Abstract
Raltegravir (MK-0518) is the first integrase (IN) inhibitor to be approved by the US FDA and is currently used in clinical treatment of viruses resistant to other antiretroviral compounds. Virological failure of Raltegravir treatment is associated with mutations in the IN gene following two main distinct genetic pathways involving either the N155 or Q148 residue. Importantly, in most cases, an additional mutation at the position G140 is associated with the Q148 pathway. Here, we investigated the viral DNA kinetics for mutants identified in Raltegravir-resistant patients. We found that (i) integration is impaired for Q148H when compared with the wild-type, G140S and G140S/Q148H mutants; and (ii) the N155H and G140S mutations confer lower levels of resistance than the Q148H mutation. We also characterized the corresponding recombinant INs properties. Enzymatic performances closely parallel ex vivo studies. The Q148H mutation ‘freezes’ IN into a catalytically inactive state. By contrast, the conformational transition converting the inactive form into an active form is rescued by the G140S/Q148H double mutation. In conclusion, the Q148H mutation is responsible for resistance to Raltegravir whereas the G140S mutation increases viral fitness in the G140S/Q148H context. Altogether, these results account for the predominance of G140S/Q148H mutants in clinical trials using Raltegravir.
INTRODUCTION
Integrase (IN) inhibitors constitute a new class of antiretroviral agents blocking HIV-1 IN activity (1). HIV-1 IN is one of the three enzymes essential for viral replication. IN is responsible for integration of the reverse-transcribed double-stranded blunt-ended DNA into the host cell DNA and, is therefore an attractive target for anti-HIV drugs. IN catalyses two reactions: 3′-end processing and strand transfer (2). During 3′-end processing, the terminal GpT dinucleotides are cleaved from the 3′-end of each long terminal repeat (LTR), producing CpA 3′-hydroxyl ends. This reaction takes place within a nucleoprotein complex known as the preintegration complex (PIC). The PIC is then transported through the nuclear pore into the nucleus, where strand transfer occurs. During this second step, IN transfers both newly exposed 3′-extremities of the viral DNA into the target DNA by a one-step transesterification reaction, resulting in full-site integration (3).
To date, only integrase strand transfer inhibitor (INSTI) have been shown to have a potent antiviral activity in vivo (4,5). Raltegravir (RAL), a drug of the INSTI group, was recently approved for therapeutic use after clinical assays demonstrated a rapid, potent and sustained antiretroviral effect in patients with advanced HIV-1 infection (6). Because of its mechanism of action, this novel antiviral agent (ARV) is likely to be active against viruses resistant to other class of antiretroviral drugs such as nucleoside reverse transcriptase inhibitors (NRTI), non-nucleoside reverse transcriptase inhibitors (NNRTI), Protease (PI) and entry inhibitors. Indeed, RAL has already proven to be active when used with an optimized regimen in patients infected with drug-resistant viruses (6). However, resistance mutations are present in the IN gene of patients who fail to respond to RAL treatment. Resistance data from a clinical study at the Hospital Pitié-Salpêtrière show that resistance to RAL develops in two main pathways, either through mutations of the N155 or the Q148 residue (7). This study also highlights two important observations: (i) the N155 pathway may shift to the Q148 pathway over time. (ii) In most cases, the Q148H mutation occurs simultaneously with the G140S mutation. The Q148H mutation may be present as a single mutation at the beginning of the treatment, but the G140S mutation appears after a few weeks of treatment. Thus, the Q148 and G140 residues clearly play a key role in the treatment failure on RAL. Other large-scale studies have shown that the Q148 pathway is the most frequently observed of the two pathways in RAL treatment failure (8). Others mutations, such as the E92 and E157Q, have also been described (7).
We investigated the impact of the two main genetic resistance pathways (N155H and G140S/Q148H), on viral replication and the catalytic properties of recombinant INs. In particular, we investigated the effect of the G140S/Q148H double mutation, by constructing both the Q148H and G140S single mutants as well as the double mutant. We also studied the effect of IN background (laboratory strain or patient) on the properties of the enzymes. We found that the Q148H mutation caused resistance to RAL when present alone. However, this mutation severely impaired viral replication kinetics in addition to the catalytic activity of the recombinant IN. The G140S mutation did not confer strong resistance, but restored the replication capability of the Q148H mutant. Accordingly, the in vitro activity of the G140S/Q148H mutant can reach a wild-type level of activity while the single mutant Q148H cannot. Our kinetic study reveals that Q148H is a catalytic mutant blocked in an inactive conformation. The G140S mutation induces a conformational transition compatible with activity. Thus, the combination of these two mutations results in a virus that is both capable of replication and highly resistant to RAL. Finally, we found that the G140S/Q148H mutant was much more resistant than the N155H mutant. These findings are consistent with the switch from the N155 to the Q148 pathway observed after a few weeks of RAL treatment.
