Abstract
The Tc1/mariner family of DNA transposons is widespread across fungal, plant and animal kingdoms, and thought to contribute to the evolution of their host genomes. To date, an active Tc1 transposon has not been identified within the native genome of a vertebrate. We demonstrate that Passport, a native transposon isolated from a fish (Pleuronectes platessa), is active in a variety of vertebrate cells. In transposition assays, we found that the Passport transposon system improved stable cellular transgenesis by 40-fold, has an apparent preference for insertion into genes, and is subject to overproduction inhibition like other Tc1 elements. Passport represents the first vertebrate Tc1 element described as both natively intact and functionally active, and given its restricted phylogenetic distribution, may be contemporaneously active. The Passport transposon system thus complements the available genetic tools for the manipulation of vertebrate genomes, and may provide a unique system for studying the infiltration of vertebrate genomes by Tc1 elements.
INTRODUCTION
Mobilization of transposons is hypothesized to contribute to the evolution of genomes by several mechanisms, including; imperfect repair after excision, insertional mutagenesis, changes in the regulation of adjacent gene expression, and gene duplication or exon shuffling (1,2). Tc1/mariner elements are widely distributed, being found in species from animal, plant and fungal kingdoms (3,4). Transposons in this family contain a transposase gene flanked by inverted terminal repeats (ITRs) and are mobilized by a cut-and-paste mechanism (3). The Tc1/mariner transposases belong to a larger family of enzymes, including bacterial transposases, retroviral integrases, and V(D)J recombinase, all of which are characterized by a DDE or in the case of mariner and mariner-like elements a DDD motif that is involved in polynucleotidyl transfer reactions (3).
Tc1/mariner elements can be active in the soma and the germline. Therefore, regulation of transposition is required for host viability, and by extension, transposon persistence (5). Evolutionary periods of transpositional activity are thus interspersed with periods of stochastic loss (6) and ‘vertical inactivation’ of transposons, wherein only defective versions are found within the genome, containing frame-shifts, deletions and missense mutations. Nonetheless, representatives of this family of transposons have been demonstrated to be active in nematodes (7,8) and arthropods (6,9,10). In contrast, the biology of Tc1/mariner elements in vertebrate cells/genomes is understudied. Despite being present at thousands of copies per genome, neither active Tc1/mariner transposition, nor functionally intact elements have been identified in vertebrate genomes. Instead, active vertebrate Tc1/mariner elements have been synthetically created by phylogeny-informed reanimation of inactive transposons. The Sleeping Beauty (SB) transposon derived from salmonid fish represents the inaugural representative of Tc1/mariner transposon reanimation in vertebrates (11), and has been subsequently engineered to hyperactivity for genetic applications including transpositional transgenesis (TnT) and gene therapy. Additional transposons from amphibians (Frog Prince) and humans (HsMar1) have been similarly reanimated (12,13).
Leaver previously described a Tc1-like transposon (PPTN) from flatfish that is present at 200–300 copies in the plaice (Pleuronectes plattessa) genome (14,15). This element belongs to the inverted repeat/direct repeat (IR/DR) group of Tc1-like elements as described by Izsvak et al. (16), and has 74–81% identity to related but distinct elements represented in the genomes of Atlantic salmon (Salmo salar) and frogs (Rana temporaria). These elements appeared to be absent in the genomes of other fish species. The fact that these related mobile elements are found in the genomes of phylogenetically distant animals yet absent in more closely related species, led Leaver to hypothesize that this transposon family was distributed by horizontal transfer (14,17). Additionally, the identification of structurally intact native transposons suggested the tantalizing possibility that these Tc1-like transposons might be active.
We have developed the native PPTN transposon as a binary non-autonomous system, henceforth referred to as ‘Passport’, and have demonstrated it as functionally competent for transposition. We have tested the Passport transposon system in multiple vertebrate cell lines and provide molecular evidence of transposition. Our data indicates that Passport is active, and when mobilized prefers to integrate into the transcriptional units of genes. An expanded analysis of the phylogeny of Passport reveals that in addition to the presence of this transposon in other flatfish, including flounder (Platichthys flesus) and turbot (Scophthalmus maximus) (18), Passport shares a high degree of similarity with other recently reported transposons, including; Eagle, Glan and Barb (19,20). Combined, these observations suggest that this structurally and functionally intact transposon family may remain active in vertebrate lineages, and given a preference for integration into genes, may be contemporaneously active in vertebrate genome evolution.
