Abstract
Methylation of RNA by methyltransferases is a phylogenetically ubiquitous post-transcriptional modification that occurs most extensively in transfer RNA (tRNA) and ribosomal RNA (rRNA). Biochemical characterization of RNA methyltransferase enzymes and their methylated product RNA or RNA–protein complexes is usually done by measuring the incorporation of radiolabeled methyl groups into the product over time. This has traditionally required the separation of radiolabeled product from radiolabeled methyl donor through a filter binding assay. We have adapted and optimized a scintillation proximity assay (SPA) to replace the more costly, wasteful and cumbersome filter binding assay and demonstrate its utility in studies of three distinct methyltransferases, RmtA, KsgA and ErmC’. In vitro, RmtA and KsgA methylate different bases in 16S rRNA in 30S ribosomal particles, while ErmC’ most efficiently methylates protein-depleted or protein-free 23S rRNA. This assay does not utilize engineered affinity tags that are often required in SPA, and is capable of detecting either radiolabeled RNA or RNA–protein complex. We show that this method is suitable for quantitating extent of RNA methylation or active RNA methyltransferase, and for testing RNA-methyltransferase inhibitors. This assay can be carried out with techniques routinely used in a typical biochemistry laboratory or could be easily adapted for a high throughput screening format.
INTRODUCTION
Post-transcriptional methylation of RNA by RNA methyltransferases (RMT) is universal and occurs to its greatest extent in transfer RNA (tRNA) and ribosomal RNA (rRNA). All of these methylations require the S-adenosyl-methionine (SAM) cofactor as methyl donor. Measurements of labeled product may be used to kinetically and enzymologically characterize the enzyme and potential inhibitors or to assess the extent and consequences of product RNA methylation.
We have been studying three different classes of rRNA methyltransferase: one is the housekeeping protein, KsgA (1); the other two, RmtA (and related RMT family members) (2) and ErmC’ (3), confer aminoglycoside and macrolide antibiotic resistance, respectively, on their bacterial hosts. KsgA converts both A1518 and A1519 (Escherichia coli numbering) of 16S rRNA in the 30S ribosomal particle to N6,N6-dimethyladenosines (4) by catalyzing the transfer of four methyl groups from SAM to each 16S rRNA molecule. RmtA and related members of its enzyme class catalyze transfer of one methyl group to the N7 position of G1405 (E. coli numbering) of 16S rRNA in the 30S ribosomal particle (5). ErmC’ catalyzes the mono- or di-methylation of A2058 (E. coli numbering) in the 23S rRNA of the 50S ribosomal particle (6).
In vitro characterization of RMT usually requires the use of 3H-methyl-SAM (radiolabeled on the methyl group) as cofactor reactant with the RNA substrate, and a method of separating 3H-methyl-RNA product from unreacted 3H-methyl-SAM. The separation is usually accomplished by filter binding (7) or with a spin column (8). While these methods are widely used, they suffer from a variety of shortcomings, including empirical determination of wash conditions, variation in deposition efficiencies, large volumes of radioactive waste and the inability to scale up to a large number of parallel measurements used in HTS. One way to circumvent these deficiencies has been to use fluorescence-based assays that measure the S-(5′-adenosyl)-l-homocysteine (SAH) product rather than the methylated RNA. This type of assay has the advantages of being adaptable for high throughput screening (HTS) and not requiring radiolabeled reagents, but it does not directly measure the methylated product, which is often the objective of this type of assay. One version of this assay used SAH antibodies with conjugate tracers to measure activity, but high cross-reactivity of the anti-SAH antibody with SAM (9) limited its use to low concentrations of SAM (3 μM). Other indirect assays using coupled enzymes to convert SAH to detectable products (10–12) are potentially ambiguous, since test compounds (e.g. inhibitor screens) could inhibit these enzymes but not the methyltransferase.
