Abstract
Virtual Screening is an increasingly attractive way to discover new small molecules with potential medicinal value. We introduce a novel strategy that integrates use of the molecular docking software Surflex with experimental validation by the method of competition dialysis. This integrated approach was used to identify ligands that selectively bind to the triplex DNA poly(dA)-[poly(dT)]2. A library containing ∼2 million ligands was virtually screened to identify compounds with chemical and structural similarity to a known triplex intercalator, the napthylquinoline MHQ-12. Further molecular docking studies using compounds with high structural similarity resulted in two compounds that were then demonstrated by competition dialysis to have a superior affinity and selectivity for the triplex nucleic acid than MHQ-12. One of the compounds has a different chemical backbone than MHQ-12, which demonstrates the ability of this strategy to ‘scaffold hop’ and to identify small molecules with novel binding properties. Biophysical characterization of these compounds by circular dichroism and thermal denaturation studies confirmed their binding mode and selectivity. These studies provide a proof-of-principle for our integrated screening strategy, and suggest that this platform may be extended to discover new compounds that target therapeutically relevant nucleic acid morphologies.
INTRODUCTION
Nucleic acid sequences that form triple helices have become a source of increasing interest as a way to interfere with DNA transcription and modulate gene expression (1–3). Several approaches attempt to use triplex nucleic acids to interfere with the transcription of genes, through either inducing the formation of triplex or stabilizing existing triplex nucleic acids. The former approach is the so-called ‘anti-gene’ approach and involves the administration of triplex forming oligonucleotides (TFOs), which are short sequences of nucleic acids that can bind to the major groove of duplex nucleic acids and promote the formation of triplex structures (4,5). TFOs have already been successful in reducing transcription of the c-myc oncogene that is located in the promoter site of genes (4,6). However, there are currently significant challenges associated with the use of TFOs and triplex structures in general. First, TFOs have significantly lower activity in cell-based systems, compared to in vitro systems (7). This has been ascribed to many factors including improper cellular localization or degradation of the oligonucleotide (7,8). A second limitation is the inherent low stability of many triplex structures (9,10). The latter limitation is the focus of this work where we demonstrate the use of a novel virtual and actual screening platform for identifying several compounds that can selectively bind to and stabilize a triplex nucleic acid structure. These newly identified small molecules could be used to target triplex structures in several ways. First, the small molecule could stabilize pre-existing triplex structures in vivo. The small molecules could be used in an adjuvant setting with TFOs to increase stability, or alternatively the small molecules can be linked to TFOs to enhance the stability of newly formed triplex structures (8). Either of these approaches could be used to control gene expression. These capabilities make these small molecules potentially clinically relevant for treating cancer and other diseases that are closely linked with abnormal gene expression.
Several small molecules are known to intercalate into and stabilize triplex nucleic acids including coralyne, benzo[e]pyridoindoles (BePI), benzo[g]pyridoindoles (BgPI), dibenzophenanthrolines, and anthraquinones (10−14). One of the most selective and extensively studied classes are the napthylquinolines, which have been shown to intercalate into the TAT DNA triplex, poly(dA)-[poly(dT)]2 (15−17). Chaires et al. performed an extensive study (18) that characterized the selectivity and affinity of 14 napthylquinoline derivatives. The ligand MHQ-12 emerged from that study as the compound with the highest affinity and greatest selectivity for the poly(dA)-[poly(dT)]2 triplex. While this approach for the discovery of triplex-selective ligands was successful, it is a laborious and time-consuming process. We propose a novel alternative approach for finding ligands that target a particular structure in which virtual screening is used to identify promising ligand candidates followed by validation using competition dialysis. We demonstrate here that this approach can identify small molecules that intercalate into poly(dA)-[poly(dT)]2 with higher selectivity and affinity than MHQ-12. A significant result of this approach is that a small molecule with a substantially different molecular scaffold was identified that has superior affinity and selectivity for triplex DNA compared to MHQ-12. This strategy thus provides a new platform for identifying promising small molecule drugs against nucleic acid targets.
Virtual screening using molecular similarity and docking methods is becoming an increasingly important and economical approach to identify small molecule drug candidates (19). While there are numerous studies using such screening methods for targeting proteins, far fewer virtual screening efforts have been performed targeting nucleic acids. The few studies that have been performed targeting nucleic acids have produced promising results. They have shown that screening methods can accurately reproduce crystallographic structures of ligand–nucleic acid complexes using a variety of docking programs including DOCK (20), Autodock (21) and Surflex (22). Our virtual screening approach uses both ligand and structure-based discovery principles to select ligands from a commercially available library that bind to poly(dA)-[poly(dT)]2 with higher affinity and selectivity than MHQ-12. Initial virtual screening is performed with Surflex-Sim, which is a ligand similarity-based software program that has superior performance compared to most traditional 2D similarity methods (23). This program is an effective tool to rapidly pre-screen large virtual compound libraries to enrich for structurally similar ligands (23). Surflex-Sim maximizes 3D morphological similarity and alignment of a test ligand to the control ligand, which in this work was MHQ-12 (23–25). The quantitative metric that is used for evaluating Surflex-Sim results is the Surflex-Sim score, which embodies an all atom comparison and alignment of the test ligand with the control ligand. The top-ranked Surflex-Sim results were used for structure-based docking studies to dock the ligand to the intercalation site and the three grooves (major–major, major–minor and minor) (18) of the triplex structure using the docking program Surflex-Dock. Surflex-Dock performs docking of test ligands to a ‘protomol’ or idealized representation of the binding site on the nucleic acid target. The ligands are docked to the target and the poses are ranked by a Surflex Raw Score (SRS) that consists of an affinity score of the ligand for the target (25). This sequential combination of Surflex-Sim followed by Surflex-Dock produced several ligands that had hypothesized higher binding affinity and selectivity for the triplex intercalation site, compared to MHQ-12.