MATERIALS AND METHODS
Vector construction
Sequence information, maps and material requests for these constructs can be found on our web site [http://primer.ansci.umn.edu/fahrenkruglab].
pPTnP-GeN
pPTnP-GeN was produced by cloning a 3.4 kb XmaI to NheI fragment of pKT2P-GeN (21), which contained the human PGK promoter and mini-intron, EGFP, the encephalomyocarditis virus internal ribosome site, neomycin phosphotransferase, and the rabbit beta-globin poly(A) signal, into pPTn2-SE.
pPTn2-SE
Using T3-rev [TCTCCCTTTAGTGAGGGTTAATT] and T7-rev [TCTCCCTATAGTGAGTCGTATTA] primers a 102-bp PCR product of pKT2-SE that provides T7 and T3 polymerase binding sites orientated towards the ITRs of the PTn transposon and separated by a short multiple cloning site was cloned into the MscI site of prePTn1(-1). prePPTn2(-1) was made by cloning a 0.65 kb BamHI to KpnI fragment of pCR4-PPTN2A into pK-A3 opened from KpnI to BamHI. pCR4-PPTN2A was created by topo cloning a 0.65 kb PCR product amplified from prePPTN2(-2) using oligos PPTN-F1 (BamHI) [AAGGATCCGATTACAGTGCCTTGCATAAGTAT] and PPTN-R2 (KpnI) [AAGGTACCGATTACAGTGCCTTGCATAAGTATTC] into pCR4-Topo (Invitrogen). prePPTN2(-2) was created by amplifying the majority of pBluKS-PPTN5 (14) with oligos PPTN-OL2 [CCATCTTTGTTAGGGGTTTCACAGTA] and PPTN-OR1 [CCAGGTTCTACCAAGTATTGACACA]. The PCR fragment was then self-ligated to produce an empty transposon with a single MscI site in its interior.
pKUb-PTs
pKUb-PTs was made by replacing the SB11 gene in pKUb-SB11 with PTs by cloning a 1.0 kb BamHI to NheI fragment from pCR4-PTs into pKUb-SB11 from NheI to BamHI. pCR4-PTs1 was made by cloning a PCR fragment of pBluKS-PPTN4 (14) amplified with primers CDS-PTs-F1 [AAAGCTAGCATGAAGACCAAGGAGCTCACC] and CDS-PTs-R1 [AAGGATCCTCAATACTTGGTAGAACC] into pCR4-Topo (Invitrogen).
pKC-PTs
The PTs coding region was placed behind the mCAGs promoter by cloning a 1.0 kb NheI to EcoRI fragment of pKUb-PTs containing the transposase into pK-mCAG opened from EcoRI to NheI. pK-mCAG was made by cloning the mCAG promoter from pSBT-mCAG (22) as a 0.96-kb SmaI to EcoRI (filled) fragment into pK-SV40(A)x2 opened with AflII (filled).
pKUb-SB11
The construction of pKUb-SB11 has previously been described (21).
pKC-SB11
pKC-SB11 was made by cloning a 1.05-kb NheI to EcoRI fragment from pKUb-SB11 into pK-mCAG (21) opened from EcoRI to NheI.
pCMV-Bgal
pCMV-Bgal is available from Clontech (Mountainview, CA, USA) as pCMVβ.
pPTnP-PTK
A 2.7 kb PvuII to PvuII fragment of pKP-PTK_TS (21)was cloned into the EcoRV site of pPTn-RV to make pPTnP-PTK. pPTn-RV was made by cloning KJC-Adapter 4 [TCTCCCTTTAGTGAGGGTTAATTGATATCTAATACGACTCACTATAGGGAGA] into the MscI site of prePPTn2(-1) creating T7 and T3 polymerase binding sites orientated out towards the ITR of the PTn transposon and separated by an EcoRV site.
Cell culture and transposition assays
HT1080, HeLa, CHO-K1, NIH-3T3 and Vero cells are available from ATCC. TT and DF1 cells were kind gift from the laboratory Dr Douglas Foster, University of Minnesota (23,24). The isolation of PEGE cells has been described previously (21). CHO-K1 cells were grown in DMEM-F12 while all other cell lines were cultured with DMEM. Both mediums were enriched with 10% FBS, 1× Penn/Strep, and 1× l-Glutamine. PEGE cells were also enriched with insulin at 10 μg/ml.
Transposition assays were carried out after seeding cells in six-well plates to achieve 60–80% confluency prior to transfection with DNA complexed with TransIT-LT1 transfection reagent (Mirus Bio Corporation, WI, USA). Transfections were carried out according to manufacturer's instructions with a ratio of 3:1 lipid:DNA. Two days after transfection, cells were isolated from their wells with trypsin and collected by centrifugation. Two replicates of 30 000 cells were plated on 100 mm dishes and selected in the appropriate selectable media. HT1080 cells were selected in 600 μg/ml of G418. For puromycin selection, HT1080, HeLa, Cho-K1, NIH-3T3, Vero, TT1, DF1 and PEGE cells were selected under 0.65, 0.4, 8.0, 1.5, 1.8, 0.35, 0.8 and 0.3 μg/ml puromycin, respectively. After colony formation, typically 9–12 days under selection, colonies were stained with methylene blue and counted.
