Abstract
Epixenosomes, ectosymbionts on hypotrich ciliates (genus Euplotidium) defend their host against the ciliate predator Litonotus lamella. Although here only Euplotidium itoi and Euplotidium arenarium from tide pools along a rocky shore near Leghorn (Ligurian sea) were studied in detail, these epibionts are certainly present on specimens of E. itoi and on other Euplotidium species in similar north coastal habitats. The complex life history of epixenosomes has two main stages. In stage I, cells with typical prokaryotic structure divide by binary fission. Stage II cells show complex organization with different cytoplasmic compartments where an extrusive apparatus within a proteinaceous matrix, although not membrane-bounded, differs from the remaining cytoplasm. The ejection process is involved in defense; extrusive apparatus is surrounded by a basket consisting of bundles of tubules. These tubules, 22 ± 3 nm in diameter, delimited by a wall made up of globular structures, are sensitive to inhibitor of tubulin polymerization (nocodazole/4°C temperature) and react positively with different antitubulin antibodies, two of which are monoclonal. The prokaryotic vs. eukaryotic nature of epixenosomes was resolved by comparative sequence analysis of amplified small subunit rRNA genes and in situ hybridization with fluorescently labeled rRNA-targeted polynucleotide probes. These unique ectosymbionts are phylogenetically related to Verrucomicrobia. Epixenosomes represent marine symbionts in this recently discovered division of the Bacteria.
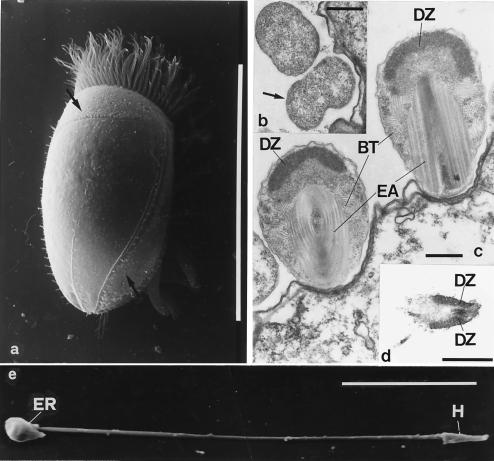
Peculiar episymbionts are located on the surface of marine ciliates of the Euplotidium genus. As these episymbionts present all the characteristics (with the exception of the internal localization) considered by Corliss (1) as being typical of the structures he called “xenosomes,” that is, from the ancient Greek, alien bodies, we refer to them as “epixenosomes,” i.e., external alien bodies. Epixenosomes of Euplotidium itoi and Euplotidium arenarium were studied in detail by microscopic and cytochemical techniques (2, 3). They possess quite similar features and have the same typical localization in a well defined cortical band on the host dorsal surface (see Fig. 1a). In the epixenosome complex life history, two main stages can be recognized. During stage I (see Fig. 1b) they are spherical (1 μm in diameter), have a simple bacteria-like cell organization, and are able to divide by direct binary fission. Stage I epixenosomes transform into stage II by gradually acquiring a more complex structure (2, 4). Fully formed stage II epixenosomes (see Fig. 1c) are larger and egg-shaped (2.5 μm long and 1.2–1.3 μm wide). Their most intriguing features are: (i) an electrondense, dome-shaped zone under the cell membranes in the upper region of the cell body. It contains DNA and basic proteins and its ultrastructure resembles of eukaryotic chromatin; (ii) a basket built up of bundles of regularly arranged tubules whose inner and outer diameters (22 ± 3 and 13 ± 1, respectively) fall into the range reported for tubulin microtubules (5). Moreover, they are delimited by a wall made up of globular subunits and are sensible to nocodazole and low temperature (4°C), two factors known to depolymerize cytoplasmic microtubules in a variety of eukaryotic cells (6); (iii) a sophisticated extrusive apparatus. In the resting position, it appears as a ribbon rolled up around a central core and immersed in a complex matrix consisting of proteins. This matrix differs in its composition from the remaining cytoplasm (4). External signals of unknown origin are detected by membrane receptors located at the top of the organism. The consequent activation of the adenylate-cyclase-cAMP system triggers the ejection of the ribbon (7). In the laboratory, it is possible to stimulate the triggering by the hormone adrenalin (7). During the ejecting process, the extrusive apparatus unrolls from the inside and forms a hollow tube, about 40 μm long. It terminates with a head consisting of the apical portion of the epixenosome (see Fig. 1 d and e). So the triggering of the extrusive apparatus leads to an ejection of a portion of the cytoplasm along with the densely packed genomic material, which is flung out of the remnants of the cell body and the host surface (4). The most similar structures to the epixenosomal extrusive apparatus so far described are the R bodies of Caedibacter teniospiralis, the bacterium that confers the killer trait to Paramecium aurelia (8). The extrusive apparatus, however, is a far more complex structure. There are also eukaryotic organelles consisting of coiled ribbons that unroll from the inside during ejection; for example, the ejectisomes of cryptoprotists (9).
