Abstract
The RNA-dependent RNA polymerase (RdRP) of double-stranded RNA (dsRNA) viruses performs both RNA replication and transcription. In order to initiate RNA polymerization, viral RdRPs must be able to interact with the incoming 3′ terminus of the template and position it, so that a productive binary complex is formed. Structural studies have revealed that RdRPs of dsRNA viruses that lack helicases have electrostatically charged areas on the polymerase surface, which might facilitate such interactions. In this study, structure-based mutagenesis, enzymatic assays and molecular mapping of bacteriophage φ6 RdRP and its RNA were used to elucidate the roles of the negatively charged plough area on the polymerase surface, of the rim of the template tunnel and of the template specificity pocket that is key in the formation of the productive RNA-polymerase binary complex. The positively charged rim of the template tunnel has a significant role in the engagement of highly structured ssRNA molecules, whereas specific interactions further down in the template tunnel promote ssRNA entry to the catalytic site. Hence, we show that by aiding the formation of a stable binary complex with optimized RNA templates, the overall polymerization activity of the φ6 RdRP can be greatly enhanced.
INTRODUCTION
Double-stranded RNA (dsRNA) viruses are intricate molecular machines that carry all the necessary component proteins required for viral RNA synthesis within their capsids. In spite of notable differences in the external structural features of these viruses and host specificity, almost all dsRNA viruses share a common replication strategy (1). Since dsRNA viruses encode only a single protein species containing conserved RNA-dependent RNA polymerase (RdRP) motifs, this protein must catalyse both transcription of genomic dsRNAs into positive-sense single-stranded RNAs (+ssRNAs) and replication of the +ssRNAs into dsRNAs. The RdRP is thus a central component in the life cycle of a dsRNA virus.
The polymerase subunit of the dsRNA bacteriophage φ6 (family Cystoviridae) is highly processive and able to replicate ssRNA molecules of practically unlimited length (2,3), making it suitable for biotechnological applications (4). The φ6 RdRP uses the two-metal ion catalysis of nucleotidyl transfer, with the metal ions being coordinated by catalytic aspartates (5,6). Structural and biochemical studies have revealed a de novo mechanism for initiation of φ6 RNA polymerization (2,3,5,7). The initiation platform (amino acids 629–632), located within the C-terminal domain, protrudes into the central cavity of the enzyme (5,7,8) and controls the switch from initiation to elongation and, to a large extent, prevents back-primed initiation by physically reducing the volume of the active site (5,8,9). In back-priming (or copy-back) initiation, the 3′ end of the template loops back to form a hairpin structure, which is subsequently extended by the polymerase.
The architecture of RdRPs resembles that of a ‘right hand’, with domains that are commonly denoted as the ‘fingers’, ‘palm’ and ‘thumb’. Viral RdRPs tend to have a compact ‘cupped right hand’ conformation as opposed to the ‘open right hand’ observed in many other polymerases. The catalytic site is contained in the palm domain, and the architecture of this region is the most conserved feature in RdRPs (10). The φ6 RdRP has two positively charged tunnels. The template tunnel and the substrate tunnel, respectively, allow access of the RNA template and NTP substrates to the internally located active site (Figure 1a). The template tunnel is wide enough to accommodate single- but not double-stranded nucleic acid. The distance from the surface of the RdRP to the active site can be spanned by a five base oligonucleotide (5,7). Out of these five nucleotides, two are relatively poorly ordered, the one in the specificity pocket (a specific binding site within the C-terminal domain to which the 3′ end of the template docks during initiation) and the one closest to the surface (Figure 1). The stabilizing interactions are most extensive at the position of the third nucleotide (5,7). In bacteriophage φ6, transcription is initiated from the 3′ terminal cytidine of the minus strand template (−ssRNA). This cytidine is strictly conserved in all three genomic segments, which explains the considerable preference of the specificity pocket for templates with a 3′ terminal cytidine (5).
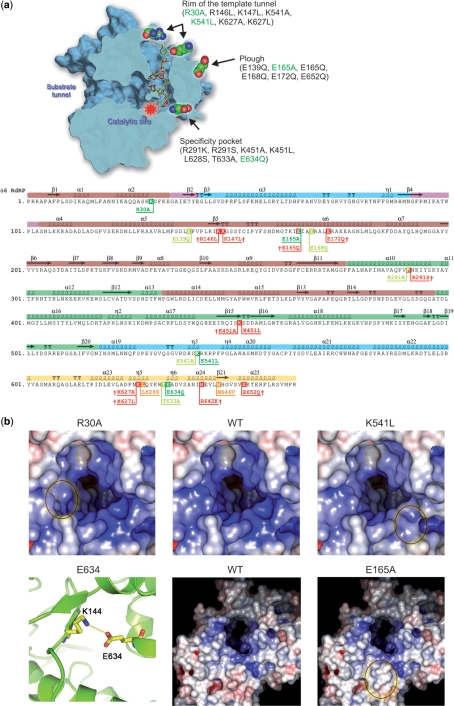
Figure 1.
Site-directed mutagenesis of φ6 RdRP. (a) Schematic presentation of the φ6 RdRP and the mutations introduced by site-directed mutagenesis. A cross section of the polymerase, exposing the catalytic site, the specificity pocket, the positively charged template tunnel (with a bound template) and the substrate tunnel, as well as the rim of the template tunnel and the plough area (top panel). Mutants marked in green were characterized in detail and the corresponding amino acids are also depicted by space-filling models in the RdRP structure. The location of the tested mutations with respect to the φ6 RdRP secondary structure elements (bottom panel), coloured according to the generic polymerase domain architecture: dark red, fingers domain (amino acids 1–30, 104–276, 333–397); dark green, palm domain (277–332, 398–517); blue, thumb domain (37–91, 518–600); light yellow, C-terminal domain (604–664); pink, connecting chains (31–36,92–103). The location of the mutated amino acids are highlighted, showing lethal mutations in red(†), mutations that could not be purified in orange and active mutations in either chartreuse (initial screening only) or green (selected for characterization). (b) Close-ups of the structural alterations of the selected mutants. Surface rendering of the rim of the template tunnel viewing towards the catalytic site (top row). The surfaces are coloured according to electrostatic charge (blue = +10 kT/e, red = −10kT/e). Introducing alanine or hydrophobic leucine in place of arginine (R30) and lysine (K541), respectively, reduces both the positive charge as well as the size and shape of the template tunnel rim. Compare the encircled areas of R30A (left) and K541L (right) to the WT (centre). R30 and K541 are located on opposite sides of the rim around the template tunnel. E634 lies within the C-terminal priming domain (specificity pocket) and forms a salt bridge with K144 (bottom row, left panel). The mutation to glutamine (E634Q) eliminates this salt bridge without major disruption of the internal packaging within the polymerase. Surface rendering of the plough area (bottom row, middle WT; right E165A). Substituting E165 to alanine reduces both the charge and size of the plough (encircled).