A critically important step after virtual screening is validation by experimental testing of the top candidates. To accomplish this, competition dialysis was employed because of its extensive use to determine the selectivity and affinity of a small molecule for single-stranded, duplex, triplex and quadruplex nucleic acid targets (13,26−36). The advantage of competition dialysis is that it is not limited to the target sequence, or a simple comparison with another form of DNA, but with as many nucleic acid forms as are included in the assay. Competition dialysis involves dialyzing solutions of an array of nucleic acid sequences and structures against a common solution containing a test ligand (26). The solution is allowed to reach equilibrium, and the amount of ligand that is bound to each nucleic acid is measured using either fluorescence or absorbance (26). Comparison of the total and relative amounts of ligand bound to each nucleic acid assesses the affinity and selectivity, respectively, of the ligand for any included nucleic acid. Competition dialysis testing is used here to validate the affinity and selectivity of the top virtual screening hits. Circular dichroism and thermal denaturation were used for further characterization of the triplex binding of the top virtual screening candidate hits.
By using this integrated approach we have identified small molecules that have higher selectivity and affinity for triplex poly(dA)-[poly(dT)]2 than the original molecule, MHQ-12, and which are among the most selective and tightest triplex binding molecules reported to date.
MATERIALS AND METHODS
Virtual library construction
The triplex-selective ligand MHQ-12 was constructed and hydrogen atoms were added using Macromodel (Schrodinger, Inc.). The ligand was energetically minimized using a sequential combination of 2000 iterations of a Steepest Descent algorithm followed by 2000 iterations of a Polak-Ribier Conjugate Gradient algorithm. AMBER ligand atom types were assigned using Sybyl (Tripos, Inc.). The program Antechamber in the software suite Amber (UCSF) was used to calculate AM1-BCC charges for the ligand and to convert to a MOL2 file format. A virtual set of 1.962 million ZINC compounds in MOL2 format were obtained from the ZINC 2007 ‘all-purchasable’ subset of ligands from the University of California San Francisco (37). The first 1.962 million ligands were downloaded out of a total of 2.7 million ligands from the 2007 ZINC ‘all-purchasable’ database. The ‘Reference’ subset of the ‘all-purchasable’ library was used directly as downloaded from the ZINC website, and was not filtered to select for ligands with any particular chemical property. The ligand 3D coordinates were generated by using detailed methods previously described (37). Other ligand preparation procedures performed were protonation of the ligand based on a ‘Reference’ pH 7 condition and assignment of partial atomic charges from AMSOL semi-empirical quantum calculations (37). The triplex nucleic acid structure poly(dA)-[poly(dT)]2 with an intercalation site was constructed using B-type parallel triplex (38) with X-ray structural intercalation site backbone fragments [Protein Data Bank entry 1p20.ent] and minimized holding the heavy atoms fixed.
Surflex methods
The program Surflex (UCSF) containing both the Surflex-Sim molecular similarity and the Surflex-Dock molecular docking programs was run on 30 AMD Opteron 246 processors (2.0 GHz) running the Linux Red Hat operating system for all virtual screening experiments. Surflex-Sim experiments were performed using the ‘align_list’ function to compare the MHQ-12 triplex selective ligand against 1.962 million compounds in the ZINC library. The top 350 ligands, ranked according to the highest Surflex-Sim score, were selected as candidates for Surflex-Dock studies and were extracted as individual MOL2 files from the library files using inhouse PERL scripts. The Surflex-Dock docking algorithm and scoring functions have been described in detail previously (39,40). Briefly, Surflex-Dock operates by a surface shape-based method, aligning each test ligand to a ‘protomol.’ The protomol consists of a series of molecular fragments that characterize the surface properties of the target active site including steric effects, hydrogen bond acceptor groups and hydrogen bond donor groups (39,40). After alignment of the ligands to the protomol, each pose is scored based on hydrophobic and polar contacts between atoms. The scoring function also includes a term for solvation although this contribution has typically been difficult to incorporate accurately in many molecular docking programs (41). Most importantly for these purposes, Surflex-Dock was previously used successfully to reproduce the crystal structures of several mono and dicationic small molecules bound to either the minor groove of nucleic acids or by intercalation between base pairs (22). This suggests that Surflex-Dock may have particular utility for finding new small molecules that target nucleic acid structures and thus is the basis for its use here. Finally, while many other programs are generally available for molecular docking, most have been used almost exclusively for protein–ligand docking, and have not been validated for nucleic acid–ligand docking. The previous published use of Surflex-Dock for successfully modeling binding of ligands to nucleic acids thus makes this program a rational selection for use in these studies (22). For the Surflex-Dock experiments, four protomols were generated to cover the major–major groove, major–minor groove, minor groove and intercalation sites of the triplex nucleic acid, using the same methods we previously described (22). The ‘proto_bloat’ function was set to accommodate all reasonable interactions of the protomols with the triplex target sites. Docking of the ligands to the target was performed using a whole molecule approach, as described previously (22,25,39). The Surflex-Dock experiments involved docking each of the ligands to all four protomols individually, in separate experiments. Surflex-Dock was operated with parameters ‘Multistart 5’ and ‘Random 5’ which we have previously shown returned accurate top-ranked docked poses for a set of small molecules to their respective nucleic acid targets (22). The Surflex-Dock poses were ranked according to the highest Surflex Raw Score. Surflex-Sim and Surflex-Dock poses were visualized using AutoDockTools (The Scripps Research Institute). The properties of compounds 1 and 2 used in the QSAR analysis were generated with QikProp (Schrodinger, Inc.).
