Abstract
Regulation of expression of the CFTR gene is poorly understood. Elements within the basal promoter of the gene do not fully explain CFTR expression patterns, suggesting that cis-regulatory elements are located elsewhere, either within the locus or in adjacent chromatin. We previously mapped DNase I hypersensitive sites (DHS) in 400 kb spanning the CFTR locus including a cluster of sites close to the 3′-end of the gene. Here we focus on a DHS at +6.8 kb from the CFTR translation end-point to evaluate its potential role in regulating expression of the gene. This DHS, which encompasses a consensus CTCF-binding site, was evident in primary human epididymis cells that express abundant CFTR mRNA. We show by DNase I footprinting and electophoretic mobility shift assays that the cis-regulatory element within this DHS binds CTCF in vitro. We further demonstrate that the element functions as an enhancer blocker in a well-established in vivo assay, and by using chromatin immunoprecipitation that it recruits CTCF in vivo. Moreover, we reveal that in primary epididymis cells, the +6.8 kb DHS interacts closely with the CFTR promoter, suggesting that the CFTR locus exists in a looped conformation, characteristic of an active chromatin hub.
INTRODUCTION
The cystic fibrosis transmembrane conductance regulator (CFTR) gene is located at chromosome 7q31.2, where it is flanked upstream by ASZ1 (ankyrin repeat, SAM and basic leucine zipper) and downstream by CTTNBP2 (cortactin binding protein 2). These neighbouring genes have very different expression profiles: CFTR is expressed primarily in specialized epithelial cells (1–3), while ASZ1 is transcribed exclusively in the testis and ovary (4), and CTTNBP2 is highly expressed in the brain, kidney and pancreas, with lower levels of expression in other tissues (5). We previously identified two enhancer-blocking insulators 5′ and 3′ to the CFTR gene that had distinct properties. First, a DNase I hypersensitive site (DHS) located at −20.9 kb with respect to the translation start site was associated with a classical CTCF-dependent insulator element (6). CTCF, a ubiquitously expressed, zinc finger DNA-binding protein (7,8) often establishes independently regulated domains of gene activity. A second element, located 3′ to CFTR, within a DHS at +15.6 kb also demonstrated enhancer-blocking activity but this was independent of CTCF-binding. The +15.6 kb DHS was marked by a peak of euchromatin-specific histone modifications, unlike the −20.9 kb DHS (6), supporting the hypothesis that these elements function by different mechanisms.
CFTR exhibits tightly regulated expression, both temporally during development, and spatially in different tissue types (1,9,10). However, somewhat paradoxically, the CFTR promoter resembles that of a house-keeping gene, in that it is CpG rich, contains no TATA box, has multiple transcription start sites and has several putative binding sites for the transcription factor Sp1 (11). Consistent with promoters of this type, the CFTR promoter demonstrates no apparent tissue-specificity, suggesting the involvement of distal regulatory elements in control of CFTR expression. It is probable that these elements are associated with DHS across 400 kb encompassing the CFTR locus (12–15). In addition to the prominent site at +15.6 kb other DHS were evident 3′ to the coding region of the gene, in particular, a complex cluster of sites at +5.4 kb, +6.8 kb, +7.0 kb and +7.4 kb from the CFTR translation end-point (13). The DHS at +5.4 kb and +7.0 kb were observed in a variety of cell types, irrespective of CFTR expression; however, the DHS at +6.8 kb and +7.4 kb were only found in a restricted number of CFTR-expressing cell-types, including primary epididymis cells, suggesting that they may contain tissue-specific regulatory elements that participate in controlling CFTR-expression (13).
Here we demonstrate that the +6.8 kb DHS is associated with a tissue-specific CTCF-binding site that displays enhancer-blocking activity comparable to that of other known insulator elements, including the one at the CFTR −20.9 kb DHS. CTCF is thought to be involved in regulating nuclear organisation and CTCF-dependent chromatin loops exist (16–18), which may depend on tethering to the nuclear matrix (19,20) and/or association with cohesins (21–23). Hence, we next evaluated the three-dimensional structure of the CFTR locus in primary cells that exhibit the +6.8 kb DHS and express CFTR. Using chromosome conformation capture (3C), we demonstrate that a region encompassing the +6.8 kb DHS shows strong interaction with the CFTR promoter in these cells. We predict that looping of CFTR, possibly induced by CTCF, enables key regulatory elements at the 3′-end of the gene to move into close proximity with the CFTR promoter, so activating cell-type-specific expression.
MATERIALS AND METHODS
Cell culture
The K562 erythroleukemia cell line (24) was cultured in RPMI 1640 supplemented with 10% fetal calf serum (FCS). The Caco2 (25) cell line was cultured in DMEM supplemented with 10% FCS. Primary human fetal male epididymis cells (26) were cultured in CMRL1066 medium, 15% FCS, supplemented with hydrocortisone, insulin and cholera toxin. Human skin fibroblasts (GM08333) were cultured in MEM medium with 15% FCS.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out following the Upstate protocol with minor modifications. Briefly, 5 × 107 cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Crosslinking was stopped by the addition of Glycine to 0.125 M. Cells were washed in cold phosphate-buffered saline (PBS) containing protease inhibitors (Roche) and lysed in 1 ml of 1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1) plus protease inhibitors. Sonication was carried out to produce fragments of 1 kb or under.
