Abstract
Chromatin immunoprecipitation (ChIP) studies were conducted in human hepatocytes treated with rifampicin in order to identify new pregnane-X receptor (PXR) target genes. Genes, both previously known to be involved and not known to be involved in drug disposition, with PXR response elements (PXREs) located upstream, within or downstream from their potentially associated genes, were identified. Validation experiments identified several new drug disposition genes with PXR binding sites. Of these, only CYP4F12 demonstrated increased binding in the presence of rifampicin. The role of PXR in the basal and inductive response of CYP4F12 was confirmed in hepatocytes in which PXR was silenced. We also assessed the association of PXR-coactivators and -corepressors with known and newly identified PXREs. Both PXR and the steroid receptor coactivator (SRC-1) were found to bind to PXREs in the absence of rifampicin, although binding was stronger after rifampicin treatment. We observed promoter-dependent patterns with respect to the binding of various coactivators and corepressors involved in the regulation of CYP4F12, CYP3A4, CYP2B6, UGT1A1 and P-glycoprotein. In conclusion, our findings indicate that PXR is involved in the regulation of CYP4F12 and that PXR along with SRC1 binds to a broad range of promoters but that many of these are not inducible by rifampicin.
INTRODUCTION
The human pregnane-X receptor (PXR, NR1I2) is an essential regulator of a large and growing array of drug disposition genes which correspond to all phases of drug metabolism. These include phase I enzymes such as cytochrome P450 (CYP), phase II enzymes such as uridine-5′-diphosphate glucuronosyltransferases (UGTs) and transporters such as the multidrug resistance protein MDR1 P-glycoprotein (Pgp) (1). A large body of literature has revealed that PXR activation by xenobiotics, in the liver and intestine, results in a significant increase in the expression of drug metabolizing enzymes and transporters (1–5). In addition to increased gene expression, PXR can repress gene expression (6), indicating that gene regulation via PXR is complex. Although PXR functions as a defense mechanism against toxic insults, it also represents the molecular basis for pharmacokinetic drug–drug interactions. For example, if one drug activates PXR, administration of this drug can promote its own elimination (autoinduction) or the elimination of other coadministered drugs that are metabolized and eliminated by PXR-target gene products, thereby reducing the efficacy of drug therapies in patients on combination therapy (7–9).
As a prototypical nuclear receptor, PXR has a DNA-binding domain (DBD) at the N-terminus and a ligand-binding domain (LBD) at the C-terminus. The DBD is responsible for binding to regulatory DNA sequences such as the AGGTCA-like direct repeats spaced by 3, 4 or 5 bases (DR3, DR4 and DR5), and the everted repeats separated by 6 or 8 bases (ER6 and ER8) located in the PXR target genes (10). The LBD is multifunctional in that it is capable of ligand binding, dimerization, transcriptional activation, and interactions with transcriptional co-factors. The C-terminal helix termed AF-2 is responsible for transcriptional activation by recruiting coactivators through conformational rearrangement, or gene repression by interactions with transcriptional corepressors (7,11).
Understanding of coactivators and corepressors of PXR in the expression of target genes is relatively limited. Recently, Moore et al. (12) reviewed some of the key coactivators and corepressors involved in the PXR regulation of drug metabolizing enzymes and transporters. Small heterodimer partner (SHP/NC0B2) and nuclear receptor corepressor 2 (NCoR2/SMRT) were identified as corepressors, while steroid receptor coactivators 1 (SRC1/NCOA1) and 2 (SRC2/GRIP1), nuclear receptor interacting protein 1 (NRIP1/RIP140), peroxisome proliferator-activated receptor-gamma coactivator (PGC-1), and Forkhead transcription factor FKHR (FOXO1) were reported as coactivators.
While NCoR, SHP and SMRT are known to be corepressors of genes, studies undertaken by several groups using transfection assays in CV1, HepG2, HEK293 and yeast cells indicate that only SHP and SMRT are involved in transcriptional repression of PXR (13–16). Ourlin et al. (13) demonstrated in HepG2 cells that SHP blocks PXR binding to DNA in a ligand-dependent manner. In addition, their studies using transient transfection assays proved that increased expression of SHP resulted in a decrease in PXR activation in the presence of rifampicin.
Tirona et al. (17) highlighted the critical involvement of hepatocyte nuclear factor 4α (HNF4α), together with PXR in the regulation of CYP3A4. Using mammalian two-hybrid assays Li and Chiang (18) showed that HNF4α and SHP compete to interact with PXR. These studies also revealed that SHP only partially blocked the PXR/SRC1 interaction and was unable to block the PXR/PGC-1α interaction. Using chromatin immunoprecipitation (ChIP) analysis, they observed that rifampicin increased the recruitment of HNF4α and SRC1 to the CYP3A4 chromatin but reduced PGC1α recruitment.
In most studies referenced above, cell lines in which proteins were overexpressed were not of hepatic origin and promoter elements were outside of their natural chromatin environment. In addition, to assess DNA–protein and protein–protein interactions researchers used electro mobility shift assays that make use of ‘naked’ DNA as a probe, whereas in living cells much of the DNA is covered by nucleosomes. Thus, many binding sites for a protein in living cells could be masked by the presence of nucleosomes, but they would be scored as positive by the biochemical assays (19).
The goals of the experiments presented in this manuscript were 3-fold: First, using ChIP, we searched for new PXR target genes in cryopreserved primary human hepatocytes treated with rifampicin. Second, using ChIP-based assays, we studied binding of PXR, as well as PXR-corepressors and -coactivators to newly identified and established PXR response elements (PXREs) before and after treatment with rifampicin. Third, we investigated whether newly identified genes in the ChIP experiments were bona fide PXR target genes by measuring mRNA levels after small interfering RNA (siRNA)-mediated knockdown of PXR.
MATERIALS AND METHODS
Materials
Cryopreserved human hepatocytes, antibiotic/antimycotic Torpedo Mix, In VitroGRO-CP Media and In VitroGRO-HI Incubation Media were purchased from In Vitro Technologies (Baltimore, MD, USA). Rifampicin, glycine, Igepal, phenylmethanesulfonyl fluoride (PMSF) and formaldehyde were purchased from Sigma-Aldrich (St Louis, MO, USA). RNeasy 96 Kit was purchased from Qiagen (Valencia, CA, USA). Anti-PXR (sc-25381), -SRC-1 (sc-8995), -SRC-3 (sc-7216), -N-CoR (sc-8994), -HNF4α (sc-8987), -GRIP-1 (sc-8996), -PGC-1α (sc-13067), -SMRT (sc-1610), -FKHR (sc-11350), -RIP140 (sc-8997) and -SHP (sc-15283) antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Anti-RNA pol II (CTD phospho Ser-2, ab5095) antibody was purchased from Abcam Inc. (Cambridge, MA, USA). TaqMan® Universal PCR Master Mix was purchased from Applied Biosystems (Foster City, CA, USA). Collagen-coated 6-well plates were purchased from BD Biosciences (San Jose, CA, USA). ON-TARGETplus GAPDH Control Pool siRNA (D-001830), ON-TARGETplus Non-Targeting Pool siRNA (D-001810), ON-TARGETplus SMARTpool human NR1I2 siRNA (L-003415) and DharmaFECT-1 transfection reagent were purchased from Dharmacon Inc. (Lafayette, CO, USA). All other reagents and chemicals were of the highest grade and purchased from Fisher Scientific (Pittsburg, PA, USA).
Cell culture and drug treatment
Cryopreserved primary human hepatocytes from three different donors (LHO: 68-year-old female Caucasian; 455: 37-year-old female Caucasian; and DMQ: 59-year-old African-American female) were used in our studies. Hepatocytes were plated in 6-well collagen-coated plates at a seeding density of 1.5 × 106 cells per well in In VitroGRO-CP Media and 2.2% antibiotic/antimycotic Torpedo Mix and placed in an incubator (37°C, 5% CO2). After 24 h the plating medium was discarded and hepatocytes were treated for 3 h with either DMSO (0.1%) or rifampicin 10 µM, prepared in In VitroGRO-HI incubation Media and 2.2% antibiotic/antimycotic Torpedo Mix. Following the 3 h incubation period, hepatocytes were fixed with 1/10 formaldehyde solution (11% formaldehyde, 0.1 M NaCl, 1 M EDTA, 50 mM HEPES pH 7.9). Fixation was stopped by adding 1/20 volume of glycine solution (2.5 M glycine, water) to the existing media. Cells were washed, centrifuged (800g for 10 min.) and resuspended in phosphate buffered saline (PBS)–Igepal solution (0.5% Igepal in PBS). Thereafter, cells were centrifuged (800g for 10 min.) and again resuspended in PBS–Igepal to which 100 µl PMSF (100 mM in ethanol) was added. Cells were then centrifuged (800g for 10 min.), pelleted and snap-frozen on dry ice and stored at −80°C for further analysis.