Figure 1.
Epixenosomes and their host. (a) Dorsal view of E. itoi at scanning electron microscope. Arrows indicate epixenosomes in the cortical band. (Bar = 100 μm.) (b) Sectioned stage I epixenosomes. Arrow indicates the dividing one. (c) Stage II epixenosomes sectioned at different levels. DZ, Apical (dome-shaped) chromatin-like zone. EA, Extrusive apparatus. BT, Microtubule-like elements forming a “basket” around EA. (d) Section through the head of an ejected epixenosome. (Bars = 1 μm.) (e) The tube at the end of ejection. ER, Epixenosome remnant. H, Head. (Bar = 10 μm.)
Even the episymbiotic association between epixenosomes and the ciliate host, although not fully understood, certainly presents peculiarities. Epixenosomes are not vital for the ciliate, at least in a noncompetitive environment. It has been proven that not only the multiplication of epixenosomes but also their differentiation from stage I to stage II is correlated with the host cell cycle and that epixenosomes themselves also influence the morphogenesis of the ciliate in some way. The behavior of the cortical region corresponding to the epixenosomal band is different whether the episymbionts are present or not (10). Moreover, it has been experimentally demonstrated (11) that by their extrusive process epixenosomes defend the ciliate host from predators. Indeed, the ciliate predator Litonotus easily ingests Euplotidium without epixenosomes, whereas it is not able to ingest Euplotidium with epixenosomes unless in the latter the ejecting capacity of the symbionts is inhibited.
The complexity and the uniqueness of the cell structure makes it difficult to assign epixenosomes to a defined group of microorganisms. Moreover, it has been demonstrated that the microtubular structures of the basket reacted with different antibodies all specific for tubulin (6) (a typical eukaryotic protein), whereas no positive reaction was observed by in situ hybridization with an oligonucleotide (12) complementary to the majority of bacterial 16S rRNAs. On this basis, even the possibility that epixenosomes, in spite of the lack of a nuclear envelope, could be of eukaryotic origin has been considered (3).
In the present study, a more thorough molecular study was carried out with the aim to reconstruct the phylogenetic affiliation of epixenosomes.
Materials and Methods
Collection and Culturing.
Specimens of E. arenarium were collected from a rocky shore in the Ligurian sea and maintained in the laboratory in artificial sea water, periodically enriched with the green alga Dunaliella salina, the diatom Pheodactylum tricornutum, and the flagellate Chilomonas for food. Cultures were maintained at 20°C and received 12 h light and 12 h darkness.
Freshly collected specimens and the cultures were repeatedly checked for the presence of epixenosomes. Clones without the symbionts could be obtained in the laboratory in the following way: specimens were isolated and maintained in sea water not enriched with food organisms for up to 12 days. They were then singly transferred to the enriched culture medium and allowed to begin their cell cycle again.
Electron Microscopy.