MATERIALS AND METHODS
Cells and viruses
MT4 cells were cultured in RPMI 1640 containing 10% fetal calf serum. 293T and HeLa-P4 cells were cultured in Dubelcco's modified Eagle medium supplemented with 10% fetal calf serum, 100 units penicillin/ml (Invitrogen), and 100 µg streptomycin/ml (Invitrogen).
HIV-1 IN mutants were generated as previously described. Briefly, the fragment encoding IN of the replication-competent pNL43 virus was digested with AgeI and EcoRI, inserted into the Bluescript vector and IN mutants were obtained by mutagenesis (Quick change mutagenesis kit, Stratagene). We studied the E92Q, G140S, Q148H, N155H and G140S/Q148H mutations in a pNL43 background. The constructs were checked by sequencing and the fragment was then inserted into pNL43. HIV-1 virus stocks of all mutants were prepared by transfecting 293T cells. Transfection assays were carried out by the calcium phosphate method. Viral supernatants, 48 h post-transfection, were filtered through a 0.45 µm-pore-size-filter and frozen at −80°C. HIV-1 p24gag antigen contents in viral supernatants were determined by enzyme-linked immunosorbent assay (Perkin-Elmer Life Sciences).
HIV infectivity assay
Single-cycle titers of the virus were determined in HeLa-P4 cells. HeLa-P4 cells are HeLa CD4 LTR-LacZ cells in which lacZ expression is induced by the HIV transactivator protein Tat, making it possible to quantify HIV-1 infectivity precisely from a single cycle of replication. Cells were infected, in triplicate, in 96-well plates, with virus (equivalent of 3 ng of p24gag antigen). The single-cycle titers of viruses were determined 48 h after infection by quantifying β-galactosidase activity in P4 lysates in a colorimetric assay (the CPRG assay) based on the cleavage of chlorophenol red-β-D-galactopyranoside (CPRG) by β-galactosidase. For IC50 determination, cells were infected with viruses and grown in the presence of increasing concentrations of RAL or the diketo-acid L731-988. The 50% inhibitory concentration (IC50) was determined as the drug concentration giving 50% inhibition of β-galactosidase levels with respect to untreated infected cells. Cell survival was also estimated with a standard MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay.
Viral infections
MT4 cells were concentrated at 2 × 106/ml and infected with viruses (50 ng of p24gag antigen per 106 cells). When required, cells were treated with 500 nM RAL inhibitor (Merck & Co.) or 25 µM zidovudine (AZT; Sigma) before infection. In the experiment with RAL, the medium containing the drug was replaced every three days (to maintain drug concentration). At various time points after infection, one to 3 million cells were harvested and dry cell pellets were frozen at −80°C until use.
DNA extraction and real-time PCR
Total cell DNA was extracted with a QIAamp blood DNA minikit (QIAGEN, Courtaboeuf, France). Quantifications of total HIV-1 DNA, 2-LTR circles and integrated HIV-1 DNA were performed by real-time PCR on a LightCycler instrument (Roche Diagnostics) using the fit point method provided in the LightCycler quantification software, version 3.5 (Roche Diagnostics) as previously described (9).
Cell equivalents were calculated based on amplification of the β-globin gene (two copies per diploid cell) with commercially available materials (Control Kit DNA; Roche Diagnostics). 2-LTR circles, total and integrated HIV-1 DNA levels were determined and expressed as copy numbers per 106 cells.
Characterization of IN protein activity in vitro
Two IN sequences, F4 and F12, obtained in a clinical study and corresponding to the IN sequences of a patient before and after treatment with RAL, respectively, were expressed, as previously described (7). The F4 polymorphism consists of eight mutations with respect to the ‘laboratory’ WT pNL43 IN: K7Q, E11D, L101I, K127R, I135V, I200L, V201I, I220L. F12 harbors these mutations together with the G140S/Q148H double mutation in the F4 background. The introduction of the entire sequence of the IN gene with the G140S/Q148H mutation from the patient into the pNL43 context did not result in a productive infection, suggesting that other viral proteins of the patient were required for efficient replication (data not shown). The comprehension of the potential influences of other viral proteins on viral replication is in progress in our laboratory.
In parallel, the E92Q, G140S, Q148H, N155H and G140S/Q148H mutations were obtained by site-directed mutagenesis from pET-15b, containing the WT sequence. The wild-type and mutant HIV-1 INs used for DNA-binding and 3′-processing assays were produced in Escherichia coli BL21 (DE3) and purified under non-denaturing conditions as previously described (10).
