Abstract
Electric signal processing has evolved to manage rapid information transfer in neuronal networks and muscular contraction in multicellular organisms and controls the most sophisticated man-built devices. Using a synthetic biology approach to assemble electronic parts with genetic control units engineered into mammalian cells, we designed an electric power-adjustable transcription control circuit able to integrate the intensity of a direct current over time, to translate the amplitude or frequency of an alternating current into an adjustable genetic readout or to modulate the beating frequency of primary heart cells. Successful miniaturization of the electro-genetic devices may pave the way for the design of novel hybrid electro-genetic implants assembled from electronic and genetic parts.
INTRODUCTION
Recent advances in synthetic biology have led to the design of engineered cells emulating basic computational functions known from semiconductor-based electronic circuits (1–3). Synthetic gene networks implanted into living systems were able to perform tight regulation of gene expression (4), fundamental logic Boolean operations (5), establish epigenetic long-term memory (6,7), provide band-detect filter characteristics (2,8) and hysteretic signal-insulating qualities (9), program time-delayed (2) or oscillating (10) gene expression and process multi-channel information within cells (11) and populations (8) as well as among different species (8,12,13). Although electronic and cell engineers are using similar circuit architectures and standardized parts (2,3) to assemble complex computing units with potentially compatible signal processing capacities, the design of electro-genetic interfaces managing mutual information processing between gene transcription in mammalian cells and electronic processing units has not yet gathered momentum. Interestingly, direct electricity-based gene expression has not been evolved as a major control theme or remains to be discovered. Previous work on electro-genetic devices mainly relied on non-specific effects of electric fields on the entire transcriptome of Escherichia coli cells (14) or on the use of electrically triggered light (15–17) to indirectly activate gene expression. In contrast to such non-specific modulation of gene expression showing genome-wide pleiotropic impact, synthetic biologists have successfully engineered mammalian transcription switches that enable reversible and adjustable activation or repression of specific trangenes in response to external stimuli, such as antibiotics (18–20), quorum-sensing molecules (21,22), (gaseous) metabolites (23–26) or cultivation temperature (27,28). All of these transcription control circuits capitalize on a generic design consisting of a synthetic transactivator (a fusion between a heterologous transcriptional repressor and a mammalian transactivation domain) which specifically binds and activates a chimeric promoter (assembled by a placing repressor-specific operator 5′ of a minimal promoter) in an inducer-responsive manner. Such a standard configuration offers optimal functional compatibility among these transcription circuits and provides a toolbox of individual transgene control modalities for assembly of complex higher order mammalian transcription networks (2,4,9,29,30).
We have designed synthetic electro-genetic devices, which enable electricity-induced expression of specific transgenes in mammalian cells as well as mammalian cell-based control of microelectronic circuits.
MATERIALS AND METHODS
Mammalian cell culture
AIRCHO-SEAP (26) transgenic for acetaldehyde-inducible expression of human placental secreted alkaline phosphatase (SEAP) was cultivated in HTS medium (Cell Culture Technology, Gravesano, Switzerland) supplemented with 5% (v/v) fetal calf serum (FCS, PAN Biotech GmbH, Aidenbach, Germany, cat. no. 3302, lot no. P251110), 100 U/ml penicillin, 100 µg/ml streptomycin, 1 µg/ml puromycin and 400 µg/ml G418 sulfate in a humidified atmosphere containing 5% CO2. All expression studies were done in triplicate using 125 000 AIRCHO-SEAP cells seeded into 2 cm2 cell culture dishes. Neonatal rat ventricular cardiomyocytes were isolated from newborn rat hearts (Wistar rats; Elevage Janvier, Le Genest Saint Isle, France) as described before (31). NRCs were cultivated in 67% (v/v) Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen), 17% (v/v) M-199 EBSS (Amimed, Allschwil, Switzerland), 10% (v/v) horse serum (Invitrogen, cat. no. 16050-122, lot. 336379), 5% (v/v) FCS and 50 µg/ml gentamycin (Sigma, St Louis, MO, USA, cat. no. G1914).