For scanning electron microscope observation, specimens were fixed in 2% OsO4 in sea water. They were then placed on poly-l- Lysine hydrobromide-coated coverslips, dehydrated in ethanol, and, after critical point drying, coated with gold and examined with a JEOL/JSM-5410. For transmission, electron microscope specimen were fixed with 2.5% glutaraldehyde and 1% OsO4 in 0.1 M cacodylate buffer. After fixation, they were dehydrated and embedded in Epon–Araldite mixture. Sections were contrasted with uranyl acetate and lead citrate (13).
DNA Extraction.
As any attempt to separate epixenosomes from their host was unsuccessful, DNA was extracted from harvested cultures of E. arenarium together with their epixenosomes. For comparison, DNA was extracted from cultures of E. arenarium without symbionts generated in the laboratory. It was performed in 4.2 M guanidine thiocyanate, 0.037 M N-laurylsarcosine, 0.1 M Tris⋅HCl (pH 8.0), 0.1% β-mercaptoethanol, followed by phenol-chloroform extraction. To minimize the possible contaminants, the cultures were repeatedly washed in artificial, sterilized sea water before the extraction.
Sequence Analyses.
The analyses of small subunit (SSU) rRNA genes from the extracted DNA were performed by using various oligonucleotide primers for the in vitro amplification by the PCR. The primers used were (W. Ludwig, personal communication): (i) the universal bacterial forward 5′-AGAGTTTGATYMTGGCTC-3′, Escherichia coli positions 8–27, and reverse 5′-CAKAAAGGAGGTGATCC-3′, E. coli positions 1529–1542; (ii) universal for Eukaryotes forward 5′-AACCTGGTTGATCCTGCCAG-3′ E. coli positions 6–25, and reverse 5′-GATCCTTCTGCAGGTTCACCTAT-3′, E. coli positions 1511–1533; (iii) designed for Archaea forward 5′-ATTCYGKTTGATCCYGSC-3′, E. coli positions 6–23 and reverse 5′-GGAGGTGATCCAGCCGCAG-3′, E. coli positions 1521–1540. PCR products were then sequenced by an automated Li-Cor (Lincoln, NE) 4000L DNA sequencing device both directly using internal primers (14) and upon cloning using a commercial available TOPO TA cloning kit (Invitrogen). Cycle-sequencing protocols based on the chain-termination technique were applied using the Thermo Sequenase fluorescent labeled primer cycle sequencing kit of Amersham. Y = (C/T); M = (A/C); K = (G/T); S = (G/C).
Phylogenetic Analyses.
The determined sequences were aligned by using the sequence editor of the arb program package (14). The sequence was incorporated in a global tree with a special parsimony program of the arb package that allows no change in the overall topology of the tree. The topology within the Verrucomicrobia was then optimized by using only alignment positions that are invariant in at least 50% of the entire set of sequences of Verrucomicrobia.
In Situ Hybridization.
Samples of E. arenarium harboring epixenosomes were fixed with saturated mercurium chloride, washed in distilled water, resuspended in 0.1% agarose, attached by air drying to glass slides, and dehydrated up to 90% ethanol. Fluorescently labeled polynucleotide probes were synthesized by in vitro transcription of a variable region of the cloned 16S rRNA gene (E. coli positions 8–296) (15). The template for the in vitro transcription was generated by PCR using the bacterial universal forward primer (5′-AGAGTTTGATYMTGGCTCAG-3′) and the reverse primer EPI277-T3 (5′-ATAGGTATTAACCCTCACTAAAGGGACAGATCAGCTACCCGTCTTA-3′) specific for a small subgroup of Verrucomicrobia. The reverse primer contained a T3 RNA polymerase promoter sequence at its 5′-end (underlined sequence). The resulting PCR fragments were transcribed in polynucleotide probes by using fluorescein-labeled UTP. The hybridization solution contained 20 ng probe/μl, 90% formamide, 75 mM NaCl, 20 mM Tris⋅HCl (pH 8.0), and 0.01% SDS. The hybridization temperature was 53°C. The hybridization with the Cy3-labeled oligonucleotide probe Eare-832-Cy3 (5′-Cy3-ATTTCGCACGCCTCCAAT-3′) specific for Euplotidium was performed subsequently by using the standard protocol with 20% formamide in the hybridization buffer (16). Specimens of E. coli were treated in the same way as controls. Micrographs were obtained with the Zeiss confocal microscope LSM 510 v.2.01.