ChIP-based assays
In order to identify PXREs present on genes in the entire human genome, cross-linked chromatin from hepatocytes treated with DMSO (0.1%) or 10 µM rifampicin were immunoprecipitated against a validated-PXR antibody. Cloning, sequencing, analysis of tags and confirmation of the PXR-binding sites using polymerase chain reaction (PCR) primers (Supplementary Table S1, Supplementary Data) targeting a region within 200 bp of each selected alignment were conducted as previously described (20). Assays to confirm that increased PXR-binding resulted in increased transcription were undertaken by immunoprecipitating cross-linked chromatin against RNA polymerase II (Pol II) antibody and the enrichment of Pol II was determined by quantitative (q)-PCR (20). To assess the involvement of coactivators and corepressors in the PXR-regulation of CYP3A4, CYP2B6, CYP4F12, UGT1A1 and MDR1 cross-linked chromatin was immunoprecipitated against the aforementioned coactivators and corepressors and the enrichment of SRC-1, SRC-3, GRIP1, NCOR, PGC1α, HNF4α, FKHR, RIP140, SMRT and SHP were determined by q-PCR.
RNA interference
Cryopreserved human hepatocytes (120 000 cells/well) were cultured in antibiotic-free In VitroGRO-HI plating medium in collagen-I coated 48-well plates. After 24 h ON-TARGETplus SMARTpool human NR1I2 siRNA (L-003415) was used to knockdown PXR. ON-TARGETplus GAPDH Control Pool siRNA (D-001830) and ON-TARGETplus Non-Targeting Pool siRNA (D-001810) were used as internal controls. Briefly, siRNA duplexes (100 nmol/well) and DharmaFect-1 (3 µl/well) were diluted in 50 µl antibiotic- and serum-free In VitroGRO-HI incubation medium separately and mixed gently and incubated for 5 min at room temperature. Afterward, PXR-siRNA and DharmaFect-1 were mixed (total volume 100 µl) and incubated at room temperature for 20 min. Then, 100 µl siRNA–DharmaFect-1 complex was diluted with 100 µl antibiotic-free medium and added to the well. Under these conditions, the transfected cells looked morphologically normal after 72 h, and they did not differ from untransfected cells in cell viability and levels of the 18S mRNA housekeeping gene (data not shown). Thereafter, hepatocytes were treated with 10 µM rifampicin or 0.1% DMSO (vehicle control) for 48 h. To ensure that functional and specific silencing was obtained, the mRNA levels of PXR and known PXR target genes (CYP3A4, CYP2B6, Pgp and UGT1A1) were compared between PXR-siRNA and SCR (scrambled) siRNA groups before and after treatments in all experiments.
Reverse-transcription Q-PCR
Total RNA was isolated using the RNeasy 96 Kit. A two-step reverse transcriptase (RT)-PCR reaction was conducted by reverse transcribing 50 ng of total RNA to cDNA using TaqMan® Reverse Transcription Reagents, according to the TaqMan® Universal PCR Master Mix protocol. PCR reactions were then prepared by adding an aliquot of cDNA (3 µl) to a reaction mixture containing the TaqMan® Fast Universal PXR Master Mix solution, primers (Supplementary Table S1, Supplementary Data), and probes for the specific gene targets. PCR-amplified cDNAs were detected by real-time fluorescence on an ABI PRISM 7900 Fast Sequence Detection System (Perkin Elmer, Wellesley, MA, USA). Gene expression values were calculated based on the comparative ΔΔCt method and normalized to values obtained for 18S ribosomal RNA.
Statistical analysis
The differences in PXR-, Pol II-, SRC-1-, SRC-3-, GRIP1-, NCOR-, PGC1α-, HNF4α-, FKHR-, RIP140-, SMRT- and SHP binding, between hepatocytes treated with 10 µM rifampicin or 0.1% DMSO (vehicle control) were analyzed using the Student's t-test. Statistical analysis of the differences in mRNA gene expression was based upon differences in the ΔCt values. Differences with P-values <0.05 were considered significant.
RESULTS
Identification of new PXR target genes
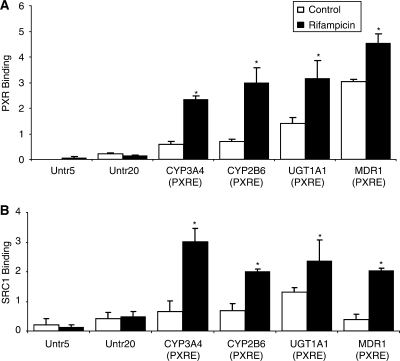
In order to identify new genes containing PXR binding sites, we performed ChIP analyses with cryopreserved human hepatocytes treated with the prototypical inducer rifampicin (10 μM, 3 h). The batches of hepatocytes used in these studies were selected for high inducibility of CYP3A4 mRNA (45- to 75-fold relative to vehicle control) after treatment with rifampicin (10 μM) for 48 h (data not shown). To validate the polyclonal anti-PXR antibody used for ChIP, the ability to detect known PXR target genes was determined (21): Enrichment of CYP3A4, CYP2B6, UGT1A1 and MDR1 PXRE were 3.9-, 4.2-, 2.2- and 1.5-fold higher in rifampicin as opposed to vehicle (control; 0.1% DMSO)-treated cells (Figure 1A). Remarkably, binding of PXR to all PXREs was also detected in the absence of rifampicin (Figure 1A), although binding was significantly lower than that after treatment with rifampicin. Measuring of PXR interaction to two untranscribed regions (Untr) served as negative controls. While ChIPs with a control IgG are often conducted as a negative control, our findings (data not shown) suggested that the values obtained using this approach provided a less reliable baseline than assaying negative control regions (Untr) with the experimental antibody. Also, we have observed that different antibodies have different levels of background (20). Our unpublished data also suggest that antibody titration experiments do not necessarily show the expected concentration-dependent increase and then plateau, but one continues to see some increase (but not linear or proportional with the antibody amount) upon increasing the antibody amount. In our studies, we could not utilize much higher antibody amounts which we believe were already in excess.
Figure 1.
PXR (A) and SRC-1 (B) binding to several known PXRE-containing promoters including CYP3A4, CYP2B6, UGT1A1, MDR1 and Untr 5 and 20. Cryopreserved human hepatocytes were treated with vehicle (Control; 0.1% DMSO) or rifampicin (10 μM) for 3 h and processed for ChIP assays as described in Materials and methods section. Studies repeated with different batches of hepatocytes yielded similar results. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05.
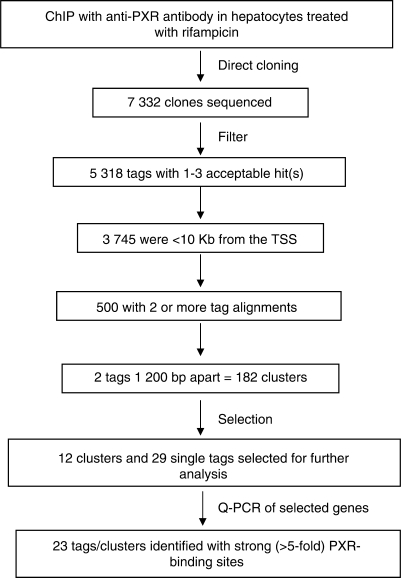
A discovery approach for the identification of PXR binding sites in the human genome was initiated (see Materials and methods section). After chromatin precipitation with the anti-PXR antibody, ∼7300 tags representing anti-PXR immunoprecipitated DNA, which were derived from rifampicin-treated cells, was sequenced (Figure 2). Approximately 5000 of these sequences produced acceptable genomic alignments from which 32.7% (or about 1/3) did not map within 10 kb of any gene. The number of genes that had at least one tag alignment within 10 kb was 3745 (using the NCBI gene database). Of these 3745 genes, 500 had two or more tag alignments (Supplementary Table S2, Supplementary Data). For these 500 genes, there were a total of 1244 aligns (within ±10 kb), of which 877 (70.5%) mapped inside genes, 190 (15.3%) upstream and 177 (14.2%) downstream. Our analysis included all of these tags since PXRE sites may be important wherever they lie within the gene and do not necessarily need to be located in the 5′ UTR. Because ChIP fragments were <600 bp, 1200 was set as the maximal distance between two tags representing the same binding site, such that ∼500 tags generated 182 clusters consisting of two or more distinct alignments within 1200 bp). Genes identified as known to contain PXR binding sites in their promoter regions were CYP3A4 and MDR1. As expected, the lack of identification of additional known PXR target genes indicated that the screen had not reached saturation.