Immunoprecipitations were performed overnight at 4°C with 10 μl of a CTCF-specific antibody (Upstate 07-729) and 200 μl of chromatin (corresponding to 1 × 107 cells), and complexes were collected with protein A agarose beads for 1 h. No antibody samples were prepared, in which chromatin was incubated with protein A agarose beads alone. Immunoprecipitations were washed, DNA eluted and cross-links reversed according to the Upstate protocol.
All immunoprecipitations were performed in duplicate or triplicate. Immunoprecipitated and 1/10 diluted input DNA samples were used as templates for Taqman qPCR. Primer and probes sets corresponding to regions of interest within the CFTR locus were designed using primer express 1.0 software (Supplementary Data) and obtained from Eurogentec or IDT. Reactions were carried out following the ABI protocol and performed in triplicate.
Enhancer-blocking activity assay
The plasmids pNI and pNI-FII were a kind gift from the Felsenfeld group (27). Enhancer-blocking assays were performed as previously described (27,28). G418-resistant colonies were counted after 2–3 weeks of selection and data subjected to statistical analysis by one-way ANOVA followed by Dunnett's multiple comparison test.
Electrophoretic mobility shift assays (EMSAs)
Caco2 nuclear extracts were prepared by standard methods (29). In vitro translated (IVT) CTCF was made by in vitro transcription and translation of pCTCF (30) using the TNT-Kit (Promega) and following manufacturer's instructions. EMSA experiments were carried out as described previously (6). Antibody supershift reactions were performed using anti-CTCF (Upstate 07-729) or anti-RARα (sc-773×).
In vitro DNase I footprinting
The +6.8 kb DHS was PCR-amplified using primers 6.8B-F (AAGAACATTATGAAAGGTGGTC; AC000061: 64235–64256) and 6.8B-R (AAGATAAAATGTCTTTGAGATT; AC000061: 64510–64489), and then cloned into pCRscript (Invitrogen). Following excision by either EcoRV/NotI or ClaI/SacII digestion, Klenow DNA polymerase fill-in with [α-32P]-dCTP was used to label either the sense or anti-sense strand, respectively. DNase I footprinting reactions were performed as described previously (31).
Chromosome conformation capture (3C)
3C was performed as described previously (32,33), with minor modifications. Briefly, 1 × 107 cells were fixed with 2% formaldehyde for 10 min at room temperature. Cells were lysed in 5 ml cold lysis buffer [10 mM Tris (pH 8), 10 mM NaCl, 0.2% NP-40, 1× protease inhibitor cocktail (Roche)] and the nuclei collected by centrifugation. Following extraction with 0.3% SDS, chromatin was digested overnight with 2000 U HindIII. Ligations were performed in a total reaction volume of 6.5 ml, using 100 U T4 DNA ligase (Roche) and incubation at 14°C for 4 h followed by 30 min at room temperature. Cross-links were reversed by proteinase K treatment at 65°C overnight. Samples were purified by phenol–chloroform extraction followed by ethanol precipitation, and then re-suspended in 150 μl H2O. The concentration of each sample was determined by SYBR green qPCR, using the B13F/B13R primer set (amplicon found within a HindIII fragment; see Supplementary Data) and comparison to a genomic DNA reference of known concentration. Samples were subsequently diluted to a concentration of 100 ng/μl.
A Taqman probe and reverse primer was designed that was specific to a HindIII fragment at the bait region of interest (i.e. the CFTR promoter). Multiple forward primers were then designed that were each specific to different HindIII fragments across the CFTR locus (see Supplementary Data for primer and probe sequences and locations). Using a dilution series of digested/re-ligated BAC DNA template, each forward primer was demonstrated to function with the ‘fixed’ Taqman probe and reverse primer to amplify with 100% efficiency. To quantify ligation events within 3C samples, 200 ng of 3C template was used per 20 μl Taqman qPCR reaction. The ligation efficiency (or ‘interaction frequency’) between each fragment and the CFTR promoter was corrected for the interaction between two HindIII fragments within the ubiquitously expressed excision repair cross-complementing rodent repair deficiency, complementation group 3 (ERCC3) locus, which has been reported to adopt the same spatial conformation in different tissues (16,34,35).
RESULTS
A cluster of four DHS was previously identified at the 3′-end of the CFTR gene, at +5.4 kb, + 6.8 kb, +7.0 kb and +7.4 kb from the CFTR translation termination site (13). The DHS at +5.4 kb and +7.0 kb were observed in a variety of cell types, irrespective of CFTR expression; however, the DHS at +6.8 kb and +7.4 kb were only found in a restricted number of CFTR-expressing cell-types, including primary human epididymis cells.
A predicted CTCF-binding site within the +6.8 kb DHS corresponds to a 60-bp DNase I footprint
An in silico search for regulatory factors that could bind to the +5.4 kb, +6.8 kb, +7.0 kb and +7.4 kb DHS regions of the CFTR locus (AC000061 62735–65135), revealed a site within the +6.8 kb DHS region that matched the consensus sequence for CTCF binding (7,30) at 13 out of 14 bp (AC000061 64412–64399) (Table 1).
Table 1.