The rim of the template tunnel entrance is studded with positively charged amino acids (5) (Figure 1). A short distance from this rim a negatively charged area flanked by a positively charged ‘groove’ forms a plough-shaped protrusion on the polymerase surface (5) (Figure 1). This area is believed to facilitate the separation of dsRNA during transcription and to guide the −ssRNA into the tunnel while the +ssRNA slides over the positively charged patch, the so called surface channel (5,7). Similar specialized features have been discovered in other RdRPs, including bovine viral diarrhoea virus, hepatitis C virus (HCV) and rabbit haemorrhagic disease virus (RHDV) (11,12).
The genome of bacteriophage φ6 consists of three segments, S (2948 nt), M (4063 nt) and L (6374 nt). In each segment, the genes are clustered into functional groups flanked by 5′-packaging and 3′-replication signals (13,14). The 3′ ends of all three φ6 transcripts form secondary structures that display considerable similarity. These structures resemble that of a clover-leaf, and they are believed to provide stability in the event of an exonuclease attack (15). These terminal regions have also been found to affect the efficacy of RNA polymerization during replication (2,16).
To date, the φ6 RdRP is one of the best-characterized viral polymerases. Our aim is to dissect the φ6 RdRP catalysed polymerization reaction into distinct steps. The mechanism of initiation and the assembly of a productive initiation complex have already been elucidated using both biochemical and structural approaches (5,7–9,17,18). We are currently probing the transition from initiation to elongation. The nature of elongation has been explicated in a few studies (2,3,17–19). Recently, we have presented a possible mechanism for the termination of the polymerization reaction (20). In this study, we focus on the pre-initiation phase of replication, namely the process leading to the formation of a productive φ6 RdRP-RNA template binary complex.
In order to gain further understanding of the formation of productive φ6 RdRP-RNA template binary complexes, we designed a set of mutations located at specific regions of the polymerase that are believed to be of structural or functional importance. These locations included the plough, the rim of the template tunnel and the specificity pocket (Figure 1), which are potentially crucial for the events preceding initiation of RNA synthesis. With the aid of in silico secondary and tertiary structure computation, we also designed a set of partially dsRNA hybrids of both endogenous and exogenous ssRNA templates to study the formation of stable φ6 RdRP-template RNA binary complexes. We were able to show that the positively charged rim around the entrance of the template tunnel interacts with the incoming template RNA and that the level of RNA synthesis can be significantly increased by stabilizing the binary complex. This was achieved by using RNA hybrids that have a readily available 3′ end, lacking secondary structure and minimal interaction with the bulk of the template. We show that the use of such templates can even compensate the disadvantageous phenotype of certain mutant polymerases to a near wild-type (WT) level.
METHODS
Designing the φ6 RdRP mutants
The mutations were inserted at structural features that were believed to be important in the formation of productive binary complexes. Furthermore, the sites of the mutations were chosen by consideration of their local environment with the aim of selecting residues where changes were likely to introduce only limited local structural changes. This analysis and the design of the four mutant structures (R30A, E165A, K541L and E634Q) were performed using Coot (21), and the most common side-chain rotamers did not cause any stereochemical clashes.
RNA secondary and tertiary structure predictions
The simulated two-dimensional RNA structure predictions were performed at desired temperature using the ‘RNAfold’-program from the Vienna RNA Package (22). The oligonucleotide binding was simulated by constraining the corresponding nucleotides from forming pairs with other nucleotides in the sequence (this was checked by hand after the computational prediction). Three-dimensional models of RNA structures were calculated using the ‘rna_2d3d’-program within the STRUCTURELAB-environment (23). The three-dimensional models were directly based on two-dimensional structures. The visualization was done using the VMD package (24).
Bacterial strains and plasmids
Escherichia coli DH5α (Gibco-BRL) was the host for plasmid propagation and molecular cloning. E. coli BL21(DE3) (Novagen) was used for protein expression and purification. Plasmid pEMG2 (20) was used for WT φ6 RdRP production. Plasmid pEM33 (17) encodes WT φ6 RdRP with a histidine tag and pEM28 (8) the RdRP gene with YKW(630–632) to GSG mutations. Point mutations were introduced into plasmid pEM33 by site-directed mutagenesis, yielding the pNL-plasmid family (Supplementary Table 1). The 6× His-tag was removed by cloning the mutated site from the pNL-plasmids using appropriate restriction endonucleases into pEMG2, giving rise to plasmids pSVe2 (E634 to Q mutation), pSVe3 (R30A) and pSVe5 (K541L) or into pEM2 (2), yielding pMH6 (E165A). Plasmids pEM15 (3), pEM19 (8), pLM659 (25) and pT7luc (26) were used as templates for ssRNA production with T7 RNA polymerase.
Site-directed mutagenesis
The point mutations were introduced into the WT φ6 RdRP cDNA backbone using either QuickChange (Stratagene) or PCR site-directed mutagenesis. QuickChange mutagenesis was performed by standard PCR (27) using PfuTurbo (Stratagene) polymerase, appropriate oligonucleotides (Supplementary Table 2) and plasmid pEM33 as a template. For PCR site-directed mutagenesis, the parental DNA was digested with DpnI restriction endonuclease. The NruI-NsiI restriction fragment of pEM33 plasmid DNA was used as a template for PCR amplification with PfuTurbo polymerase and appropriate oligonucleotides (Supplementary Table 2) (27). The PCR products were digested with NruI and NsiI restriction endonucleases and ligated with similarly cut pEM33 vector.
