Abstract
Genetic screens performed in model organisms have helped identify key components of the RNA interference (RNAi) pathway. Recessive genetic screens have recently become feasible through the use of mouse embryonic stem (ES) cells that are Bloom's syndrome protein (Blm) deficient. Here, we developed and performed a recessive genetic screen to identify components of the mammalian RNAi pathway in Blm-deficient ES cells. Genome-wide mutagenesis using a retroviral gene trap strategy resulted in the isolation of putative homozygous RNAi mutant cells. Candidate clones were confirmed by an independent RNAi-based reporter assay and the causative gene trap integration site was identified using molecular techniques. Our screen identified multiple mutant cell lines of Argonaute 2 (Ago2), a known essential component of the RNAi pathway. This result demonstrates that true RNAi components can be isolated by this screening strategy. Furthermore, Ago2 homozygous mutant ES cells provide a null genetic background to perform mutational analyses of the Ago2 protein. Using genetic rescue, we resolve an important controversy regarding the role of two phenylalanine residues in Ago2 activity.
INTRODUCTION
The RNA interference (RNAi) pathway is a highly conserved, sequence-specific gene silencing pathway that functions in most eukaryotes (1–5). In mammals, RNAi can be triggered by 20–23 nt double-stranded small interfering RNAs (siRNAs) or by short hairpin RNAs (shRNAs); shRNAs expressed from transgenes are produced by RNA polymerase II/III in the nucleus (6,7) and then exported from the nucleus via exportin-5 (3,8). In the cytoplasm, the RNAse III enzyme, Dicer, cleaves the shRNA into an siRNA (1–3,5,9). The siRNA is then unwound through an unknown mechanism and one strand of the siRNA, the guide strand, is incorporated into an Argonaute 2 (Ago2) containing RNA induced silencing complex (RISC) that targets the complementary mRNA for cleavage and degradation (3–5,10).
RNAi serves important functions in diverse biological processes including development, differentiation, cell division, apoptosis and defense against transposons and viruses (1,2,5,10). A greater understanding of the RNAi pathway will enhance our ability to more effectively use RNAi for genetic analyses and therapeutics (5,7). Our knowledge of the RNAi mechanism and machinery has been greatly increased through genetic, biochemical and structural biology studies. Forward genetic screens in several model organisms have been instrumental in uncovering key players and roles of RNAi. For example, Argonaute proteins, major RNAi components, were discovered in genetic screens performed in Caenorhabditis elegans (11). Argonautes were also identified in other forward genetic screens for quelling in Neurospora crassa and posttranscriptional gene silencing (PTGS) in Arabidopsis thaliana, thus linking these three related pathways (12,13). Other components of the C. elegans RNAi pathway, RNA-dependent RNA polymerases (RdRPs) and RecQ proteins, were also identified using forward genetic screens (14–17). A screen performed in Drosophila demonstrated that Dicer1 and Dicer2 serve distinct functions in the miRNA and siRNA pathways, respectively (18).
Components identified in these genetic screens have helped direct our analyses of RNAi in mammals and highlighted the fact that several species-specific differences exist for this pathway (8). For instance RdRPs, important components of plant and worm RNAi, have not been found to play a role in RNAi in flies or mammals (6,8,10). RecQ proteins are important for RNAi in C. elegans and N. Crassa, yet the mammalian homologs, Werner and Bloom, are not involved in RNAi (19). Thus, a screen for components of the RNAi pathway in a mammalian system has the potential to identify RNAi components that may be unique to mammals.
One obstacle to performing a recessive genetic screen in mammalian cells is the diploid nature of the genome, which requires that any recessive mutation be rendered homozygous in order to observe a mutant phenotype. This challenge has recently been partially overcome by the use of Bloom's syndrome protein (Blm)-deficient mouse embryonic stem (ES) cells. The Bloom gene encodes a homolog of the bacterial RecQ helicase and loss of this gene leads to genome instability. An 18- to 27-fold higher loss-of-heterozygosity (LOH) has been reported for Blm-deficient ES cells compared with wild-type ES cells (20,21). Therefore, these cells cause heterozygous mutations to be rendered homozygous at a much higher frequency. Several pioneering studies have shown that Blm-deficient ES cells are amenable to forward phenotypic screens (21–23). A screen performed in Blm-deficient ES cells for mismatch repair genes identified a novel component, Dnmt1 (22).
In this study, we took advantage of the high rate of LOH that occurs in Blm-deficient ES cells and developed a drug resistance selection scheme to enrich for RNAi mutants. Using retroviral gene traps, we successfully performed a genome-wide screen for mammalian RNAi mutants. This screen led to the isolation of multiple gene trap mutants homozygous for Ago2. Thus, the identification of Ago2, a known essential component of RNAi, validates the utility of our screening strategy. With future improvements in insertion mutagenesis and an increase of genomic coverage, our system has the potential to identify other unknown components in the mammalian RNAi pathway.
MATERIALS AND METHODS
Cell culture
Mouse ES cells were grown on feeder cells and maintained in M15 as previously described (24). For the various drug selections used in the study, the following drugs and concentrations were used: 250–350 μg/ml Geneticin [G418 (Invitrogen)], 1 × HAT Supplement (Invitrogen), 10 μM 6-TG [6-thioguanine (Sigma)], 1–2 μg/ml Puromycin (Invivogen), 5–10 μg/ml Blasticidin (Invitrogen).