We have developed a scintillation proximity assay (SPA) for the assay of RMT using commercially available yttrium silicate scintillant (YSi) beads that capture DNA and RNA by non-specific electrostatic binding. In our application, radiolabeled, methylated RNA product bound to the bead activates the scintillant whose emission can be measured and quantitated. The intrinsic physical separation of the radiolabeled, methylated RNA product from radiolabeled SAM reactant permits the accurate direct quantitation of methylated RNA product without the need for a separation step. This method should be generally applicable to assay of RNA-methyltransferases and their products and adaptable to HTS.
MATERIALS AND METHODS
Protein expression and purification
KsgA was expressed and purified as described (7). RmtA and ErmC’ were expressed from cloned, synthetic genes inserted into pET15b plasmids, rmtA in Rosetta 2 (DE3) cells (Novagen) and ermC in BL21 (codon+ DE3-RIL) cells (Stratagene). All three enzymes were purified on a HiTrap Ni-chelating column (Amersham) to >95% homogeneity on SDS gel.
30S purification
Wild type and ksgR strains of E. coli were provided by Dr Heather O’Farrell and 30S subunits were prepared according to a previously described method (7). Concentration of 30S was determined by multiplying OD260 by 6 pmol/ml.
30S particle methylation reaction
A standard 50 μl reaction contained 10 pmol enzyme (RmtA or KsgA), 10 pmol 30S (wild type for RmtA, unmethylated ksgR for KsgA), 0.02 mM 3H-methyl-SAM (780 cpm/pmol) (Perkin Elmer) and reaction buffer incubated at 37°C for a specified time. Prior to mixing the reactants, 30S particles were heated at 42°C for 5 min to anneal subunits into a homogenous conformation. Reactants were mixed and the reaction initiated by addition of SAM. Background reactions were performed in the same way without enzyme. In reactions with RmtA and for its background measurement, reaction buffer (buffer R) consisted of 40 mM Tris, pH 7.2, 40 mM NH4Cl, 8 mM MgOAc and 1 mM DTT. Reaction buffer for KsgA reactions and for its background measurement was 40 mM Tris at pH 7.4, 40 mM NH4Cl, 4 mM MgOAc, 6 mM 2-mercaptoetanol.
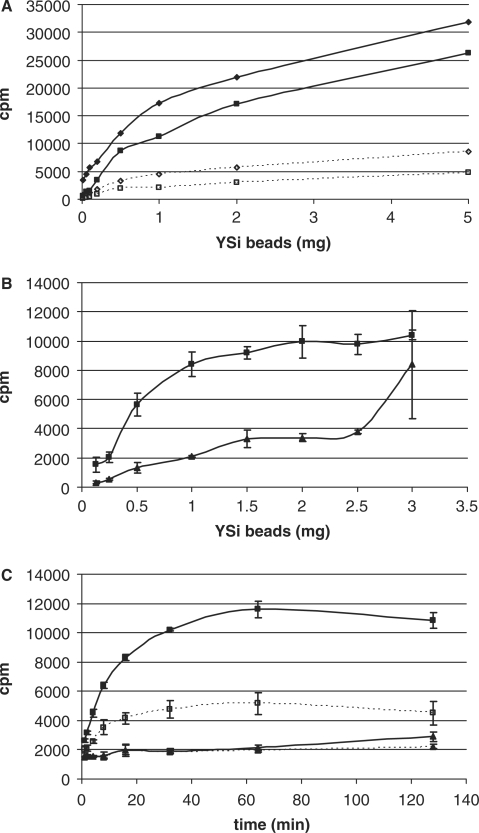
At the end of the reaction time, the 50 μl reaction volume was removed and added to clear 1.5 ml vials containing 180 μl deionized water, 10 μl (100 mg/ml) YSi binding beads (cat. RPNQ0013, GE Life Sciences) and 10 μl 100 mM unlabeled SAM (Sigma) and mixed thoroughly by pipette. Vials were then incubated in the dark for 40 min with mixing by inversion at 20 and 40 min and then centrifuged at 13 000 rpm for 3 min, placed in the mouth of a 15 ml scintillation vial, and counted in a Packard 1500 Tri-Carb Liquid Scintillation Analyzer. For experiments to optimize bead concentration (Figure 1A) and water dilution (Figure 1B), sample/bead mixtures were pipetted into the bottom of the scintillation vial and allowed to settle for an additional 20 min in the dark without mixing before being counted. Time-course assays of samples before and after centrifugation (Figure 1C) were done in one vial in volumes sufficient for eight samples. Samples of 50 μl were taken at each of the eight designated time points and measured by the standard SPA method with the additional step of counting samples just prior to centrifugation. Signal and background reactions were measured in triplicate at 1, 2, 4, 8, 16, 32, 64 and 128 min.