Compounds for biophysical testing
The highest ranked candidates identified by virtual screening were the ligands with ZINC identification numbers 632255 and 4623551, which will be referred to hereafter as compound 1 and compound 2, respectively. Compound 1 (42), is 4-(4-methylpiperazino-2-(2-naphthyl)quinoline and was obtained from Sigma-Aldrich (Milwaukee, WI, USA). Compound 2 is 1-phenyl-4-pyrrolidino-2,3-dihydro-1/H/-pyrrolo[2,3-/b/]quinoline and was obtained from Chemical Block (N.D. Zelinsky Institute of Organic Chemistry, Moscow, Russia). As positive controls, the known triplex selective ligands MHQ-15 and OZ-85H were synthesized as previously described (18).
Competition dialysis method
Competition dialysis experiments were done as previously described (18,27,28,43). The array of oligonucleotides used is given in Supplementary Table S1. The methods used to construct the oligonucleotides are elaborated in detail by Ren and Chaires (26). Briefly, natural DNA including calf thymus, Micrococcus lysodeikticus and Clostridium perfringens were obtained from Sigma (St Louis, MO, USA) and sonicated, phenol extracted and purified using methods previously described (26,44). The remaining 30-nt long oligonucleotides including poly(dA), poly(dT), poly (dAdT), poly (dAdT)-(dAdT) and poly (dGdC)-(dGdC) were synthesized and obtained from Pharmacia Biotech (Piscataway, NJ, USA). Poly(rA), poly(rU) and quadruplex sequences were obtained from Integrated DNA Technologies (Coralville, IA, USA). The Z-DNA structure was produced by bromination of poly (dGdC)-(dGdC), as previously described (45). Remaining duplex nucleic acids such as poly(rArU) were produced by mixing poly(rA) and poly(rU) in a 1:1 equimolar ratio, heating at 90°C for 2 min and subsequently cooling to room temperature. Similarly, triplex nucleic acids were made by mixing poly (dAdT) with poly(dT) in a 1:1 equimolar ratio, heating at 90°C for 2 min and subsequently cooling to room temperature. Quadruplex nucleic acid solutions were heated at 90°C, cooled to room temperature and left at 4°C for 48 h prior to use in competition dialysis. All nucleic acid solutions were extensively dialyzed against Na2HPO4 (6 mM), NaH2PO4 (2 mM), NaCl (185 mM), EDTA (0.1 mM), pH 7 using Pierce (Rockford, IL, USA) 3500 Da molecular weight cutoff dialysis cassettes. All nucleic acid structures were characterized by circular dichroism spectra, UV absorbance spectra and thermal denaturation, as previously described (26). All nucleic acid samples were of identical concentration of 75 μM, expressed in terms of monomeric unit (base pairs for duplex DNA, triplets for triplex DNA and tetrads for quadruplex DNA). The monomer concentration scale was used to alleviate possible problems arising from length differences among the polynucleotide and natural DNA samples. At the end of the dialysis equilibration period, ligand concentrations were determined by fluorescence. A volume of 180 μl of each sample was carefully transferred into one well of a 96-well microtiter plate (Costar® cat 3915; Corning Inc., Corning, NY, USA). To each sample, 20 μl of a 10% (w/v) sodium dodecyl sulfate (SDS) stock solution was added to give a final concentration of 1% (w/v) SDS. The technique of adding SDS has been used extensively by many research groups for dissociating ligands from nucleic acid structures (46). Dissociation of the ligand from the DNA is critical to ensure that there are no complexities arising from differences in the optical properties of free and bound ligands. The total ligand concentration (Ct) within each dialysis well was determined using a fluorescence standard curve for each tested ligand. Appropriate corrections were made for the small dilution resulting from the addition of the SDS stock solution. The free ligand concentration (Cf) was determined from an aliquot of the dialysate solution, which typically did not vary significantly from the initial 1 μM concentration. Fluorescence measurements were made using a Safire microplate reader (Tecan US, Durham, NC, USA), with the following parameters: excitation and emission bandwidth, 10 nm, gain: 100. Compound OZ-85H: exc./emission 260/494 nm, compound MHQ-15: exc./emission 260/437 nm, compound 1 exc./emission 260/490 nm, compound 2: exc./emission 348/446 nm. The bound ligand concentration (Cb) was then determined by:
| 1 |
Binding constants, specificity sums (SS) and the ratio Cb/SS were calculated as previously described (46) and are provided in Supplementary Equation (S1).