Molecular cloning
To construct mouse U6-expressed shRNA vectors, the following oligos were phosphorylated, annealed and ligated into a BbsI and XhoI digested mU6 vector. For Hprt (hypoxanthine phosphoribosyl transferase), shRNA Hprt5 sense: 5′-TTT GCA CTG AAT AGA AAT AGT GAT TTA TTA ATA TCA CTA TTT CTA TTC AGT GCT TTT TC-3′; shRNA Hprt5 antisense: 5′-TCG AGA AAA AGC ACT GAA TAG AAA TAG TGA TAT TAA TAA ATC ACT ATT TCT ATT CAG TG-3′. For firefly luciferase (F-luc), shRNA luc1 sense: 5′-TTT GTG TCG CTC TGC CTC ATA GAA TTA TTA TTT CTA TGA GGC AGA GCG ACA CTT TTT C-3′ and shRNA luc1 antisense: 5′-TCG AGA AAA AGT GTC GCT CTG CCT CAT AGA AAT AAT AAT TCT ATG AGG CAG AGC GAC A-3′. For miRNA version of Has-mir-30a, 2264: 5′-TTT GCT GCA AAC ATC CTC GAC TGG AAG TTA TTA ATC TTT CAG TCG GAT GTT TGC AGC TTT TTC-3′ and 2265: 5′-TCG AGA AAA AGC TGC AAA CAT CCG ACT GAA AGA TTA ATA ACT TCC AGT CGA GGA TGT TTG CAG-3′.
To construct PGGV5 gene trap, a 110-bp loxP-containing unique DNA fragment was inserted between NheI and SacI of PGGV2's 3′LTR. For PGGV6 and PGGV7, pPolII was used as a shuttling vector to introduce the pPolII promoter and SA2 into PGGV5. A segment of the PGGV5 virus (the LacZ region between two Hpa sites used for Southern analysis) was amplified and inserted into the pPolII vector by HindIII and ClaI digestion to create pPolII-Hpa. To construct PGGV6, the pPolII-Hpa vector was cut with EcoRI and ClaI and the insert containing the polII promoter and Hpa region was ligated into EcoRI/ClaI digested PGGV5. To create the SA2, the following oligos: 1961 (5′-CGA TAG GGT TTC CTT GAC AAT ATC ATA CTT ATC CTG TCC CTT TTT TTT CCA CAG-3′), 1962 (5′-GAT CCT GTG GAA AAA AAA GGG ACA GGA TAA GTA TGA TAT TGT CAA GGA AAC CCT AT-3′), were annealed, phosphorylated and ligated into pPolII-Hpa by BamHI/ClaI digestion. To construct PGGV7, the pPolII-Hpa-SA2 was cut with EcoRI and ClaI and the insert containing the polII promoter-Hpa-SA2 was inserted into EcoRI/ClaI digested PGGV5. All primers were obtained from Integrated DNA Technologies.
Retroviral infection
To produce recombinant gene trap retroviruses, phoenix cells were transfected with the gene trap retroviral constructs using the Calcium–Phosphate precipitation method (Clontech) as described before (22). To infect the reporter cell lines, ES cells were plated at 3 × 106 cells per 10 cm plate and infected with retroviruses in the presence of polybrene the following day.
Transfection of dual luciferase reporters and effectors
For transfection, ES cells were passaged at least twice onto feeder-free gelatinized plates in M15 with LIF to remove feeders. ES cells were plated at ∼1–2 × 105 cells per well of a gelatinized 24 well plate. The following day, cells were transfected using lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For the shRNA cleavage assay the following constructs were transfected: 0.1 μg of CMV-Firefly luciferase, 0.02 μg of CMV-Renilla luciferase and 0.25 μg of the effector DNA (U6-hprt or U6-luc1). For the miRNA assay, the following constructs were transfected: 0.1 µg CMV-Firefly-mir30x6, 0.02 µg CMV-Renilla luciferase and 0.25 µg of the effector DNA (U6-hprt or U6-mir30A).
Splinkerette PCR
The proviral junction fragment was identified through Splinkerette PCR, described in (25). Genomic DNA was digested with Sau3AI and ligated with splinkerette adaptors. The initial PCR was performed with primer 1092 5′-GCT AGC TTG CCA AAC CTA CAG GTG G-3′ and 1094 5′-CGA AGA GTA ACC GTT GCT AGG AGA GAC C-3′ (for V2) or 1748 (5′-TAG GTC ACT CGA CCT GCA GAC C-3′) and 1094 (for V5–V7). The PCR product was diluted 1/1000 and used for a second round of PCR with primers 1093 5′-GCC AAA CCT ACA GGT GGG GTC TTT-3′and 1095 5′-GTG GCT GAA TGA GAC TGG TGT CGA C-3′ (for V2) or 1749 5′-TCG ACC TGC AGA CCA AGA TCG CT-3′ and 1095 (for V5–V7). As a control, DNA from noninfected reporter cells was used in the PCR. Specific PCR fragments obtained only in the virus-infected mutant was isolated, gel purified and either TA-cloned or directly used for sequencing. All primers were obtained from Integrated DNA Technologies.
Western, northern and Southern blot analysis
Antibodies against the following proteins were used for western blotting: rabbit anti-Hprt polyclonal antibody (Santa Cruz Biotechnology, Inc.), mouse monoclonal antibody against HA (12CA5) (Developmental Studies Hybridoma Bank, DHSB) mouse monoclonal anti-β-Tubulin antibody (DHSB) and rabbit monoclonal antibody against Ago2 (Cell Signaling Technology). For northern blotting of Hprt, a cDNA probe was PCR amplified from the PGK-Hprt vector. For Southern blot analyses, a 470 bp fragment of LacZ sequence was labeled with α-32P-dCTP and used to detect proviral junction fragments.