Figure 1.
Optimization of assay conditions. (A) Dilution of samples of 3H-methyl-SAM with water: beads mixed with 50 μl of sample (buffer R, 20 μM SAM 780 cpm/pmol) before adding water (closed diamond); beads mixed with 50 μl of sample (buffer R, 20 μM SAM 780 cpm/pmol, 10 pmol 30S) before adding water (closed square); beads mixed with 50 μl of sample (buffer R, 20 μM SAM 780 cpm/pmol) followed by the addition of 3 ml of water (open diamond); beads mixed with 50 μl of sample (buffer R, 20 μM SAM 780 cpm/pmol, 10 pmol 30S) after adding 3 ml of water (open square). Data shown were from single samples. (B) Optimizing bead concentration. Beads mixed with quenching SAM and 50 μl of signal reaction (10 pmol RmtA, 10 pmol 30S, buffer R, 20 μM SAM 780 cpm/pmol, 37°C for 1 h) and diluted to 250 μl with water (closed square); beads mixed with quenching SAM and 50 μl of background reactions (10 pmol 30S, buffer R, 20 μM SAM 780 cpm/pmol, 37°C for 1 h) and diluted to 250 μl with water (closed triangle). (C) Centrifuging samples from time courses of RmtA + wild-type 30S. Signal reactions before centrifugation (open square); signal reactions after centrifugation (closed square); background reactions before centrifugation (open triangle); background reactions after centrifugation (closed triangle). Assays from (B) and (C) were done in triplicate, plotting the average with error bars of ± 1 SD.
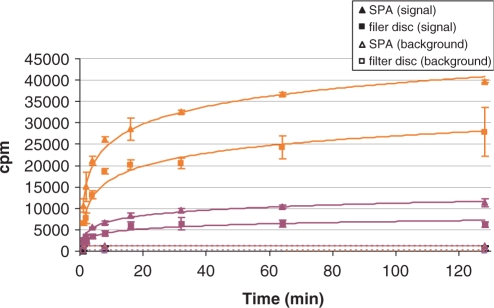
Parallel time course assays (Figure 2) were done as above except the reaction volume was sufficient for 16 samples for signal reactions and 6 samples for background reactions. Samples of 100 μl were removed at each time point, quenched with 20 μl 100 mM unlabeled SAM, and divided into two 60 μl aliquots for determination by the SPA and filter binding methods. Signal reactions were measured at 1, 2, 4, 8, 16, 32, 64 and 128 min, while background reactions were measured at 1, 8 and 128 min, all in triplicate.
Figure 2.
Parallel time-course assays measured with SPA and filter binding methods. Maroon are RmtA + wild-type 30S; orange are KsgA and ksgR 30S. Solid symbols indicate signal reactions (10 pmol 30S, 10 pmol enzyme, 20 μM SAM 780 cpm/pmol); empty symbols indicate background reactions (10 pmol 30S, 20 μM SAM 780 cpm/pmol). Measurements by YSi SPA beads (closed triangle) and the corresponding filter binding measurements (closed square). Assays were done in triplicate, plotting the average with error bars of ± 1 SD.
Excel was used to obtain a linear fit of data in Figures 3, 6 and background data in Figure 2, and a logarithmic fit of signal data in Figures 2 and 4.