Results
Electron Microscopical Observations.
A dorsal view of E. itoi by scanning electron microscope is shown in Fig. 1a. Epixenosomes have a precise localization along a cortical band. Sections of epixenosomes at the two main stages of their life cycle are visible in Fig. 1 b and c. Epixenosomes following the ejecting process are seen in Fig. 1 d and e. Fig. 1d shows a section of the head of an ejected extrusive apparatus, and Fig. 1e is a picture of the whole formed tube.
Sequence Analyses.
DNA of E. arenarium with and E. arenarium without epixenosomes was amplified. No amplification products of the expected size were obtained when PCR was performed with the standard primers routinely used to amplify almost complete bacterial 16S rRNA genes. On the contrary, primers specific for most known eukaryotic 18S rRNA genes gave a distinct band. This PCR product was homogeneous in both samples and, upon direct sequencing, that obtained from E. arenarium with epixenosomes was identical with the 18S rRNA gene amplicon derived from the E. arenarium laboratory clone lacking symbionts. Using the pair of SSU rRNA gene primers originally designed for Archea, a faint PCR product with the expected size could only be retrieved when DNA was prepared from the host colonized with the symbionts. This PCR product was purified and directly sequenced. Based on the obtained partial sequence, it was possible to optimize the PCR by replacing the archaeal forward by the bacterial forward primer. Applying this bacterial/archaeal primer combination, the PCR yielded a distinct amplification product suitable for cloning and direct sequencing. The almost complete sequences of the cloned SSU rRNA genes were identical to each other and to the directly determined sequence of the amplicon, indicating that the obtained PCR product was homogeneous and most likely originated from the epixenosomes.
Phylogeny.
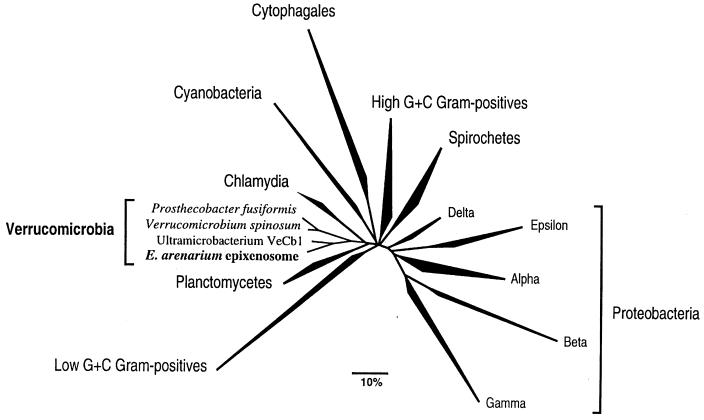
Comparative sequence analyses of the retrieved 16S rRNA with about 10,000 other known 16S rRNA sequences available in the arb database (14) revealed an affiliation to the recently discovered bacterial division Verrucomicrobia (17). In Fig. 2, a phylogenetic tree based on 16S rRNA sequences showing the position of the sequenced gene within the major lineages of bacteria is shown. To make the tree clearer, the less known divisions of Bacteria and environmental sequences were removed. The closest relatives of epixenosomes are recently cultivated ultramicrobacteria indigenous to anoxic rice paddy soil (18). However, the similarity of the determined sequence to the 16S rRNA sequences of the cultured ultramicrobacteria was around 84%, excluding any close taxonomic relationship.
Figure 2.
Phylogenetic tree based on 16S rRNA sequences showing the position of epixenosomes within the major lineages of Bacteria. The shown tree is derived from an optimized global tree reconstructed from more than 10,000 aligned SSU rRNA sequences belonging to all three domains using various treeing programs included in the arb package.
In Situ Hybridization.