RESULTS
Development of a recessive genetic screen for RNAi mutants in Blm-deficient ES cells
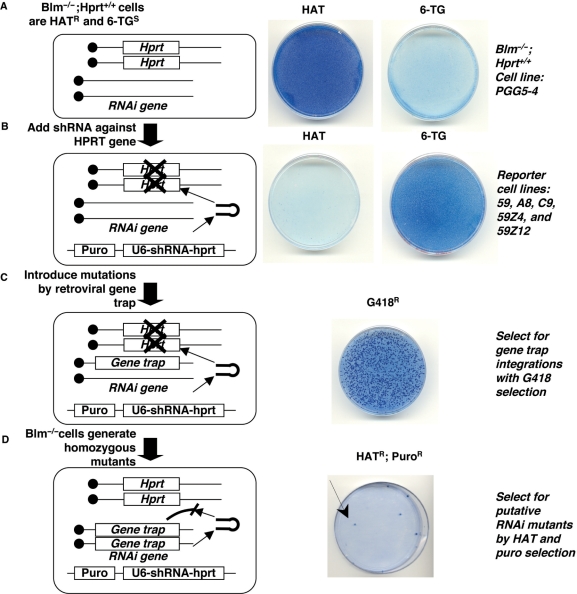
Previous studies indicated that, on average, one out of every 5000 Blm-deficient ES cells could become homozygous for a mutation (20–22). Therefore, a genetic screen performed in Blm-deficient ES cells depends on a powerful selection scheme to distinguish mutant cells from the rest of the population. To establish a selection system for RNAi mutants in Blm-deficient ES cells, we chose Hprt as the reporter gene because it allows for both positive and negative selection. When stably expressed in the cells, Hprt confers resistance to the drug HAT [hypoxanthine aminopterin thymidine, HAT resistant (HATR)] and sensitivity to the drug 6-TG [6-thioguanine, 6-TG sensitive (6-TGS)]. By introducing an shRNA against the Hprt gene, we can select for cells that silence the reporter gene (becoming 6-TG resistant, 6-TGR) through a working RNAi pathway. More importantly, after mutagenesis the Hprt reporter gene allows for positive selection (HATR) of cells that become RNAi deficient.
We constructed our selection system using a Blm-deficient ES cell line, PGG5-4, that contains two copies of the Hprt gene (Blm-deficient Hprt+/+ ES cells) (22). This cell line was originally engineered from NM5 ES cells that are Hprt-deficient and Blm-deficient. Two copies of the PGK-Hprt minigene were sequentially targeted at the mouse gdf7 locus in order to maintain homozygosity for the Hprt gene required in an LOH-based recessive genetic screen. The expression of Hprt in the cells was confirmed by drug selection with HAT and 6-TG (Figure 1A). We electroporated the cells with a U6-promoter driven shRNA to silence the Hprt gene through RNAi. The shRNA contained a puromycin (puro) marker (puro::shRNA) to select for cells that stably incorporated the transgene. Blm-deficient cells expressing the puro::shRNA effectively silenced the Hprt gene and the cells became puromycin resistant (puroR), HAT sensitive (HATS) and 6-TGR (Figure 1B). These reporter cell lines were expanded and used as the cell lines for screening. Although our preliminary results showed that one copy of shRNA was sufficient to knock-down Hprt expression to an undetectable level, the use of two independent shRNA transgenes can compensate for the loss of a single transgene through mitotic recombination. Therefore, reporter cell lines were created that contained a second copy of Hprt shRNA, selectable with zeocin (zeo::shRNA) in addition to the puro::shRNA (not depicted in the diagram for simplicity). In the presence of Hprt shRNA, RNAi competent cells were selected with 6-TG and then expanded to establish several cell lines including 59, a8, c9 (containing puro::shRNA) 59Z4 and 59Z12 (containing both puro::shRNA and zeo::shRNA) (Figure 1B).
Figure 1.
A recessive genetic screen to isolate RNAi mutants in Blm-deficient ES cells. The screen is diagrammed on the left and the drug resistance phenotype of the cells is shown on the right. Viable ES cells appear dark blue when stained with methylene blue. (A) Blm−/− Hprt+/+ cells are HATR and 6-TGS. (B) After the addition of a puromycin-linked U6-shRNA-Hprt to silence the Hprt gene, cells become HATS and 6-TGR. (C) Gene trap mutagenesis generates mutations in the cells and can be selected with the drug G418. (D) The Blm-deficient gene trapped ES cells accumulate homozygous gene trap mutations. Mutations that inactivate the RNAi pathway lead to Hprt expression and these cells are HATR and PuroR.
We carried out our screen on several different RNAi-competent reporter cell lines (59, a8, c9, 59Z4 and 59Z12) because the locations of the stably incorporated shRNAs are unknown and the shRNAs might be expressed at different levels. The location of the shRNAs may also affect the probability of isolating an RNAi essential gene. For example, if a retroviral integration occurs on the opposite chromatid of the same chromosome in which the puro::shRNA resides, a homozygous mutant for this integration site is less likely to be retained in our screen because we preferentially select for the chromatid containing the puro::shRNA over the sister chromatid containing the retroviral integration site.