Lentiviral vectors
Lentivectors pCD20 and pCD22, enabling acetaldehyde-responsive expression of human bone morphogenetic protein 2 (BMP-2), have been described previously (32). In brief, pCD20 (5′LTR-oriSV40-cPPT-RRE-PAIR-bmp-2-3′LTRΔU3) encodes the bmp-2 gene under control of the acetaldehyde-responsive promoter PAIR and pCD22 (5′LTR-oriSV40-cPPT-RRE-PhEF1α-alcR-3′LTRΔU3) encodes constitutive expression of the acetaldehyde-dependent transactivator AlcR (26). pBP253 (5′LTR-oriSV40-cPPT-RRE-PhEF1α-bmp-2-3′LTRΔU3) is the control lentivector for constitutive bmp-2 expression (32). Vesicular stomatitis virus-pseudotyped lentiviral particles were produced and titrated as described before (33). Abbreviations: cPPT, central polypurine tract; LTR, long-terminal repeat; oriSV40, simian virus 40-derived origin of replication; PhEF1α, human elongation factor 1α promoter; RRE, rev-response element.
Standard input device
A custom-designed electrolysis chamber (Febikon Labortechnik, Wermelskirchen, Germany, cat. no. E-pb0) containing 250 ml 150 mM succinic acid supplemented with 1% (v/v) ethanol was used as standard input device. The chamber was placed in an airtight 3.6 l polypropylene box (Migros, Zurich, Switzerland) containing a humidified atmosphere with 5% CO2. In a typical experimental set-up, a direct current (DC) source (PowerPac HC, Bio-Rad, Hercules, CA, USA) or an alternating current (AC) generator (Lapp & Co., Zurich, Switzerland, model Th) were used to power the standard input interface for 1 h, which was then incubated for 23 h before SEAP expression was profiled using the output interface. Acetaldehyde and ethanol were quantified using Gastec tubes (26).
At high currents Joule heating (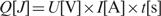 ) of the cellular processing unit (CPU) was observed, corresponding to a temperature increase of 49 K at 500 mA (28.5 V) assuming a specific heat capacity of c = 4.18 kJ kg−1 K−1 for the buffer. Acetaldehyde concentrations up to 150 p.p.m. do not compromise cell viability and SEAP production (26).
) of the cellular processing unit (CPU) was observed, corresponding to a temperature increase of 49 K at 500 mA (28.5 V) assuming a specific heat capacity of c = 4.18 kJ kg−1 K−1 for the buffer. Acetaldehyde concentrations up to 150 p.p.m. do not compromise cell viability and SEAP production (26).
Miniaturized input device
Modified PVDF-based hollow fibers (1 mm inner diameter, 25 mm long; Spectrum Laboratories, Rancho Dominguez, CA, USA, cat. no. M138615) were filled with 16.7 µl of a 1.7% (w/v) agarose-based hydrogel containing 125 mM succinic acid and 17% (v/v) ethanol. Both ends were equipped with platinum electrodes (diameter 1 mm), sealed and connected to a DC power source.
Output device
SEAP activity was converted into an electric signal using a coupled enzymatic–optic device. SEAP-containing cell culture supernatant was incubated for 5 min with p-nitrophenylphosphate as described previously (34) and the resulting p-nitrophenolate was quantified optically at 405 nm using a Novaspec II photometer (Pharmacia, Freiburg, Germany) equipped with an output port providing an electric signal (millivolts) proportional to absorbance.
Complementary metal-oxide semiconductor high-density microelectrode arrays
High-density microelectrode arrays (HD-MEA) consist of 11 016 metal electrodes and 126 channels, each of which contains recording and stimulation electronics for bidirectional communication with electrogenic cells. The HD-MEAs were manufactured as described previously (32,35). The arrays were coated with 20 µg/ml laminin for 3 h prior to seeding 105 neonatal rat cardiomyocytes (NRCs) in 1 ml medium. After incubation for 24 h, the cells were transduced with 106 pCD20/pCD22- or pBP253-derived lentiviral particles engineered for acetaldehyde-inducible or constitutive BMP-2 expression, respectively. After 24 h of transduction, the medium was exchanged and the cell-containing HD-MEAs were connected to the input interface which was set to different power levels for 1 h. The cell-containing HD-MEAs were then incubated for 48 h at 37°C before NRC beating was recorded for 20 s. The error bars represent the standard deviation from three readings.