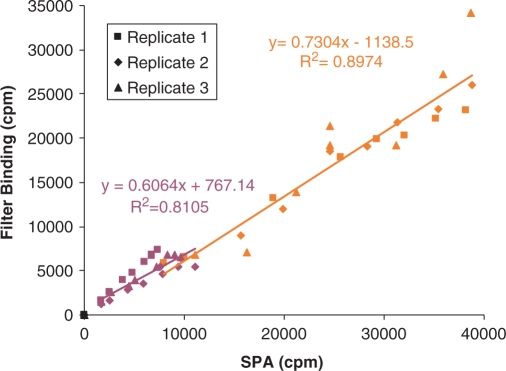
Figure 3.
Correlation of SPA and filter binding measurements. Net counts from time points in Figure 2 are plotted with SPA counts along the X-axis and filter binding counts along the Y-axis. RmtA + wild-type 30S (maroon); KsgA and ksgR + 30S (orange). Combined replicates were used to fit the least squares trend line.
Figure 6.
Dependence on enzyme concentration. 30S ribosomal particles (10 pmol) were incubated with RmtA (maroon) for 8 min and 10 pmol ksgR 30S ribosomal particles were incubated with KsgA (orange) for 3.5 min. Samples were done in triplicate and the average plotted with error bars of ± 1 SD.
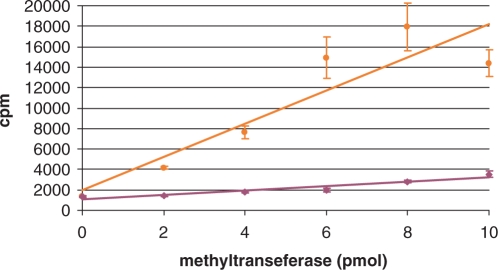
Figure 4.
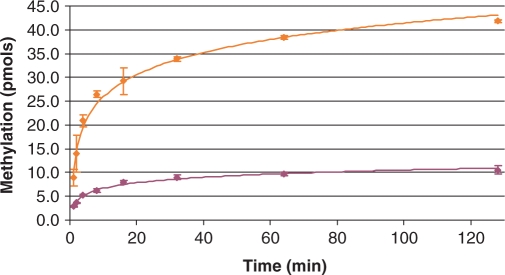
Extent of methylation. Time courses measured by SPA in this figure are expressed in number of methyl groups incorporated into RNA. Formulas derived from Figure 3 were used to convert SPA measurements into ‘filter binding equivalent’ values for quantitation of methyl group incorporation. RmtA + wild-type 30S (maroon); KsgA + ksgR 30S (orange).
Inhibition studies were done using RmtA and wild-type 30S in a triple volume reaction. Inhibitor or solvent was added at a specified concentration prior to the addition of SAM to initiate the reaction. Reactions were stopped after 8 min and dispensed in equal volumes into three vials with YSi beads. Sinefungin (Sigma) was dissolved in deionized H2O, SAH (Sigma) was dissolved in DMSO (American Bioanalytical). Data in Figure 7A and B were fitted with a sigmoidal dose–response curve provided in Sigma Plot 8.0; IC50 values were calculated as described (13).
Figure 7.
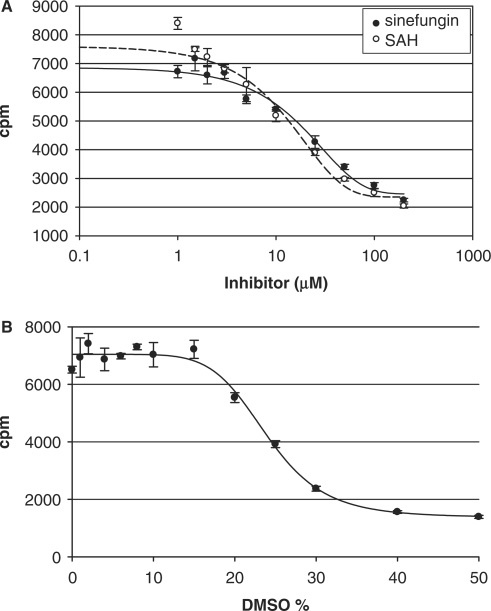
Inhibitors of methylation. For each concentration of inhibitor, inhibitor/DMSO was added to 30 pmol RmtA + 30 pmol wild-type 30S ribosomal particles + 20 μM SAM 780 cpm/pmol for an 8 min reaction. One-third of the reaction volume was added to each of the three vials of SPA beads and the average plotted with error bars of ± 1 SD. (A) Reactions with sinefungin (closed circle) and with SAH (open circle). (B) Reactions with DMSO alone.