To screen for RNAi mutant cells, we used a recombinant retroviral gene trap strategy to mutagenize the reporter cell lines. Retroviral gene trap mutagenesis has been successfully used in recessive genetic screens performed in Blm-deficient ES cells (22,23). The major advantage of this approach is the ability to use the gene trap provirus as a molecular tag to identify the integration site. Integration within a gene results in splicing of upstream exons to the gene trap, and a polyadenylation signal in the gene trap prematurely terminates the mRNA leading to a loss of downstream exons (Figure 2A). The gene trap contains a neomycin resistance marker to allow for Geneticin (G418) selection of the stably integrated and expressed gene traps. After integration, the gene trap contains two loxP sites, and Cre-expression removes the gene trap leaving behind a single long terminal repeat (LTR) in the locus. In some cases, a gene trap event can be reverted after Cre excision provided that the remaining viral LTR does not interfere with the endogenous locus.
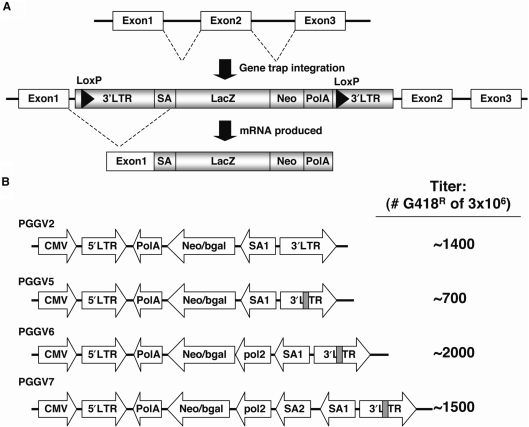
Figure 2.
Diagram of retroviral gene trap constructs. (A) Schematic of how gene traps create mutations after integration. As an example, integration of a proviral gene trap between exons 1 and 2 leads to loss of exons 2 and 3. (B) Diagrams of PGGV2-PGGV7 are depicted. Grey bars in PGGV5-7 represent unique insertion in LTR. The titer for each construct is shown to the right. SA, splice acceptor; LacZ, beta-galactosidase; Neo, neomycin resistance marker; PolA, polyadenylation.
To increase the genomic coverage of our screen, four different retroviral gene trap constructs were used (Figure 2B). The PGGV2 retroviral gene trap vector was used in previous recessive screens employing Blm-deficient ES cells (22,23). Several modifications to PGGV2 were incorporated to create the following retroviral gene trap vectors: PGGV5, PGGV6 and PGGV7 (Figure 2B). PGGV5 contained a unique sequence inserted into the LTR of PGGV2 to facilitate subsequent molecular cloning of the integration. PGGV6 was a modified version of PGGV5 containing an additional promoter to drive neomycin resistance independent of the gene trap integration site. PGGV7 was a modified version of PGGV6 with an additional splice-acceptor site.
The reporter ES cell lines were infected with gene trap retroviruses produced from a phoenix packaging cell line. ES cells containing retroviral integrations that were active gene traps were selected by G418 resistance (Figure 1C). After distinct G418R colonies had formed, the cells were propagated twice to allow accumulation of homozygous recessive mutations for the gene trap. The putative RNAi mutant cells were selected with HAT for their ability to regain Hprt expression. HATR colonies were also selected with puromycin to ensure that the puro::shRNA was retained (Figure 1D). For PGGV2 and PGGV5, we cumulatively screened 80 000 gene trap clones and for PGGV6 and PGGV7, we cumulatively screened 80 000 gene trap clones. For each G418R clone, on average we screened 10 000–40 000 cells. Through this drug selection strategy, we successfully isolated HATR putative RNAi mutant ES cells.
Validation of RNAi mutants
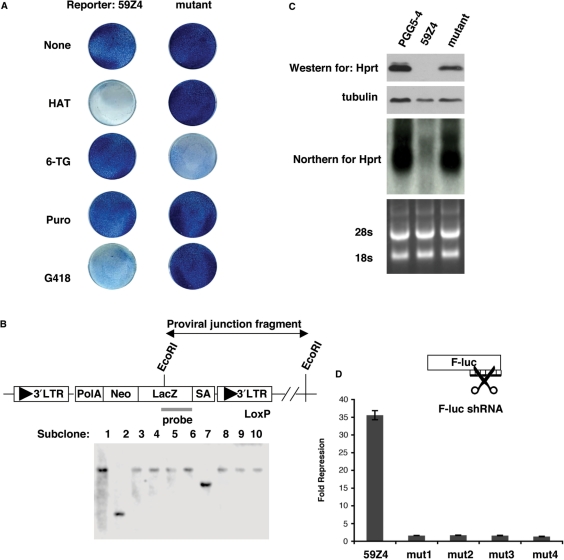
We initially performed three assays to eliminate false positives and facilitate analysis of unique mutants. First, to verify the drug-resistance profile, colonies were transferred to 96-well plates and replica plated for selection by HAT, puro and G418 (Figure 3A). RNA and protein samples from mutant clones that passed all drug resistance tests were collected for further analyses. Second, we identified unique gene trap events among candidate clones isolated from the same plate. The process of expanding the gene-trapped cells to generate homozygous mutations could potentially result in identical mutant clones being isolated multiple times. To avoid redundant analysis of identical mutant clones, a Southern blot was performed on clones isolated from the same plate. Genomic DNA was digested with EcoRI, which cuts once in the provirus and then hybridized with a probe for a unique region of the gene trap (Figure 3B). Southern blot analyses revealed that clones isolated off the same plate contained a similar proviral–host genome junction fragment based on size, most likely caused by daughter clones arising from passaging (Figure 3B). In addition, Southern blot analyses revealed a single band for the majority of the clones, which verified that the retroviral gene trap did not integrate more than once in the genome. Lastly, Hprt expression in the mutant clones was examined by northern blot and western blot to confirm that the Hprt gene was not being silenced. Hprt expression was negligible in reporter cell lines containing the puro::shRNA but was re-expressed in the gene trap mutant clones (Figure 3C).