Southern hybridization
Genomic DNA from independent clones derived after transfection with Passport transposons (pPTnP-PTK) and Passport transposase (pKC-PTs) was isolated using standard methods. Approximately 10 μg of DNA was digested with AseI and run on a 0.7% agarose gel. The DNA was transferred to a positively charged nylon membrane using 10X SSC and standard methods. The membrane was hybridized with a random primed fragment of pKP-PTK-TS isolated after digestion with XmaI. This probe contains the bulk of the puromycin–thymidine kinase gene, about 1.5 kb.
Cloning junction fragments
Blocked linker-mediated PCR was performed as described (21) except that DNA was obtained from colonies of cells that had been dried and stained with methylene blue. Briefly, genomic DNA was digested with a cocktail of restriction enzymes, including XbaI, NheI, AvrII and SpeI. The DNA was ligated to a blocked linker made by annealing the oligos primerette-long [CCTCCACTACGACTCACTGAAGGGCAAGCAGTCCTAACAACCATG] and blink-XbaI [5′P-CTAGCATGGTTGTTAGGACTGCTTGC-3′P]. Nested PCR was performed on the ligated DNA to specifically amplify junctions between the Passport transposon and genomic DNA. The transposon-specific primers for the primary PCR included PTn-IRDR(L)-O1 [GTGTTGGTCCATTACATAAACTCACGATGAA] or PTn-IRDR(R)-O1 [GGGTGAATACTTATGCACCCAACAGATG], transposon-specific primers for the secondary PCR reactions included PTn-IRDR(L)-O2 [GCATGACAAAATGTAGAAAAGTCCAAAGG] or PTn-IRDR(R)-O2 [CAGTACATAATGGGAAAAAGTCCAAGGG].
Phylogenetic analysis
The 1626 bp DNA sequence of PPTN (Passport) was used to query the entire ENSEMBL (www.ensembl.org) genome database using BLASTN. Consensus DNA sequences were derived, as described by Leaver (14), from a minimum of seven of the most similar sequences from each genome. Deduced consensus transposase amino acid sequences were aligned using ClustalW and phylogenetic trees generated as described (14). The Atlantic salmon (Salmo salar) and rainbow trout (Oncorhyncus mykiss) EST and tentative consensus cDNA databases (http://compbio.dfci.harvard.edu/tgi/) were also interrogated with PPTN using BLASTN and sequences assembled into consensus polypeptides as described for genome sequences.
RESULTS
Native Passport is competent for transposition in cells of diverse vertebrate origin
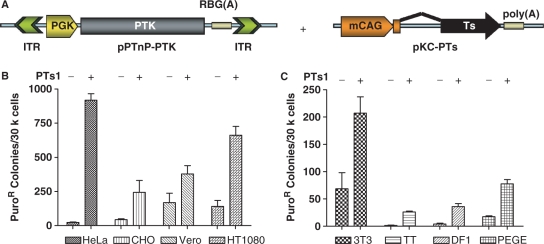
The SB, Frog Prince and HsMar1 transposon systems are active in a wide array of vertebrate cells (11–13), although to differing degrees. In order to assess the ability and ubiquity of Passport function, we undertook an analysis of TnT in human (HeLa, HT1080), monkey (Vero), pig (PEGE), hamster (CHO), mouse (3T3), chicken (DF1) and turkey (TT) cells using a Passport transposon containing a puromycin thymidine kinase fusion protein (25) driven by the mouse PGK promoter (pPTnP-PTK). Cells were transfected with the pPTnP-PTK transposon and a Passport transposase expression construct (pKC-PTs) at a Tn:Ts molar ratio of 1:0.5, or with the molar equivalent of pCMV-βgal. Following transfection, replicates of ∼30 000 cells were plated and selected in puromycin, fixed, stained and enumerated. In all cases, Passport-dependent TnT resulted in the generation of a number of puromycin-resistant colonies far exceeding that observed for controls lacking transposase, in the case of HeLa cells reflecting at least a 40-fold enhancement (Figure 1). As with other transposon systems (12,26), TnT varied between cell types (as did background-resistant colony formation), although comparing relative transpositional activity across cell lines may be confounded by the fact that transfections were conducted under identical conditions that may be suboptimal for some cell lines. Nonetheless, native Passport is functional in cells from a broad sampling of vertebrate species.
Figure 1.
Passport functions in cells from a wide variety of vertebrate sources. (A) A Passport transposon that expresses Puromycin phosphotransferase was co-transfected with a source of Passport transpoase, pKC-PTs1 (+PTs) or pCMV-Bgal (–PTs). Cells were selected in puromycin and stable colonies were counted. (B) HeLa, CHO, Vero and HT1080 cells displayed an increase in stable colony formation with the addition of Passport transposase. (C) 3T3, TT, DF1 and PEGE cells produced fewer colonies under these transfection conditions; however, the addition of Passport transposase significantly improved colony formation. The addition of transposase rather than beta-galactosidase significantly increased colony formation in all cell types (P < 0.05, except CHO where P = 0.07).