Steady-state fluorescence anisotropy-based assay
Steady-state fluorescence anisotropy values (r) were recorded on a Beacon 2000 Instrument (Panvera, Madison, WI), in a cell maintained at 25°C or 37°C under thermostatic control. The principle underlying the anisotropy-based assay was published elsewhere for DNA-binding (11,12) and 3′-processing (13,14), respectively. Briefly, IN binding to fluorescein-labeled DNA (double-stranded 21-mer oligonucleotide (ODN) mimicking the U5 viral DNA end) increases the r value, making it possible to calculate the fractional saturation function: ([DNA*IN]/[DNA]0). DNA-binding step was recorded at 25°C, using ODNs fluorescein-labeled at the 3′-terminal GT nucleotide. The percentage of complexes was then calculated according to:
| 1 |
where rfree and rsat are the anisotropy values characterizing the free and bound oligonucleotides, respectively.
Following the DNA-binding step, the sample was then shifted to a permissive temperature for the recording of 3′-processing activity (37°C). As the fluorophore is linked to the released dinucleotide, 3′-processing activity significantly decreases the r value with respect to that for the non-processed DNA. Activity can be calculated in fixed-time experiments, by disrupting all the IN/DNA complexes with SDS (0.25% final). The fraction of dinucleotides released is given by:
| 2 |
where rNP and rdinu are the anisotropy values for pure solutions of non-processed double-stranded ODN and dinucleotide, respectively. The formation of IN/DNA complexes and the subsequent 3′-processing reaction were performed by incubating fluorescein-labeled ODNs (4 nM) with IN in 20 mM Hepes pH 7.2, 1 mM dithiothreitol, 30 mM NaCl and 10 mM MgCl2. Standard 3′-processing and strand transfer activity tests based on gel electrophoresis were performed as previously described (10).
RESULTS
Resistance of viral mutants to RAL
Sequence analysis of clinical isolates obtained during RAL treatment led to the identification of various mutations at specific positions. Two main profiles were identified, one based on the N155H mutation and the other based on the G140S/Q148H mutations, which seemed to be present together in most cases. After RAL administration, only mutations in the IN gene were observed (7).
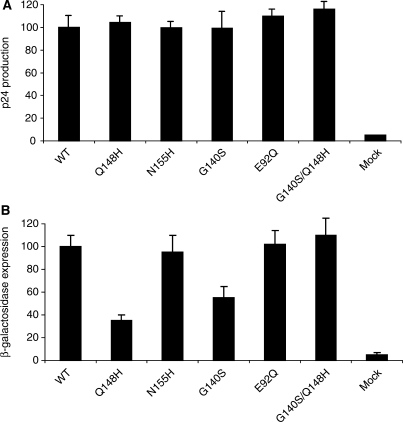
We introduced IN mutations (i.e. N155H or E92Q or G140S/Q148H) into a WT pNL43 background. Two other constructs with single G140S or Q148H mutations were analyzed in parallel. Levels of p24 production were determined 48 h after transfection. They were found to be similar for all mutations (Figure 1A). We can therefore conclude that these mutations do not significantly impair virus assembly or the release step. HeLa-P4 cells were then infected for 48 h to study the early replication step. The mutations clearly had differential effects on viral infectivity (Figure 1B). The two single mutations, N155H and E92Q, neither disrupt the early steps of replication nor expression of integrated DNA, as shown by the corresponding β-galactosidase levels, which were similar to the WT. However, the Q148H and G140S mutations significantly decreased viral infectivity (3-fold and 2-fold less β-galactosidase expression for Q148H and G140S, respectively, as compared with the WT level), probably because of a defect in the integration process. Interestingly, the combination of these two mutations in the same virus resulted in levels of viral infectivity similar to those of the WT and much higher than obtained with the Q148H and G140S single mutations. These findings suggest that these two point mutations individually impair the viral infectivity but that their combination results in a wild-type level of infectivity.
Figure 1.
p24 and infectivity of IN mutant viruses. (A) Quantification of p24 protein, 48 h after transfection of 5 µg of each virus DNA. (B) Viral infectivity for WT and mutants. Viral infectivity was determined in a single-cycle replication assay using HeLa p4 indicator cells and 3 ng of p24 antigen for each virus. Cells were exposed to virus during 48 h. Early steps of infections were assessed by measuring β-galactosidase activity in cell extracts by the CPRG method. For panels A and B, the results are expressed as percentages of the value obtained for the WT. The data shown are the means of three independent experiments.
We investigated the mechanism underlying the effects of these mutations on resistance to RAL, by determining the IC50 for each IN mutant (Figure 2A). The IC50 value (10 nM) obtained for the WT virus confirmed the potency of RAL as an inhibitor of HIV-1. At 24 or 72 h, no cytotoxic effects from the concentrations used in this experiment were observed in the MTT assay after infection (Figure 2B). G140S and E92Q mutants had slightly higher IC50 values (30 nM) than the WT. In sharp contrast, the N155H, Q148H and G140S/Q148H mutants had much higher IC50 values, at 130, 450 and >1000 nM, respectively. Thus, all the mutants identified in clinical trials using RAL were resistant to this compound, but to different extents. The same experiment was conducted with the strand transfer inhibitor L,731-988, a diketo acid which, similar to RAL, belongs to the INSTI group. The resistance profiles observed with this drug followed the same pattern as RAL (data not shown) but the values obtained were in low micromolar range for L,731-988, rather than in the nanomolar range as seen with RAL. Thus DKA, a well-characterized drug and RAL, both of which share the same mechanism of action, probably bind to the same binding site, inhibiting the strand transfer reaction by interfering with the binding of target DNA to IN.