Extraction of 16S and 23S rRNA from 30S and 50S particles
Methylated 16S rRNA after reaction with KsgA or RmtA was extracted with buffer saturated phenol (Invitrogen) and precipitated with ethanol before dilution and counting. 23S rRNA was similarly extracted from 50S particles with phenol prior to methylation by incubation with ErmC’.
RESULTS AND DISCUSSION
Optimization of SPA assay conditions
Bead concentration affects the signal to noise ratio
To optimize the YSi bead concentration, we tested signal and background reactions with different amounts of YSi beads (Figure 1A). We found that for our assay conditions, 1 mg of YSi beads gave the highest signal:noise ratio with a signal greater than 6000 cpm. These experiments also suggest that the apparent failure to reach saturation of the beads with bound, methylated 30S ribosomal particles is due to the non-proximity effect (NPE) as bead concentration increases (Figure 1A). The NPE arises from excitation of beads by radiolabeled substrate that is within the path length of the isotope's β-particle (1.5 μm for 3H; GE Health Sciences), but is either free in solution or bound to other nearby beads. Bead concentrations should be optimized for specific applications.
Minimizing background from 3H-methyl-SAM
3H-methyl-SAM alone in reaction buffer produced surprisingly high counts (Figure 1B) that would raise background levels in the assay. Addition of cold 30S (10 pmol) ribosomal particles to the incubation of 3H-methyl-SAM with YSi beads lowered the counts, particularly for vials with smaller amounts of YSi beads, indicating that SAM and 30S ribosomal particles compete for sites on the YSi beads.
A significant diminution of background counts from 3H-methyl-SAM binding to the beads was achieved by dilution of the sample with 3 ml of water (Figure 1B). This indicated that the major contributor of 3H-methyl-SAM to the background counts is through the NPE and not through binding to the beads. Dilution of the reaction solution with water to a volume of ∼250 μl (the well volume of a typical 96-well plate) on addition to the YSi beads almost entirely eliminated this background. Attempts to lower background counts by dilution with solutes (adenosine, unlabeled SAM, spermine, HCl or MeOH) showed that only unlabeled SAM lowered background without lowering signal. We conclude that background contribution from non-specific binding of 3H-methyl-SAM to YSi beads is intrinsically optimized because the higher affinity 30S ribosomal particles in the sample and the cold SAM used in the quenching step reduce bound 3H-methyl-SAM to an inconsequential level.
Sample centrifugation increases the signal to noise ratio
We found that centrifuging samples just prior to scintillation counting improved the signal:noise significantly (>5:1). Counts from signal reactions more than doubled, while those of background reactions increased only slightly (Figure 1C). This phenomenon may also be due to the NPE, arising in this case from more tightly packed radiolabeled 30S particle-bound beads exciting proximal YSi beads within the 1.5 μm path length of the tritium β-particles. Background counts did not increase significantly on centrifugation, consistent with the low level of bound radiolabeled substrate. More importantly, centrifugation of the beads after product binding also reduced variability among replicate samples, possibly by detaching beads bound to the walls of the vial and packing them more uniformly. A centrifugation step can be incorporated into HTS experiments by centrifuging 96-well plates in a special rotor as described in a similar SPA HTS assay (14).