Passport is sensitive to overproduction inhibition
Overproduction inhibition, where excess transposase reduces the rate of transposition, is a hallmark of Tc1/mariner elements (5). We thus undertook an analysis of this effect for Passport and compared its sensitivity to that of the well-characterized SB transposon system (27,28). A series of transfections was performed with varying ratios of transposase to transposon vector to measure the effect of increasing transposase concentration on the rate of transposition. In addition, two promoters [human UbC (29) and mCAG (22,30)] were used to drive expression of the Passport transposase across a broad range of transposase levels (Figure 2). In HT1080 cells, gene expression from the mCAGs promoter is 5- to 10-fold higher than from the UbC promoter (data not shown). A constant amount of transposon (pPTnP-GeN, 75 fmol) was co-transfected with transposase vector containing either the UbC or mCAGs promoter (pKC-PTs or pKUb-PTs at a Tn:Ts molar ratio of 1:0.2, 1:0.5, 1:1, 1:2 or 1:5 corresponding to 15, 37.5, 75, 150 and 375 fmol of transposase plasmid). The total amount of transfected DNA was kept at 2 µg by supplementing with pCMV-βgal DNA. To compare to the SB transposon system, identical reactions were performed with an SB transposon (pKT2P-GeN) and SB11 transposase expressed from the UbC and mCAGs promoters (pKUb-SB11 and pKC-SB11). Following transfection, two replicates of ∼30 000 cells were plated and selected in G418 for 10–14 days, fixed, stained and the resulting colonies enumerated. Our previous studies indicated that a molar ratio of 1:1 SB transposon to SB transposase expressed from the human UbC promoter resulted in near-optimal transposition rates for the SB transposon system. Therefore to correct for any variation in transfection or selection, a 1:1 ratio of pKT2P-GeN:pKUb-SB11 was included as in internal standard for each day of transfection. The relative sensitivity of the two transposon systems to overproduction inhibition is presented in Figure 2B and C, where colony formation is expressed relative to the contemporary pKT2P-GeN:pKUb-SB11 internal standard. As shown in Figure 2B, the hyperactive SB system resulted in more than twice as many colonies as the native Passport system (Figure 2C) at their respective optimal Tn:Ts ratios. As expected, the SB transposon system is sensitive to overproduction inhibition. The peak transpositional activity for Passport was observed using a 1:5 ratio of pPTnP-GeN:pKUb-PTs1 or a 1:0.2 ratio of pPTnP-GeN:pKC-PTs1, beyond which increasing transposase expression resulted in reduced transposition, indicating that Passport is indeed susceptible to overproduction inhibition.
Figure 2.
Examination of overproduction inhibition. (A) To examine the effect of transposase dose on transposition rates, a constant amount of pTnP-GeN (75 fmol) was co-transfected with five different molar ratios of transposase expression vector driven by either the human UbC promoter (pKUb-Ts) or the mCAGs promoter (pKC-Ts), where T and Ts generically refer to either SB or Passport components. In all cases the total amount of DNA transfected was adjusted to 2 µg by the addition of the appropriate amount of pCMV-Bgal. After transfection and selection in G418, colonies were counted and the data compared to an internal reference transfection of SB at a ratio of 1:1U. The raw data for the internal reference transfection came from a total of 30 replicates and ranged from 68 to 324, with a median of 150 and a mean of 170 (data not shown). The relative transposition efficiencies confirm overproduction inhibition of (B) the SB transposon system and (C) demonstrate overproduction inhibition of the Passport transposon system. Error bars represent the SE.
Molecular characterization of Passport transposition
To validate transposition we examined the number of integration events per cellular clone by Southern analysis. Transposition is supported by hybridizing fragments of varying lengths, corresponding to genomic restriction sites at varying distances from the transposon insertion-sites (Figure 3A). Non-transpositional DNA integration results in the formation of multi-copy concatemers (31) that are expected to result in a predictable restriction enzyme fragment derived from sites within the transposon vector (Figure 3B). The Southern analysis of DNA isolated from 15 HT1080 clones revealed that Passport indeed had transposed into the human genome, with one to four integrations per cellular clone (Figure 3C). Clones 4, 5 and 9 also contain a hybridizing band near the predicted size of a concatemer, although low signal intensity suggests low copy inserts not inconsistent with transposition.
Figure 3.