RESULTS AND DISCUSSION
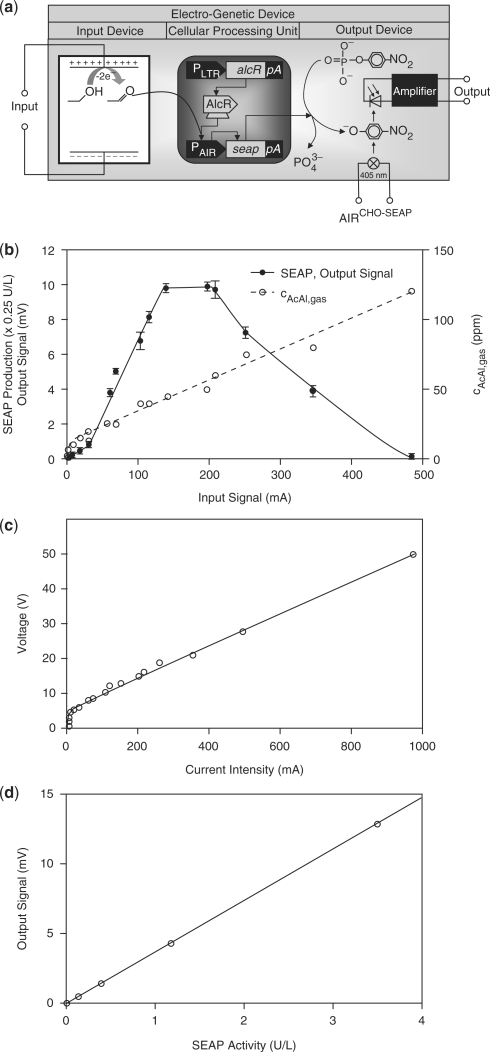
The generic electro-genetic input device enabling electronic transcription control in engineered mammalian cells was designed by linking electrochemical oxidation of ethanol to acetaldehyde with acetaldehyde-inducible transgene expression (26). Electrochemical oxidation of ethanol was performed using a platinum anode and cathode in an electrolysis chamber containing 1% (v/v) ethanol. Acidic pH was chosen to favour higher acetaldehyde production compared to basic pH at which the major oxidation end product is acetic acid (36). For acidification of the electrolyte, we replaced the standard perchloric acid (37) that is electrolysed to toxic chlorine with non-volatile and biocompatible succinic acid. As central cellular processing unit (CPU), we used Chinese hamster ovary (CHO-K1)-derived cells engineered for constitutive expression of the Aspergillus nidulans-derived acetaldehyde-dependent transactivator AlcR which binds and activates the acetaldehyde-inducible promoter PAIR in the presence of acetaldehyde and so triggers expression of the human placental SEAP (26). SEAP production was scored by an enzymatic–optical process, consisting of a photodiode converting SEAP-catalysed dephosphorylation of p-nitrophenyl-phosphate to coloured p-nitrophenolate into a dose-dependent electric signal thereby providing a standard gene-electronic output device (Figure 1a). Input and output devices could either be operated as standalone electronic-cell interfaces or be sequentially linked to use engineered mammalian cells as CPUs plugged into an integrated electronic circuit.
Figure 1.