CD titration and thermal denaturation methods
CD titrations were done as previously described (47), using a Jasco 810 spectropolarimeter. Instrument settings were: wavelength range (220–500 nm), scan rate (100 nm min−1), averaging time (0.125 s), bandwidth (1 nm), number of scans (2), temperature (20°C). The effect of ligands on the thermal denaturation of triplex DNA was studied using the exact protocol described previously (46).
RESULTS AND DISCUSSION
Virtual screening
The initial step in virtual screening was performing Surflex-Sim to determine which of the ligands in the library were most structurally similar to the known, triplex selective intercalator MHQ-12. Of the approximately 2 million ligands screened for similarity against MHQ-12, 350 ligands had a Surflex-Sim score of greater than 0.70 (range 0.875–0.704) and were selected for Surflex-Docking studies. A cutoff Surflex-Sim score of 0.70 was selected based on previous studies which suggested that this is the lowest score where the ligand structure–function relationship is typically maintained (25). The next step in the virtual screening process involved performing Surflex-Dock studies with the top 350 ranked Surflex-Sim ligands using the intercalation site and the three grooves (major–major, major–minor and minor) of the triplex as individual docking targets. Interestingly, MHQ-12 has the top Surflex Raw Score out of all 350 ligands that were docked to the intercalation site, which directly supports the ability of Surflex-Dock to successfully dock and rank a known selective triplex intercalator. We propose a new metric to evaluate the Surflex-Dock results, the ‘Normalized Surflex Raw Score (NSRS)’. The rationale behind the normalization of the Surflex Raw Score is that the score for a ligand binding to a single site on a target measures only the interaction with that one site. However, a ligand may have multiple interaction sites on a particular target. Therefore, for selectivity for a particular mode of binding, it is crucial to determine the binding of the ligand to the site of interest relative to the binding to other potential sites on the target. Since ligands interact with nucleic acids typically through either the groove-binding or intercalation, protomols were constructed at the three grooves and the intercalation site (48). Binding of the ligand to the intercalation site relative to binding in the three grooves embodies the ‘normalized’ affinity and specificity of the ligand for triplex intercalation. The following metric determines the NSRS for the intercalation site for each of the top 350 Surflex-Dock results:
 |
2 |
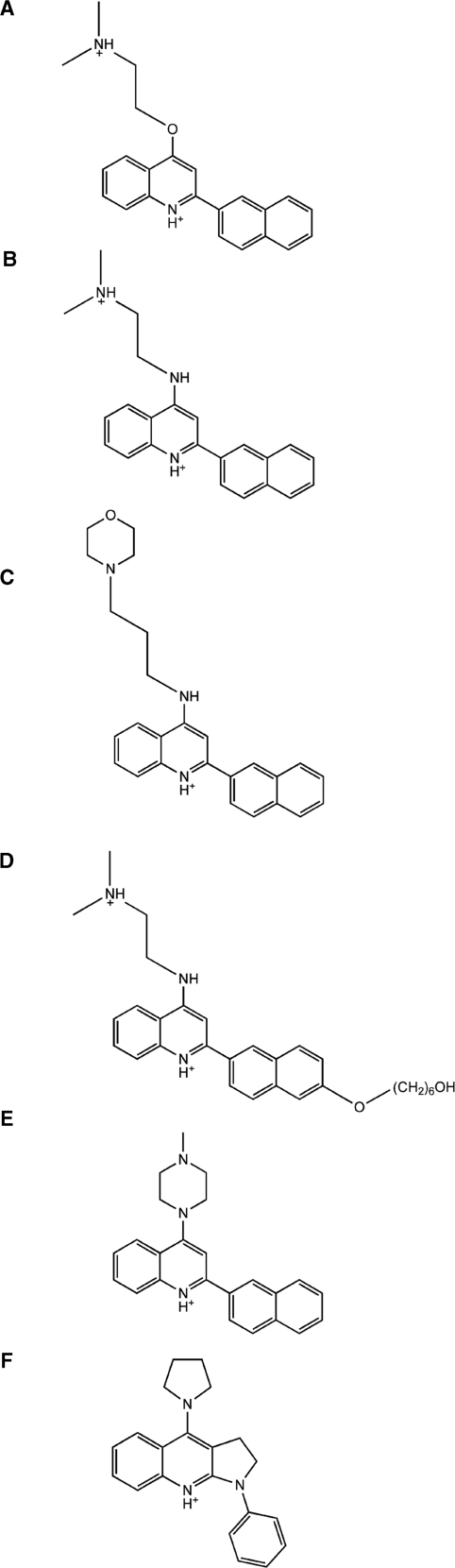
Ranking of the 350 intercalation site Surflex-Dock results by NSRS shows that only three ligands have a higher NSRS score than MHQ-12 (NSRS of 6.8) (Figure 1A). The ligands are LS-08 (49) (Figure 1B), compounds 1 (Figure 1E) and 2 (Figure 1F) and have NSRS values of 7.03, 7.34 and 7.39, respectively (Figure 1). Interestingly, LS-08 (Figure 1B) which was identified by our virtual screening methodology, was previously tested by Chaires (46) and shown to be highly triplex selective, which adds validity to our virtual screening approach used to identify triplex selective ligands. Based on the NSRS values, compounds 1 (Figure 1E) and 2 (Figure 1F) were hypothesized to have superior affinity and selectivity for binding to the triplex nucleic acid, and were tested by competition dialysis. Two known triplex selective compounds, MHQ-15 (Figure 1C) and OZ-85H (Figure 1D) served as positive controls, as these compounds have been extensively studied and characterized (18). Biophysical characterization was performed by circular dichroism and thermal denaturation to assess the ability of the compounds to intercalate into the DNA triplex.