Figure 2.
Resistance of IN mutants to RAL. (A) HeLa p4 cells were infected, in triplicate, with 3 ng of each virus, in the presence of various RAL concentrations. β-Galactosidase production was quantified by the CPRG assay. Data from a representative experiment (performed three times) is shown. The IC50 was determined as the concentration of RAL inhibiting β-galactosidase production by 50% with respect to untreated infected cells. (B) MTT assay. The MTT assay was performed 48 and 72 h after infection for all viruses. For the WT and G140S/Q148H mutant, the assay was performed with and without 500 nM RAL. The data shown are the means of three independent experiments.
Effects of mutations on viral DNA forms during replication
We investigated the effects of mutations during viral replication, by real-time PCR (qPCR) analysis, on the different viral nucleic acid species (total viral DNA, 2-LTR circles and integrated viral DNA) in cells infected with WT virus or resistant mutants.
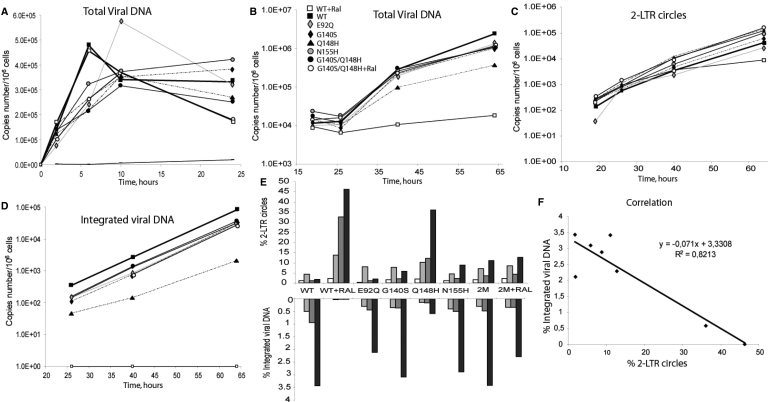
Total viral DNA synthesis during the first 24 h after infection was similar for all mutants studied, and the kinetics of DNA synthesis were similar to those of the WT (Figure 3A). Furthermore, for all mutant and WT viruses, DNA synthesis peaked 10 h after infection, as typically reported in such studies (15), indicating that none of the mutations significantly affected the timing of the reverse transcription step. As a control, no viral DNA synthesis was detected with the WT in the presence of 25 µM AZT. We performed the same experiment for the WT and the G140S/Q148H mutant in the presence of 500 nM RAL (a concentration 50 times higher than the IC50 value for the WT). Viral DNA synthesis levels were similar in the presence and absence of RAL for both the WT and the G140S/Q148H mutant (Figure 3A), confirming the absence of RAL effect on the reverse transcription step.
Figure 3.
Kinetics of viral DNA synthesis. CEM cells were infected with 40 ng of p24 antigen and levels of intracellular HIV-1 DNA species were monitored by qPCR. (A) Dynamics of total viral DNA during the first 24 h of infection. WT+RAL (open square), WT (filled square), E92Q (gray diamond), G140S (filled diamond), Q148H (filled triangle), N155H (gray circle), G140S/Q148H (filled circle), G140S/Q148H+Ral (open circle), WT + AZT (straight line). (B-D) Quantification of viral DNA over a 3-day period. (B) Total Viral DNA (C) 2-LTR circles (D) integrated HIV-1 DNA. The scales in panels A to D are logarithmic. Depicted results were obtained from a representative experiment. (E) The percentage of 2-LTR circles and integrated viral DNA were determined by calculating the ratio of 2-LTR circles copy number and integrated DNA levels, respectively, over total viral DNA. (F) Correlation between the percentages of 2-LTR circles and integrated viral DNA. The data shown are the means of three independent experiments.
We quantified total viral DNA synthesis at a later time point to assess the ability of mutants to replicate in infected cells. qPCR quantification showed the kinetics of total viral DNA accumulation to be similar for the WT and all mutants, with the exception of the Q148H virus, for which smaller amount of viral DNA were detected, i.e. 5- and 10-fold less than for the WT virus, 39 and 64 h after infection, respectively. The apparent discrepancy between β-Gal assays and integration quantification of the G140S mutant will be further discuss in the next section. For WT infection in the presence of RAL, only a faint viral DNA signal was detected after the reverse transcription step: viral DNA, was reduced by 10- and 100-fold, 39 and 64 h after infection, respectively, when compared with WT infection in the absence of RAL, demonstrating the efficacy of this compound for blocking viral replication. In sharp contrast, RAL had only a slight effect on the DNA synthesis of the G140S/Q148H mutant (DNA synthesis was reduced by 1.2-fold at 64 h after infection), confirming the strong resistance of this double mutant. Moreover, Figure 3B clearly shows the better fitness of G140S/Q148H in comparison to the single-mutant Q148H.