Preparation of RNA templates
Synthetic ssRNAs were produced by run-off transcription in vitro with T7 RNA polymerase (2). Templates for the T7 transcription were prepared by either cutting the plasmid DNA with restriction endonucleases or by PCR amplification. The full-length transcript of the S genome segment (s+) was transcribed from the PCR amplified fragment of plasmid pLM659 (oligos 41 and 47; Supplementary Table 2). RNA templates sΔ+13 and sΔ+HP were transcribed from SmaI digested pEM15 and pEM19 plasmids, respectively. The template specificity RNAs (sΔ+, sΔ+A, sΔ+C, sΔ+G, sΔ+U and luc, lucA, lucC, lucG, lucU) were transcribed from DNA following PCR amplification of pEM15 or pT7luc using DyNAzyme II polymerase (Finnzymes Ltd.) and oligonucleotides 41–51 (Supplementary Table 2) (3,28). The template (s+rep) for the electrophoretic mobility shift assays (EMSAs) was transcribed from the 300 bp PCR-amplified fragment of plasmid pLM659 (oligos 52 and 47). For the template ssRNA-oligonucleotide hybrids, RNA oligonucleotides complementary to the 3′ end region of the s+ ssRNA (nos. 53–65 for s+, 66–72 for luc; Supplementary Table 2) were purchased from Biomers.Net (Ulm, Germany). The hybridization was performed as described previously (8,29) with some modifications: 8.9 pmol ssRNA and 24 pmol oligonucleotide were incubated in 50 mM HEPES–KOH (pH 7.0) and 0.1 M KCl at 65°C for 1 min and cooled to 30°C over a period of 30 min. Excess oligonucleotides were removed by purification in G50 spin columns (GE Healthcare), and the purified RNA hybrids were stored in 25 mM Tris–HCl (pH 8.0), 25 mM NaCl. Genomic dsRNA templates were obtained from WT φ6 bacteriophage infections (30). All RNAs were dissolved in sterile 18 Ω milli-Q water (Millipore Corporation), and the concentrations were determined by spectrophotometry (A260). The quality of each preparation was monitored by electrophoresis in 1% agarose (TBE) gels (27).
φ6 polymerase assay
Both the WT and mutated φ6 RdRPs were expressed in E. coli BL21(DE3) cells containing the appropriate expression plasmid at room temperature over night and purified to near homogeneity as previously described (2). The replication (ssRNA template) and transcription (dsRNA template) assays were carried out using standard reaction conditions (8), with 10 mM ATP and GTP and 2 mM CTP and UTP. All optimizations were carried out using the s+ template. After a 1-h incubation period, the reactions were stopped by the addition of 2× loading buffer (31) or 10× loading dye (RNA hybrids) (27). Analysis of the reaction products was performed using standard 0.8% agarose gel (TBE) electrophoresis (27). The signals were collected by autoradiography on BAS1500 image plates (Fujifilm), which were scanned using a Fuji BAS-1500 phosphorimager (Fujifilm). Digital image analysis (densitometry) was performed using AIDA Image Analyser v. 3.44 software (Raytest Isotopenmeβgeräte GmbH, Germany), measuring the band intensities in 1D Evaluation mode using Lane and Peak Determination.
Elongation rate
A modification of the heparin trap assay (17) was applied to analyse elongation separately from the initiation phases of the polymerization reaction. Providing limited nucleotides (10 mM ATP and GTP), the WT and mutant polymerases were allowed to extend 10 nt from the 3′ end of the s+ template at standard reaction conditions. Following a 20 min incubation at 30°C, heparin sodium (Sigma-Aldrich, H3393) was added to a final concentration of 6.25 mg/ml, and incubation was continued for an additional 5 min. The reactions were subsequently supplemented with 2 mM CTP and UTP, as well as 1 μCi [α32P]-UTP (Amersham Biosciences, Inc., 3000 Ci/mmol). Aliquots sampled at different time points were processed and analysed as described for the φ6 polymerase assay. A negative control was supplemented with heparin sodium and a positive control with an equal volume of sterile 18 Ω milli-Q water already in the pre-incubation mixture. The elongation rates were obtained by graphical extrapolation, as previously described (2).
Electrophoretic mobility shift assay
A modification of the φ6 polymerase assay using standard reaction conditions (8) and 0.25 μg s+rep template with a trace amount of [γ32P]-labelled s+rep ssRNA (T4 polynucleotide kinase, Promega, [γ32P]-ATP, Amersham Biosciences, Inc., 3000 Ci/mmol) was incubated with 0.04–0.16 mg/ml of WT or mutant polymerase on ice for 20 min. A corresponding volume of sterile 18 Ω milli-Q water was used in the control reactions. Prior to loading, 10× loading dye (27) was added to the reaction mixtures and the RNA-polymerase complexes were resolved by electrophoresis in 8% native polyacrylamide gels (Tris–glycine buffer) (27) at 60 V and 20 mA for 24 h at 4°C. Following electrophoresis, the gels were dried on Whatman 3 filter paper, and signal collection and data processing were performed as described earlier.
RESULTS
Site-directed mutagenesis of the φ6 RdRP
Based on the existing structural data (5,7), we designed a set of mutant polymerase proteins that targeted three key structural features within the enzyme, the plough, the rim of the template tunnel and the specificity pocket (Figure 1; Supplementary Table 1). Out of the planned 22 histidine-tagged φ6 RdRP mutations, nine functional mutants were obtained (Figure 1a). This subset of functional mutants was assayed for polymerase activity using the positive-strand of φ6 genome segment S as a template (2,8). The polymerase activity of all the mutants was either low or moderate in comparison to the histidine-tagged WT RdRP (Supplementary Table 1). Due to the possible adverse effect of the tag, point mutants R30A, E165A, K541L and E634Q (Figure 1, Supplementary Table 1), which displayed the most favourable qualities (polymerase activity, expression level and purification characteristics), were recloned without the tag and purified. The selected enzymes were representatives for all the functional mutations at their respective structural sites within the protein.