Potential CTCF-binding site within the +6.8 kb DHS region
 |
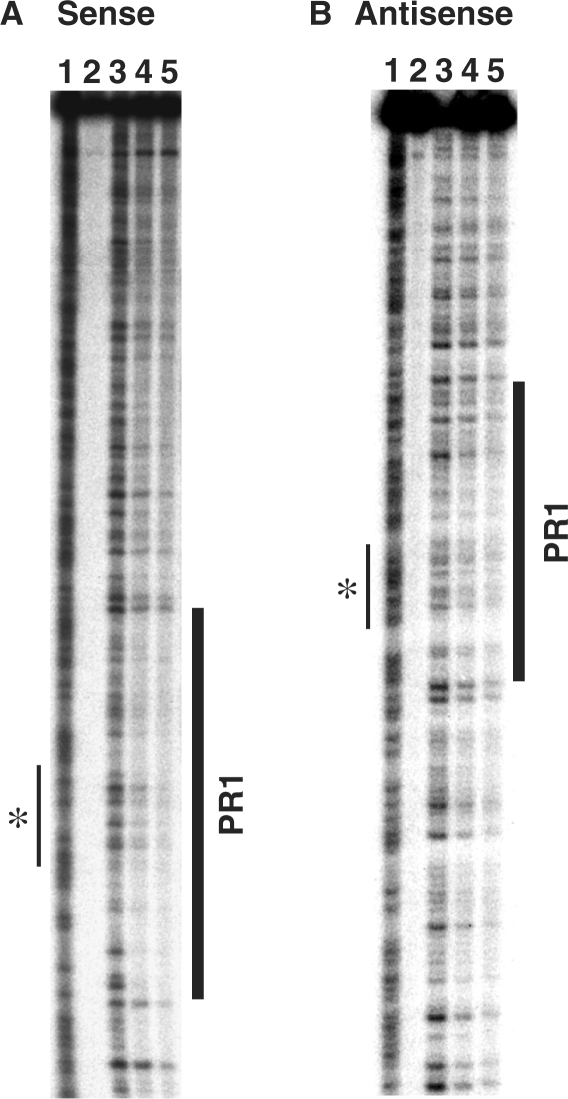
A 275-bp fragment spanning the +6.8 kb DHS (AC000061 64235–64510) was labelled at the 3′-end of either the sense or the anti-sense strand and used as a template for DNase I footprinting. In the presence of Caco2 cell nuclear extract, the +6.8 kb DHS sense strand exhibited protection from DNase I digestion (PR1) over 60 bp, at AC000061 64373–64436 (Figure 1A and Supplementary Figure 1). A weaker protected region was also evident at the corresponding location on +6.8 kb DHS anti-sense strand (Figure 1B). Extensive DNase I footprints are characteristic of those produced by CTCF binding (36), and furthermore, the putative site of interaction with CTCF identified in silico was centrally positioned within PR1 (Figure 1).
Figure 1.
In vitro DNase I footprinting of the +6.8 kb DHS probe. Experiments using (A) sense and (B) anti-sense strands are shown. Both gel images are labelled as follows: 1, AG ladder; 2, No DNase I; 3, No Caco2 nuclear extract; 4 and 5, 40 μg and 80 μg Caco2 cell nuclear extract, respectively. For both panels, protected region 1 (PR1) is highlighted by a bold vertical line. Narrow vertical line marked with an asterisk shows position of putative CTCF-binding site.
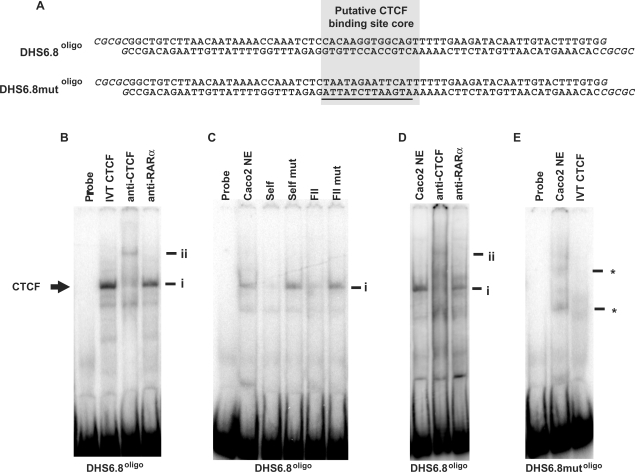
CTCF binds to the +6.8 kb DHS sequence in vitro
In vitro binding of CTCF at the putative +6.8 kb DHS-binding site (PR1), was investigated by EMSA. A 78-bp oligonucleotide probe, DHS6.8oligo, was generated that spanned the putative +6.8 kb DHS CTCF-binding site (Figure 2A and Supplementary Figure 1). DHS6.8mutoligo, had the same sequence with the exception of alterations in the 14-bp putative CTCF-binding site core which was positioned centrally within DHS6.8oligo (Figure 2A). EMSA experiments with the DHS6.8oligo probe demonstrated that this sequence binds IVT CTCF (Figure 2B, i) and that the complex formed was supershifted with an antibody specific to CTCF (Figure 2B, ii), but not an isotype control antibody (anti-RARα). DHS6.8oligo also formed a complex with Caco2 nuclear extract that was of the same mobility as that formed with IVT CTCF (Figure 2C, i). The complex formed between DHS6.8oligo and Caco2 nuclear extract was competed by unlabelled self, but not by DHS6.8mutoligo (Figure 2C). The complex was also competed by FII (a known CTCF-binding site from the chicken β-globin locus), but not by a mutant version of this probe (FII mut) (Figure 2C). Furthermore, the complex formed between DHS6.8oligo and Caco2 nuclear extract was also supershifted by an antibody specific to CTCF (Figure 2D, ii), but not by an isotype control antibody. When the DHS6.8mutoligo probe was used in EMSA experiments, the CTCF complex was not observed with either Caco2 nuclear extract (Figure 2E) or IVT CTCF (data not shown). In the presence of Caco2 nuclear extract, DHSmutoligo exhibited two weak complexes (*), possibly due to CTCF site mutation creating novel transcription factor binding sites. However, given the lack of interaction between DHS6.8mutoligo and IVT CTCF, these novel complexes are likely to involve proteins other than CTCF.