Figure 2.
Construction of the PXR-binding site library. Outline of the strategy for ChIP-cloning and filtering of sequences. ‘Filter’: short sequences (<40 bp), highly repetitive sequences and those with <90% identity to the genome were eliminated. ‘Selection’: 12 clusters with strong PXR binding (>5-fold binding relative to the negative control region) were selected for further testing. In addition, 29 tags were selected which were in the proximity of genes known to be involved in different metabolic processes including drug metabolism, or recently identified as PXR target genes by gene profiling in studies with the colon carcinoma cell line LS180 (Table 1) (2).
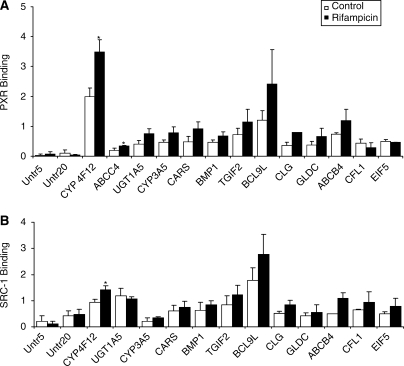
The binding of PXR to identified genomic locations in cryopreserved hepatocytes treated with either vehicle or rifampicin was confirmed by q-PCR using PXR primers encompassing ∼200 nt of the putative PXR binding sites (Supplementary Table S1, Supplementary Data). Of the 182 clusters, 12 clusters with strong PXR binding (>5-fold binding relative to the negative control region) were selected for further testing (Table 1). Several of these were not in the proximity of genes known to be involved in drug metabolism or transport but were involved in apoptosis, electron transport, hormone catabolic processes, response to chemical stimuli and steroid, estradiol, linoleic acid, retinol, tryptophan, arachidonic acid, fatty acid and lipid metabolism. In addition, 29 tags were selected which were in the proximity of genes known to be involved in different metabolic processes including drug metabolism, or recently identified as PXR target genes by gene profiling in studies with the colon carcinoma cell line LS180 (Table 1) (2). Of the 41 tags/clusters selected, 23 showed elevated PXR binding (>5-fold increased binding compared to the negative control regions) (Table 2). Interestingly, of the 23 identified PXR-binding sites only 11 showed a trend towards increased PXR-binding in the presence of rifampicin compared to the vehicle (Table 2 and Figure 3A). However, statistically significant differences were only observed for CYP4F12 and ABCC4.
Table 1.
Tags/Clusters from discovery approach selected for further analysis
| Genomic region | Tag/Cluster name | Gene/Proteome description |
|---|---|---|
| CYP3A4 (PXRE) | Control (literature) | |
| CYP2B6 | Control (literature) | |
| UGT1A1 | Control (literature) | |
| MDR1 | Control (literature) | |
| CARS | GP0059_02_B04-frag008 | Encodes a class 1 aminoacyl-tRNA synthetase, cysteinyl-tRNA synthetase. Each of the 20 aminoacyl-tRNA synthetases catalyzes the aminoacylation of a specific tRNA or tRNA isoaccepting family with the cognate amino acid. Alterations in this region have been associated with the Beckwith–Wiedemann syndrome, Wilms tumor, rhabdomyosarcoma, adrenocortical carcinoma, and lung, ovarian and breast cancer. |
| OSBP2 | GP0059_06_B06-frag009 | Oxysterols are byproducts of cholesterol that can have cytotoxic effects on many cell types. The membrane-bound protein encoded by this gene contains a pleckstrin homology domain and an oxysterol-binding region. It binds oxysterols such as 7-ketocholesterol and may inhibit their cytotoxicity. |
| PFAS | GP0059_03_A11-frag004 | The enzyme encoded by this gene catalyzes the fourth step of inosine monophosphate biosynthesis. |
| CD59 | GP0059_06_C01-frag008 | This gene encodes a cell surface glycoprotein that regulates complement-mediated cell lysis, and it is involved in lymphocyte signal transduction. This protein is a potent inhibitor of the complement membrane attack complex, whereby it binds complement C8 and/or C9 during the assembly of this complex, thereby inhibiting the incorporation of multiple copies of C9 into the complex, which is necessary for osmolytic pore formation. This protein also plays a role in signal transduction pathways in the activation of T cells. Mutations in this gene cause CD59 deficiency, a disease resulting in hemolytic anemia and thrombosis, and which causes cerebral infarction. |
| CEACAM6 | GP0059_05_C06-frag011 | Carcinoembryonic antigen is one of the most widely used tumor markers in serum immunoassay determinations of carcinoma. An apparent lack of absolute cancer specificity for CEA probably results in part from the presence in normal and neoplastic tissues of antigens that share antigenic determinants with the 180 kDa form of CEA. |
| DOCK1 | GP0059_08_D04-frag004 | Dedicator of cytokinesis 1, member of the CDM family, contains an SH3 domain, binds phosphatidylinositol-3,4,5-triphosphate, integrin receptor-mediated complex formation with BCAR1 and CRK leads to RAC1 activation and phagocytosis of apoptotic cells. |
| FLJ10986 | GP0059_06_D06-frag004 | |
| CYP4F12 | Cluster 19 153 (GP0059_07_F05-frag007; GP0059_06_D09-frag005) | Cytochrome P450 family 4 subfamily F polypeptide 12, metabolizes arachidonic acid by omega 3-hydroxylation, metabolizes docosahexanoic acid, docosapentaenoic acid, leukotrienes and the antihistamine drug ebastine. |
| CYP3A5 | GP0059_02_H05-frag007 | This protein localizes to the endoplasmic reticulum and its expression is induced by glucocorticoids and some pharmacological agents. The enzyme metabolizes drugs such as nifedipine and cyclosporine as well as the steroid hormones testosterone, progesterone and androstenedione. This gene is part of a cluster of cytochrome P450 genes on chromosome 7q21.1. This cluster includes a pseudogene, CYP3A5P1, which is very similar to CYP3A5. This similarity has caused some difficulty in determining whether cloned sequences represent the gene or the pseudogene. |
| CYP3A5P2/CYP3A4 | Cluster 7 170 (GP0059_04_A01-frag006; GP0059_01_D05-frag007) | |
| ABCC4 | GP0059_10_C11-frag004 | The protein encoded by this gene is a member of the superfamily of ATP-binding cassette (ABC) transporters. This protein is a member of the MRP subfamily which is involved in multi-drug resistance. The specific function of this protein has not yet been determined; however, this protein may play a role in cellular detoxification as a pump for its substrate, organic anions. |
| SULT1B1 | GP0059_01_B02-frag007 | Sulfotransferase enzymes catalyze the sulfate conjugation of many hormones, neurotransmitters, drugs and xenobiotic compounds. These cytosolic enzymes are different in their tissue distributions and substrate specificities. The gene structure (number and length of exons) is similar among family members. |
| SLC40A1 | Cluster 2 215 (GP0059_10_F11-frag006; GP0059_01_C04-frag005) | Solute carrier family 40 (iron-regulated transporter) member 1, an iron transporter involved in iron homeostasis and regulation of translation initiation, possibly involved in development; gene mutations are a cause of autosomal dominant hemochromatosis. |
| KCNK5 | Cluster 6 129 (GP0059_03_F09-frag006; GP0059_10_A04-frag005) | Potassium channel subfamily K member 5, mediates a pH and acid sensitive, noninactivating, outwardly rectifying K+ conductance, involved in potassium ion transport, inhibited by amide anesthetics. |