Both efficient IN inhibitors and mutations in the catalytic triad DD35E of IN have been reported to cause a decrease in the amount of integrated DNA, with a concomitant increase of 2-LTR circles (16,17), suggesting that 2-LTR circles molecules results from a defect in the integration process. We then analyzed the kinetics of 2-LTR accumulation and quantified integrated viral DNA, to obtain greater insight into the effects of the mutations and of RAL on the integration process (Figure 3C and D). The percentages of 2-LTR circles and integrated DNA viral forms are shown in Figure 3E. Again, our results clearly demonstrate that the Q148H mutant was more strongly affected than other mutants, as shown by the significant decrease in the amount of integrated DNA forms and the simultaneous accumulation of 2-LTR circles. A qualitatively similar but more pronounced effect was observed with the WT in the presence of RAL. No integrated DNA was detected, despite the sensitivity of the qPCR approach (9), demonstrating the efficiency of RAL as an inhibitor of the integration process. Remarkably, RAL had only a small effect on the levels of 2-LTR circles and integrated forms with the G140S/Q148H double mutant (Figure 3E). Figure 3F shows the correlation between accumulation of the episomic viral genome and the decrease in the amount of integrated DNA. This correlation supports the idea that the accumulation of 2-LTR circles results principally from an integration defect.
We assayed the supernatant of infected cells for the presence of viral particles, by quantifying p24 48 and 72 h after infection, to determine whether viral production was compatible with the amount of integrated DNA. Consistent with the quantification results obtained for the different DNA forms (Figure 3), only the production of p24 by the WT in the presence of RAL and by the Q148H mutant was severely impaired (p24 production decreased by 25- and 6-fold, respectively) (Figure 4). Figure 4 also confirms the resistance of G140S/Q148H as shown by the limited effect of RAL on the p24 production of the double mutant. The viral particles obtained from all IN mutants were infectious (data not shown).
Figure 4.
p24 production. Viral particles released in the supernatant were determined 48 and 72 h after infection by quantification of the p24 protein (see Materials and methods section). The data shown are the means of four independent experiments.
Thus, IN mutations have no crucial consequences for the dynamics of viral replication, except the for the Q148H mutant, which displayed high levels of 2-LTR circle accumulation related to an integration defect. Therefore, the similar profile of this mutant as compared to mutants of the catalytic triad strongly suggests a defect of IN at the catalytic level (18). We then studied the effect of the Q148H and/or G140S mutations on 3′-processing and strand transfer activities using recombinant proteins.
G140S rescues the catalytic defect due to the Q148H mutation
The G140S/Q148H double mutation is one of the main profiles identified in patients resistant to RAL. These mutations seem to appear simultaneously or over a very short period of time in patients treated with this compound. We investigated the role of the G140 and Q148 residues in the resistance, by producing recombinant INs harboring either the G140S or the Q148H mutation, and the G140S/Q148H double mutant. In parallel, we also obtained recombinant proteins encoding the entire IN sequence, as found in patients with the double G140S/Q148H mutation.
A plot of 3′-processing activity (corresponding to 3 h of incubation at 37°C) as a function of IN concentration gave a characteristic bell-shaped curve, with activity peaking at a concentration of about 200 nM, for both WT IN, expressed in NL-43 context (Figure 5A, left panel) or in patients context before RAL treatment (Figure 5A, right panel) in accordance with previous results (13,14). Under the same conditions, the activities of the Q148H, G140S and G140S/Q148H were severely impaired, with the degree of impairment as follows: Q148H < < G140S/Q148H ≈ G140S < WT (Figure 5). It is important to note that both proteins, WT and G140S/Q148H, in the patient background displayed stronger activity than INs in the NL-43 context. We are currently investigating the effects of the polymorphism on intrinsic IN activity.
Figure 5.
Comparative study of the DNA-binding and catalytic properties of wild-type and RAL-resistant INs. (A) 3′-Processing activity—after 3 h of incubation at 37°C—as a function of IN concentration. 3′-Processing activities were quantified as described in Materials and methods section, using a 21-mer DNA substrate (4 nM), with MgCl2 as a cofactor (10 mM) in 20 mM Hepes (pH 7.2), 1 mM DTT and 30 mM NaCl. (B) DNA binding of wild-type and mutant INs. The DNA-binding step was assessed by steady-state fluorescence anisotropy as described in Materials and methods section. Experimental conditions were similar to those described in A. IN and DNA were incubated together for 15 min before recording steady-state anisotropy. (C) Kinetics of 3′-processing for the different proteins. IN concentration was 200 nM. The same symbols were used in panels A, B and C: (open square) wild-type NL-43; (filled square) G140S/Q148H NL-43; (filled triangle) Q148H NL-43; (open triangle) G140S NL-43; (open circle) wild-type patient; (filled circle) G140S/Q148H patient. Strand transfer products are depicted (middle panel). The percentages of strand transfer (shown besides the gel) were obtained after the normalization by the 3′-processing activity.