Figure 3.
Confirmation of RNAi mutants. (A) Methylene blue staining of viable cells showing the drug resistance of a putative RNAi mutant as compared to the reporter cell line, 59Z4. (B) A Southern blot of EcoRI digested DNA using a probe unique to the gene trap identifies distinct gene-trap integration events by size. (C) Western and northern blots show that Hprt protein and RNA expression returns to levels similar to the parental cell line, PGG5-4. For the western blot, β-tubulin is shown as a loading control. For the northern blot, the 28S and 18S rRNAs are shown as a loading control. (D) A luciferase assay was used to confirm that HATR clones are RNAi-defective. The fold repression of F-luc by the luc1-shRNA is shown for the reporter (59Z4) and mutant cell lines. The data shown are the average of six independent luciferase assays, and error bars represent the standard error of the mean. The F-luc data was normalized to R-luc and the fold repression of F-luc by the control shRNA was set to 1 (not plotted). The reporter cell line represses the F-luc reporter 35-fold while the mutants repress 1- to 2-fold.
Alternative reasons could account for the Hprt expression in the isolated HATR clones. For instance, a recombination event could result in loss of the shRNA while still retaining the purmoycin marker. To eliminate false positives, we performed a secondary functional assay to confirm the RNAi mutant phenotype of the isolated clones. A F-luc reporter was used to measure the repression by shRNA against F-luc. Renilla luciferase (R-luc) served as an internal control for the transfection efficiency. Reporter cell lines were transfected with F-luc, R-luc and either control shRNA or shRNA against F-luc. An ∼35-fold repression of F-luc was observed in the original reporter cell line (59Z4) transfected with the F-luc shRNA, whereas a dramatic loss of F-luc repression was seen in several isolated mutants (Figure 3D). Thus, the luciferase assay confirmed that the isolated HATR clones were truly RNAi-defective.
Identification of Ago2 mutants validates the utility of the screen
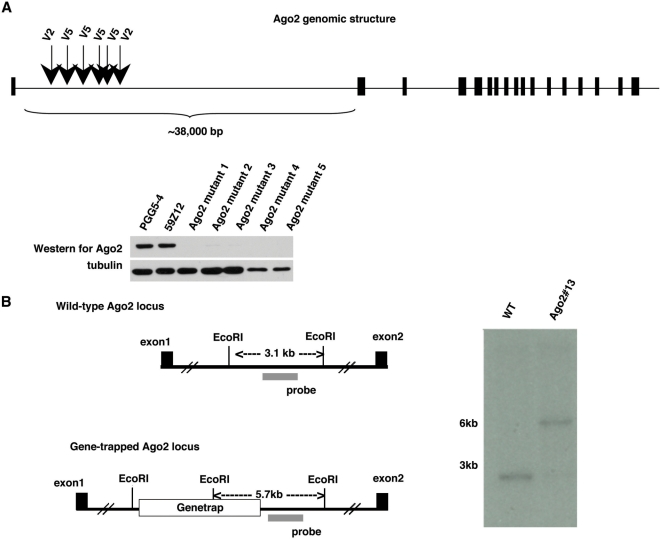
After confirmation of the RNAi mutant phenotype by luciferase assay, we performed Splinkerette PCR to identify the gene trap integration site. Six independent clones had gene trap integrations within the 38-kb intron 1 of Ago2 (Figure 4A). Loss of Ago2 was confirmed by western blot on several of the mutants (Figure 4A). Other clones, which were isolated once, included VIG (by PGGV6), and CA150 (by PGGV7). To confirm that the Ago2 clones were indeed homozygous for the gene trap, we performed Southern blot analysis. For Ago2 clone #13, genomic DNA was digested with EcoRI and hybridized with a probe detecting a region flanking the gene trap. Southern blot analysis showed that the Ago2 mutant cells are homozygous for the gene trap (Figure 4B). However, two other mutant clones (VIG and CA150) were found to be heterozygous for the gene trap (data not shown). The significance of these heterozygous gene trap clones remains to be determined.
Figure 4.
Isolation of Ago2 mutants. (A) All Ago2 gene-trap mutants were identified in the first intron of the Ago2 locus, six are depicted in the diagram with the gene trap construct that was used in the screen shown above the arrowheads. A western blot for Ago2 shows that endogenous Ago2 was below detection in the mutant cells. (B) To confirm that the isolated Ago2 mutant was homozygous for the gene trap, a Southern blot was performed on EcoRI digested DNA from the reporter cell line and one of the Ago2 mutants, Ago2# 13. A probe for a region flanking the Ago2 gene trap was used to detect the wild-type locus as a 3.1 kb fragment and the gene trap locus as a 5.7 kb fragment. The Ago2 mutant is homozygous for the gene trap as shown by the detection of only the 5.7 kb fragment.