| UGT1A5 | GP0059_07_C01-frag002 | Encodes a UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway that transforms small lipophilic molecules, such as steroids, bilirubin, hormones and drugs, into water-soluble, excretable metabolites. |
| AKR1C1 | GP0059_01_B10-frag003 | Aldo-keto reductase family 1 member C1 acts in xenobiotic and progesterone metabolism, increased expression is observed in lung cancer, glaucoma and polycystic ovary syndrome. |
| AKR1C2 | GP0059_02_C12-frag007 | Aldo-keto reductase family 1 member C2 (dihydrodiol dehydrogenase), functions in bile transport, steroid metabolism and xenobiotic metabolism, expression is decreased in prostate and breast cancer but upregulated in esophageal squamous cell carcinoma. |
| CYP2C8 (gene) | GP0059_03_D11-frag004 | Cytochrome P450 family 2 subfamily C polypeptide 8, a member of heme-binding mono-oxygenase superfamily, metabolizes steroids, fatty acids and xenobiotics, altered expression may contribute to colon cancer development. |
| CYP2C8 (downstream) | GP0059_05_C04-frag010 | |
| PAPSS2 | GP0059_01_A08-frag002 | 3′-phosphoadenosine 5′-phosphosulfate synthase 2, involved in skeletal development; mutation in the corresponding gene correlates with spondyloepimetaphyseal dysplasia, mouse Papss2 gene is associated with brachymorphism. |
| HDAC7A | GP0059_06_H02-frag002 | Histone acetylation/deacetylation alters chromosome structure and affects transcription factor access to DNA. The protein encoded by this gene has sequence homology to members of the histone deacetylase family. This gene is orthologous to mouse HDAC7 gene whose protein promotes repression mediated via the transcriptional corepressor SMRT. |
| TEAD2 | GP0059_01_B06-frag007 | TEA domain family member 2, a member of the TEA DNA-binding domain family of transcription factors, regulates transcription, may be involved in organ morphogenesis and central nervous system development. |
| HIST1H1E | GP0059_06_E12-frag008 | The linker histone, H1, interacts with linker DNA between nucleosomes and functions in the compaction of chromatin into higher order structures. This gene is intronless and encodes a member of the histone H1 family. Transcripts from this gene lack polyA tails but instead contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6. |
| BMP1 | GP0059_06_G08-frag002 | Encodes a protein that is capable of inducing formation of cartilage in vivo. Although other bone morphogenetic proteins are members of the TGF-β superfamily, this gene encodes a protein that is not closely related to other known growth factors. |
| CFL1 | GP0059_09_F09-frag0005 | Cofilin 1 (non-muscle), plays a role in actin filament depolymerization and lymphocyte chemotaxis, involved in the G protein-coupled receptor protein and Rho protein signaling pathways. |
| EIF5 | GP0059_05_G08-frag003 | |
| TGIF2 | GP0059_03_G06-frag003 | This gene is a DNA-binding homeobox protein and a transcriptional repressor. The encoded protein appears to repress transcription by recruiting histone deacetylases to TGF-β-responsive genes. This gene is amplified and overexpressed in some ovarian cancers, and mutations in this gene can cause holoprosencephaly. |
| BBC3 | Cluster 19-76 | Bcl-2 binding component 3, regulates p53 (TP53)-dependent and independent apoptosis, increased expression of BBC3 plays a role in chronic lymphocytic leukemia and lung neoplasms and is used in the treatment of glioma and colorectal neoplasms. |
| CLG | Cluster 19-185 | |
| GLDC | Cluster 9-228 | Glycine decarboxylase (glycine dehydrogenase, P-protein), part of the glycine cleavage system that catalyzes the decarboxylation of glycine; mutation of the corresponding gene causes nonketotic hyperglycinemia. |
| ABCB4 | GP0059_06_D02-frag005 | This protein is a member of the MDR/TAP subfamily. Members of the MDR/TAP subfamily are involved in multidrug resistance as well as antigen presentation. This gene encodes a full transporter and member of the P-glycoprotein family of membrane proteins with phosphatidylcholine as its substrate. The function of this protein has not yet been determined; however, it may involve transport of phospholipids from liver hepatocytes into bile. |
| CENTD2 | Cluster 11-186 | Centaurin delta 2, functions as a GTPase activating protein (GAP) for both Arf and Rho family proteins, mediates PIP3 phosphoinositide-dependent Golgi apparatus membrane changes, filopodia formation and cell spreading. |
| ZNF281 | Cluster 1-52 | Zinc finger protein 281, a transcriptional repressor, member of the ZBP family, contains four Kruppel-type zinc fingers. |
| BCL9L | Cluster 11-267 | B-cell CLL/lymphoma 9-like, a BCL9-related protein that acts as a transcriptional regulator, induces β-catenin (CTNNB1) nuclear translocation epithelial to mesenchymal transition, enhances cell migration, expression is upregulated in colorectal tumors. |
| ARNT2 | Cluster 15-179 | Aryl-hydrocarbon receptor nuclear translocator 2, a protein that heterodimerizes with HIF-1alpha (HIF1A) and is involved in hypoxia-induced erythropoietin (EPO) expression. |
| ASS1 | Cluster 9-86 | The protein encoded by this gene catalyzes the penultimate step of the arginine biosynthetic pathway. There are approximately 10–14 copies of this gene including the pseudogenes scattered across the human genome, among which the one located on chromosome 9 appears to be the only functional gene for argininosuccinate synthetase. Mutations in the chromosome 9 copy of ASS cause citrullinemia. Two transcript variants encoding the same protein have been found for this gene. |
| CTNNA1 | GF0059_10_B02-frag003 | |
| ELK1 | GP0059_02_G12-frag003 | This gene is a member of the Ets family of transcription factors and of the ternary complex factor (TCF) subfamily. Proteins of the TCF subfamily form a ternary complex by binding to the the serum response factor and the serum reponse element in the promoter of the c-fos proto-oncogene. The protein encoded by this gene is a nuclear target for the ras-raf-MAPK signaling cascade. |
| MTL5 | GP0059_04_G12-frag010 | Metallothionein proteins are highly conserved low-molecular-weight cysteine-rich proteins that are induced by and bind to heavy metal ions and have no enzymatic activity. They may play a central role in the regulation of cell growth and differentiation and are involved in spermatogenesis. This gene encodes a metallothionein-like protein which has been shown to be expressed differentially in mouse testes and ovary. |
| WT | GP0059_05_B10-frag005 | |
| XRCC5 | GP0059_01_D07-frag005 | The protein encoded by this gene is the 80 kDa subunit of the Ku heterodimer protein which is also known as ATP-dependant DNA helicase II or DNA repair protein XRCC5. Ku is the DNA-binding component of the DNA-dependent protein kinase, and it functions together with the DNA ligase IV-XRCC4 complex in the repair of DNA double-strand break by non-homologous end joining and the completion of V(D)J recombination events. |
Table 2.