Synthetic mammalian cell-based electro-genetic device. (a) Circuit diagram of the electro-genetic device. DC applied to the input device results in the electrochemical conversion of ethanol into acetaldehyde, which enables the acetaldehyde-dependent transactivator AlcR to bind and induce transcription from its cognate promoter PAIR which triggers transcription of the human placental SEAP. SEAP subsequently catalyses the production of coloured p-nitrophenolate, which is quantified photometrically at 405 nm by a photodiode and converted into an electric output signal. AlcR, acetaldehyde-inducible transactivator; pA, polyadenylation signal; PAIR, AlcR-responsive promoter; PLTR, murine stem cell virus 5′ long terminal repeat-derived promoter. (b) Characterization of the CPU. The CPU was connected to DC of different intensities for 1 h, and the resulting acetaldehyde concentration as well as the electric output signal were quantified after 24 h. (c) Correlation between DC input and corresponding voltage. (d) Characterization of the output interface. SEAP activity was plotted against the electric signal measured by the output interface.
When input devices and CPUs were operated with DC < 30 mA no significant SEAP production could be observed. Between 30 mA and 140 mA there was a direct correlation between input current, acetaldehyde production and transgene expression (Figure 1b and c). Between 140 mA and 200 mA DC SEAP expression reached a plateau since acetaldehyde-inducible transgene expression became saturated although the input device continued to produce increasing acetaldehyde levels. Beyond 200 mA DC SEAP production decreased as a consequence of a current-induced temperature increase, which steadily reduced CPU viability (Figure 1b; Materials and Methods section). Such thermal destruction is a characteristic the electro-genetic device shares with any electronic circuitry.
The photodiode-based output device was shown to provide electric signals, which were proportional to SEAP production in a range of 0–30 U/l (corresponding to an output signal of 0–1000 mV), which enabled full coverage of the transgene expression dynamics of mammalian cells (Figure 1b and d). Consolidating the dose-response characteristics of input and output devices as well as the CPU, the entire synthetic electro-genetic circuitry shows an overall dynamic range of 30–140 mA for the input current and 0.7–10 mV for the output signal.
A mammalian cell-based integrator
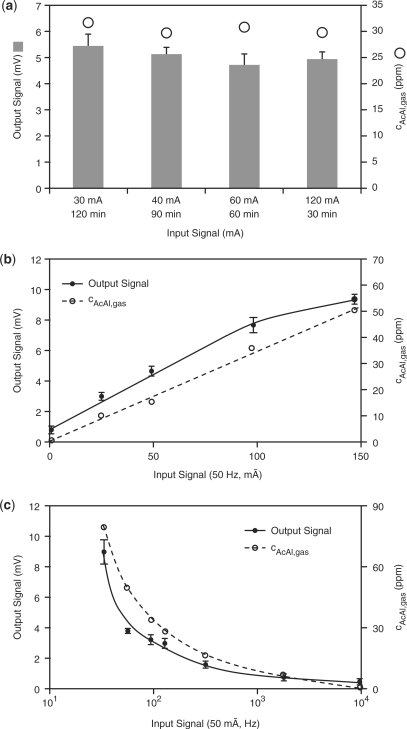
Within its linear operation range (30 mA DC ≤ input current ≤ 140 mA DC, see above), the electro-genetic device functions as a cell-based electronic integrator mimetic since electrochemical acetaldehyde production is a direct function of the exposure time and the intensity of the current. For validation of the integrator characteristics, the electro-genetic device was connected for different periods of time to currents with varying intensities, while the overall amount of electron flux was kept constant (I × t = constant). The observation that acetaldehyde levels as well as the electric output were identical for all time profiles, suggests that the device has the capacity to process electronic signals in an integrator-like manner (Figure 2a).
Figure 2.
Electro-genetic circuits. (a) Mammalian cell-based integrator. The CPU was connected to DC of different intensities and for various periods of time; the product of time and current intensity was kept constant (t × I = constant). The resulting acetaldehyde levels and electric output signals were scored after 24 h. (b) Mammalian cell-based AM detector. The CPU was connected to an AC of 50 Hz and different intensities for 1 h and the acetaldehyde levels as well as the electric output current were quantified after 24 h. (c) Mammalian cell-based FM detector. The CPU was connected to an AC of 50 mA and different frequencies for 1 h and the acetaldehyde levels as well as the electric output current were quantified after 48 h.