As the increase in the IN concentration did not compensate for the defect in 3′-processing activity of the mutants (i.e. increasing the concentrations of mutant proteins did not result in wild-type levels of activity, as shown in Figure 5A), the defect was probably not because of a decrease in the overall affinity for the DNA substrate, being instead due to a catalytic defect. Accordingly, quantification of IN–DNA complexes by steady state anisotropy (Figure 5B), indicated that complex formation was not altered by any type of mutation. Our activity data after 3 h of incubation highlight some discrepancies between in vitro and ex vivo results, particularly for the G140S/Q148H double mutant virus, which replicated as well as the WT virus, despite the lower activity of the corresponding recombinant IN.
To understand this apparent discrepancy, we next studied the entire 3′-processing kinetics of WT and mutants. Interestingly, the kinetics of processing for the G140S and G140S/Q148H, although delayed in time as compared to the WT (t1/2 = 20.8 h for mutants, t1/2 = 4.1 h for WT), showed that these two mutants were able to reach WT levels of activity (Figure 5C). Again, this result was independent of the context (NL-43 or patients) (compare left and right panels). The sigmoidal nature of the kinetics observed strongly suggests that IN must undergo a conformational transition to shift from a catalytically inactive state to an active state (INI⇔INA). 3′-Processing activity as analyzed by gel-electrophoresis with standard procedures (10) gave similar results (data not shown). Moreover, quantification of the product of the strand transfer reaction indicated that, under conditions in which the 3′-processing levels of the G140S/Q148H reached the WT levels, the yields of strand transfer are similar for the WT and the double mutant. In fact, after normalization by the 3′-processing activity, we found that the strand transfer efficiency was 6.5%, 2.2% and 5.5% for WT, Q148H and G140S/Q148H mutants, respectively (Figure 5C). In contrast to G140S and G140S/Q148H, the single mutant Q148H appears to be more severely impaired than the other mutants, with a 3′-processing activity increasing only slightly to 35%, after up to 50 h of incubation, with concomitant decrease in the yield of strand transfer (Figure 5C). Most likely, the INI⇔INA transition accounts for the slow single turn-over rate constant characterizing IN (12). The effect of the Q148H mutation which strongly impairs this transition is reversed by adding the G140S mutation. These two residues belong to the catalytic loop 140–149, the flexibility of which was previously described to be essential for activity (19,20). The Q148 residue was shown to be essential to establish specific contacts with viral DNA (21,22). Our results, showing that overall affinity of the IN-DNA complex was not influenced by the Q->H mutation at this position, do not exclude a fine repositioning of the catalytic loop relative to the viral DNA end, leading to a non-competent catalytic complex. Interestingly, two other mutations of the catalytic loop (G149A and G149A/G140A) display similar post DNA-binding defects (20). The residue G140 participates in catalytic loop hinge formation and its mutation could restore specific contacts compatible with catalysis between the loop of the double mutant and the viral DNA end. Simulation and molecular dynamics (MD) studies on the catalytic loop of IN is under progression in our laboratory. We have found that the Q148 residue belongs to a Ω-shaped hairpin (144–148) that can move in a gate-like manner toward the active site as a rigid body. The G140 residue is not directly involved in the flexibility of the catalytic loop but plays a critical role in controlling the overall motion of the loop and finally in controlling its precise positioning relative to the phosphodiester bound to be cleaved (‘In Silico study suggests that raltegravir-resistant mutations modify the DNA recognition properties of HIV-1 Integrase’ by Tchertanov et al., Third International Conference on Retroviral Integrase; September 14–18, 2008).
DISCUSSION
To date, only INSTIs have been shown to be true inhibitors of HIV-1 DNA integration. These compounds specifically inhibit strand transfer and target the pre-integration complex in vivo. RAL (or MK-0518) is a member of the INSTI family. This IN inhibitor was the first to be approved by the FDA for the treatment of AIDS. RAL is generally well tolerated although a recent study suggested an association between RAL and rhabdomyolysis (23). It is currently undergoing late-stage clinical trials with patients infected with multidrug-resistant HIV-1 viruses. However, several mutations occur very rapidly, within 11 weeks of beginning treatment. These mutations are E92Q, N155H and the G140S/Q148H double mutation, which seems to appear preferentially (8).