Genetic rescue of Ago2 mutant ES cells
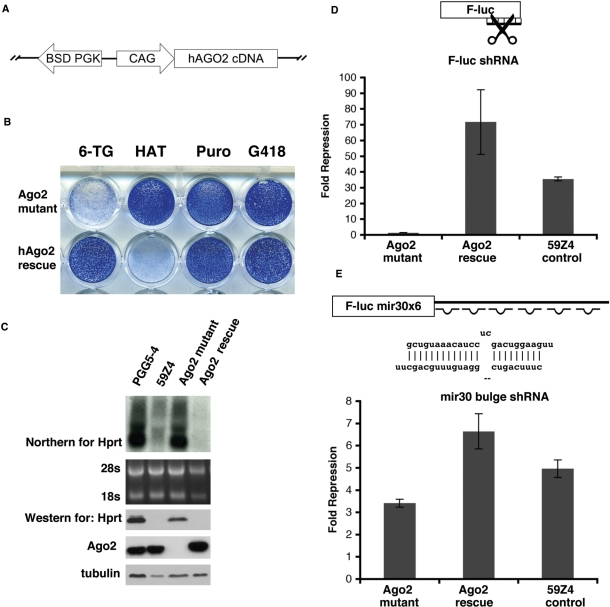
To confirm Ago2 is the cause of the RNAi mutant phenotype, we performed a genetic rescue with a human Ago2 (hAgo2) transgene. A blasticidin (BSD)-linked hemagglutinin-tagged hAgo2 (BSD-HA-hAgo2) transgene was electroporated into the mutant cells and selected with BSD (Figure 5A). Following BSD selection, cells were selected with 6-TG to identify Hprt-silent revertants (Figure 5B). Introduction of the hAgo2 transgene was able to revert the HATR/6-TGS phenotype to the reporter HATS/6-TGR phenotype (Figure 5B). Western and northern analysis of Hprt levels confirmed that the Hprt gene was silenced in the rescued clones (Figure 5C). The 6-TGR clones of hAgo2 were pooled and further analyzed by luciferase assay. The hAgo2-rescued cells regained their ability to efficiently silence the F-luc reporter as compared to the original reporter cell line, 59Z4 (Figure 5D). Therefore, the hAgo2 transgene rescued the RNAi defect in these cells.
Figure 5.
Rescue of the gene trap Ago2 mutant by a hAgo2 transgene. (A) Schematic of the hAgo2 rescue construct. (B) Rescue of the Ago2 mutant reverts the drug resistance phenotype to HATS and 6-TGR (compare to the drug resistance phenotype of the Ago2 mutant). (C) Rescue of the Ago2 mutant also reverts the levels of Hprt back to reporter cell (59Z4) levels as shown by northern and western blots. An Ago2 western blot performed on parental (PGG5-4), reporter (59Z4), mutant and rescued cell lines showing similar levels of Ago2 in the rescued cells. (D) A luciferase assay for RNAi-directed mRNA cleavage confirms the rescue of Ago2 mutant by hAgo2 transgene. (E) A luciferase assay was used to examine miRNA repression and shows that the Ago2 mutant exhibits a slight decrease in miRNA repression compared to the original reporter cell line 59Z4. Calculations for data shown in (D) and (E) were performed as in Figure 3D.
Of the four mammalian Argonautes, Ago2 has been shown to be the only Argonaute capable of performing mRNA cleavage (26,27). In contrast, all mammalian Argonautes are capable of binding miRNAs (26). To test whether the Ago2 mutant would also show a defect in the miRNA pathway, we performed a miRNA repression assay using a F-luc reporter with six miRNA binding sites for mir30 (F-luc mir30 × 6). The Ago2 mutant cells exhibited only a mild reduction in miRNA repression compared to the control cells (Figure 5E). This result corroborates previous reports that Ago2 is essential for siRNA-mediated target mRNA cleavage but not required for miRNA mediated translational repression (26,27).
Mutations in the Ago2 mid-domain disrupt its cleavage activity
The luciferase-reporter assay revealed that the homozygous Ago2 gene trap cell line has a nearly complete loss of RNAi-directed cleavage activity. Thus, these cells provide an Ago2 null genetic background to examine the effects of specific Ago2 mutations. The Ago2-deficient cell line also allows testing of genetic rescue by drug selection, as expression of wild-type hAgo2 reverted Ago2 mutant cells to 6-TGR. An assay based on drug selectable rescue in mutant cells would be useful for rapid structure–function analysis of RNAi pathway components. To test this possibility, we asked whether this method could be used to resolve a controversy regarding Ago2 activity. Ago2-F2V2 is an Ago2 mutant in which two highly conserved phenylalanines (F470 and F505) in the mid-domain are mutated to valines. Recent reports on this Ago2 mutant have had contradictory findings. One group found that the Ago2-F2V2 mutations disable the ability of hAgo2 to bind to m7G caps, while they observed no effect on the ability of hAgo2-F2V2 to perform RNAi-directed mRNA cleavage in an in vitro assay (28). A second group discovered that the equivalent mutations disrupt the ability of Drosophila Ago1 and Ago2 to repress mRNA through a tethering assay, however they did not observe a loss of binding ability to m7G caps (29). The F2V2 mutations could globally affect the Ago2 protein, which might account for the loss of translational repression observed in this mutant.