Binding of methylated 30S ribosomal particles to YSi beads is instantaneous
Incubation time for YSi beads with product mix was optimized to ensure maximum capture of 30S ribosomal particles. A large batch (70 pmol) of 30S rRNA particles was methylated by RmtA for 1 h and separated into individual 10 pmol portions to test the effect of different incubation times (0, 20, 40, 60, 80, 100 and 120 min). YSi beads (density = 4.1 g/cm3) quickly settle in solution, so beads were thoroughly mixed with a pipette on addition of the reaction sample (time 0) and then mixed by inversion every 20 min for the specified time. The signal from time 0 was approximately the same as the other incubation times (data not shown), indicating that 30S binding occurs almost instantaneously and is complete with the first mixing. For convenience, all succeeding samples were incubated for 40 min; i.e. mixed by pipette initially and again at 20 and 40 min before centrifugation.
With these optimized conditions, we attained minimum count levels of ∼3000 cpm and signal:noise ratio of ∼5:1, which are the recommended benchmarks for this bead-based assay (GE Health Sciences). While these conditions are useful as a guide to implementing the assay, optimization should be done for applications to different assay systems.
SPA vs. filter-binding method in measurement of time courses and extent of methylation
Our standard filter-binding assay for measurement of methyltransferase activity is based on an earlier method (15) in which reaction solutions are pipetted onto cationic filter discs, washed twice with ∼200 ml of 5% TCA and rinsed with ∼3 ml ethanol. After drying for an hour, filter discs are placed in 3 ml of scintillation fluid for counting.
We compared this method with the SPA method in parallel time-course experiments. Using our standard optimized reaction conditions, we ran time courses of reaction for RmtA and KsgA, including separate background measurements (Figure 2). For each time point, double the normal amount was removed from the reaction, quenched and divided so that equal amounts were measured by either the SPA method or the traditional filter binding technique. Both sets of experiments showed the expected exponential curve with an early linear phase extending up to about 5 min and an extended asymptotic plateau. Although the SPA produced a higher background than the filter binding method, signals from the SPA were greater than for filter binding after backgrounds were subtracted. The consistent values of SPA background indicate that the high background does not compromise the results. The SPA background increased slightly over time in a linear fashion. This could be due to degradation of 3H-methyl SAM to a radiolabeled product with higher affinity for SPA beads.
The data from these parallel time courses allowed us to calculate the amount of 3H-methyl transferred to RNA from the SPA measurements. In the filter binding method, the extent of methylation can be easily determined from knowledge of the amount of RNA substrate in the assay and the measurable specific activity of the 3H-methyl-SAM used. However, in the SPA format, the amount of 3H-methyl-SAM used cannot be directly measured with a specific-activity test, because the different binding affinities of 3H-methyl SAM and 3H-methylated RNA to the SPA beads will yield a different response for the same amount of 3H-methyl group.
This problem is circumvented by plotting the SPA and filter binding responses from the same samples and obtaining the linear equation that relates them (Figure 3). SPA measurements can then be converted into ‘filter binding equivalent’ values, from which RNA methylation is quantified (Figure 4) using the specific activity of 3H-methyl-SAM. The number of methyl groups incorporated into RNA by KsgA or RmtA determined from the SPA assay is consistent with full methylation of the 10 pmol of 30S substrate by the two enzymes: 10 pmol of methyl group for RmtA and 40 pmol for KsgA.
Performing a parallel filter binding assay may be needed only when quantifying methyl group incorporation. In this case, once a correlation of SPA and filter binding measurements is established for a particular enzyme reaction, subsequent SPA measurements could be quantified by use of the previously obtained SPA to filter binding conversion factor and the measured specific activity of the 3H-methyl-SAM.