List of identified strong PXR binding sites
| Binding site ID | Nearest gene, mRNA, EST, or genomic location | GenBank ID | Also known as | Distance to start of gene | Cluster information |
Normalized binding |
Binding ratios | ||
|---|---|---|---|---|---|---|---|---|---|
| No. of tags | Length, nt | Control ± SD | Rifampicin ± SD | Rifampicin/ control | |||||
| SLC40A1 | Solute carrier family 40 (iron-regulated transporter), member 1 | NM_014585 | FPN1; HFE4; MTP1; IREG1; MST079; MSTP079; SLC11A3 | −2914, −3986 | 2 | 1113 | 0.30 ± 0.10 | 0.39 ± 0.17 | 1.29 |
| KCNK 5 | Potassium channel, subfamily K, member 5 | NM_003740 | FLJ11035, K2p5.1, TASK-2, TASK2 | 18 682, 18 670 | 2 | 44 | 0.66 ± 0.07 | 0.85 ± 0.14 | 1.29 |
| SULT 1B1 | Sulfotransferase family, cytosolic, 1B, member 1 | NM_014465 | ST1B2; SULT1B2; MGC13356 | −271 | 0.41 ± 0.16 | 0.34 ± 0.03 | 0.82 | ||
| AKR1C1/2 (gene) | Aldo-keto reductase family 1, member C1 | NM_001354 | 2-ALPHA-HSD, 20-ALPHA-HSD, C9, DD1, DDH, DDH1, H-37, HAKRC, MBAB, MGC8954 | 4088 | 0.39 ± 0.13 | 0.40 ± 0.22 | 1.03 | ||
| AKR1C1/2 (downstream) | NM_001354 | 18 979 | 0.45 ± 0.13 | 0.40 ± 0.07 | 0.90 | ||||
| EIF5 | Eukaryotic translation initiation factor 5 | NM_001969 | EIF-5A | −1198 | 0.50 ± 0.05 | 0.47 ± 0.01 | 0.93 | ||
| CYP2C8 (gene) | Cytochrome P450, family 2, subfamily C, polypeptide 8 | NM_000770 | CPC8, P450 MP-12/MP-20 | 18 489 | 0.30 ± 0.10 | 0.25 ± 0.01 | 0.86 | ||
| CD59 | CD59 molecule, complement regulatory protein | NM_000611 | 16.3A5, EJ16, EJ30, EL32, G344, MGC2354, MIC11, MIN1, MIN2, MIN3, MSK21, PROTECTIN, p18-20 | 36 417 | 0.89 ± 0.14 | 1.08 ± 0.30 | 1.21 | ||
| CYP4F12a | Cytochrome P450, family 4, subfamily F, polypeptide 12 | NM_023944 | F22329_1 | 8204, 8197 | 2 | 46 | 1.99 ± 0.29 | 3.49 ± 0.41 | 1.75 |
| ABCC4a | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | NM_005845 | RP11-74A12.1, EST170205, MOAT-B, MOATB, MRP4 | 90 518 | 0.20 ± 0.06 | 0.34 ± 0.02 | 1.69 | ||
| UGT1A5a | UDP glucuronosyltransferase 1 family, polypeptide A5 | NM_019078 | UDPGT, UGT1*5, UGT1E | −3758 | 0.41 ± 0.13 | 0.75 ± 0.18 | 1.84 | ||
| CYP3A5a | Cytochrome P450, family 3, subfamily A, polypeptide 5 | NM_000777 | CP35, P450PCN3, PCN3 | 7277 | 0.47 ± 0.08 | 0.79 ± 0.19 | 1.70 | ||
| CARSa | Cysteinyl-tRNA synthetase | NM_001751 | CARS1, CYSRS, MGC:11246 | 15 820 | 0.48 ± 0.19 | 0.92 ± 0.23 | 1.92 | ||
| BMP1a | Bone morphogenetic protein 1 | NM_006131 | FLJ44432, PCOLC, PCP, TLD | 4711 | 0.46 ± 0.08 | 0.69 ± 0.14 | 1.48 | ||
| TGIF2a | TGF-β-induced factor homeobox 2 | AF055012 | 5′-TG-3′ interacting factor 2; TGF-β-induced transcription factor 2; TGF-β-induced factor 2 (TALE family homeobox); homeobox protein TGIF2; transcription growth factor-β-induced factor 2 | 1373 | 0.72 ± 0.21 | 1.15 ± 0.41 | 1.61 | ||
| BCL9La | B-cell CLL/lymphoma 9-like | NM_182557 | BCL9-2, DLNB11 | −23 391, −23 240, −23 141 | 3 | 291 | 1.20 ± 0.33 | 2.42 ± 1.14 | 2.01 |
| CLGa | Pleckstrin homology domain containing, family G (with RhoGef domain) member 2 | NM_022835 | PLEKHG2, FLJ00018, FLJ22458 | −14 177, −15 081, −15 054 | 3 | 933 | 0.37 ± 0.11 | 0.81 ± 0.27 | 2.20 |
| GLDCa | Glycine dehydrogenase (decarboxylating) | NM_000170 | GCE, GCSP, HYGN1, MGC138198, MGC138200, NKH | −109 084, −109 083, −109 566 | 3 | 524 | 0.37 ± 0.12 | 0.66 ± 0.27 | 1.77 |
| ABCB4a | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | NM_018849 | MDR3; PGY3; ABC21; MDR2/3; PFIC-3 | −3078 | 0.74 ± 0.05 | 1.19 ± 0.39 | 1.60 | ||
| CFL1 | Cofilin 1 (non-muscle) | NM_005507 | CFL | −1874, −1879, −1882 | 3 | 47 | 0.44 ± 0.13 | 0.29 ± 0.16 | 0.66 |
| CYP2C8 (downstream) | NM_000770 | 33 678 | 0.51 ± 0.27 | 0.36 ± 0.15 | 0.71 | ||||
| GCKR | Glucokinase (hexokinase 4) regulator | NM_001486 | GKRP | 27 665 | 0.32 ± 0.05 | 0.19 ± 0.13 | 0.60 | ||
| GSTT1 | Glutathione S-transferase theta 1 | NM_000853 | −567 | 0.40 ± 0.20 | 0.25 ± 0.11 | 0.63 | |||
aTrend towards increased PXR binding in the presence of rifampicin compared to vehicle (Control; 0.1% DMSO).
Binding strength was determined by q-PCR signal >5-fold above the background signal obtained for the Untr5.
>5-fold binding relative to the negative control region was defined as ‘strong PXR binding’.
Figure 3.
PXR (A) and SRC-1 (B) binding to 11 of 23 genes identified as PXRE-containing promoters with a trend towards increased binding in the presence of rifampicin. Cryopreserved human hepatocytes were treated with vehicle (Control; 0.1% DMSO) or rifampicin (10 µM) for 3 h and processed for ChIP assays as described in the Materials and methods section. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05. PXR-binding to CLG and SRC-1-binding to ABCB4 in rifampicin-treated cells are based upon n = 2.
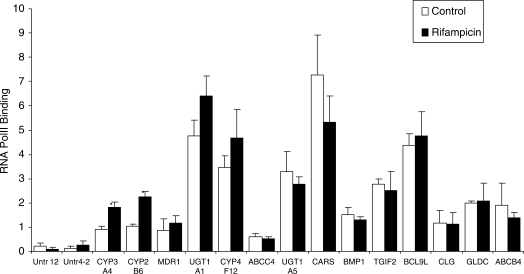
In addition to assessing increased PXR-binding, we also determined whether increased PXR binding at the newly identified sites, as well as the previously identified literature controls, did result in increased binding of DNA-dependent RNA Pol II to these genes (Figure 4). Determinations were performed in a ChIP-type assay using an antibody against RNA Pol II (20). CYP4F12 showed a trend towards increased Pol II binding in the presence of rifampicin. However, ABCC4, UGT1A5, CARS, BMP1, TGIF2, BCL9L, CLG, GLDC and ABCB4 showed no change. As expected, both CYP3A4 and CYP2B6 showed a ∼2-fold increase in binding of Pol II (Figure 4). The increase observed for UGT1A1 and MDR1 was not significant.
Figure 4.
RNA Pol II binding at or near previously known and unknown PXR binding sites. Chromatin from vehicle or 10 µM rifampicin-treated hepatocytes (3 h) were immunoprecipitated with anti-Pol II antibodies, and binding of Pol II to the candidate PXR-binding sites or nearby gene sequences were determined by q-PCR. Sequences amplified by PCR were the same as for Figure 3. Signals were normalized for the β-actin housekeeping gene. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05.
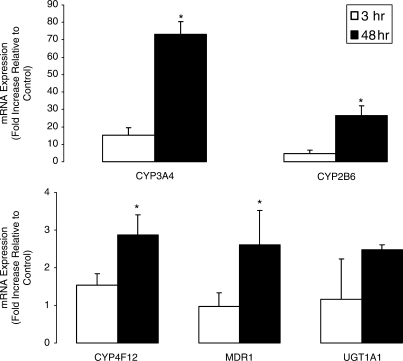
In order to study the correlation between Pol II binding by ChIP and mRNA levels, we also conducted quantitative mRNA analysis using TaqMan following treatment of hepatocytes with rifampicin for 3 or 48 h (Figure 5). While increases in mRNA expression were observed as early as 3 h, the expression was significantly higher after 48 h of drug treatment. CYP3A4, CYP2B6, CYP4F12, MDR1 and UGT1A1 demonstrated 4.8-, 5.6-, 1.9-, 2.7- and 2.1-fold higher levels of mRNA expression after 48 h compared to 3 h drug treatment. Differences in mRNA levels between genes did not correlate with the Pol II ChIP experiments. The antibody used in these analyses is against phospho-Ser 2 in the C-terminal domain of Pol II, which is the elongating form of the polymerase. The analysis was not done at the transcription start site of the genes of interest as it is known that Pol II may be bound at that area but not transcribing (22). Interestingly, the relative difference in mRNA accumulation after 3 and 48 h of treatment were most pronounced for CYP3A4 and CYP2B6.