Mutations at the putative template interaction sites alter the overall polymerization activity
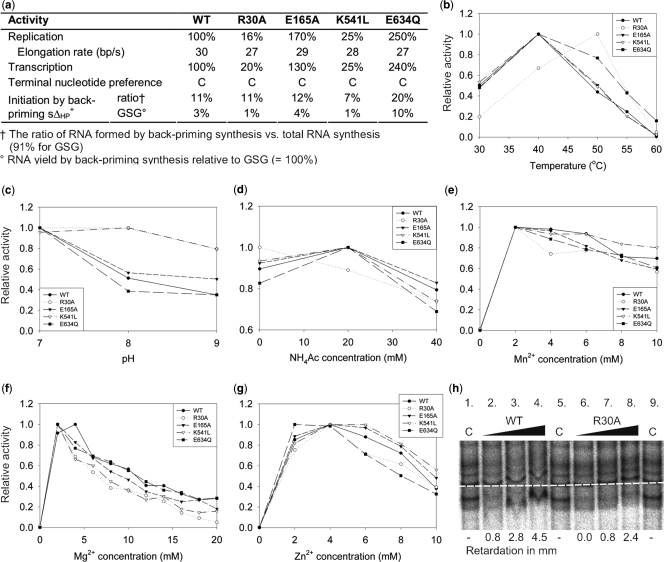
The polymerase reaction conditions preferred by the mutants were generally quite similar to those of the WT polymerase, showing only minor deviations in terms of temperature, pH, salt concentration and divalent cation concentrations (Figure 2a-g). Despite strong similarities between the optimal conditions, the template tunnel mutant R30A and the specificity pocket mutant E634Q retained polymerization activity at temperatures up to 55–60°C, i.e. high enough temperatures to cause RNA hydrolysis, whereas the other mutants retained WT characteristics with temperature optima of roughly 40°C (Figure 2b). The template tunnel rim mutants R30A and K541L displayed a significantly reduced level of RNA synthesis for both replication and transcription, whereas the specificity pocket mutant E634Q showed an increased overall activity (Figure 2a).
Figure 2.
Biochemical characterization of φ6 RdRP mutants. (a) Summary of the RNA polymerization activities of the φ6 RdRP mutants in comparison to the WT or YKW(630–632)GSG mutant (initiation by back-priming) (8). (b–g) Screening of the optimal reaction conditions for the mutant polymerases in terms of temperature, pH, ammonium acetate concentration and divalent cation concentrations (Mn2+, Mg2+, Zn2+). All results have been normalized against the highest attained polymerization activity for each RdRP. (h) Electrophoretic mobility shift assay (EMSA) of the φ6 RdRP mutants. The effect of an increasing concentration (0.04, 0.08 and 0.16 mg/ml) of WT (lanes 2–4) and R30A (lanes 6–8) polymerases on the migration of the s+rep template in 8% native polyacrylamide gel. The baseline (white dotted line) determines the position of the control (s+rep without polymerase; lanes 1, 5 and 9). The numbers below lanes 2–4 and 6–8 indicate the retardation of the s+rep template in millimetres.
A mutation in the template specificity pocket increases back-priming activity
Interestingly, the elimination of the salt bridge between E634 and K144 introduced by the E634Q mutation yielded a polymerase with increased back-priming activity (Figure 2a). Although the E634Q mutant did synthesize ∼20% of the total dsRNA product by means of back-priming initiation when using the sΔ+HP template (8) that features a terminal hairpin loop, de novo initiation was still preferred. All other φ6 RdRP mutants synthesized on average ∼10% of their total dsRNA yield by back-priming initiation (comparable with WT). In the case of the GSG mutant (positive control), where changing YKW(630–632) to GSG causes back-priming to become the major mode of initiation of RNA synthesis (8), this ratio was found to be as high as ∼90% (Figure 2a). Moreover, all tested polymerases except GSG showed a clear reduction in overall synthesis activity when using the sΔ+HP template as opposed to the equally long non-looped sΔ+13 template (data not shown). The poor performance of the WT and other mutant polymerases was expected, since their initiation sites are not spacious enough to efficiently encompass a dsRNA structure such as the 3′ hairpin loop in sΔ+HP.
Mutations at the plough region are well tolerated
In vitro transcription activity of the recombinant φ6 RdRP requires that the polymerase itself is able to melt blunt-ended dsRNA without the assistance of a separate helicase. The purified polymerase does indeed possess this activity (3), although transcription proceeds more than one order of magnitude less efficiently than replication. The negatively charged protrusion near the template tunnel on the molecular surface of the polymerase (the plough-area) has been proposed to facilitate strand displacement. Contrary to the expectations, the plough mutant E165A did not reduce the transcriptional activity but enhanced it by up to ∼30% compared to the WT (Figure 2a). In the initial screening, similar characteristics were observed for the histidine-tagged functional mutants located at the plough region (E139Q, E168Q) (data not shown).
The elongation rates are not compromised by the mutations
In order to elucidate at which stage the polymerization reaction was affected by the mutations, we applied a heparin trap assay (see Materials and methods section). Heparin is a competitive inhibitor of RNA synthesis affecting initiation but not elongation (18,32). This allows analysis of elongation separately from the initial stages of the polymerization reaction. It appears that the elongation rates of the WT and mutant polymerases are almost identical, occurring at a rate of 27–30 bp/s (Figure 2a). This suggests that the changes observed in the overall polymerization activity reflect alterations in the events preceding elongation.
Mutations at the rim of the template tunnel reduce the affinity of the RdRP for RNA
To determine possible differences in the pre-initiation phase of the RNA polymerization reaction of the tested polymerases, a RNA-binding assay was performed in order to monitor the formation of φ6 RdRP-RNA template binary complexes. This was visualized by EMSAs using the s+rep template (terminal 300 nt of the s+ template). The plough mutant E165A (data not shown), the specificity pocket mutant E634Q (data not shown) and the WT polymerase (Figure 2h) had identical binding characteristics, displaying a retardation of the RNA template migration that was proportional to the polymerase concentration. The mutants at the template tunnel rim, R30A (Figure 2h) and K541L (data not shown), showed a less pronounced shift with the template migrating in close proximity to the control RNA at all polymerase concentrations. This indicates that the rim of the template tunnel plays a role in the formation of productive φ6 RdRP-RNA template binary complexes.