SPA analysis of extracted 3H-methyl RNA
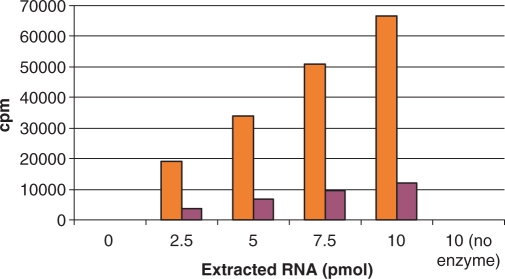
The experiments described above measure methylation of 30S subunit particles that bind intact to YSi beads. We also tested the SPA assay format on methylated RNA extracted with phenol to remove proteins. Phenol-extracted, methylated 16S rRNA from RmtA or KsgA catalyzed reactions was tested for binding to YSi beads in the SPA assay. Signals for both reaction products increased linearly with amount of extracted 3H-methyl-RNA up to 10 pmol (Figure 5). The counts measured for 10 pmol of extracted RNA were higher than those for the same amount of 30S particles, suggesting that free RNA has a higher affinity for YSi beads than RNA bound to protein in the 30S subunit or that the NPE is larger for bound RNA in the absence of protein. The background for the extracted RNA measurements was essentially nil, since all 3H-methyl SAM is washed away in the extraction process.
Figure 5.
Free 3H-methyl-RNA measured with YSi SPA beads. RNA extracted from 30S subunits following a 2-h reaction with RmtA + wild-type 30S (maroon) or KsgA + ksgR 30S (orange), and incubated with different amounts of YSi beads. RNA concentration was determined by measuring the OD260 following extraction. RNA (10 pmol) extracted from reactions without enzyme were used as a negative control. Data are for single samples.
SPA assay for methylation of free RNA substrate
To verify that this SPA procedure can measure methylation of an RNA substrate alone, parallel time course experiments were performed as above using the ErmC’ enzyme and phenol-extracted 23S rRNA from 50S ribosomal particles. The kinetics of this reaction differ from those for KsgA and RmtA, but with 10 pmol of E. coli 23S rRNA substrate, we measure ErmC’-catalyzed incorporation of 8 pmols of methyl group (data not shown), in close agreement with the results of Denoya and Dubnau (16) for this reaction. Furthermore, the correlation of SPA and filter-binding values for the ErmC’-catalyzed reaction is closely similar to that obtained for the KsgA and RmtA assays.
Use of a similar YSi SPA bead to monitor methylation of tRNA has been reported (17) for considerably different assay conditions, suggesting that this method can be applied widely to both methylated RNA and RNA–protein complexes.
Dependence on enzyme concentration
Based on the time course, we took early samples (8 min for RmtA, 3.5 min for KsgA) of reactions carried out with varying amounts of RmtA or KsgA (Figure 6) to determine the dependence on enzyme concentration. Under the reaction conditions used, the activity measured from the SPA assay is linear for enzyme concentrations up to at least 40 nM (10 pmol enzyme in the reaction), validating the use of the assay for determining enzyme concentrations.
Inhibitor assays
We tested the SPA assay for RmtA inhibition by the known methyltransferase inhibitors sinefungin and SAH (Figure 7A). Both compounds inhibited RmtA in a concentration-dependent manner with calculated IC50 values of 19.95 ± 0.09 μM for sinefungin and 9.33 ± 0.07 μM for SAH. We tested the effect of DMSO in the inhibition reactions, since this solvent is often used in HTS assays to dissolve test compounds (Figure 7B). RmtA in the presence of DMSO appeared to be fully active up to 15% (V/V), but the response from SPA beads dropped off at ≥20% DMSO. Whether the drop in signal was from the effect of DMSO on enzyme activity or on the binding of methylated RNA product to, or response of, the SPA beads was not determined.
CONCLUSIONS
Yttrium silicate nucleic-acid-binding SPA beads are shown here to be a useful method for measuring methylation of rRNA and almost certainly for other RNA and RNA–protein complexes. This assay method gives signal:noise ratios >5:1 under our optimized conditions and can be used to measure methylation of RNA alone or in 30S ribosomal particle complex with protein. While optimization of conditions for a specific assay and its products is advisable, all assays of RNA methylation will involve the common step of non-specific binding of the RNA product to the YSi beads. Therefore, the conditions described here are likely to be a good starting point for any further optimization. Through the use of a standard reference curve relating SPA counts to counts from the traditional filter binding assay, the SPA assay can give reproducible and accurate measures of the stoichiometry of methyl group incorporation. While the creation of this standard reference curve adds a step to the assay when methylation stoichiometry is to be determined, it is not necessary for routine assays of relative activities, and for high throughput applications need only be done once. This SPA method can be used in time course assays for kinetic measurements, for quantitation of methyltransferases and for assay of inhibitors in DMSO concentrations up to 15% by volume. It is easily adapted to HTS processes and for manual procedures offers the advantages over the filter binding assay of less time, materials, radioactive waste and lower overall cost.