In this report, we analyzed the effect of mutations introduction in both the viral context and in the recombinant IN, focusing our studies on the double mutant. Our data demonstrate that each mutation described conferred resistance, but the extent of that resistance differed between mutations. The G140S/Q148H double mutation confers strong resistance to the drug and viral replication levels similar to those of the WT virus. Importantly, the G140S displays weak resistance (IC50 = 30 nM) while the Q148H is strongly resistant to RAL (IC50 > 700 nM). Taken together, these data suggest that resistance results principally from the Q148H mutation. In the viral context, all mutants, including G140S, displayed replication kinetics similar to those to the WT virus, with the exception of the Q148H mutant which is characterized by a slower kinetic. Consistently, all mutants are weakly impaired in the synthesis of viral DNA as well as in their propensity to integrate their genome and consequently to produce viral particles with the notable exception of the Q148H mutant. Its integration efficiency is 7-fold less in comparison to the WT. These data demonstrate that, in the case of the G140S/Q148H double mutant, the resistance to RAL is due to the Q148H and that the G140S mutation rescues the integration deficiency and thus the kinetic of replication.
It is important to note that β-Gal assays using the HeLa-P4 cells suggests that the G140S mutant was impaired in the viral integration process, in accordance with results of Nakahara et al. (24). However, we found that the integration efficiency of the G140S was similar to those found for the WT and G140S/Q148H double mutant. To date, the reason of such discrepancy is not clear but the type of cells used for integration quantification could be crucial in modulating the mutation impact, especially in the case of the G140S mutation. Indeed, Nakahara et al. (24) have found that the viral replication was delayed in Jurkat cells but not in PBMC cells. Such a differential response of the G140S mutation depending on the cellular context could account for the lack of correlation between β-Gal assays (Figure 1) and integration quantification (Figure 3).
These data were confirmed by in vitro activity experiments. We observed that G140S/Q148H IN, although kinetically affected as compared to the WT IN, displayed WT levels of activity if the incubation time was increased (kinetic mutant). The Q148H IN was much more severely impaired and cannot reach the WT level of activity (thermodynamic mutant). Our activity data—at long incubation time—highlight a correlation between in vitro activity of IN and viral replication. Nevertheless, the slower kinetic of either G140S or G140S/Q148H, as seen in vitro, had no effect on the replication cycle of corresponding viruses. It is possible that the slow conformational transition involving the flexibility of the catalytic loop (see Results section) may not be kinetically limiting in the PIC context due to interactions with protein partners. Taken together, we hypothesized that the Q148H mutation primarily confers resistance to RAL. However, this mutation severely impaired viral replication. The G140S mutation counteracted this detrimental effect for the virus and increased viral fitness. These data clearly account for the high frequency of the G140S/Q148H mutant in clinical trials. It is important to note that the Q148H/R single mutation may occur 8 weeks after starting RAL treatment. However, 8–10 weeks later, the G140S mutation is also detected in patients. Our data of the compensation of the Q148H replication efficiency by the G140S explain why very rapidly after the administration of RAL, the G140S/Q148H mutant is detected. In addition, our data demonstrate that the Q148H mutant is more resistant to RAL in comparison to the N155H mutant. This observation explains why the N155 pathway often switched to the Q148 pathway. It is important to note that, during the course of our study, similar results were obtained by Nakahara et al. (24). The authors have studied the Q148K/R and G140S mutations as well as the G140S/Q148R and G140S/Q148K double mutants. Mutants Q148K/R displayed reduced viral fitness whatever the cell line studied (PBMC or Jurkat cells). In the case of the G140S mutant, the viral fitness is only delayed in Jurkat cells. In accordance with our results, Nakahara et al. have shown that the G140S mutation rescues the fitness of the Q148K/R mutant. Taken together, results from Nakahara et al. and our study show that the G140S mutation rescues the ability of the Q148H/K/R mutants to replicate in cells while mutation of the Q148 residue is responsible for the resistance to the drug. Note that the discrepancy concerning the nature of the mutation (K, R in the Nakahara's experiments and H in our study) is only apparent since resistance mutation were not obtained against the same anti-IN compound: S-1360 and GSK-364735 were used by Nakahara et al. while, in this study, we used RAL.
The development of strand transfer-specific inhibitor classes is an important achievement for the IN drug design. However, continued drug development is required as the ability of HIV to replicate under therapeutic pressure will undoubtedly lead to the emergence of IN drug-resistant viral strains characterized by comparable fitness to WT such as the double mutant G140S/Q148H.
To prevent from the appearance of cross-resistance, other sites of IN must be targeted as already done for the development of RT inhibitors. For example, sites responsible for the oligomerization of IN or/and interactions with cellular partners as LEDGF constitute good candidates. The combination of several IN inhibitors in optimal regimen will undoubtedly lead to optimal treatment for patients.
FUNDING
This work was supported in part by the Agence Nationale de Recherche sur le SIDA (ANRS) and by the Fondation pour la Recherche Médicale (FRM). Funding for open access charge: The French Agence Nationale de Recherche contre le SIDA (ANRS).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Françoise Simon for technical assistance and Hervé Leh for helpful discussions. We thank Ede Osemwota for reviewing the manuscript.