Design and in silico analysis of RNA templates
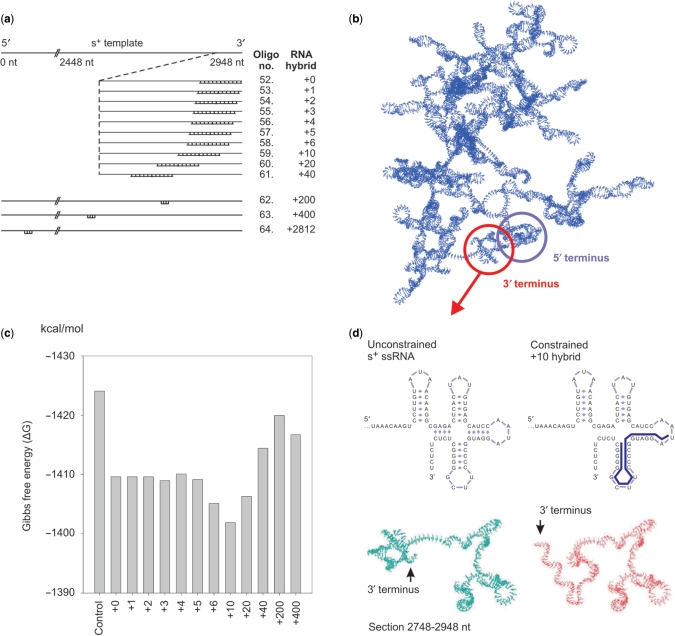
To assess the nature of the interaction between the template RNA and the φ6 RdRP, we designed a set of partially dsRNA hybrids of both endogenous (genomic s+ segment) and exogenous (firefly luciferase gene, luc) RNA templates. Complementary 20-nt long RNA oligonucleotides (Supplementary Table 2) were annealed in silico to various positions of the s+ template (Figure 3a) or the luc template (Supplementary Figure 1) to yield single-stranded 3′ overhangs of varying length. These hybridized ssRNA template-oligonucleotide complexes are referred to as ‘+n hybrid’, where n indicates the distance from the template 3′ end, which also corresponds to the length of the single-stranded 3′ overhang (0 = blunt). The changes induced by these oligonucleotides to the native structure of the template RNAs were first assessed by in silico secondary and tertiary structure calculations (Figure 3b-d; Supplementary Figure 1). A global fold of the s+ template (Figure 3b) and luc template (Supplementary Figure 1b) with the different RNA hybrids revealed differences in terms of Gibbs free energy. In the case of the s+ template, the template alone possessed the lowest and the +10 hybrid the highest free energy (Figure 3c). The same observation holds true for the luc ssRNA with the unconstrained template yielding the lowest and the luc +10 hybrid the highest free energy (Supplementary Figure 1c).
Figure 3.
In silico template RNA modelling. (a) Schematic representation of the φ6 s+ ssRNA and a close-up view of the +0 to +2812 hybrids following annealing of the complementary RNA oligonucleotides 52–64 (Supplementary Table 2) to the s+ ssRNA. (b) Predicted tertiary structure of the complete genomic φ6 ssRNA segment s+ at the RdRP reaction conditions. The 5′ (blue) and 3′ (red) termini have been encircled. (c) Changes in Gibbs free energy (▵G) of the RNA hybrids as a function of the complementary oligonucleotide location. (d) Schematic representation of the secondary structure of the unconstrained 3′ termini of the ssRNA (top-left panel) and the constrained +10 hybrid (top right panel). Local visualization of the tertiary structure of the last 200 nucleotides of the 3′ region of the s+ ssRNA (unconstrained) versus the constrained RNA (+10 hybrid) (bottom panel). Only the coding strand is depicted without the complementary oligonucleotide.
Although these global energy differences were moderate for both templates, significant local structural changes were predicted for the s+ ssRNA, especially when the +10 (Figure 3d) and +20 hybrids (data not shown) were studied. It appeared that the differences in Gibbs free energy reflected these regional changes and were not caused by a global rearrangement of the RNA. When analysing the tertiary structure prediction of the s+ ssRNA and the +10 hybrid, changes in the local fold were observed throughout the first 300 (data not shown) and last 200 bases (Figure 3d) of the 5′ and 3′ regions, respectively. Notably, in the unhybridized form the 3′ region was partially buried within the overall fold and the 5′ and 3′ termini were predicted to be spatially located close to each other without specifically interacting (Figure 3b). However, the 3′ termini of the +10 (Figure 3d) and +20 hybrids (data not shown) were predicted to protrude outwards of the main fold, reducing possible sterical clashes between the RNA and the polymerase. In contrast to the s+ template, the predicted overall tertiary structure of the unconstrained luc ssRNA was markedly different from the luc hybrids (Supplementary Figure 1b). For all luc templates tested, the first 200 nt of the 5′ terminus always retained its native fold, whereas the last 40 nt of the 3′ terminus regained it once the oligonucleotide was moved outside this region (data not shown).
Assembly of a productive φ6 RdRP-template RNA binary complex
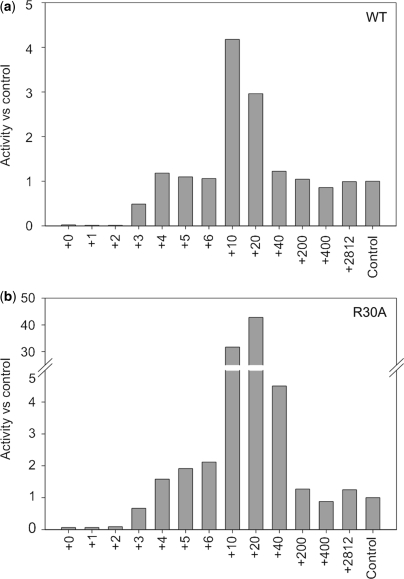
To biochemically evaluate the process of binary complex formation, polymerase assays were performed with the above-described RNA hybrids in standard φ6 RdRP reaction mixtures. Assaying the RNA hybrids of the biologically relevant endogenous template (s+) with the WT polymerase revealed that almost no dsRNA was formed with the +0 to +2 hybrids. The +3 hybrid gave rise to a dsRNA synthesis of ∼50% compared to the control (the dsRNA yield for each polymerase mutant using the genomic s+ segment as a template), and moving the oligonucleotides further downstream (+4 to +6 hybrids) resulted in a product yield equalling that of the control (Figure 4a). The same observations also apply to mutants E165A and E634Q (Supplementary Figure 2a and c). Similar characteristics were observed for polymerase mutants R30A (Figure 4b) and K541L (Supplementary Figure 2b), although their dsRNA yield slightly exceeded that of the control with the +4 to +6 hybrids.