Figure 2.
In vitro binding of CTCF at the +6.8 kb DHS region. (A) The DHS6.8oligo and DHS6.8mutoligo probes. The putative CTCF-binding site core is highlighted. Oligonucleotides were designed to form BssHII sticky ends when annealed (shown in italics), facilitating cloning into the AscI site of pNI. Mutated bases in DHS6.8mutoligo are underlined. (B) IVT CTCF; (C, D), Caco2 nuclear extracts. (B) EMSA using DHS6.8oligo. Supershift was performed with an anti-CTCF antibody and anti-RAR β was used as isotype control. (C) EMSA with DHS6.8oligo. Competition reactions were performed with 100× excess of unlabelled DHS6.8oligo (self), DHS6.8mutoligo (self mut), FII (known CTCF-binding site from chicken β-globin locus) and mutant (FIImut). (D) EMSA with DHS6.8oligo. Supershift reactions were performed as in (B). (E) EMSA with DHS6.8mutoligo. Complex marked i represents CTCF in complex with DHS6.8oligo and ii the antibody-supershifted CTCF complex. Undefined interactions formed between 6.8mutoligo and Caco2 nuclear extract are marked by *.
These data conclusively demonstrate a strong in vitro interaction between the +6.8 kb DHS region and CTCF.
CTCF binds to the +6.8 kb DHS in vivo
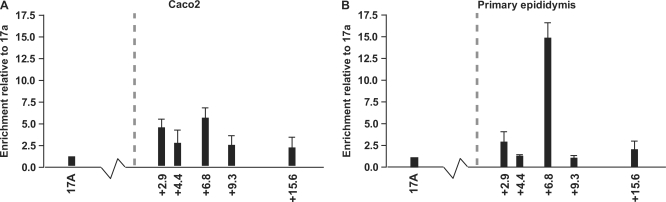
Since CTCF showed a strong interaction with DHS6.8oligo in vitro, ChIP with an antibody specific for CTCF followed by Taqman quantitative PCR analysis was used to investigate in vivo binding at this site. Chromatin from two cell types was evaluated: Caco2 colon carcinoma cells and fetal male primary epididymis cells, both of which express abundant CFTR. For Caco2 chromatin, ChIP with an antibody specific to CTCF gave an ∼5-fold enrichment of the +6.8 kb DHS relative to a region within CFTR intron 17a where there is no predicted CTCF-binding site (Figure 3A). The +15.6 kb DHS of CFTR (located ∼9 kb 3′ to the +6.8 kb DHS), that was previously demonstrated to possess CTCF-independent enhancer-blocking activity, showed no CTCF-specific enrichment, consistent with our earlier work (6). In contrast, using primary epididymis chromatin, the CTCF-specific antibody enriched the +6.8 kb DHS region by about 15-fold relative to CFTR intron 17a (Figure 3B). Approximately 2.5 kb either side of the +6.8 kb DHS, at +4.4 kb and +9.3 kb relative to the CFTR translation end-point, CTCF-specific enrichment in primary epididymis cells returned to baseline levels. The +15.6 kb DHS again showed no CTCF-specific enrichment in epididymis chromatin. For both Caco2 and primary epididymis chromatin, negative control ChIP experiments in which chromatin was immunoprecipitated with protein A beads alone (no antibody) resulted in baseline levels of enrichment at all regions (data not shown). Taken together, these results demonstrate strong in vivo binding of CTCF at the +6.8 kb DHS in primary epididymis cells, with a much lesser interaction between CTCF and the +6.8 kb DHS in the Caco2 cell line. It is noteworthy that CTCF-specific enrichment of the +6.8 kb DHS in primary epididymis cells correlates with the presence of the +6.8 kb DHS in this cell type (13).
Figure 3.
In vivo binding of CTCF at the +6.8 kb DHS region. Immunoprecipitations were performed with a CTCF-specific antibody and chromatin from (A) Caco2 cells and (B) Primary epididymis cells. No antibody control samples were also prepared, in which chromatin was incubated with Protein A agarose beads alone (data not shown). Samples were subjected to Taqman quantitative PCR analysis using probes specific for intron 17a and regions of interest 3′ to CFTR. CTCF-specific enrichment of each of these regions is shown relative to levels at intron 17a (which contains no predicted CTCF-binding sites). Vertical dashed line represents location of CFTR translation end-point, and x-axis on the right of this is drawn to scale (i.e. each data point accurately reflects the relative positions of Taqman amplicons). Immunoprecipitations were repeated at least twice. PCRs were performed in triplicate and Ct values averaged. Error bars denote S.E.M.
In addition to the CTCF-binding site that we determined experimentally at the +6.8 kb DHS, another CTCF-binding site was predicted in silico at the 3′-end of the CFTR locus, (7). This site is located at +2.9 kb from the CFTR translation end-point (AC000061:60371–60384) and has a 12 out of 14-bp match with the CTCF consensus. To evaluate CTCF-specific enrichment of this region in Caco2 and primary epididymis cells, an additional Taqman primer/probe set was designed. Both Caco2 and primary epididymis cells showed only modest CTCF-specific enrichment of the +2.9 kb region (∼4- and 5-fold, respectively) (Figure 3A and B) in comparison to 15-fold enrichment at the +6.8 kb site in epididymis chromatin. Moreover, the lack of any known DHS at this region in these cell types brings into question the biological significance of the +2.9 kb predicted CTCF-binding site.