Evaluation of diversity and number of Passport genomic integrations. (A) Transposase-mediated recombination into the genome should result in transposon fragments of variable length after digestion with AseI. The sizes of the fragments are dependent on the proximity of AseI recognition sites in the neighboring chromatin, and can be observed following Southern hybridization. (B) Commonly, when DNA integrates without the enzymatic activity of transposase, head-to-tail concatemers of variable length are formed and integrate into the genome by non-homologous end-joining. In this case, the size of this internal high-representative fragment (∼5.1 kb) is predictable based on the location of AseI sites within the transposon donor plasmid. (C) An image of our Southern hybridization of 15 independent HT1080 clones is shown. The paired head-to-tail arrows indicate the expected position of pPTnP-PTK concatemers that could potentially form during integration by non-homologous end joining. The line with outward facing arrowheads represents the size of the transposon and therefore the minimal expected size of a hybridizing fragment integrated by TnT. The asterisks mark two bands present in the HT1080 DNA that hybridize weakly with the PTK probe used here.
To further verify TnT by Passport, and to characterize the insertion target sites and preferences within HT1080 cells, junction fragments between the transposon and host genome were cloned and sequenced. Passport, like other Tc1 transposons, is expected to integrate into a TA dinucleotide and cause target-site duplication of the TA sequence at the ITR boundary. Table 1 lists 27 independent insertion events identified in HT1080 cells, demonstrating integration of the transposon into a TA within the human genome, and validating genuine transposition. The genomic location of each transposon insertion was determined by comparison of the cloned junction sequence to the human genome using Blastn (32). Insertions were dispersed across the human genome (Table 1). However, insertions did not appear to be completely random as chromosome 1, which is twice as long as chromosome 12, has no integrations, whereas chromosome 12 has six integration events. In addition, the cloned junctions were found in transcription units in 63% of the cases, which is inconsistent with a completely random integration profile.
Table 1.
Passport junctions from integration into the human genome
 |
The integration sites show the sequence outside the left ITR (L), the TA that is duplicated upon integration, and the sequence outside the right ITR (R). The first sequence indicates the sequence found in the donor plasmid (red), while the remaining represent 27 Passport integrations sites all of which occurred by transposition as indicated by the exact junction at the ITR with a TA dinucleotide from the genome. In each case the sequence represented in CAPS was cloned by blocked LM-PCR and the sequence in lower case was derived from genome sequence data. In many cases, the Passport transposon integrated into known or (predicted) genes (Locus). The transposon integrations targeted a wide variety of chromosomal positions (Chrm Pos).
Passport-like transposons are present in other fish and amphibian genomes
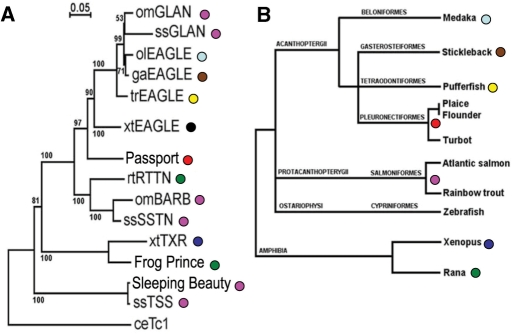
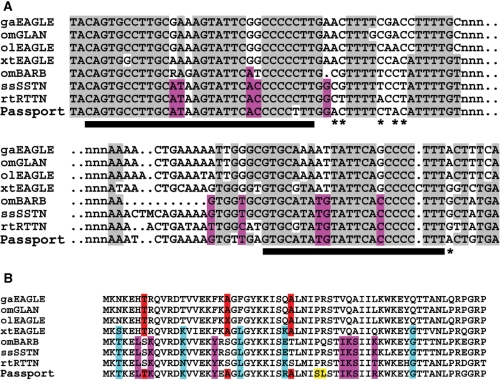
The availability of sequenced genomes provides an opportunity to compare and categorize all transposons within a species and derive consensus sequences with a minimum of experimental bias. While Passport elements were originally isolated from plaice, nearly identical elements have been identified in other flatfish, including flounder and turbot (99% and 98% DNA identity). A recent search of ENSEMBL revealed the presence of additional related transposases with high nucleotide identity (>80%) to Passport in the genomes and EST collections of Xenopus tropicalis, and pufferfish (Takafugu rubripes), stickleback (Gasterostreus aculeatus), medaka (Oryzis latipes), Atlantic salmon (Salmo salmar) and rainbow trout (Oncorhynchus mykiss). Passport-like transposons were absent from all other ENSEMBL genomes, including those of the zebrafish (Danio rerio), despite the wide range and high copy number of other Tc1-like elements in this species. Comparison of the encoded transposase amino acid sequences show that relatives of Passport form a distinct family of Tc1-like transposons that is further divided into two subfamilies, including Eagle/Glan and Barb/SSTN/RTTN (Figure 4). The salmonids (salmon and rainbow trout) contain members of both subfamilies, whilst X. tropicalis, pufferfish, stickleback and medaka contain only the Eagle/Glan subfamily. The structure of Passport is somewhat intermediate between that of Eagle/Glan and Barb/SSTN/RTTN, in that its ITRs bear a strong resemblance to Barb/SSTN/RTTN (Figure 5A) whereas its transposase-coding region seems to bear more resemblance to the Eagle/Glan subfamily. Intriguingly, alignment of the DNA-binding domains of the transposases demonstrates a distinction between Eagle/Glan and Passport/Barb/SSTN/RTTN (Figure 5B), a difference that may be functionally connected to the ITRs of these elements.