Figure 4.
RNA polymerization activity with RNA hybrids. RNA polymerization activity of WT φ6 RdRP (a) and R30A mutant (b), using different RNA hybrids. The results have been normalized against an internal control (polymerization activity with the s+ ssRNA template).
A significant increase in the dsRNA yield was observed with the +10 and +20 hybrids. The WT RdRP (Figure 4a), as well as mutants E165A and E634Q (Supplementary Figure 2a and c), reached a fourfold increase in dsRNA synthesis in comparison to the control level. However, the R30A (Figure 4b) and K541L (Supplementary Figure 2b) mutations displayed a staggering 40-fold increase in comparison to the control. Hence, the addition of an oligonucleotide at 10 or 20 nt past the 3′ end suppresses the phenotype of the template tunnel rim mutants R30A and K541L, reinstating dsRNA synthesis to the native WT level.
The exogenous luc +10 hybrid also yielded a significant increase in dsRNA synthesis for the WT and mutant polymerases (Supplementary Figure 1), although the template tunnel rim mutants R30A and K541L did not display the dramatic changes seen with the s+ hybrids. Similarly, the luc +0 hybrid was unable to create a stable binary complex, whereas the luc +5 hybrid attained a level of RNA synthesis equivalent to the native luc template (control). Additionally, for both the endogenous and the exogenous templates, the RNA synthesis stimulating effect was lost if the oligonucleotide was placed further away than 40 nt from the 3′ end (data not shown).
DISCUSSION
To date, RdRPs from dsRNA viruses such as bluetongue virus (33), birnavirus (34), reovirus (35,36), rotavirus (37–39) and bacteriophages φ6, φ8, φ12 and φ13 (2,3,5,18,40,41) have been purified and biochemically characterized to varying degrees. Previous structural analyses of the φ6 RdRP have focused on initiation of polymerization and the role of the structural Mn2+ ion (8,9,17). In this study, we analysed the events preceding initiation by monitoring the assembly of productive φ6 RdRP-RNA template binary complexes. This has involved in silico RNA modelling, template RNA modification and mutagenesis of the φ6 RdRP at areas that are believed to be of functional importance during pre-initiation, namely the plough, the rim of the template tunnel and the specificity pocket.
The mutagenesis of φ6 RdRP revealed that even relatively conservative mutations in external side chains can have a significant impact on the folding or stability of the enzyme (Figure 1). Of the four enzymes selected for further characterization, the specificity pocket mutant E634Q has especially interesting characteristics, including a substantially elevated level of RNA synthesis, an increased back-priming activity (Figure 2a) as well as a higher thermal stability of the polymerization reaction (Figure 2b). As the elongation rate of E634Q was unaffected, the elevated level of RNA synthesis indicates an enhancement of the initiation (or pre-initiation) step. The higher thermal stability of the polymerization reaction most likely reflects changes in the initial phases of the reaction. Indeed, it has been shown that the temperature sensitive phase in the φ6 RdRP catalysed reaction is de novo initiation, whereas elongation may well proceed at temperatures exceeding 50°C (18). Apparently, the disruption of the salt bridge between E634 (C-terminal domain) and K144 (fingers domain) increases the volume of the specificity pocket, causing changes in the mode and control of initiation.
The template tunnel of the φ6 RdRP (5) is not wide enough to allow the entry of dsRNA. During semi-conservative transcription, the parental +ssRNA strand has to be displaced prior to the entry of the −ssRNA strand to the template tunnel (Figure 5), a function that has been loosely assigned to the plough area (Figure 1). Similar structural features have been described for other RdRPs, including bovine viral diarrhoea virus, hepatitis C virus and rabbit haemorrhagic disease virus (11,12), suggesting that the plough area might be a common feature of viral RdRPs. Surprisingly, the phenotype of the plough mutants E165A, E139Q and E168Q were similar to the WT enzyme, and the mutations did not reduce the transcription activity. Contrary to earlier suggestions, this indicates that the plough may not be critical in breaking open dsRNA. The role of this negatively charged area remains unclear, but it might constitute a simple electrostatic barrier that forces the parental +ssRNA strand to slide along the surface channel. Mutations E165A and E634Q did not have a significant effect on the efficiency of template binding (data not shown). Instead, the template tunnel mutants R30A (Figure 2h) and K541L (data not shown) seem to have a decreased RNA affinity, suggesting that the reduced charge around the template tunnel entrance may interfere with the correct guidance of the template, thus preventing it from entering the template tunnel.
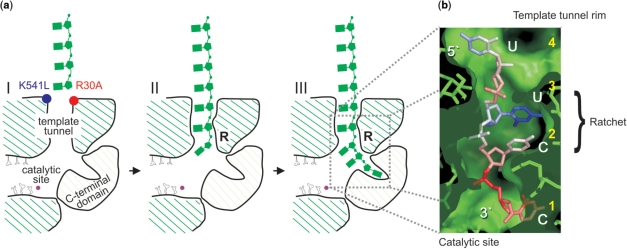
Figure 5.