Figure 1.
Chemical structures of the ligands used in virtual screening and competition dialysis experiments. (A) MHQ-12, (B) LS-08, (C) MHQ-15, (D) OZ-85H, (E) compound 1 and (F) compound 2.
Competition dialysis
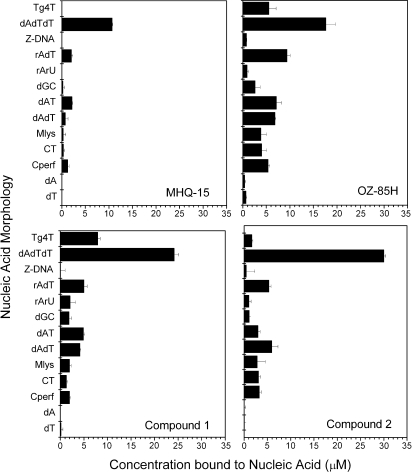
The results of the competition dialysis experiments are shown in Figure 2. It is visually apparent that compounds 1 and 2 have a much higher affinity for the TAT triplex than the two positive control reference compounds, MHQ-15 and OZ-85H. The competition dialysis results for MHQ-12 have previously been described in detail (18), and this compound has an SS of 1.32 and a Cmax/SS ratio of 8.93. Determination of the SS (Table 1) for compounds 1 and 2 demonstrates superior triplex selectivity compared to OZ-85H but slightly less selectivity than MHQ-12 and MHQ-15. However, the significantly higher binding affinities of compounds 1 and 2 translate to much higher Cmax/SS values than MHQ-12, MHQ-15 or OZ-85H. The Cmax/SS ratio for compounds 1 and 2 is significant as it suggests that compounds 1 and 2 have a superior combination of binding affinity and selectivity compared to the reference compounds. These results validate the virtual screening approach, and show that the method can be used to identify compounds with high affinity and selectivity for a target nucleic acid, in this case the DNA TAT triplex.
Figure 2.
Competition dialysis results for MHQ-15, OZ-85H, compounds 1 and 2. The concentration of bound ligand to each nucleic acid structure in the array is shown.
Table 1.
Competition dialysis metric results for the positive controls, MHQ-15, OZ-85H and the virtual screening top results, compounds 1 and 2
| Test ligand | Cb (μM) | Kapp/105 (M−1) | SS | Cmax/SS (μM) |
|---|---|---|---|---|
| MHQ-12a | 11.8 | 1.87 | 1.32 | 8.93 |
| MHQ-15 | 10.7 | 1.7 | 1.66 | 6.44 |
| OZ-85H | 17.6 | 3.1 | 3.69 | 4.77 |
| Compound 1 | 24.2 | 4.8 | 2.30 | 10.47 |
| Compound 2 | 30.0 | 6.7 | 1.92 | 15.63 |
aThese data are included from a previous publication (18).
Circular dichroism
The interaction of compounds 1 and 2 with DNA was studied by circular dichroism (Figure 3). Both compounds show pronounced induced circular dichroism (ICD) in the presence of triplex DNA. The ICD is in a spectral range where the compounds absorb light but the DNA does not. This ICD is unambiguous proof of the ligand binding to triplex DNA. For both compounds 1 and 2, the ICD is negative in sign, and relatively weak in magnitude. Such behavior is consistent with an intercalative binding mode, although the mode of binding can only be definitively established by high-resolution experimental structural analysis (50).
Figure 3.
Induced circular dichroism results for (A) compound 1 and (B) compound 2. (A) Spectra are shown for a ligand concentration of 45 μM in the presence of triplex DNA ranging from 5 μM to 450 μM triplets. (B) Spectra are shown for a ligand concentration of 22.5 μM in the presence of triplex DNA ranging from 2.25 μM to 225 μM triplets.
Thermal denaturation studies
Figure 4 shows the effects of compounds 1 and 2 on the thermal denaturation of the TAT triplex. In the absence of added ligand, two transitions are seen, corresponding to the melting of the third strand (∼30°C) and the duplex (∼70°C). Titration with both ligands results in a clear elevation of the first transition, indicating stabilization of the triplex. The effect is maximal at saturating concentrations of ligand (1:1, ligand:triplet), where melting of the triplex coalesces with duplex melting. Melting of the triplex is stabilized by ∼40°C indicating tight binding of both compounds. Neither compound 1 nor compound 2 alters the transition temperature of the duplex form to any appreciable extent, an observation that is fully consistent with the weak binding to duplex seen in competition dialysis experiments (Figure 2).
Figure 4.