To prove that the amplified and cloned sequences really originated from the epixenosomes and were not artifacts from contaminating bacteria, an in situ hybridization was done. The fluorescent polynucleotide probe Epi277-T3 was obtained by in vitro transcription of a highly variable region at the 5′ end of the cloned 16S rRNA gene (15). A polynucleotide probe was chosen for the in situ detection because they usually give stronger signals than oligonucleotide probes (19). Using this probe, epixenosomes ejected from the host as well as those associated with the host could be specifically detected. When the Cy3-labeled probe Eare-832-Cy3, specific for Euplotidium, was used along with the fluorescein-labeled polynucleotide probe for the symbionts, both organisms were each specifically labeled. Whereas the ciliate was stained in red by the Cy3-labeled probe, the green fluorescence could be seen only in correspondence to the epixenosomal band on the dorsal surface of E. arenarium or on the heads of the ejected tubes (Fig. 3). Labeling was never observed in control specimens.
Figure 3.
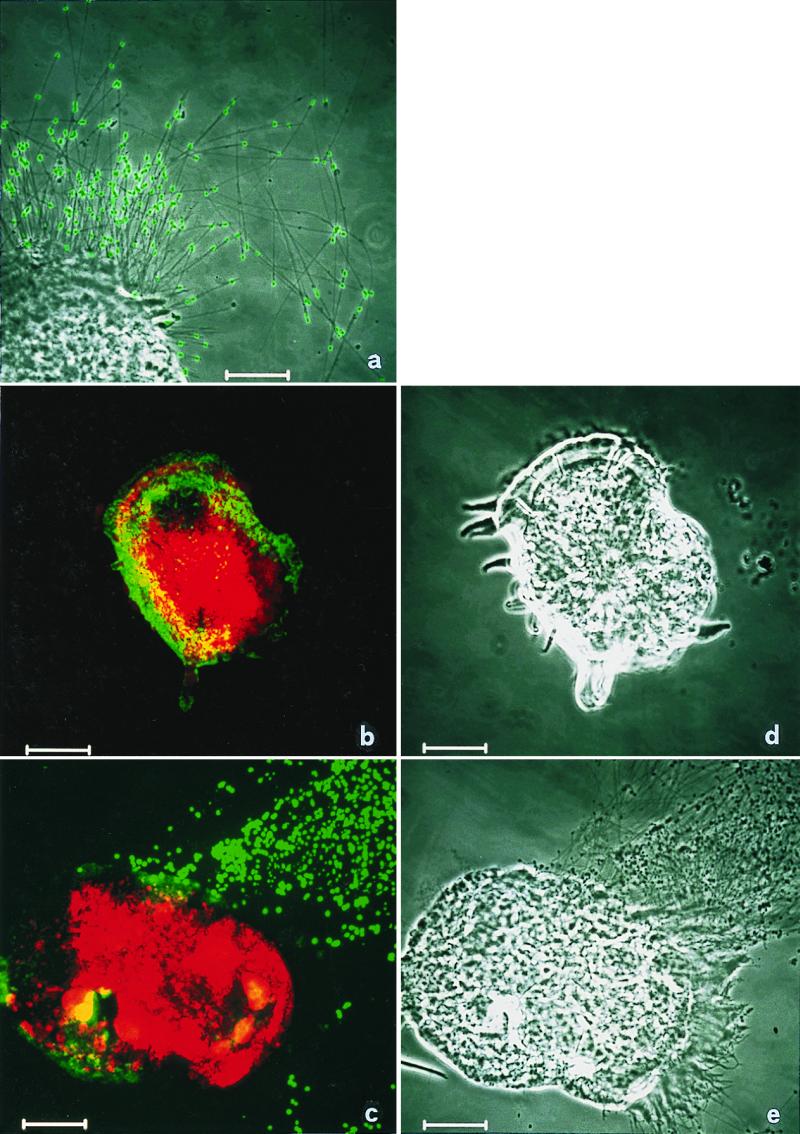
Detection of epixenosomes on E. arenarium. (a) Detail on ejected extrusive processes in phase contrast superimposed with the green signal of epixenosome specific probe. The heads of ejected epixenosomes are clearly labeled. (b and c) Double hybridization with the Cy3-labeled oligonucleotide probe Eare-832-Cy3 specific for Euplotidium (red signal), and with the fluorescein-labeled polynucleotide probe specific for epixenosomes (green signal). Both organisms are specifically labeled. The green fluorescence is visible only at the level of the epixenosomal band or on the heads of ejected tubes. (d and e) Corresponding phase contrast of b and c. (Bars = 10 μm.)