Figure 5.
Comparison of CYP3A4-, CYP2B6-, CYP4F12-, MDR1- and UGT1A1-mRNA expression following 3 and 48 h treatment with rifampicin 10 µM. Cryopreserved human hepatocytes were treated with vehicle (Control; 0.1% DMSO) or 10 µM rifampicin for either 3 or 48 h. Thereafter, total RNA was isolated and subjected to TaqMan analysis as described in the Materials and methods section. Gene expression values were calculated based on the comparative ΔΔCt method and normalized to values obtained for 18S ribosomal RNA. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05.
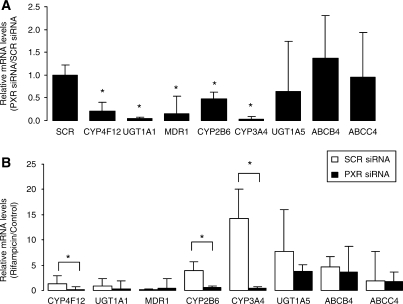
PXR knockdown in human hepatocytes
We conducted siRNA studies to further discern the involvement of PXR in the regulation of CYP4F12, UGT1A5, ABCB4 and ABCC4 which were identified as having strong PXR binding sites using ChIP analysis. We observed 79% knockdown in PXR mRNA levels in hepatocytes transfected for 72 h with PXR-siRNA relative to hepatocytes that were transfected with SCR-siRNA (data not shown). The basal expression levels of known PXR target genes, such as CYP3A4, CYP2B6, MDR1 and UGT1A1 were significantly lower (54–97%) in hepatocytes in which PXR was knocked down using PXR-siRNA compared to non-specific SCR-siRNA (Figure 6A). A statistically significant (∼80%, P < 0.05) decrease was also observed in the basal expression of CYP4F12. While ∼30% decrease was observed in the basal expression of UGT1A5, this was not statistically significant when compared to hepatocytes transfected with the non-specific SCR-siRNA. To confirm the specificity of the PXR-siRNA we also determined whether basal expression levels of the housekeeping genes 18S and GAPDH were affected. We did not observe a statistically significant decrease in the expression of these genes (data not shown) following PXR-siRNA transfection.
Figure 6.
Effect of PXR knockdown on the basal expression and inductive response of CYP4F12, CYP3A4, CYP2B6, MDR1 and UGT1A1 in human hepatocytes. Hepatocytes were transfected with ON-TARGETplus SMARTpool human NR1I2 siRNA (PXR-siRNA) or the non-specific ON-TARGETplus Non-Targeting Pool siRNA (scrambled, SCR-siRNA) as described in the Materials and methods section. (A) The effect of PXR silencing on basal expression of genes was determined relative to hepatocytes transfected with non-specific or SCR-siRNA. (B) The effect of PXR silencing on the inductive response was determined in hepatocytes transfected with PXR-siRNA or non-specific SCR-siRNA treated with rifampicin 10 µM. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05.
A comparison of fold changes in gene expression in hepatocytes transfected with either SCR-siRNA or PXR-siRNA demonstrated a statistically significant decrease in the inductive response of CYP3A4 and CYP2B6 when treated with the prototypical PXR activator rifampicin (Figure 6B). We also observed a statistically significant decrease in the inductive response of CYP4F12 in hepatocytes transfected with SCR-siRNA or PXR-siRNA following treatment with rifampicin. No significant difference was observed in the expression of UGT1A1, MDR1, UGT1A5, ABCB4 and ABCC4 in SCR- versus PXR-transfected siRNA. The inductive response that we observed for UGT1A5, ABCB4 and ABCC4 in rifampicin-treated hepatocytes transfected with SCR-siRNA relative to DMSO control without subsequent knockdown in PXR-siRNA-transfected hepatocytes may be related to the regulation of these genes by other transcription factors, such as the vitamin-D receptor (VDR), farsenoid-X receptor (FXR) and aryl hydrocarbon receptor (AHR) (23–25).
Association of corepressors and coactivators with PXR binding sites
It was previously demonstrated that the steroid receptor complex protein SRC1 associates with PXR after ligand binding (26). Using a similar approach as used to determine PXR-binding to PXR gene promoters, we assessed whether the level of binding of SRC1 was increased after the treatment of cells with rifampicin. A very similar binding pattern to that observed with the anti-PXR antibody was observed in which increased binding was found for all four genes analyzed (Figure 1B). In addition, we assessed the association of SRC1 with the 11 genes showing increased binding to PXR after exposing the cells to rifampicin (Table 2). Again, binding patterns were very similar to those found for PXR (Figure 3B). In most cases, lower binding was detectable in cells not treated with rifampicin. Of the 11 genes tested, only CYP4F12 demonstrated significantly increased binding for both PXR and SRC1 after rifampicin treatment in three batches of hepatocytes.
We extended our studies to assess the involvement of other previously published coactivators and corepressors of PXR on the regulation of CYP3A4, CYP2B6, CYP4F12, UGT1A1 and MDR1. ChIP analysis was performed with antibodies specific for the coactivators GRIP1, SRC3, PGC1α, FKHR and RIP140, and the corepressors NCoR, SMRT and SHP. All antibodies used have been evaluated by others before (see Material and methods section) and specificity was confirmed using known target genes (data not shown).
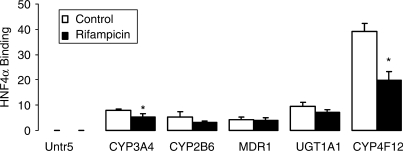
Compared to the coactivators and corepressors that we assessed, HNF4α showed a significantly higher binding to the promoter elements of CYP3A4, CYP2B6, CYP4F12, UGT1A1 and MDR1 (Figures 7 and 8), but no increased binding of HNF4α was detectable after treatment of cells with rifampicin. In fact, the converse was true in that of the five genes evaluated, two (CYP3A4 and CYP4F12) showed a significantly decreased binding in the presence of rifampicin (Figure 7).
Figure 7.
Identification of HNF4α binding to CYP3A4, CYP2B6, MDR1, UGT1A1 and CYP4F12. Chromatin from cryopreserved human hepatocytes treated with vehicle (Control; 0.1% DMSO) or rifampicin (10 µM) for 3 h were immunoprecipitated with anti-HNF4α antibody, and the enrichment of HNF4α was determined by q-PCR. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05.
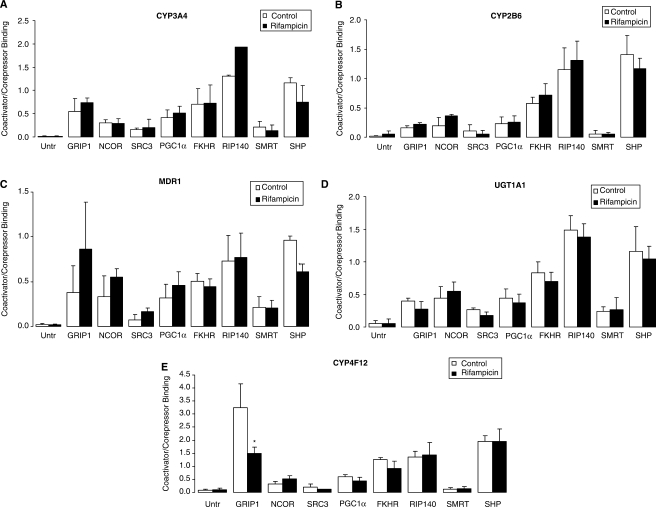
Figure 8.
Identification of PXR coactivators/corepressors binding to (A) CYP3A4, (B) CYP2B6, (C) MDR1, (D) UGT1A1 and (E) CYP4F12. Chromatin from cryopreserved human hepatocytes treated with vehicle (Control; 0.1% DMSO) or rifampicin (10 µM) for 3 h were immunoprecipitated with anti-GRIP1, -NCOR, -SRC3, -PGC1 α, -FKHR, -RIP140, -SMRT or -SHP antibody, and the enrichment of candidate coactivators/corepressors were determined by q-PCR. The error bars represent the SD from the mean of triplicate assays of an individual experiment, n = 3, *P < 0.05. RIP140-binding to CYP3A4 in vehicle (Control; 0.1% DMSO)-treated cells is based upon n = 2.