FUNDING
This work was funded from grants from the Commonwealth Health Research Board (to J.P.R. and H.T.W.) and the National Institutes of Health (to J.P.R., GM66900). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Heather C. O’Farrell for providing purified KsgA and wild-type and ksgR strains of E. coli, Dr Aiye Liang for her help with Sigma Plot, and Dr Gordona Maravic Vlahovicek for providing us with a plasmid containing the ErmC’ gene.
REFERENCES
- 1.Sparling PF. Kasugamycin resistance: 30S ribosomal mutation with an unusual location on the E. coli chromosome. Science. 1970;167:56–58. doi: 10.1126/science.167.3914.56. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet. 2003;362:1888–1893. doi: 10.1016/S0140-6736(03)14959-8. [DOI] [PubMed] [Google Scholar]
- 3.Lai CJ, Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus Aureus. Proc. Natl Acad. Sci. USA. 1971;68:856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helser TL, Davies JE, Dahlberg JE. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat. New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 5.Liou GF, Yoshizawa S, Courvalin P, Galimand M. Aminoglycoside resistance by armA-mediated ribosomal 16S methylation in human bacterial pathogens. J. Mol. Biol. 2006;359:358–364. doi: 10.1016/j.jmb.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 6.Skinner R, Cundliffe E, Schmidt FJ. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 1984;258:12706–12708. [PubMed] [Google Scholar]
- 7.O'Farrell HC, Pulicherla N, Desai PM, Rife JP. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA. 2006;12:725–733. doi: 10.1261/rna.2310406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob. Agents Chemother. 2006;50:178–184. doi: 10.1128/AAC.50.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graves TL, Zhang Y, Scott JE. A universal competitive fluorescence polarization activity assay for S-adenosylmethionine utilizing methyltransferases. Anal. Biochem. 2008;373:296–306. doi: 10.1016/j.ab.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal. Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Leffler S, Thompson DH, Hrycyna CA. A general fluorescence-based coupled assay for S-adenosylmethionine-dependent methyltransferases. Biochem. Biophys. Res. Commun. 2005;331:351–356. doi: 10.1016/j.bbrc.2005.03.170. [DOI] [PubMed] [Google Scholar]
- 13.Henry BL, Monien BH, Bock PE, Desai UR. A novel allosteric pathway of thrombin inhibition: Exosite II mediated potent inhibition of thrombin by chemo-enzymatic, sulfated dehydropolymers of 4-hydroxycinnamic acids. J. Biol. Chem. 2007;282:31891–31899. doi: 10.1074/jbc.M704257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Triantafyllou I, Cable M, Palermo R. High-throughput assays for yeast RNA 5′ triphosphatase (Cet1p) Anal. Biochem. 2008;372:89–95. doi: 10.1016/j.ab.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Poldermans B, Roza L, Van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16 S ribosomal RNA of Escherichia coli. III. purification and properties of the methylating enzyme and methylase-30 S interactions. J. Biol. Chem. 1979;254:9094–9100. [PubMed] [Google Scholar]
- 16.Denoya CD, Dubnau D. Site and substrate specificity of the ErmC 23S rRNA methyltransferase. J. Bacteriol. 1987;169:3857–3860. doi: 10.1128/jb.169.8.3857-3860.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Dwyer K, Watts JM, Biswas S, Ambrad J, Barber M, Brulé H, Petit C, Holmes DJ, Zalacain M, Holmes WM. Characterization of streptococcus pneumoniae TrmD, a tRNA methyltransferase essential for growth. J. Bacteriol. 2004;186:2346–2354. doi: 10.1128/JB.186.8.2346-2354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]