Figure 4.
Phylogeny of Passport-like transposons and their hosts. (A) Neighbor-joining plot of multiply aligned transposase consensus amino acid sequences. Sequences were aligned with ClustalW, and plotted with NJplot. Numbers represent the percentage frequencies with which the tree topology was returned after 1000 iterations. The tree is rooted to Tc1 from C. elegans. Transposon designation is prefixed by host species identifier; om, rainbow trout; ol, medaka; ga, stickleback; tr, pufferfish; ss, Atlantic salmon; ce, C. elegans; rt, Rana temporaria (frog); xt, Xenopus tropicalis. Passport, Frog Prince and Sleeping Beauty were isolated from Pleuronectes platessa, Rana sylvestris, and a variety of salmonid species, respectively. (B) Phylogeny of host species adapted from Nelson (50). The colored dots assist in pairing the transposons shown in A with the species from B.
Figure 5.
Comparison of repeat sequences and transposase DNA-binding domains of Passport-like transposons. (A) Comparison of terminal and internal 5′ repeats of the left ITR. Host species identifier as for Figure 4 prefixes transposon designation. Gray-shaded residues are common to the majority of sequences. Bars below the line indicate the conserved repeat units from within the inverted repeats. Pink highlighted sequences delineate differences between the Eagle/Glan and SSTN/Barb families. For the ITR sequences, Passport clearly aligns more closely with the SSTN/Barb families within the direct repeats, but evidence of convergence towards Eagle/Glan sequences are observed just outside of these direct repeats as indicated by the asterisks. (B) Comparison of the putative DNA-binding domains of Passport and related transposases. Pink-shaded residues indicate the amino acids that distinguish members of the Eagle/Glan family from SSTN/Barb. Residues shaded red indicate the convergence of the active Passport sequence towards the Eagle/Glan family to which it is more closely related over the length of the entire protein, whereas blue residues show some convergence of the X. tropicalis Eagle element towards the SSTN/Barb subfamily. Residues shaded in yellow seem to be unique within Passport and may be important for its activity.
DISCUSSION
We have shown for the first time that a vertebrate-derived transposon from the Tc1 family is active in its native form. Passport promotes TnT into vertebrate cells, with up to a 40-fold increase in HeLa cells and a 20-fold increase in HT1080 cells when compared to transgenesis without transposase. This corresponds to a rate of transposition up-to half the level we observed for SB11, itself a hyperactive mutant that is about 3-fold more active than the originally reanimated SB10 (27). Reanimation of SB, Frog Prince and HsMar1 relied on phylogeny-informed reconstruction of extinct elements (11–13). Efforts to develop hyperactive transposases for application to TnT and gene therapy have applied both structure-based (33,34) and phylogenetics-informed (27,35,36) approaches. Indeed, the native Passport transposase sequence has been considered in improvements to SB and it contains several residues that have been synthetically introduced to generate hyperactive SB mutants, including; L205 and VR207/8 (35), and R130 and Q243 (27). Changes have also been made in the cis-acting ITR (36,37), as well as the spacer sequence between the ITRs of the SB transposon (26,36), resulting in the development of improved transposons, and evidence that only flanking IR/DR are required to constitute an effective transposon, a finding recapitulated in our study.
We demonstrated that Passport transposons display overproduction inhibition analogous to other Tc1/mariner elements (5). Interestingly, despite using identical promoters in the SB and Passport transposase expression constructs, optimal transposition and the emergence of overproduction inhibition for Passport occurred under conditions expected to correspond to significantly higher levels of transposase expression. We can estimate that optimal transposition for Passport requires more than double the amount of transposase expression as SB, since their maximal transposition occurred at Tn:Ts molar equivalents of 1:5 and 1:2, respectively. This could result from differences in the translational efficiency or stability of the encoded transposases, differences in the affinities of the transposases for their corresponding transposons, or from innate variance in transpositional activity (disparities not unexpected when comparing native and hyperactive transposon systems).