REFERENCES
- 1.Pommier Y, Johnson AA, Marchand C. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 2005;4:236–248. doi: 10.1038/nrd1660. [DOI] [PubMed] [Google Scholar]
- 2.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J. Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, Mizuuchi M, Burke T.R., Jr., Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeJesus E, Berger D, Markowitz M, Cohen C, Hawkins T, Ruane P, Elion R, Farthing C, Zhong L, Cheng AK, et al. Antiviral activity, pharmacokinetics, and dose response of the HIV-1 integrase inhibitor GS-9137 (JTK-303) in treatment-naive and treatment-experienced patients. J. Acquir. Immun. Defic. Syndr. 2006;43:1–5. doi: 10.1097/01.qai.0000233308.82860.2f. [DOI] [PubMed] [Google Scholar]
- 5.Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, et al. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 6.Grinsztejn B, Nguyen BY, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet. 2007;369:1261–1269. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- 7.Malet I, Delelis O, Valantin MA, Montes B, Soulie C, Wirden M, Tchertanov L, Peytavin G, Reynes J, Mouscadet JF, et al. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 2008;52:1351–1358. doi: 10.1128/AAC.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller MD, Danovich RM, Ke Y, Witmer M, Zhao J, Harvey C, Nguyen BY, Hazuda D. Longitudinal analysis of resistance to the HTV-1 integrase inhibitor raltegravir: results from P005 a Phase II study in treatment-experienced patients. Antiviral Therapy. 2008;13:A8. [Google Scholar]
- 9.Brussel A, Delelis O, Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol. Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 10.Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochon JC, LeCam E, Coulaud D, Auclair C, Mouscadet JF. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 11.Agapkina J, Smolov M, Barbe S, Zubin E, Zatsepin T, Deprez E, Le Bret M, Mouscadet JF, Gottikh M. Probing of HIV-1 integrase/DNA interactions using novel analogs of viral DNA. J. Biol. Chem. 2006;281:11530–11540. doi: 10.1074/jbc.M512271200. [DOI] [PubMed] [Google Scholar]
- 12.Smolov M, Gottikh M, Tashlitskii V, Korolev S, Demidyuk I, Brochon JC, Mouscadet JF, Deprez E. Kinetic study of the HIV-1 DNA 3'-end processing. FEBS J. 2006;273:1137–1151. doi: 10.1111/j.1742-4658.2006.05139.x. [DOI] [PubMed] [Google Scholar]
- 13.Delelis O, Carayon K, Guiot E, Leh H, Tauc P, Brochon JC, Mouscadet JF, Deprez E. Insight into the integrase-DNA recognition mechanism: a specific DNA-binding mode revealed by an enzymatically labeled integrase. J. Biol. Chem. 2008;283:27838–27849. doi: 10.1074/jbc.M803257200. [DOI] [PubMed] [Google Scholar]
- 14.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J. Biol. Chem. 2006;281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 15.Maroun M, Delelis O, Coadou G, Bader T, Segeral E, Mbemba G, Petit C, Sonigo P, Rain JC, Mouscadet JF, et al. Inhibition of early steps of HIV-1 replication by SNF5/Ini1. J. Biol. Chem. 2006;281:22736–22743. doi: 10.1074/jbc.M604849200. [DOI] [PubMed] [Google Scholar]
- 16.Svarovskaia ES, Barr R, Zhang X, Pais GC, Marchand C, Pommier Y, Burke, T.R., Pathak VK. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J. Virol. 2004;78:3210–3222. doi: 10.1128/JVI.78.7.3210-3222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 18.Wiskerchen M, Muesing MA. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asante-Appiah E, Skalka AM. HIV-1 integrase: structural organization, conformational changes, and catalysis. Adv. Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 20.Greenwald J, Le V, Butler SL, Bushman FD, Choe S. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry. 1999;38:8892–8898. doi: 10.1021/bi9907173. [DOI] [PubMed] [Google Scholar]
- 21.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AA, Santos W, Pais GC, Marchand C, Amin R, Burke T.R., Jr., Verdine G, Pommier Y. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J. Biol. Chem. 2006;281:461–467. doi: 10.1074/jbc.M511348200. [DOI] [PubMed] [Google Scholar]
- 23.Zembower TR, Gerzenshtein L, Coleman K, Palella F.J., Jr. Severe rhabdomyolysis associated with raltegravir use. AIDS. 2008;22:1382–1384. doi: 10.1097/QAD.0b013e328303be40. [DOI] [PubMed] [Google Scholar]
- 24.Nakahara K, Wakasa-Morimoto C, Kobayashi M, Miki S, Noshi T, Seki T, Kanamori-Koyama M, Kawauchi S, Suyama A, Fujishita T, et al. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 2008 doi: 10.1016/j.antiviral.2008.10.007. [Epub ahead of print, Nov 21, 2008] [DOI] [PubMed] [Google Scholar]