None of the other coactivators that we assessed showed a statistically significant increase in binding in cells treated with rifampicin versus vehicle control. Interestingly, GRIP1 showed a decrease in binding to CYP4F12 in the presence of rifampicin. Of the corepressors tested, while a trend towards decreased binding of SHP was observed for CYP3A4 (Figure 8A), CYP2B6 (Figure 8B) and UGT1A1 (Figure 8D), we observed a statistically significant decrease only for MDR1 (Figure 8C).
DISCUSSION
Here we report the identification of new genes containing PXR binding sites using ChIP-based assays in cryopreserved primary human hepatocytes. The use of ChIP can detect the direct association of PXR and coactivators/corepressors with promoter elements. This is different from gene profiling experiments with cells treated with PXR-activators where it cannot be excluded that the induction of genes is a secondary effect of PXR activation. Our studies indicate that PXR binds to PXREs in the presence and absence of rifampicin, and that binding of PXR to many of the newly identified genes does not increase after treatment with rifampicin. The ability of PXR to regulate the basal expression and inducibility of one of these new genes, CYP4F12, was confirmed by hepatocytes depleted of PXR by siRNA. Finally, we extended our studies and also found that binding of SRC1 correlates with transcriptional activation of PXR target genes, whereas this was not found for other coactivators or corepressors studied.
Using a ChIP-based discovery approach, we were able to identify genes containing PXR binding sites which were mapped against the entire human genome (Supplementary Table S2, Supplementary data). Tags were identified which were localized large distances upstream from, within or downstream of their potentially associated genes. The identified tags were located on all 23 chromosomes and included drug metabolizing and transporter genes as well as genes not involved in drug disposition. The presence of PXR-binding sites on genes not involved in drug disposition is not unexpected given that recent evidence suggests that PXR plays a role in hepatic lipid metabolism (27). For instance, PXR binds to FOXA2 (Supplementary Table S2, Supplementary Data) and represses its’ activity and the genes encoding the key enzymes in energy metabolism. Furthermore, Dai et al. (28) observed that ACC1 (Acetyl CoA carboxylase 1), which is a lipogenic enzyme, was consistently lower in PXR-null mice than wild-type mice. Their data indicated that PXR is involved in the regulation of ACC-1 gene expression during liver regeneration.
Using the data obtained in the ChIP-based discovery approach we were able to confirm that several previously unidentified drug disposition genes contain PXREs (Table 2). These included phase I (CYP4F12) and phase II (SULT1B1 and UGT1A5) drug metabolizing enzymes, as well as transporters (SLC40A1 and ABCB4). In addition, our data also confirmed previous studies which indicated that CYP3A5, CYP2C8 and GSTT1 were PXR-target genes (Table 2) (1,2,29). Similar to what we observed for the genes with known PXR-binding sites (Figure 1A), PXR binding to the PXREs on 11 genes analyzed in Figure 3A also occurred in the absence of rifampicin. To ensure that our findings were not due to the high inducibility of PXR in the cells we used in our experiments, we have looked at binding to several targets in two to three batches of hepatocytes. The data indicated that results were consistently in agreement. While our discovery approach identified many new genes binding to PXR, the number of tags sequenced likely did not reach saturation, and therefore more PXR target genes may be identified in the future.
As expected, in our PXR antibody validation studies with known PXR-binding sites, all four genes showed significantly higher PXR binding than was observed for the control regions (Figure 1A). Similar to what we observed during the discovery approach, binding of PXR to these PXREs was also detected in the absence of rifampicin in which the binding to MDR1 was much higher than that to UGT1A1, CYP2B6 and CYP3A4. Our PXR knockdown studies add additional evidence of PXR binding to PXREs in the absence of ligand and the subsequent reduction in the basal expression of CYP3A4, CYP2B6, UGT1A1, Pgp and CYP4F12 (Figure 6). This is in line with findings by Squires et al. (30) that PXR was present in the nucleus of mouse liver cells prior to the addition of a ligand. Similarly, in primary human hepatocytes in which PXR was knocked down using the hPXR-siRNA–adenovirus expression system, Kojima et al. (31) observed a reduction in the basal expression of CYP3A4 and CYP2B6 mRNA levels.
Our studies did confirm that addition of rifampicin significantly increased the binding of PXR to the PXREs of CYP3A4, MDR1, UGT1A1 and CYP2B6. Interestingly, the fold increase of PXR binding to PXRE was several-fold higher for CYP3A4 than was observed for MDR1 (Figure 1A). This may explain why minimal induction of MDR1 was observed after treatment with rifampicin compared to CYP3A4 (Figure 5). Also of interest was the finding that PXR binding to PXREs did not always significantly increase in the presence of rifampicin (Figure 3) which subsequently did not result in an increase in gene transcription (Figure 4). This phenomenon is difficult to explain and could be related to the fact that PXR binding to PXREs occurs in the absence of the ligand which could lead to saturation of PXREs on specific genes (30). It should also be noted that PXR binding to PXREs is not the only determinant of an increase in gene transcription but involves numerous corepressors and coactivators (12).
Using ChIP we identified CYP4F12 as having PXR-binding sites. We confirmed the involvement of PXR in the regulation of CYP4F12 by assessing its’ basal expression and inducibility in human hepatocytes in which PXR was knocked down. CYP4F12 mRNA has been detected in several major tissues involved in drug disposition including the liver, kidney and small intestine (32,33). It is the only member of the CYP4F family involved in xenobiotic metabolism. In particular, CYP4F12 has been shown to be highly efficient toward metabolism of antihistaminic compounds, such as ebastine and terfenadine (32,34,35). In addition to xenobiotics, CYP4F12 is also involved in the oxidation of arachadonic acid and stable prostaglandin H2 (PGH2) analogs (32,34).
Previous studies have highlighted the importance of the coactivator SRC-1 in PXR activation and the subsequent increase in gene transcription (36). Our studies confirmed that the SRC-1 binding pattern mirrors that observed with PXR (Figures 1B and 3B). Similar to PXR, SRC-1 binding was increased after treatment of hepatocytes with rifampicin. This is in line with crystallographic studies which demonstrated that SRC-1 binds to PXR and stabilizes the PXR-LBD and together with the ligand exerts an additive effect on the stability of the receptor (36). Binding of SRC-1 to the surface of PXR limits the ability of the receptor to continuously change its conformation and helps trap a single, active conformation of the ligand.
In addition to SRC-1, several other PXR-coactivators and -corepressors have been implicated in PXR-mediated gene regulation. These include the coactivators GRIP1, PGC1α, FKHR and RIP140, and the corepressors SMRT and SHP (12). Analysis of these coactivators and corepressors did, in most cases, show significant binding to the promoter elements of the target genes in the absence of rifampicin compared to the control Untr (Figure 8). Treatment of the hepatocytes with rifampicin did not result in a consistent increase in binding in the case of coactivators or decreased binding in the presence of corepressors. This may be related to our studies being conducted in human hepatocytes and not in a recombinant in vitro system. In comparison, studies by others were conducted in cell lines in which proteins were overexpressed and were outside their natural chromatin environment (13–16). Overall, however, our data suggests that binding of the coactivators and corepressors studied were involved in the regulation of gene expression as binding was higher than to the negative control and, with few exceptions, the overall pattern of binding to the various promoter elements studied were similar. An important point to consider is that all our analyses were done after treatment of hepatocytes with rifampicin and we cannot exclude that different binding behavior could be found in the presence of other PXR activators.
HNF4α has been shown to cooperate at the −7.8 kb XREM in the PXR-dependent induction of CYP3A4 expression (17). Even though HNF4α binding was detectable in the presence and absence of rifampicin, it was unexpected that we observed a decrease in binding of HNF4α in the presence of rifampicin (Figure 7). The role of HNF4α in PXR-mediated gene activation is not clear as Li and Chiang (18) and Tegude et al. (37) have shown that binding of HNF4α to the XREM-DR1 element is not required for gene activation of the CYP3A4 promoter. This was confirmed in a study by Kamiyama et al. (38) in which HNF4α was knocked down using an adenovirus expressing system in human hepatocytes. They observed that hHNF4α-siRNA did not significantly affect the fold-induction of CYP2B6, CYP2C8, CYP2C9 or CYP3A4 mRNA levels following treatment with CYP inducers, such as phenobarbital, rifampicin and dexamethasone. There is strong evidence, however, that HNF4α is involved in regulating basal expression of CYP3A4 (37,38).