We conducted a limited survey of Passport integration sites and observed a significant preference for integration into genes (likelihood ratio >5000:1). This characteristic has also been observed for the piggyBac transposon system, a non-Tc1 element (38), but contrasts sharply with the random integration site preferences for the SB transposon system (39), suggesting Passport may be especially suitable for functional genomics applications. Indeed, there are a variety of transposons useful in vertebrate cells, including; SB (11), Frog Prince (12), Tol2 (40), minos (41,42), piggyBac (43,44), Ac/Ds (45,46), Tol1 (47), HsMar1 (13), Harbinger (48) and now Passport. The unique transposon ITRs and preferences for target sites provides a toolbox that can be implemented in response to a variety of needs. Particularly relevant applications include the serial introduction of multiple transgenes into cells/animals without disturbing previously integrated transposons, and enhanced insertional mutagenesis screens that capitalize on differences in integration site preferences (49).
An examination of genome-sequence data for diverse organisms shows that Tc1 elements related to Passport are also present in X. tropicalis and in other fish species. In X. tropicalis these transposons have been termed Eagle (20) and in rainbow trout Glan and Barb (19). Members of the Eagle/Glan family are phylogenetically widespread and their distribution is generally in agreement with the accepted phylogeny for these species (50), indicating vertical transmission. On the other hand, Barb/SSTN/RTTN family members are restricted to salmonid fish and Rana frogs (14), likely representing horizontal transmission.
Passport transposons appear to be an intermediate between the Eagle/Glan group and the Barb/SSTN/RTTN group of transposons, with ITRs bearing a strong resemblance to Barb/SSTN/RTTN but a transposase more akin to the Eagle/Glan subfamily. Perhaps transposon ‘hybridization’ has resulted in the genesis of Passport in Pleuronectid genomes. Regardless, the correspondence of differences in the DNA-binding domains of these closely related transposases and their cognate ITRs provides a rationale for future investigations focused on the evolution and engineering of transposase specificity.
Passport represents the first opportunity to study the biology of a native functionally intact vertebrate Tc1 element. As such, it may provide important information about transposon function and regulation that could have been lost or modified during synthetic reanimation or subsequent hyperactivity mutagenesis. Provided suitable culture conditions can be developed for plaice cells, Passport may also provide the seminal opportunity to study evolved regulatory or accommodating interactions between vertebrate Tc1 transposons and the host genome, perhaps missing when elements are artificially introduced into a naive genome.
The functional integrity and phylogenetic restriction of Passport sequences suggest contemporaneous activity. Further study of Passport integration sites by high throughput sequencing could provide important information about the history of transposition in flatfish, contributing to our understanding of the roles of DNA transposons in vertebrate genome evolution (51). The prevalence and genomic location of Passport transposons among geographically diverse plaice and related natural fish populations may be helpful in determining the time course and extent of Passport infiltration. Of particular interest is the potential to address the hypothesis that transposon activity may increase in response to stress (52), a phenomenon recently supported by observations in other teleosts (19). Although speculative, perhaps Passport activity could prove a useful indicator for managing geographically disperse flatfish fisheries in the face of ecological and fishing pressures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work and funding for open access publication was supported by grants from the USDA National Research Initiative (2008-35205-18852 to S.C.F.).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev. Genet. 1999;25:291–296. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 4.Robertson HM. The Tcl-mariner superfamily of transposons in animals. J. Insect Physiol. 1995;41:99–105. [Google Scholar]
- 5.Hartl DL, Lozovskaya ER, Nurminsky DI, Lohe AR. What restricts the activity of mariner-like transposable elements. Trends Genet. 1997;13:197–201. doi: 10.1016/s0168-9525(97)01087-1. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson JW, Medhora MM, Hartl DL. Molecular structure of a somatically unstable transposable element in Drosophila. Proc. Natl Acad. Sci. U.S.A. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins J, Forbes E, Anderson P. The Tc3 family of transposable genetic elements in Caenorhabditis elegans. Genetics. 1989;121:47–55. doi: 10.1093/genetics/121.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moerman DG, Waterston RH. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics. 1984;108:859–877. doi: 10.1093/genetics/108.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry EG, Witherspoon DJ, Lampe DJ. A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics. 2004;166:823–833. doi: 10.1534/genetics.166.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muñoz-López M, Siddique A, Bischerour J, Lorite P, Chalmers R, Palomeque T. Transposition of Mboumar-9: identification of a new naturally active mariner-family transposon. J. Mol. Biol. 2008;382:567–572. doi: 10.1016/j.jmb.2008.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 12.Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, Izsvak Z, Ivics Z. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leaver MJ. A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission. Gene. 2001;271:203–214. doi: 10.1016/s0378-1119(01)00530-3. [DOI] [PubMed] [Google Scholar]