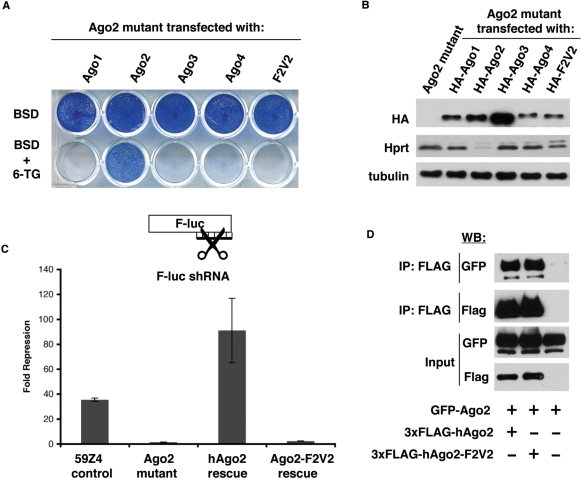
To further examine the effect of the F2V2 mutation on cleavage activity, we attempted genetic rescue of the Ago2 mutant using the Ago2-F2V2 mutant. A BSD-linked HA-tagged Ago2-F2V2 transgene (BSD-HA-Ago2-F2V2) was electroporated into the Ago2 mutant and cells were selected with BSD. For these studies, BSD-linked HA-tagged constructs of the noncleavage competent Argonautes (Ago1, 3 and 4) served as negative controls and BSD-HA-hAgo2 served as a positive control (Figure 6A). The transfected cells were selected with either BSD alone or BSD with 6-TG. Only the hAgo2 construct was capable of rescuing the Ago2 mutant cell line by 6-TG selection (Figure 6A). A western blot for HA confirms that the levels of the BSD-HA-hAgo2 and BSD-HA-hAgo2-F2V2 were comparable, demonstrating that the lack of rescue was not due to a lower level of expression of the hAgo2-F2V2 (Figure 6B). Consistent with drug selection, western blots showed that only Ago2 could repress the Hprt gene through a working RNAi pathway, whereas Ago1, 3, 4 and Ago2-F2V2 were unable to do so (Figure 6B). In addition, we performed an siRNA-based luciferase assay to examine cleavage activity and did not observe any ability of the Ago2-F2V2 mutant protein to repress the luciferase reporter (Figure 6C). Thus, our data are in agreement with Eulalio and colleagues (29), demonstrating that the F2V2 mutations may produce global defects in Ago2's function.
Figure 6.
Ago2 mutant ES cells can serve as a tool to perform mutational analysis. (A) The mutant Ago2 cell line was transfected with transgenes expressing a BSD-linked, HA-tagged cDNA construct for Ago1, Ago2, Ago3, Ago4 and Ago2-F2V2. Cells were selected with BSD or BSD with 6-TG. Only stable transfection of WT-Ago2 can revert the 6-TGS phenotype of the mutant cells. (B) Western blot for HA shows the expression levels of different Ago proteins. Western blot for Hprt shows that only Ago2 expression silences Hprt expression. Tubulin is shown as a loading control. (C) A luciferase assay for RNAi-directed cleavage activity shows that Ago2-F2V2 is deficient for cleavage activity. (D) Coimmunoprecipitation performed in 293T cells shows that GFP-Ago2 is associated with Flag-tagged Ago2 as well as Flag-tagged Ago2-F2V2.
Previously, it was shown that immunoprecipitated Ago2-F2V2 was capable of cleavage in vitro (28). In order to rationalize the contradiction between our data and this finding, we reasoned that the immunoprecipitated Ago2-F2V2 might have also coimmunoprecipitated WT-Ago2. To test this idea, we transfected 293T cells with WT-GFP-Ago2 and either 3xFLAG-WT-Ago2 or 3xFLAG-Ago2-F2V2. Immunoprecipitation of Flag-tagged Ago2 proteins revealed that both WT-Ago2 and Ago2-F2V2 were associated with WT-GFP-Ago2 (Figure 6D). Thus, the previous observation of cleavage activity associated with Ago2-F2V2 may have been due to the presence of coimmunoprecipitated endogenous Ago2. Our finding that Ago2-F2V2 is deficient in RNAi-directed cleavage, together with the finding that F2V2 substitution abolishes translational repression (29) suggests a critical role of the mid-domain for overall function or structural integrity of Ago2.
DISCUSSION
Key components of the RNAi pathway have been identified through genetic screens in model organisms. Performing a recessive genetic screen in a mammalian system is feasible through the use of Blm-deficient ES cells. Here, we show the utility of screening for RNAi components in Blm-deficient cells by isolating an essential component of RNAi, Ago2. Several Ago2 mutants were isolated through our screen. One interpretation of this result is that Ago2 is the most essential component for siRNA-mediated RNAi. Interestingly, minimal RISC has been shown to be composed solely of Ago2 and siRNA (30). In addition, the identification of six independent Ago2 gene traps within intron 1 could indicate a hot spot for retroviral integration. Consistent with this, a similar Ago2 gene trap ES cell line was described in the gene trap consortium (www.genetrap.org).