Model for the formation of φ6 RdRP binary complex. (a) Schematic representation of the φ6 RdRP with an ssRNA reaching the template tunnel (panel I). The rim and lining of the template tunnel are positively charged. This electrostatic steering positions the incoming ssRNA template (green) for entry into the template tunnel (panel I). However, mutations R30A (red) and K541L (blue) locally disrupt the charge of the rim (panel I). The incoming template strand is stabilized by interactions between the RNA and specific residues in the template tunnel, the so-called ratchet area (panel II and III, marked R). All the steps leading up to the formation of the productive binary complex (panel III) are passive energy events. (b) RNA oligonucleotide bound in the template tunnel of φ6 RdRP. The four ordered RNA nucleotides (fifth at the template tunnel entrance, not depicted) are coloured according to temperature factor (low- to high-coloured blue to red), showing that the central two nucleotides are the best ordered (the 3′ cytidine approaches the catalytic site and the 5′ uridine is at the molecular surface). The amino acids and molecular surface of φ6 RdRP are rendered as green sticks and a semi-transparent envelope, respectively. The sequence and respective ends of the oligonucleotide are labelled in white, whereas the order of the template nucleotides is labelled in yellow. Note the close fit of the RNA within the polymerase tunnel, especially in the central portion, reflecting the molecular engagement that occurs in this region (ratchet area).
Furthermore, it was observed that annealing a primer to the very 3′ end of the template significantly reduced the dsRNA yield for all polymerases studied (Figure 4a and b). This reduction was most prominent when the oligonucleotide was complementary to the last three nucleotides of the 3′ end of the template (+0 to +2 hybrids). However, all polymerases reached their normal level of RNA synthesis once the oligonucleotide was placed at four to five nucleotides from the 3′ end (+4 or +5 hybrid; Figure 4a,b) an overhang corresponding to that at the 3′ terminus of the φ6 s+ ssRNA template (Figure 3b,d). This overhang also coincides with the distance from the surface of the φ6 RdRP to the catalytic site. In the crystal structures of the complex of φ6 RdRP with different oligonucleotides, only four 3′ terminal nucleotides of the template can be visualized in the template tunnel (Figure 5) (5,7). Our biochemical analyses show that a 3-nt overhang is the minimum capable of yielding significant dsRNA synthesis (Figure 4a and b). These observations match closely with the structural results (5,7), where the second and the third nucleotide are the best-defined (Figure 5), suggesting that these are the points of engagement of RNA within the template tunnel.
Overall, the computational secondary and tertiary structure predictions of the RNA hybrids (Figure 3; Supplementary Figure 1b) closely match the biochemical data (Figure 4a and b; Supplementary Figure 1, 2). Previous in silico structure predictions have proposed that the last ∼80 nt of the genomic s+ ssRNA form a knot-like clover-leaf fold (Figure 3b) (42). These structures are believed to provide both a stabilizing function in the case of an exonuclease attack, and in the case of viral genomes to constitute an in vivo replication signal (2,42). Annealing an oligonucleotide within this region (+10 or +20 hybrid) efficiently destroyed these secondary and tertiary structures, leaving a sufficiently long single-stranded ‘overhang’ (Figure 3d). This unfolding of the 3′ terminus is also reflected in the Gibbs free energy calculations. Interestingly, the highest free energy (Figure 3c, Supplementary Figure 1c) and the highest replication activity (Figure 4, Supplementary Figure 1d–h) coincide. Notably, the severe phenotypic effects of the R30A and K541L mutations were suppressed using the +10 and +20 hybrids, such that WT levels of activity were restored (Figure 4, Supplementary Figure 2).
The tertiary structure predictions (Figure 3) suggest that the partially buried fold of the 3′ terminus of the s+ template could cause sterical clashes between the template and the RdRP, possibly hampering the initial melting of the template. The outward protruding single stranded 3′ ends of the +10 to +20 hybrids apparently minimize these structural obstructions, thus making the 3′ end of the template readily available for the polymerase. This allows suppression of the phenotype of the mutations at the rim of the template tunnel that have a reduced ability to productively bind RNA templates.
Our studies on the formation of productive binary complexes allow us to propose a model for the pre-initiation of polymerization (Figure 5). This model for pre-initiation constitutes stochastic interactions that occur to form productive RdRP complexes without the input of external energy via ATP-hydrolysis. Initially, when an incoming ssRNA template comes in contact with the φ6 RdRP, weak electrostatic interactions occur with the positively charged residues lining the rim of the template tunnel (Figure 5a-I), allowing the RNA to bind in the template tunnel. The efficiency at which this occurs is dependent on the structure of the 3′ end of the ssRNA template. Freely available template 3′ ends with no secondary structures and/or sterical clashes facilitate the entry into the template tunnel, whereas highly structured templates with buried 3′ ends are less efficient in forming productive polymerase-template binary complexes. Furthermore, disrupting the charges along the rim of the template tunnel, as in mutants R30A and K541L, causes a significant reduction in the RNA-binding affinity of the polymerase (Figure 2h). Our results also indicate that the binary complex becomes productive once three nucleotides enter the template tunnel (Figures 4 and 5a-II). Specific interactions between the φ6 RdRP and RNA are largely ‘blind’ to the sequence of the bases (Figure 5a-II) and guide the template down towards the catalytic site (Figure 5a-III). At the site of the ratchet, located at the second and third nucleotide from the specificity pocket (Figure 5b), the interactions are most defined. Further stabilization of the binary complex arises from cytidine-specific interactions in the specificity pocket (Figure 5a-III). Once this occurs, nucleotides that enter the active site, via the nucleotide tunnel, can sense and engage with the bases of the terminal nucleotides and initiate polymerization.
The mutagenesis approach used in this study has yielded further insight into the structure-related functions of the φ6 RdRP, demonstrating how, for this intricate molecular machine, even minor changes can yield unexpected results. It is acutely sensitive to mutagenesis; most of the designed mutants were lethal and different amino acid substitutions within the same residue can give rise to either functional or inactive mutants. So, although it might be possible to enhance the thermostability and processivity of the RdRP, by mutations on the rim of the template tunnel and in the specifity pocket, the outcome from such mutations is difficult to predict and would need to be biochemically assessed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Finnish Centre of Excellence Programme 2006–2011 (Academy of Finland grant 1213467 to D.H.B.) as well as SPINE2–COMPLEXES project LSHG–CT–2006–031220. L.P.S., LSHG-CT-2006-031220 to D.I.S., M.R.L.K. and A.P.A. are fellows of the Helsinki Graduate School in Biotechnology and Molecular Biology. D.I.S. is supported by the Medical Research Council, UK. Funding for open access charge: Finnish Centre of Excellence Programme 2006-2011, Academy of Finland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Riitta Tarkiainen, Anna Latva-Käyrä, Soile Storman and Janne Turunen for their excellent technical assistance as well as Sampo Vehma and Martin Heger for constructing plasmids pSVe2, pSVe3, pSVe5 and pMH6, respectively.