Enhancer-blocking activity at the +6.8 kb DHS
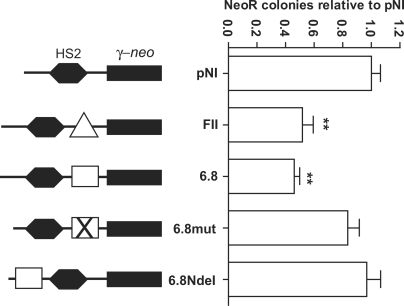
Binding of CTCF is necessary for enhancer-blocking activity at the majority of vertebrate insulator elements (37,38). Since the +6.8 kb DHS binds CTCF, and is located in an intergenic region, we used a well-established assay (27,28) to investigate whether this element possessed enhancer-blocking activity. This test uses the pNI plasmid, in which the mouse HS2 enhancer of the β-globin LCR is positioned upstream of a neor gene driven by the human γ-globin promoter. Sequences to be assayed for insulator activity are cloned into a unique AscI site located between the enhancer and promoter. Transfection of these constructs into K562 human erythroleukemia cells yields G418-resistant colonies, with a frequency dependent on the level of enhancer-promoter communication (and hence enhancer-blocking activity of the intervening fragment).
The DHS6.8oligo and DHS6.8mutoligo oligonucleotides were designed with BssHII sticky ends (Figure 2A), facilitating their direct cloning into the AscI site of pNI. An enhancer-blocking activity assay was performed, with the number of neomycin-resistant colonies obtained for each construct normalized to the empty pNI plasmid. Results were subjected to statistical analysis by one-way ANOVA followed by Dunnett's multiple comparison test. When inserted into the enhancer-blocking position of pNI, FII, the CTCF-binding core from the known 5′HS4 chicken β-globin insulator (28,37), significantly reduced the number of colonies (2- to 3-fold; P < 0.01) (Figure 4). The pNI-DHS6.8oligo construct also gave a significantly lower number of colonies compared to pNI (2- to 3-fold, P < 0.01). In contrast, although the pNI-DHS6.8mutoligo construct gave a slightly lower number of colonies than pNI, this was not significant (P > 0.05). This suggests that CTCF binding at the +6.8 kb DHS is responsible, at least in part, for the enhancer-blocking activity of this element.
Figure 4.
Enhancer-blocking activity at the +6.8 kb DHS region. Each construct used in the enhancer-blocking assay is depicted as follows: pNI is the empty pNI plasmid and pNI-FII (FII) contains a known insulator from the chicken β-globin locus (represented by a triangle). pNI-6.8 (6.8) contains the wild-type DHS6.8oligo (represented by a rectangle) and pNI-6.8mut (6.8mut) contains the mutant version DHS6.8mutoligo (represented by a rectangle with a cross in it). pNI-6.8NdeI (6.8NdeI) contains the wild-type DHS6.8oligo sequence cloned upstream of the HS2 enhancer (as opposed to between HS2 and the γ-neo reporter). The number of NeoR colonies obtained for empty pNI was given a value of 1, and the number of colonies obtained with all other constructs was expressed relative to this value. Error bars denote S.E.M. of triplicate experiments carried out on three separate occasions, except for 6.8NdeI which was performed in triplicate on one occasion. Different plasmid DNA preparations were used for each experiment within the triplicates. **P < 0.01.
As some CTCF-binding sites are associated with silencer activity (39), we investigated this possibility for the CFTR +6.8 kb DHS CTCF-binding site. A modified version of DHS6.8oligo was generated with NdeI sticky ends (as opposed to BssHII), enabling DHS6.8oligo to be cloned into an NdeI site upstream of the HS2 enhancer (as shown in Figure 4). However, in this arrangement, DHS6.8oligo had no significant effect upon colony number (P > 0.05) (see 6.8NdeI, Figure 4). This demonstrates that the enhancer-blocking activity of the CFTR +6.8 kb DHS is directional (in that it only has an effect when positioned between an enhancer and promoter), consistent with this element being an insulator rather than a silencer (27).
Looping of the CFTR locus in CFTR-expressing cell types
CTCF sites that flank the β-globin locus interact with the promoters of actively expressed globin genes, bringing them within proximity of the β-globin ‘active chromatin hub’ (ACH) (16–18). Within the CFTR locus, in addition to the 3′ CTCF-binding site at +6.8 kb investigated here, we previously characterized a CTCF-binding site 5′ to the locus in the −20.9 kb DHS region (6). The presence of these two DHS flanking the locus suggested similarities to β-globin. Hence we investigated whether either of the CTCF sites that flank CFTR interacts with the CFTR promoter region using chromosome conformation capture (3C) (33).