Thermal melting results for (A) compound 1 and (B) compound 2. Derivative melting curves were obtained using 32 μM triplex DNA and ligand concentrations ranging from 0 to 16 μM (A) or 0 to 32 μM (B). The peak near 30°C is for the melting of the third stand, while that near 70°C is for melting of the duplex.
Validation of QSAR
In the previous study of naphthylquinoline binding to triplex DNA (18), a QSAR was derived from competition dialysis binding data. The best three-term QSAR to emerge was:
 |
3 |
In this relationship, log Kapp is the logarithm of the apparent binding constant (Table 1), SASA is the total solvent accessible surface area in Å2, EA is electron affinity in eV and HBa is the number of hydrogen bond acceptors. The physical meaning of this is as follows. As SASA increases, log Kapp increases in magnitude, indicating higher affinity for triplex DNA. Increases in the magnitudes of EA and HBa result in decreasing binding affinity. Increasing the solvent-accessible surface areas of naphthylquinoline compounds results in higher affinity for the triplex. Greater electron affinity and more hydrogen bond acceptors reduce the affinity of naphthylquinolines for triplex DNA.
Binding data obtained for compounds 1 and 2 in this study validate the published QSAR. The molecular descriptors SASA, EA and HBa were calculated using QikProp, and substituted into Equation (3). For compound 1, log Ka = 5.07 was predicted, compared to a measured value of log Kobs = 5.68. For compound 2, log K-values of 5.18 and 5.82 were calculated and observed, respectively. The differences in calculated and observed values correspond to a factor of about four in binding constants, an acceptable agreement for predictions from a QSAR.
Triplex-binding ligands
While these newly reported compounds have some of the tightest binding properties found to date by competition dialysis, it is worth considering limitations to this platform. One consideration is that the array of nucleic acids used in the assay is not all-encompassing and represents only a broad number of structural and sequence variants. Certain morphologies of nucleic acids to which ligands may bind may not be present in the array. To address this, current efforts are underway to expand the assay to include a larger number of nucleic acid solutions. This limitation also highlights however that a key benefit of competition dialysis is the ability to customize any number of targets for the assay. As new potentially therapeutic nucleic acid structures are discovered, they can be added to the assay and screened using this platform. Another consideration from these results is that the assay remains particularly relevant as an in vitro test, but may not necessarily reflect in vivo conditions and ligand binding behavior. This is particularly true when considering differences in the abundance of various forms of nucleic acids. For example, because of the abundance of duplex DNA, the observed approximately 5-fold difference in binding of compound 2 to the TAT triplex may be insufficient to overcome non-specific binding of the ligand to duplex DNA in vivo. However, this may be irrelevant if ligands are discovered that bind with true selectivity and high affinity to only a single nucleic acid structure. While these are some relevant considerations when using this platform, the approach as demonstrated here is valuable for identifying potential small molecules with desirable binding properties for a single nucleic acid structure.
CONCLUSION
This work demonstrates a novel strategy for discovering small molecules that can selectively bind to the triplex nucleic acid, poly(dA)-[poly(dT)]2. Through the combination of virtual screening by Surflex and experimental validation by competition dialysis, compounds 1 and 2 were discovered. These compounds have the highest overall affinities and selectivities reported for triplex binders as determined by competition dialysis. Further biophysical characterization by circular dichroism and thermal melting confirmed the mechanism of action of these new compounds and verified the predictive nature of the virtual screening methodologies. Several aspects of the virtual screening results are noteworthy. First, the combination of a ligand-based (Surflex-Sim) with a structure-based approach (Surflex-Dock) proved to be a powerful and highly computationally efficient way to identify triplex selective small molecules, as Surflex-Sim is two orders of magnitude faster than Surflex-Dock. Second, our development of the NSRS metric, which can predict a particular mode of binding of triplex selective ligands with both similar and different (scaffold hopping) chemical scaffolds. This is significant as it has the potential to identify new classes of small molecules that may have much higher affinity and selectivity for a given nucleic acid target. Future work will focus on extending this integrated virtual and actual screening platform to target other nucleic acid structures that may hold medicinal value and physiological relevance.