Discussion
Although epixenosomes have been extensively studied from different points of view, their real nature was obscure up to now. To investigate their phylogenetic affiliation, the comparative sequence analysis of genes coding for the RNA part of the small subunit of the ribosome (SSU rRNA) was chosen because this molecule is suitable for the phylogenetic identification of a wide range of organisms belonging to any of the three domains known as Bacteria, Archaea, and Eukarya (20). From the results reported here, it can be affirmed that epixenosomes, the peculiar episymbionts of ciliates of Euplotidium genus, are prokaryotes phylogenetically related to Verrucomicrobia, a recently discovered division of the Bacteria (17). Indeed, the specificity of the label obtained by in situ hybridization with the polynucleotide probe Epi277-T3 clearly demonstrates that we had amplified, cloned, and sequenced the 16S rRNA gene of epixenosomes. This 16S rRNA gene, like that of other Verrucomicrobia so far sequenced, has two mutations in the target region of the routinely used universal bacterial probe EUB338 (12). It may prevent the binding of this probe to the 16S rRNA molecule of epixenosomes. Therefore, it is not surprising that in previous experiments (3) carried out at the electron microscopical level, the oligonucleotide probe usually used for the in situ detection of bacteria did not react with epixenosomes. In the light of the present data, obtained with specifically constructed probes, the positive results obtained with some eukaryotic polynucleotide probes (3) were unspecific or artifactual.
It is noteworthy that no amplification product of epixenosomes was obtained with the primer set generally used for bacterial 16S rRNA genes, but with an atypical combination of the forward bacterial primers and the reverse archaeal primer. Consequently, the use of universal bacterial primers could have led to an underestimation of members of the Verrucomicrobia in studies of microbial ecology involving clone libraries. The phylogenetic depth of this bacterial division is nevertheless mainly represented by sequences retrieved from environmental samples and only two type species are validly described (17). The cultivated representatives of the Verrucomicrobia were isolated from different habitats and differ significantly in their morphological features. Verrucomicrobium spinosum and Prosthecobacter species were originally isolated from freshwater and are typical prosthecate (appendaged) bacteria, whereas the ultramicrobacteria (dwarf-cells, ranging in volume from 0.03 to 0.04 μm3) were found in wet soil (18). Uncultured representatives of the Verrucomicrobia seem to be particularly abundant in soil (21). Epixenosomes are to date the first report of symbionts among the Verrucomicrobia. In contrast, most other bacterial endo- and ectosymbionts of protists belong to the Proteobacteria (22). Epixenosomes appear to have unique morphological features among prokaryotes. Their complex structure involves, at least during stage II, the overproduction of a protein that is sensible to tubulin inhibitors, binds tubulin antibodies, and is organized in stable tubular structures (6), as well as of unidentified specialized proteins forming the extrusive apparatus. The presence of tubulin microtubules has never been demonstrated in prokaryotes (23). A tubulin-like protein, e.g., FtsZ, widespread among bacteria, is involved in the septum formation during cell division (24). However, although it is able to form tubules, in vitro (25), the formation of a basket-like structure as in the case of stage II epixenosomes has never been described. Moreover, the extrusion apparatus represents a unique feature. Interestingly, it is involved in the defensive role played by epixenosomes (11). A role that in other ciliates is played by cellular organelles, namely extrusomes of the trichocyst type (26–28). One may even speculate whether some trichocysts or other extrusomes originated from internalized epixenosome-like bacteria.