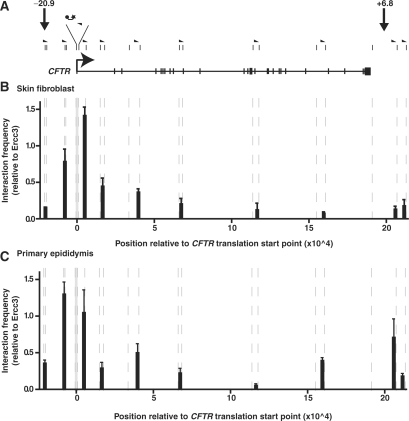
Formaldehyde-crosslinked nuclei from human primary skin fibroblasts (CFTR non-expressing) and human primary epididymis epithelial cells (CFTR-expressing) were subjected to HindIII digestion and subsequent 3C analysis. A fixed Taqman probe and reverse primer were designed within a HindIII fragment at the CFTR promoter, and multiple forward primers were generated within distal regions across the CFTR locus (Figure 5A and Supplementary Data). These forward primers were located within HindIII fragments encompassing the −20.9 kb, +6.8 kb and +15.6 kb DHS, and within specific intronic HindIII fragments. The assay fragments were positioned ∼25 to 50 kb apart, such that they would give a good overall representation of the structure of the CFTR locus (Figure 5A). Real-time PCR reactions using the ‘fixed’ reverse probe/primer and each of the ‘variable’ forward primers enabled quantification of ligation events (subsequently referred to as ‘interaction frequency’) between the CFTR promoter and specific distal regions within each 3C sample.
Figure 5.
3C analysis of the CFTR locus. (A) Scale figure of the CFTR gene with exons marked by vertical bars and the translation start site represented by a bent arrow. Small vertical lines above the gene denote HindIII sites and half arrow heads show the locations of 3C primers. Due to space constraints, primers are not drawn to scale, and the fixed CFTR promoter HindIII fragment is expanded, showing the location of Taqman probe (joined ball and star) and the reverse primer. The –20.9 kb DHS and +6.8 kb DHS locations are also indicated. (B) 3C data from primary human skin fibroblast cells. (C) 3C data from primary human epididymis cells. The x-axis for each chart is drawn to scale, with units representing base pairs relative to the CFTR translation start point (0). Both charts are aligned with the CFTR gene figure above. Vertical dashed lines represent HindIII sites. The interaction frequency between a fixed HindIII fragment at the CFTR promoter (shaded grey) and HindIII fragments at various regions across the CFTR gene was measured by Taqman quantitative PCR. The interaction frequency at each point is expressed relative to the interaction between two HindIII fragments within the ubiquitously expressed ERCC3 gene. Data shown represent the average of two independent experiments. Each real-time PCR reaction was performed three times and averaged. Error bars denote SEM.
Within primary human skin fibroblasts, which do not express CFTR, interaction frequency with the CFTR promoter decreased as a function of distance from the promoter, with no significant interaction between the CFTR promoter and either the −20.9 kb or +6.8 kb DHS (Figure 5B). Furthermore, there were no peaks of interaction at any of the intronic locations assayed. These data suggest that within CFTR-negative primary skin fibroblasts, the CFTR locus exists in a relaxed structure, or at least one that does not contain stable loops that bring distal regulatory elements close to the promoter.
In contrast to the skin fibroblast data, in CFTR-expressing primary epididymis cells, the CFTR locus exhibited a 3C profile that suggested specific interactions between the promoter and distal regions (Figure 5C). For example, a moderate interaction was detected between the CFTR promoter and a region within intron 19, possibly indicating the presence of an intronic regulatory element that interacts with the promoter. However, most significantly, a strong interaction frequency was detected between the CFTR promoter region and a HindIII site at the 3′-end of the CFTR locus encompassing the +6.8 kb DHS. Indeed, despite being located more than 200 kb from the CFTR promoter region, this 3′ fragment demonstrated an interaction frequency with the CFTR promoter that was greater than a HindIII fragment within intron 1 of the CFTR, <10 kb from the CFTR promoter (Figure 5C). For the adjacent 3′ fragment (∼210 kb from the CFTR promoter), interaction frequency fell dramatically to an apparently baseline level, suggesting that the CFTR promoter interacts specifically with elements in the HindIII fragment encompassing the +6.8 kb DHS.
Unlike the +6.8 kb DHS, in primary epididymis cells, no significant interaction was detected between the CFTR promoter and the −20.9 kb DHS CTCF-binding site (Figure 5C). The lack of interaction between these two sites was also confirmed using a −20.9 kb DHS-specific Taqman probe and reverse primer in combination with a forwards primer specific to the CFTR promoter region (data not shown). Furthermore, using the −20.9 kb DHS probe, no interaction was detected with the +6.8 kb DHS.
These data indicate that within CFTR-expressing primary epididymis cells, the CFTR locus exists within a looped structure, with a HindIII fragment encompassing the +6.8 kb DHS CTCF site being in close proximity to the CFTR promoter. In contrast, at the opposite end of the locus, no interaction is apparent between the −20.9 kb DHS CTCF site and the CFTR promoter.
DISCUSSION
Expression of the CFTR gene is tightly controlled, both temporally during development and spatially in different tissue types. However, the underlying genetic mechanisms responsible for conferring these complex CFTR expression patterns are poorly understood. The CFTR promoter is weak and demonstrates little tissue specificity, suggesting that key regulatory elements must be located elsewhere within the gene or in adjacent genomic regions. To identify potential regulatory elements, we previously performed DHS mapping in a variety of cell types, across 400 kb spanning the CFTR locus. Of the DHS that were found, a subset showed some correlation with CFTR expression. These included two DHS at the 3′-end of the gene, at +6.8 kb and +7.4 kb from the CFTR translation end-point.