We found that Ago2 mutant ES cells display only a slight defect in miRNA-mediated repression. In contrast, the gene trap Ago2 mutants display a dramatic loss of RNAi-directed cleavage activity. The nearly cleavage-deficient background of the Ago2 mutant provides a useful tool to examine the cleavage activity of Ago2 proteins with specific mutations. We transfected an Ago2 containing mid-domain mutations, Ago2-F2V2, into our Ago2 mutant cell line to assess the ability of Ago2-F2V2 to perform RNAi-directed cleavage. Through this method, we have shown that the two mutations in the mid-domain of Ago2 abolish cleavage activity. The recent crystal structure of the Thermus thermophilus Argonaute bound to a guide-strand reveal that the 5′-phosphate of the guide strand is bound in a pocket in the mid-domain (31). It is possible that the F2V2 mutations might have disrupted the structural conformation of the binding pocket of the mid-domain, thus disabling its ability to bind siRNA. Our system can be further used to characterize the importance of newly identified posttranscriptional modifications of Ago2 on RNAi-directed cleavage activity (32,33).
Although this screen was successful in isolating Ago2, there appears to be limitations of this system. For example, the screen did not pull out other core components of the RNAi pathway such as Dicer. One reason for this could be the growth defect exhibited by Dicer−/− ES cells (34). Slower growing Dicer−/− cells may be lost during several passages of Blm-deficient ES cells to create homozygous mutations. The screen also identified heterozygous gene traps, whose homologs have been identified in previous studies for RNAi components in other model systems. VIG and CA150 have been isolated previously as RNAi components (35–37). However, these clones were proven to be heterozygous for their gene traps, thus making it difficult to determine whether they are the causative mutations. One possible explanation for the gene trap heterozygotes is the accumulation of point mutations in our reporter cell lines over time. These point mutations may rest on the sister allele of the gene trapped allele or a completely independent mutation. The accumulation of point mutations may be avoided by the use of an inducible Blm-deficient cell line to control the mutagenic rate of the cells (21).
Improvements to our system can be implemented to potentially identify more components of the RNAi pathway. A new method to mutagenize ES cells, the Piggy-Bac transposon, has recently been reported (38). Similar to gene traps, the Piggy-Bac transposon can be molecularly analyzed for the site of integration, and can potentially circumvent the problems associated with the nonrandom bias of retroviral gene trap mutagenesis. In addition, transposons can be removed by expressing transposase, facilitating analysis of revertants. Another way to improve the screen might be to use a different observable phenotype to select for RNAi mutants. For example, the screen could be set up to detect RNAi mutants using a GFP reporter combined with FACS analysis. The use of a different selectable phenotype has greatly improved other screens for RNAi pathway genes. For instance, a screen for RNAi components in C. elegans based on embryonic lethality pulled out three genes (39). In contrast, a screen in C.elegans based on silencing of GFP in seam cells led to the identification of 90 genes (35). Therefore, the use of various selectable phenotypes can be advantageous for pulling out more RNAi components.
Understanding the repertoire of mammalian RNAi components has many potential benefits. A new class of endogenous siRNAs (endo-siRNAs) has recently been identified in mice and flies (40). A better understanding of the mammalian RNAi pathway can help elucidate the RNAi mechanism of these newly identified endo-siRNAs. In addition, RNAi-based therapies can be improved by a more comprehensive knowledge of the RNAi pathway in a mammalian system. We have shown that a recessive genetic screen for mammalian RNAi genes can be performed in Blm-deficient ES cells to pull out a key component of the siRNA pathway. Therefore, with improvements like a new insertional mutagenesis tool, the transposon-mediated gene trap, this screening system has the potential to identify as-yet-unknown mammalian RNAi components.
FUNDING
National Institute of Health (5R21GM079528); Illinois Department of Public Health (to X.W.); CMBD training grant from the National Institute of Health T32 GM008061 (to M.T., partial). Funding for open access charge: 5R21GM079528.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank A. Bradley and G. Guo for PGG5-4 ES cells and PGGV2 gene trap construct and G. Guo and W. Wang for technical advice. We thank A. Dudley and D. Trombly for critical reading and comments on the article.
REFERENCES
- 1.Mello CC, Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 4.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe D. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 6.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 7.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 8.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 9.Huppi K, Martin SE, Caplen NJ. Defining and assaying RNAi in mammalian cells. Mol. Cell. 2005;17:1–10. doi: 10.1016/j.molcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 10.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 11.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 12.Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- 13.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl Acad. Sci. USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 15.Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 16.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 17.Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways.[see comment] Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 19.Stein P, Svoboda P, Stumpo DJ, Blackshear PJ, Lombard DB, Johnson B, Schultz RM. Analysis of the role of RecQ helicases in RNAi in mammals. Biochem. Biophys. Res. Commun. 2002;291:1119–1122. doi: 10.1006/bbrc.2002.6578. [DOI] [PubMed] [Google Scholar]
- 20.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 21.Yusa K, Horie K, Kondoh G, Kouno M, Maeda Y, Kinoshita T, Takeda J. Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom's syndrome gene. Nature. 2004;429:896–899. doi: 10.1038/nature02646. [DOI] [PubMed] [Google Scholar]
- 22.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Bradley A. A recessive genetic screen for host factors required for retroviral infection in a library of insertionally mutated Blm-deficient embryonic stem cells. Genome Biol. 2007;8:R48. doi: 10.1186/gb-2007-8-4-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 25.Devon RS, Porteous DJ, Brookes AJ. Splinkerettes–improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. [see comment] Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 27.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 30.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 33.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 36.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caudy AA, Hannon GJ. Induction and biochemical purification of RNA-induced silencing complex from Drosophila S2 cells. Methods Mol. Biol. 2004;265:59–72. doi: 10.1385/1-59259-775-0:059. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl Acad. Sci. USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol. Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]