FUNDING
National Institutes of Health [1R01GM077422]; James Graham Brown Foundation. Funding for open access charge: National Institutes of Health [1R01GM077422].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Chin JY, Schleifman EB, Glazer PM. Repair and recombination induced by triple helix DNA. Front. Biosci. 2007;12:4288–4297. doi: 10.2741/2388. [DOI] [PubMed] [Google Scholar]
- 2.Seidman MM, Puri N, Majumdar A, Cuenoud B, Miller PS, Alam R. The development of bioactive triple helix-forming oligonucleotides. Ann. NY Acad. Sci. 2005;1058:119–127. doi: 10.1196/annals.1359.020. [DOI] [PubMed] [Google Scholar]
- 3.Kalota A, Shetzline SE, Gewirtz AM. Progress in the development of nucleic acid therapeutics for cancer. Cancer Biol. Ther. 2004;3:4–12. doi: 10.4161/cbt.3.1.517. [DOI] [PubMed] [Google Scholar]
- 4.Seidman MM, Glazer PM. The potential for gene repair via triple helix formation. J. Clin. Invest. 2003;112:487–494. doi: 10.1172/JCI19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchini S, Leumann CJ. Recent improvements in antigene technology. Curr. Opin. Chem. Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Kim HG, Reddoch JF, Mayfield C, Ebbinghaus S, Vigneswaran N, Thomas S, Jones DE, Jr., Miller DM. Inhibition of transcription of the human c-myc protooncogene by intermolecular triplex. Biochemistry. 1998;37:2299–2304. doi: 10.1021/bi9718191. [DOI] [PubMed] [Google Scholar]
- 7.Carbone GM, McGuffie E, Napoli S, Flanagan CE, Dembech C, Negri U, Arcamone F, Capobianco ML, Catapano CV. DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene. Nucleic Acids Res. 2004;32:2396–2410. doi: 10.1093/nar/gkh527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacoste J, Francois JC, Helene C. Triple helix formation with purine-rich phosphorothioate-containing oligonucleotides covalently linked to an acridine derivative. Nucleic Acids Res. 1997;25:1991–1998. doi: 10.1093/nar/25.10.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchand C, Bailly C, Nguyen CH, Bisagni E, Garestier T, Helene C, Waring MJ. Stabilization of triple helical DNA by a benzopyridoquinoxaline intercalator. Biochemistry. 1996;35:5022–5032. doi: 10.1021/bi952908l. [DOI] [PubMed] [Google Scholar]
- 10.Moraru-Allen AA, Cassidy S, Asensio Alvarez JL, Fox KR, Brown T, Lane AN. Coralyne has a preference for intercalation between TA.T triples in intramolecular DNA triple helices. Nucleic Acids Res. 1997;25:1890–1896. doi: 10.1093/nar/25.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keppler M, Zegrocka O, Strekowski L, Fox KR. DNA triple helix stabilisation by a naphthylquinoline dimer. FEBS Lett. 1999;447:223–226. doi: 10.1016/s0014-5793(99)00284-7. [DOI] [PubMed] [Google Scholar]
- 12.Baudoin O, Marchand C, Teulade-Fichou M-P, Vigneron J-P, Sun J-S, Garestier T, Helene C, Lehn J-M. Stabilization of DNA triple helices by crescent-shaped dibenzophenanthrolines. Chem. Eur. J. 1998;4:1504–1508. [Google Scholar]
- 13.Ren J, Bailly C, Chaires JB. NB-506, an indolocarbazole topoisomerase I inhibitor, binds preferentially to triplex DNA. FEBS Lett. 2000;470:355–359. doi: 10.1016/s0014-5793(00)01335-1. [DOI] [PubMed] [Google Scholar]
- 14.Strekowski L, Hojjat M, Wolinska E, Parker AN, Paliakov E, Gorecki T, Tanious FA, Wilson WD. New triple-helix DNA stabilizing agents. Bioorg. Med. Chem. Lett. 2005;15:1097–1100. doi: 10.1016/j.bmcl.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Cassidy SA, Strekowski L, Fox KR. DNA sequence specificity of a naphthylquinoline triple helix-binding ligand. Nucleic Acids Res. 1996;24:4133–4138. doi: 10.1093/nar/24.21.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keppler MD, James PL, Neidle S, Brown T, Fox KR. DNA sequence specificity of triplex-binding ligands. Eur. J. Biochem. 2003;270:4982–4992. doi: 10.1046/j.1432-1033.2003.03901.x. [DOI] [PubMed] [Google Scholar]
- 17.Strekowski L, Say M, Zegrocka O, Tanious FA, Wilson WD, Manzel L, Macfarlane DE. Bis-4-aminoquinolines: novel triple-helix DNA intercalators and antagonists of immunostimulatory CpG-oligodeoxynucleotides. Bioorg. Med. Chem. 2003;11:1079–1085. doi: 10.1016/s0968-0896(02)00525-4. [DOI] [PubMed] [Google Scholar]
- 18.Chaires JB, Ren J, Henary M, Zegrocka O, Bishop GR, Strekowski L. Triplex selective 2-(2-naphthyl)quinoline compounds: origins of affinity and new design principles. J. Am. Chem. Soc. 2003;125:7272–7283. doi: 10.1021/ja034181r. [DOI] [PubMed] [Google Scholar]
- 19.Kapetanovic IM. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem. Biol. Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Shafer RH, Kuntz ID. Structure-based discovery of ligands targeted to the RNA double helix. Biochemistry. 1997;36:11402–11407. doi: 10.1021/bi970756j. [DOI] [PubMed] [Google Scholar]
- 21.Evans DA, Neidle S. Virtual screening of DNA minor groove binders. J. Med. Chem. 2006;49:4232–4238. doi: 10.1021/jm0601957. [DOI] [PubMed] [Google Scholar]
- 22.Holt PA, Chaires JB, Trent JO. Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J. Chem. Inf. Model. 2008;48:1602–1615. doi: 10.1021/ci800063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain AN. Ligand-based structural hypotheses for virtual screening. J. Med. Chem. 2004;47:947–961. doi: 10.1021/jm030520f. [DOI] [PubMed] [Google Scholar]