A mammalian cell-based amplitude-modulation (AM) detector
The CPU's electronic signal integration capacity is not limited to DC but can also be operated to score the amplitude of an AC power source. Upon connection of the CPU to an AC of 50 Hz, the resulting acetaldehyde production as well as the electric signal generated by the output interface were monitored and were shown to directly correlate with the amplitude of the electric input signal thereby confirming the ability of the electro-genetic device to function as an AM detector (Figure 2b).
A cell-based frequency modulation detector
The efficacy of AC-based electrolysis is frequency dependent—with increasing frequencies, polarization phenomena at the electrodes occur and re-oxidation/re-reduction of the reaction products further decrease the electrochemical production of acetaldehyde in a frequency-dependent manner (Figure 2c). In order to characterize the ability of the electro-genetic device to detect the frequency of an AC and to convert it into a DC output signal, the input interface of the electro-genetic device was connected to AC with constant amplitude (50 mA) and increasing frequencies (50–10 000 Hz). With increasing AC frequencies, decreasing acetaldehyde production was dose-dependently translated into decreasing SEAP expression, which converted into a linear electric signal by the enzymatic–optical output interface thereby validating the electro-genetic device as a frequency modulation (FM) detector (Figure 2c).
A cell-based frequency generator
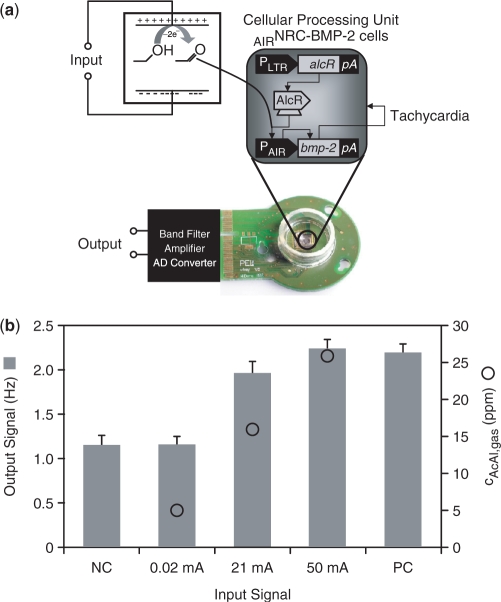
Electric signals linked to complex intracellular signalling cascades are well-known to manage muscular contraction in specialized mammalian cells (38). For example, cardiac ventricular contraction frequency of NRCs is modulated by BMP-2 that induces receptor-mediated activation of the myocyte-specific enhancer factor 2A via phosphatidylinositol 3-kinase in a dose-dependent manner (39). By transducing NRCs cultivated on complementary metal-oxide semiconductor (CMOS)-based HD-MEAs with lentiviral particles (32) engineered for electro-inducible acetaldehyde-responsive expression of BMP-2, we were able to convert DC into an oscillating electronic signal with a defined frequency (Figure 3a). Challenging this cell-based frequency generator with increasing input current resulted in elevated BMP-2 expression which stimulated NRCs to beat at higher frequency as scored by the HD-MEA (Figure 3b–e).
Figure 3.