The +6.8 kb DHS was only observed in fetal epididymis cells and adult lung tissue, both CFTR-expressing primary cell types. Here we demonstrate that the +6.8 kb DHS contains a predicted CTCF-binding site with a close match to the consensus. Moreover, this region shows a strong in vivo association with CTCF in chromatin extracted from primary epididymis cells, correlating with the presence of the DHS in these cells. Although we did not previously observe the +6.8 kb DHS in Caco2 colon carcinoma cells experiments with Caco2 chromatin showed slight CTCF-specific enrichment at this site. It is thus possible that within a large population of Caco2 cells, a small number of cells exhibit the +6.8 kb DHS.
While most CTCF-binding sites throughout the human genome appear to be cell-type invariant, a fraction are cell-type specific (7). This cell-type specificity may be conferred by DNA methylation at CpG dinucleotides, an epigenetic mark that is known to affect CTCF binding (38,40). However, within the CFTR +6.8 kb DHS, there is an absence of CpG dinucleotides, with the nearest CpG located about 200 bp 5′ to the CTCF-binding site core. It therefore seems unlikely that tissue-specific control of CTCF binding at the +6.8 kb DHS is controlled by DNA methylation, suggesting the use of a different regulatory mechanism. For example, it is possible that events leading to the formation of the +6.8 kb DHS, such as nucleosome remodelling, may regulate CTCF binding.
Genome-wide analysis of CTCF binding in primary human fibroblast IMR90 cells revealed ∼14 000 genomic regions flanked by CTCF-binding sites, so-called CTCF-pair-defined domains (CPDs) (7). Within these CPDs, the average distance between CTCF-binding sites was found to be 212 090 bp. It is therefore of interest that the CFTR gene is contained within an apparent CPD of 214 000 bp, considering the distance between the +6.8 kb DHS and the previously described −20.9 kb DHS CTCF-dependent insulator element (6).
At the β-globin locus, CTCF sites that flank the locus associate with actively transcribed globin promoters within the β-globin active chromatin hub (ACH) (16–18). In the present study, we demonstrated that there is no association between the CFTR promoter and the −20.9 kb DHS CTCF-binding element in the upstream flanking region, irrespective of CFTR expression. In contrast, in CFTR-expressing primary epididymis cells, we observed a strong interaction between the CFTR promoter and a region at the 3′-end of the CFTR gene encompassing the +6.8 kb DHS CTCF-binding site. This interaction was not seen in skin fibroblast cells, which do not express CFTR, suggesting that ‘looping’ of the CFTR locus correlates with active expression of the gene.
In addition to the +6.8 kb DHS, the 3′ HindIII fragment demonstrated here to interact with the CFTR promoter encompasses a number of other DHS. These include the cluster of adjacent DHS (+5.4 kb, +7.0 kb and +7.4 kb) (13) and the previously characterized +15.6 kb DHS enhancer-blocking element (6). Within primary epididymis cells, the +6.8 kb, +7.0 kb and +15.6 kb DHS were all evident (13). We therefore cannot rule out the possibility that looping of the CFTR locus in primary epididymis cells is induced by interaction of either the +7.0 kb or the +15.6 kb DHS with the CFTR promoter. However, given that in other cell types the presence of these DHS shows poor correlation with CFTR expression, it seems highly likely that an important role is performed by the +6.8 kb DHS, which was only observed in a subset of CFTR-expressing primary cell types.
It seems probable that the induction and maintenance of looping of the CFTR locus is dependent on the interaction of multiple regulatory elements located in different DHS regions, both intronic and flanking the gene (Figure 6). Indeed we now have evidence that several intronic DHS also contribute to the loops (Ott et al., 2008. Journal of Cellular and Molecular Medicine, in Press). Hence, though a single element, such as that within the DHS at the 3′-end of the gene is unlikely to be solely responsible for the looping phenomenon, it may play a pivotal role in primary epididymis. We predict a co-ordinated interplay of the regulatory elements located within DHS at the 3′-end of the CFTR gene and note similarities between this cluster of DHS and the human β-globin locus control region (LCR) (41): (i) Both comprise five DHS (+5.4 kb, +6.8 kb, +7.0 kb, +7.4 kb and +15.6 kb in the case of CFTR; 5′HS1-5 in the case of β-globin). In the case of the CFTR 3′ region, these DHS are spread over 11 kb, while the β-globin LCR is about 16 kb in size; (ii) both contain CTCF-binding elements (+6.8 kb DHS in the case of CFTR; 5′HS5 in the case of β-globin); (iii) when their corresponding genes are actively transcribed, both regions contain peaks of euchromatin-specific histone modifications (at the +15.6 kb DHS in the case of CFTR) (6); and (iv) both interact with the promoters of their corresponding genes, inducing genomic loops and the formation of an ACH. Future experiments will determine whether the DHS cluster 3′ to CFTR does indeed represent an LCR. Understanding the precise mechanisms by which CFTR expression is regulated is likely to be of direct practical relevance in the design of effective gene therapy vectors for cystic fibrosis.
Figure 6.
Looping model for CFTR gene. In CFTR-expressing cell types, such as primary epididymis cells, elements in the CFTR 3' flanking region are in close proximity with the CFTR promoter. This 3' flanking region includes the tissue-specific +6.8-kb DHS, shown here to bind CTCF, as well as other previously described DHS (13,6). Protein factors bound at each of these sites interact with the promoter-bound transcription machinery, thus forming an active chromatin hub (ACH) and helping regulate expression of the CFTR gene. In addition to DHS from the 3' flanking region, intronic DHS such as the intestine-specific intron 1 element (31,42,43) and others (represented by the elements marked ‘?’), may contribute to the CFTR ACH in a tissue-specific manner.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Cystic Fibrosis Foundation USA; NIH R01 HL094585; the Cystic Fibrosis Trust UK; Children's Memorial Research Center; and a Medical Research Council scholarship (to N.P.B., partial).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Douglas Vernimmen (Weatherall Institute of Molecular Medicine, University of Oxford) for help with establishing chromosome conformation capture experiments. We thank Dr Gary Felsenfeld for pNI constructs and Dr Jeannie Lee for pCTCF.