In conclusion, in cryopreserved primary human hepatocytes we identified several new genes containing PXR binding sites. We confirmed the involvement of PXR in the regulation of CYP4F12 using siRNA. We also found that the SRC-1 binding pattern is very similar to PXR and that both bind to promoter elements in the absence of ligand. Finally, we did not observe a pattern correlating with rifampicin treatment with respect to other PXR coactivators/corepressors involved in gene regulation in the presence of rifampicin.
FUNDING
Funding for open access charge: Merck & Co., Inc.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane X receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- 2.Hartley DP, Dai X, Yabut J, Chu X, Cheng O, Zhang T, He YD, Roberts C, Ulrich R, Evers R, et al. Identification of potential pharmacological and toxicological targets differentiating structural analogs by a combination of transcriptional profiling and promoter analysis in LS-180 and Caco-2 adenocarcinoma cell lines. Pharmacogenet. Genomics. 2006;16:579–599. doi: 10.1097/01.fpc.0000220561.59972.7a. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld JM, Vargas R, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol. Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- 4.Staudinger J, Liu YP, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab. Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- 5.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, Latour A, Liu YP, Klaassen CD, Brown KK, Reinhard J, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl Acad. Sci. USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, Chiang JY. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G74–G84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 7.Ding X, Lichti K, Staudinger JL. The mycoestrogen zearalenone induces CYP3A through activation of the pregnane X receptor. Toxicol. Sci. 2006;91:448–455. doi: 10.1093/toxsci/kfj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 2001;276:14581–14587. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- 9.Lin JH. CYP induction-mediated drug interactions: in vitro assessment and clinical implications. Pharm. Res. 2006;23:1089–1116. doi: 10.1007/s11095-006-0277-7. [DOI] [PubMed] [Google Scholar]
- 10.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 11.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 12.Moore DD, Kato S, Xie W, Mangelsdorf DJ, Schmidt DR, Xiao R, Kliewer SA. International Union of Pharmacology. LXII. The NR1H and NR1I receptors: constitutive androstane receptor, pregnene X receptor, farnesoid X receptor alpha, farnesoid X receptor beta, liver X receptor alpha, liver X receptor beta, and vitamin D receptor. Pharmacol. Rev. 2006;58:742–759. doi: 10.1124/pr.58.4.6. [DOI] [PubMed] [Google Scholar]
- 13.Ourlin JC, Lasserre F, Pineau T, Fabre JM, Sa-Cunha A, Maurel P, Vilarem MJ, Pascussi JM. The small heterodimer partner interacts with the pregnane X receptor and represses its transcriptional activity. Mol. Endocrinol. 2003;17:1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- 14.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 15.Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR) Proc. Natl Acad. Sci. USA. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeshita A, Koibuchi N, Oka J, Taguchi M, Shishiba Y, Ozawa Y. Bisphenol-A, an environmental estrogen, activates the human orphan nuclear receptor, steroid and xenobiotic receptor-mediated transcription. Eur. J. Endocrinol. 2001;145:513–517. doi: 10.1530/eje.0.1450513. [DOI] [PubMed] [Google Scholar]
- 17.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat. Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Chiang JY. Rifampicin induction of CYP3A4 requires pregnane X receptor cross talk with hepatocyte nuclear factor 4alpha and coactivators, and suppression of small heterodimer partner gene expression. Drug Metab. Dispos. 2006;34:756–764. doi: 10.1124/dmd.105.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson KD, Bresnick EH. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods. 2002;26:27–36. doi: 10.1016/S1046-2023(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 20.Labhart P, Karmakar S, Salicru EM, Egan BS, Alexiadis V, O’Malley BW, Smith CL. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc. Natl Acad. Sci. USA. 2005;102:1339–1344. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- 22.Wu JQ, Snyder M. RNA polymerase II stalling: loading at the start prepares genes for a sprint. Genome Biol. 2008;9:220. doi: 10.1186/gb-2008-9-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finel M, Li X, Gardner-Stephen D, Bratton S, Mackenzie PI, Radominska-Pandya A. Human UDP-glucuronosyltransferase 1A5: identification, expression, and activity. J. Pharmacol. Exp. Ther. 2005;315:1143–1149. doi: 10.1124/jpet.105.091900. [DOI] [PubMed] [Google Scholar]
- 24.Oude Elferink RP, Paulusma CC. Function and pathophysiological importance of ABCB4 (MDR3 P-glycoprotein) Pflugers Arch. 2007;453:601–610. doi: 10.1007/s00424-006-0062-9. [DOI] [PubMed] [Google Scholar]
- 25.Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, et al. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J. Biol. Chem. 2001;276:39411–39418. doi: 10.1074/jbc.M106340200. [DOI] [PubMed] [Google Scholar]
- 26.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- 27.Konno Y, Negishi M, Kodama S. The roles of nuclear receptors CAR and PXR in hepatic energy metabolism. Drug Metab. Pharmacokinet. 2008;23:8–13. doi: 10.2133/dmpk.23.8. [DOI] [PubMed] [Google Scholar]
- 28.Dai G, He L, Bu P, Wan YJ. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology. 2008;47:1277–1287. doi: 10.1002/hep.22129. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol. Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- 30.Squires EJ, Sueyoshi T, Negishi M. Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J. Biol. Chem. 2004;279:49307–49314. doi: 10.1074/jbc.M407281200. [DOI] [PubMed] [Google Scholar]
- 31.Kojima K, Nagata K, Matsubara T, Yamazoe Y. Broad but distinct role of pregnane X receptor on the expression of individual cytochrome p450s in human hepatocytes. Drug Metab. Pharmacokinet. 2007;22:276–286. doi: 10.2133/dmpk.22.276. [DOI] [PubMed] [Google Scholar]
- 32.Bylund J, Bylund M, Oliw EH. cDna cloning and expression of CYP4F12, a novel human cytochrome P450. Biochem. Biophys. Res. Commun. 2001;280:892–897. doi: 10.1006/bbrc.2000.4191. [DOI] [PubMed] [Google Scholar]
- 33.Stark K, Schauer L, Sahlen GE, Ronquist G, Oliw EH. Expression of CYP4F12 in gastrointestinal and urogenital epithelia. Basic Clin. Pharmacol. Toxicol. 2004;94:177–183. doi: 10.1111/j.1742-7843.2004.pto940404.x. [DOI] [PubMed] [Google Scholar]
- 34.Hashizume T, Imaoka S, Hiroi T, Terauchi Y, Fujii T, Miyazaki H, Kamataki T, Funae Y. cDNA cloning and expression of a novel cytochrome p450 (cyp4f12) from human small intestine. Biochem. Biophys. Res. Commun. 2001;280:1135–1141. doi: 10.1006/bbrc.2000.4238. [DOI] [PubMed] [Google Scholar]
- 35.Hashizume T, Imaoka S, Mise M, Terauchi Y, Fujii T, Miyazaki H, Kamataki T, Funae Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J. Pharmacol. Exp. Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- 36.Watkins RE, Davis-Searles PR, Lambert MH, Redinbo MR. Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J. Mol. Biol. 2003;331:815–828. doi: 10.1016/s0022-2836(03)00795-2. [DOI] [PubMed] [Google Scholar]
- 37.Tegude H, Schnabel A, Zanger UM, Klein K, Eichelbaum M, Burk O. Molecular mechanism of basal CYP3A4 regulation by hepatocyte nuclear factor 4 alpha: evidence for direct regulation in the intestine. Drug Metab. Dispos. 2007;35:946–954. doi: 10.1124/dmd.106.013565. [DOI] [PubMed] [Google Scholar]
- 38.Kamiyama Y, Matsubara T, Yoshinari K, Nagata K, Kamimura H, Yamazoe Y. Role of human hepatocyte nuclear factor 4alpha in the expression of drug-metabolizing enzymes and transporters in human hepatocytes assessed by use of small interfering RNA. Drug Metab. Pharmacokinet. 2007;22:287–298. doi: 10.2133/dmpk.22.287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.