- 24.Cleves AE, Jain AN. Robust ligand-based modeling of the biological targets of known drugs. J. Med. Chem. 2006;49:2921–2938. doi: 10.1021/jm051139t. [DOI] [PubMed] [Google Scholar]
- 25.Cleves AE, Jain AN. Effects of inductive bias on computational evaluations of ligand-based modeling and on drug discovery. J. Comput. Aided Mol. Des. 2008;22:147–159. doi: 10.1007/s10822-007-9150-y. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 27.Chaires JB. Competition dialysis: an assay to measure the structural selectivity of drug-nucleic acid interactions. Curr. Med. Chem. Anticancer Agents. 2005;5:339–352. doi: 10.2174/1568011054222292. [DOI] [PubMed] [Google Scholar]
- 28.Ragazzon PA, Garbett NC, Chaires JB. Competition dialysis: a method for the study of structural selective nucleic acid binding. Methods. 2007;42:173–182. doi: 10.1016/j.ymeth.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrasco C, Rosu F, Gabelica V, Houssier C, De Pauw E, Garbay-Jaureguiberry C, Roques B, Wilson WD, Chaires JB, Waring MJ, et al. Tight binding of the antitumor drug ditercalinium to quadruplex DNA. Chembiochem. 2002;3:1235–1241. doi: 10.1002/1439-7633(20021202)3:12<1235::AID-CBIC1235>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Shi X, Chaires JB. Sequence- and structural-selective nucleic acid binding revealed by the melting of mixtures. Nucleic Acids Res. 2006;34:e14. doi: 10.1093/nar/gnj012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy PM, Phillips VA, Jennings SA, Garbett NC, Chaires JB, Jenkins TC, Wheelhouse RT. Biarylpyrimidines: a new class of ligand for high-order DNA recognition. Chem. Commun. 2003;10:1160–1161. doi: 10.1039/b301554h. [DOI] [PubMed] [Google Scholar]
- 33.Chaires JB, Ren J, Hamelberg D, Kumar A, Pandya V, Boykin DW, Wilson WD. Structural selectivity of aromatic diamidines. J. Med. Chem. 2004;47:5729–5742. doi: 10.1021/jm049491e. [DOI] [PubMed] [Google Scholar]
- 34.Xing F, Song G, Ren J, Chaires JB, Qu X. Molecular recognition of nucleic acids: coralyne binds strongly to poly(A) FEBS Lett. 2005;579:5035–5039. doi: 10.1016/j.febslet.2005.07.091. [DOI] [PubMed] [Google Scholar]
- 35.Ren J, Qu X, Dattagupta N, Chaires JB. Molecular recognition of a RNA:DNA hybrid structure. J. Am. Chem. Soc. 2001;123:6742–6743. doi: 10.1021/ja015649y. [DOI] [PubMed] [Google Scholar]
- 36.Arya DP, Xue L, Tennant P. Combining the best in triplex recognition: synthesis and nucleic acid binding of a BQQ-neomycin conjugate. J. Am. Chem. Soc. 2003;125:8070–8071. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 37.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trent JO. Molecular modeling of drug-DNA complexes: an update. Methods Enzymol. 2001;340:290–326. doi: 10.1016/s0076-6879(01)40428-9. [DOI] [PubMed] [Google Scholar]
- 39.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 40.Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput. Aided Mol. Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 41.Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil CR. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br. J. Pharmacol. 2008;153(Suppl 1):S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strekowski L, Gulevich Y, Baranowski TC, Parker AN, Kiselyov AS, Lin SY, Tanious FA, Wilson WD. Synthesis and structure-DNA binding relationship analysis of DNA triple-helix specific intercalators. J. Med. Chem. 1996;39:3980–3983. doi: 10.1021/jm9603734. [DOI] [PubMed] [Google Scholar]
- 43.Chaires JB. Structural Selectivity of Drug-Nucleic Acid Interactions Probed by Competition Dialysis. Heidleberg: Springer-Verlag GMBH & Co.; 2005. [DOI] [PubMed] [Google Scholar]
- 44.Chaires JB, Dattagupta N, Crothers DM. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on interaction of daunomycin with deoxyribonucleic acid. Biochemistry. 1982;21:3933–3940. doi: 10.1021/bi00260a005. [DOI] [PubMed] [Google Scholar]
- 45.Moller A, Nordheim A, Kozlowski SA, Patel DJ, Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984;23:54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- 46.Chaires JB. A competition dialysis assay for the study of structure-selective ligand binding to nucleic acids. In: Beaucage SL, editor. Current Protocols in Nucleic Acid Chemistry. New York, NY: John Wiley & Sons, Inc.; 2003. Chapter 8. Unit 8.3. [DOI] [PubMed] [Google Scholar]
- 47.Garbett NC, Ragazzon PA, Chaires JB. Circular dichroism to determine binding mode and affinity of ligand-DNA interactions. Nat. Protoc. 2007;2:3166–3172. doi: 10.1038/nprot.2007.475. [DOI] [PubMed] [Google Scholar]
- 48.Chaires JB. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Wilson WD, Zhao M, Patterson SE, Wydra RL, Janda L, Strekowski L, Schinazi RF. Design of RNA interactive anti-HIV agents: unfused aromatic intercalators. Med. Chem. Res. 1992;2:102–110. [Google Scholar]
- 50.Eriksson M, Norden B. Linear and circular dichroism of drug-nucleic acid complexes. Methods Enzymol. 2001;340:68–98. doi: 10.1016/s0076-6879(01)40418-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.