REFERENCES
- 1.Crawford I, Maloney PC, Zeitlin PL, Guggino WB, Hyde SC, Turley H, Gatter KC, Harris A, Higgins CF. Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc. Natl Acad. Sci. USA. 1991;88:9262–9266. doi: 10.1073/pnas.88.20.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelhardt JF, Yankaskas JR, Ernst SA, Yang Y, Marino CR, Boucher RC, Cohn JA, Wilson JM. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat. Genet. 1992;2:240–248. doi: 10.1038/ng1192-240. [DOI] [PubMed] [Google Scholar]
- 3.Kreda SM, Mall M, Mengos A, Rochelle L, Yankaskas J, Riordan JR, Boucher RC. Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol. Biol. Cell. 2005;16:2154–2167. doi: 10.1091/mbc.E04-11-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan W, Rajkovic A, Viveiros MM, Burns KH, Eppig JJ, Matzuk MM. Identification of Gasz, an evolutionarily conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-alpha motif, and a basic leucine zipper. Mol. Endocrinol. 2002;16:1168–1184. doi: 10.1210/mend.16.6.0864. [DOI] [PubMed] [Google Scholar]
- 5.Cheung J, Petek E, Nakabayashi K, Tsui LC, Vincent JB, Scherer SW. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics. 2001;78:7–11. doi: 10.1006/geno.2001.6651. [DOI] [PubMed] [Google Scholar]
- 6.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broackes-Carter FC, Mouchel N, Gill D, Hyde S, Bassett J, Harris A. Temporal regulation of CFTR expression during ovine lung development: implications for CF gene therapy. Hum. Mol. Genet. 2002;11:125–131. doi: 10.1093/hmg/11.2.125. [DOI] [PubMed] [Google Scholar]
- 10.Trezise AE, Chambers JA, Wardle CJ, Gould S, Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum. Mol. Genet. 1993;2:213–218. doi: 10.1093/hmg/2.3.213. [DOI] [PubMed] [Google Scholar]
- 11.Yoshimura K, Nakamura H, Trapnell BC, Dalemans W, Pavirani A, Lecocq JP, Crystal RG. The cystic fibrosis gene has a “housekeeping”-type promoter and is expressed at low levels in cells of epithelial origin. J. Biol. Chem. 1991;266:9140–9144. [PubMed] [Google Scholar]
- 12.Smith AN, Wardle CJ, Harris A. Characterization of DNASE I hypersensitive sites in the 120kb 5′ to the CFTR gene. Biochem. Biophys. Res. Commun. 1995;211:274–281. doi: 10.1006/bbrc.1995.1807. [DOI] [PubMed] [Google Scholar]
- 13.Nuthall HN, Moulin DS, Huxley C, Harris A. Analysis of DNase I hypersensitive sites at the 3′ end of the cystic fibrosis transmembrane conductance regulator gene. Biochem. J. 1999;341:601–611. [PMC free article] [PubMed] [Google Scholar]
- 14.Smith DJ, Nuthall HN, Majetti ME, Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics. 2000;64:90–96. doi: 10.1006/geno.1999.6086. [DOI] [PubMed] [Google Scholar]
- 15.Phylactides M, Rowntree R, Nuthall H, Ussery D, Wheeler A, Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur. J. Biochem. 2002;269:553–559. doi: 10.1046/j.0014-2956.2001.02679.x. [DOI] [PubMed] [Google Scholar]
- 16.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 17.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 19.Dunn KL, Zhao H, Davie JR. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 2003;288:218–223. doi: 10.1016/s0014-4827(03)00185-x. [DOI] [PubMed] [Google Scholar]
- 20.Yusufzai TM, Felsenfeld G. The 5′-HS4 chicken beta-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl Acad. Sci. USA. 2004;101:8620–8624. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 23.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 25.Fogh J, Wright WC, Loveless JD. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl. Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 26.Harris A, Coleman L. Ductal epithelial cells cultured from human foetal epididymis and vas deferens: relevance to sterility in cystic fibrosis. J. Cell Sci. 1989;92(Pt 4):687–690. doi: 10.1242/jcs.92.4.687. [DOI] [PubMed] [Google Scholar]
- 27.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 28.Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken beta-globin insulator. Proc. Natl Acad. Sci. USA. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 31.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, Harris A. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 32.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 33.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 34.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 35.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 37.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 38.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 39.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 41.Mahajan MC, Karmakar S, Weissman SM. Control of beta globin genes. J. Cell Biochem. 2007;102:801–810. doi: 10.1002/jcb.21507. [DOI] [PubMed] [Google Scholar]
- 42.Paul T, Li S, Khurana S, Leleiko NS, Walsh MJ. The epigenetic signature of CFTR expression is coordinated via chromatin acetylation through a complex intronic element. Biochem. J. 2007;408:317–326. doi: 10.1042/BJ20070282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowntree R, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, Huxley C, Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum. Mol. Genet. 2001;11:1455–1464. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.