- 15.Leaver MJ, Wright J, George SG. Structure and expression of a cluster of glutathione S-transferase genes from a marine fish, the plaice (Pleuronectes platessa) Biochem. J. 1997;321(Pt 2):405–412. doi: 10.1042/bj3210405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izsvak Z, Ivics Z, Hackett PB. Characterization of a Tc1-like transposable element in zebrafish (Danio rerio) Mol. Gen. Genet. 1995;247:312–322. doi: 10.1007/BF00293199. [DOI] [PubMed] [Google Scholar]
- 17.Robertson HM. The mariner transposable element is widespread in insects. Nature. 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 18.Pocwierz-Kotus A, Burzynski A, Wenne R. Family of Tc1-like elements from fish genomes and horizontal transfer. Gene. 2007;390:243–251. doi: 10.1016/j.gene.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Krasnov A, Koskinen H, Afanasyev S, Molsa H. Transcribed Tc1-like transposons in salmonid fish. BMC Genomics. 2005;6:107. doi: 10.1186/1471-2164-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinzelle L, Pollet N, Bigot Y, Mazabraud A. Characterization of multiple lineages of Tc1-like elements within the genome of the amphibian Xenopus tropicalis. Gene. 2005;349:187–196. doi: 10.1016/j.gene.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Clark KJ, Carlson DF, Foster LK, Kong BW, Foster DN, Fahrenkrug SC. Enzymatic engineering of the porcine genome with transposons and recombinases. BMC Biotechnol. 2007;7:42. doi: 10.1186/1472-6750-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohlfest JR, Frandsen JL, Fritz S, Lobitz PD, Perkinson SG, Clark KJ, Nelsestuen G, Key NS, McIvor RS, Hackett PB, et al. Phenotypic correction and long-term expression of factor VIII in hemophilic mice by immunotolerization and nonviral gene transfer using the Sleeping Beauty transposon system. Blood. 2005;105:2691–2698. doi: 10.1182/blood-2004-09-3496. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer-Klein J, Givol I, Barsov EV, Whitcomb JM, VanBrocklin M, Foster DN, Federspiel MJ, Hughes SH. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 24.Kong BW, Foster LK, Foster DN. Establishment of an immortal turkey turbinate cell line suitable for avian metapneumovirus propagation. Virus Res. 2007;127:106–115. doi: 10.1016/j.virusres.2007.03.028. [DOI] [PubMed] [Google Scholar]
- 25.Abuin A, Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol. Cell. Biol. 1996;16:1851–1856. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 27.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 28.Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol. Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 29.Gill DR, Smyth SE, Goddard CA, Pringle IA, Higgins CF, Colledge WH, Hyde SC. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1alpha promoter. Gene Therapy. 2001;8:1539–1546. doi: 10.1038/sj.gt.3301561. [DOI] [PubMed] [Google Scholar]
- 30.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 31.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zayed H, Izsvak Z, Khare D, Heinemann U, Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 36.Zayed H, Izsvak Z, Walisko O, Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the Sleeping Beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 38.Wilson MH, Coates CJ, George AL., Jr piggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 39.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hori H, Suzuki M, Inagaki H, Oshima T, Koga A. An active Ac-like transposable element in teleost fish. J. Marine Biotechnol. 1998;6:206–207. [PubMed] [Google Scholar]
- 41.Klinakis AG, Zagoraiou L, Vassilatis DK, Savakis C. Genome-wide insertional mutagenesis in human cells by the Drosophila mobile element Minos. EMBO Rep. 2000;1:416–421. doi: 10.1093/embo-reports/kvd089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franz G, Savakis C. Minos, a new transposable element from Drosophila hydei, is a member of the Tc1-like family of transposons. Nucleic Acids Res. 1991;19:6646. doi: 10.1093/nar/19.23.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 45.Emelyanov A, Gao Y, Naqvi NI, Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClintock B. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. U.S.A. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koga A, Shimada A, Kuroki T, Hori H, Kusumi J, Kyono-Hamaguchi Y, Hamaguchi S. The Tol1 transposable element of the medaka fish moves in human and mouse cells. J. Human Genet. 2007;52:628–635. doi: 10.1007/s10038-007-0161-2. [DOI] [PubMed] [Google Scholar]
- 48.Sinzelle L, Kapitonov VV, Grzela DP, Jursch T, Jurka J, Izsvak Z, Ivics Z. Transposition of a reconstructed Harbinger element in human cells and functional homology with two transposon-derived cellular genes. Proc. Natl Acad. Sci. U.S.A. 2008;105:4715–4720. doi: 10.1073/pnas.0707746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 50.Nelson JS. Fishes of the World. 4th. Hoboken, NJ: John Wiley; 2006. [Google Scholar]
- 51.Wessler SR. Transposable elements and the evolution of eukaryotic genomes. Proc. Natl Acad. Sci. U.S.A. 2006;103:17600–17601. doi: 10.1073/pnas.0607612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClintock B. The significance of responses of the genome to challenge. Science (New York, N.Y.) 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.