REFERENCES
- 1.Mertens P. The dsRNA viruses. Virus Res. 2004;101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Makeyev EV, Bamford DH. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 2000;19:124–133. doi: 10.1093/emboj/19.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makeyev EV, Bamford DH. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aalto AP, Sarin LP, van Dijk AA, Saarma M, Poranen MM, Arumae U, Bamford DH. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage φ6 RNA-dependent RNA polymerase. RNA. 2007;13:422–429. doi: 10.1261/rna.348307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 6.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 7.Salgado PS, Makeyev EV, Butcher SJ, Bamford DH, Stuart DI, Grimes JM. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure. 2004;12:307–316. doi: 10.1016/j.str.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Laurila MR, Makeyev EV, Bamford DH. Bacteriophage φ6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 2002;277:17117–17124. doi: 10.1074/jbc.M111220200. [DOI] [PubMed] [Google Scholar]
- 9.Laurila MR, Salgado PS, Stuart DI, Grimes JM, Bamford DH. Back-priming mode of φ6 RNA-dependent RNA polymerase. J. Gen. Virol. 2005;86:521–526. doi: 10.1099/vir.0.80492-0. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006;16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Bruenn JA. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 2003;31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi KH, Groarke JM, Young DC, Kuhn RJ, Smith JL, Pevear DC, Rossmann MG. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl Acad. Sci. USA. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage φ6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Poranen MM, Tuma R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. doi: 10.1016/j.virusres.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Mindich L, Qiao X, Onodera S, Gottlieb P, Strassman J. Heterologous recombination in the double-stranded RNA bacteriophage φ6. J. Virol. 1992;66:2605–2610. doi: 10.1128/jvi.66.5.2605-2610.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frilander M, Gottlieb P, Strassman J, Bamford DH, Mindich L. Dependence of minus-strand synthesis on complete genomic packaging in the double-stranded RNA bacteriophage φ6. J. Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poranen MM, Salgado PS, Koivunen MR, Wright S, Bamford DH, Stuart DI, Grimes JM. Structural explanation for the role of Mn2+ in the activity of φ6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang H, Gottlieb P, Wei H, Bamford DH, Makeyev EV. Temperature requirements for initiation of RNA-dependent RNA polymerization. Virology. 2003;314:706–715. doi: 10.1016/s0042-6822(03)00460-4. [DOI] [PubMed] [Google Scholar]
- 19.Vilfan ID, Candelli A, Hage S, Aalto AP, Poranen MM, Bamford DH, Dekker NH. Reinitiated viral RNA-dependent RNA polymerase resumes replication at a reduced rate. Nucleic Acids Res. 2008;36:7059–7067. doi: 10.1093/nar/gkn836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poranen MM, Koivunen MR, Bamford DH. Nontemplated terminal nucleotidyltransferase activity of double-stranded RNA bacteriophage φ6 RNA-dependent RNA polymerase. J. Virol. 2008;82:9254–9264. doi: 10.1128/JVI.01044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures (The Vienna RNA Package) Mon. Chem. 1994;125:167–188. [Google Scholar]
- 23.Shapiro BA, Kasprzak W. STRUCTURELAB: a heterogeneous bioinformatics system for RNA structure analysis. J. Mol. Graph. 1996;14:194–205. doi: 10.1016/s0263-7855(96)00063-x. 222–224. [DOI] [PubMed] [Google Scholar]
- 24.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb P, Strassman J, Qiao X, Frilander M, Frucht A, Mindich L. In vitro packaging and replication of individual genomic segments of bacteriophage φ6 RNA. J. Virol. 1992;66:2611–2616. doi: 10.1128/jvi.66.5.2611-2616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolb VA, Makeyev EV, Spirin AS. Co-translational folding of an eukaryotic multidomain protein in a prokaryotic translation system. J. Biol. Chem. 2000;275:16597–16601. doi: 10.1074/jbc.M002030200. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 28.Makeyev EV, Bamford DH. Primer-independent RNA sequencing with bacteriophage φ6 RNA polymerase and chain terminators. RNA. 2001;7:774–781. doi: 10.1017/s1355838201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 30.Bamford DH, Ojala PM, Frilander M, Walin L, Bamford J.KH. In: Methods in Molecular Genetics. Adolph KW, editor. Vol. 6. San Diego: Academic Press; 1995. pp. 455–474. [Google Scholar]
- 31.Pagratis N, Revel HR. Detection of bacteriophage φ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology. 1990;177:273–280. doi: 10.1016/0042-6822(90)90480-f. [DOI] [PubMed] [Google Scholar]
- 32.Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276:39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 33.Boyce M, Wehrfritz J, Noad R, Roy P. Purified recombinant bluetongue virus VP1 exhibits RNA replicase activity. J. Virol. 2004;78:3994–4002. doi: 10.1128/JVI.78.8.3994-4002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan J, Vakharia VN, Tao YJ. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc. Natl Acad. Sci. USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starnes MC, Joklik WK. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 36.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage-structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 37.Patton JT. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng CQ, Wentz MJ, Cohen J, Estes MK, Ramig RF. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J. Virol. 1996;70:2736–2742. doi: 10.1128/jvi.70.5.2736-2742.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H, Makeyev EV, Bamford DH. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 2001;75:11088–11095. doi: 10.1128/JVI.75.22.11088-11095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H, Makeyev EV, Butcher SJ, Gaidelyte A, Bamford DH. Two distinct mechanisms ensure transcriptional polarity in double-stranded RNA bacteriophages. J. Virol. 2003;77:1195–1203. doi: 10.1128/JVI.77.2.1195-1203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage φ6. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.