Acknowledgments
We thank Markus Schmid for his help with the confocal microscope and Simone Gabrielli for photographic work. G.P. is grateful to European Molecular Biology Organization for a short-time fellowship.
Abbreviation
- SSU
small subunit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the EMBL nucleotide sequence database [accession nos. Y19166 (E. arenarium) and Y19169 (epixenosomes of E. arenarium)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.030438197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.030438197
References
- 1.Corliss J O. J Protozool. 1985;32:373–376. [Google Scholar]
- 2.Verni F, Rosati G. J Protozool. 1990;37:337–343. [Google Scholar]
- 3.Rosati G. Symbiosis. 1999;26:1–23. [Google Scholar]
- 4.Rosati G, Verni F, Lenzi P. Eur J Protistol. 1993;29:238–245. doi: 10.1016/S0932-4739(11)80278-6. [DOI] [PubMed] [Google Scholar]
- 5.Margulis L. Symbiosis in Cell Evolution. 2nd Ed. San Francisco: Freeman; 1993. pp. 217–261. [Google Scholar]
- 6.Rosati G, Lenzi P, Verni F. Micron. 1993;24:465–471. [Google Scholar]
- 7.Rosati G, Giambelluca M A, Grossi M, Morelli A. Protoplasma. 1997;197:57–63. [Google Scholar]
- 8.Preer J R, Preer L B, Jurand A. Bacteriol Rev. 1974;38:113–163. doi: 10.1128/br.38.2.113-163.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kugrens P, Lee R E, Corliss J O. Protoplasma. 1994;181:164–190. [Google Scholar]
- 10.Giambelluca M A, Rosati G. Eur J Protistol. 1995;32:77–80. [Google Scholar]
- 11.Rosati G, Petroni G, Quochi S, Modeo L, Verni F. J Eukaryotic Microbiol. 1999;46:278–282. [Google Scholar]
- 12.Amann R I, Binder B J, Olson R I Y, Chisholm S W, Devereux R, Stahl D A. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulin J J. In: Protocols in Protozoology. Lee J J, Soldo A T, editors. Lawrence: Soc. Protozool.; 1992. , C-17.1. [Google Scholar]
- 14.Ludwig, W. & Strunk, O. (1997) arb: A Software Enviroment for Sequence Data (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps). [DOI] [PMC free article] [PubMed]
- 15.Spring S, Lins U, Amann R, Schleifer K-H, Ferreira L C S, Esquivel D M S, Farina M. Arch Microbiol. 1998;169:136–147. doi: 10.1007/s002030050553. [DOI] [PubMed] [Google Scholar]
- 16.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 17.Hedlund B P, Gosink J J, Staley J T. Antonie Leeuwenhoek. 1997;72:29–38. doi: 10.1023/a:1000348616863. [DOI] [PubMed] [Google Scholar]
- 18.Janssen P H, Schuhman A, Morschel E, Rainey F A. Appl Environ Microbiol. 1997;63:1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trebesius K, Amann, Ludwig W, Müllegger K, Schleifer K-H. Appl Environ Microbiol. 1994;60:3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woese C R, Kandler O, Wheelis M L. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felske A, Akkermans A D L. Lett Appl Microbiol. 1998;26:219–223. doi: 10.1046/j.1472-765x.1998.00322.x. [DOI] [PubMed] [Google Scholar]
- 22.Amann R, Ludwig W, Schleifer K H. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bermudes D, Hinkle G, Margulis L. Microbiol Rev. 1994;58:387–400. doi: 10.1128/mr.58.3.387-400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson H P. Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 25.Bramhill D, Thompson C M. Proc Natl Acad Sci USA. 1994;91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harumoto T, Miyake A. J Exp Zool. 1991;260:84–92. [Google Scholar]
- 27.Knoll G, Haacke-Bell B, Plattner H. Eur J Protistol. 1991;27:381–385. doi: 10.1016/S0932-4739(11)80256-7. [DOI] [PubMed] [Google Scholar]
- 28.Miyake A, Harumoto T. Eur J Protistol. 1996;32:128–133. [Google Scholar]