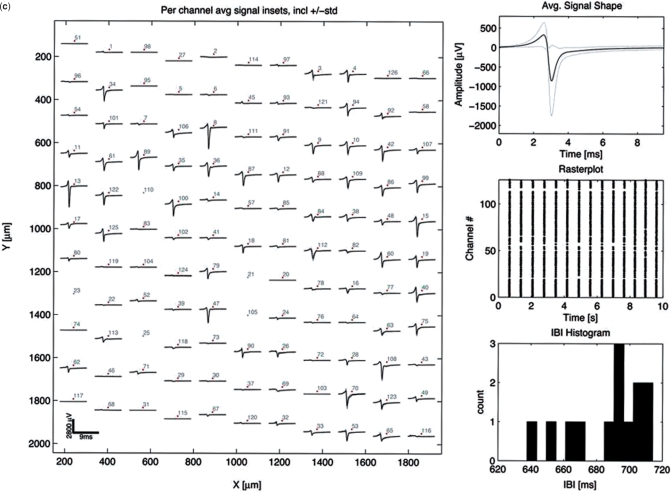
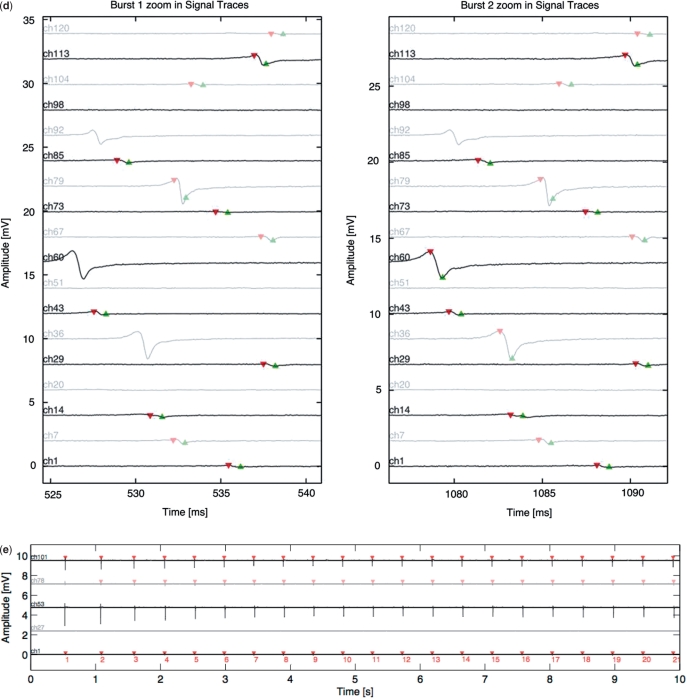
Mammalian cell-based frequency generator. (a) Circuit diagram of the cell-based frequency generator. DC power converts ethanol into acetaldehyde, which dose-dependently triggers expression of the BMP-2 in engineered rat cardiomyocytes (AIRNRC-BMP-2) and increases the contraction frequency (tachycardia). (b) The beating frequency of cardiomyocytes is recorded as a function of the input current and acetaldehyde concentration using a CMOS-based HD-MEA. NC: negative control, mock-transduced cardiomyocytes; PC: positive control, cells transduced for constitutive BMP-2 expression. (c) HD-MEA-based analysis of the electrogenic behaviour of NRCs engineered for electro-inducible acetaldehyde-responsive BMP-2 expression. The dataset shown as example was recorded at a direct input current of 50 mA corresponding to a beating frequency of 2.1 Hz. Detailed activation map illustrating the average signal shape of the 121 selected electrodes during 10 s. Average signal shape over all 121 channels. Raster plot showing a dot for each contraction on each channel over time. Inter-burst interval or inter-beat interval used to calculate the average beating frequency and beating frequency variation. (d) Zoom-in of two selected bursts/beats on different electrodes. (e) Long signal trace showing the synchronized contraction frequency on five selected electrodes or channels.
Miniaturization of the input interface
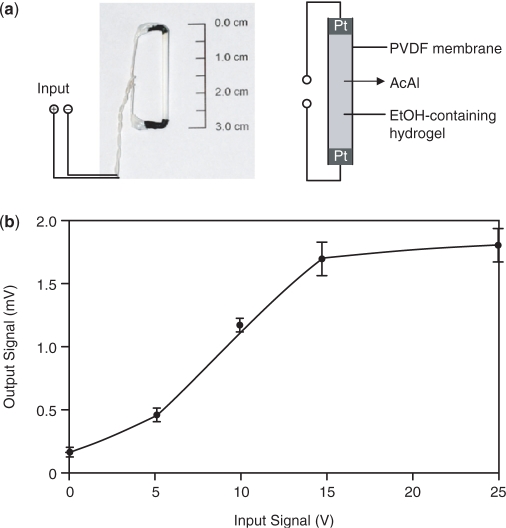
Akin to electronic equipment, which is continuously miniaturized, we have reduced the size of the initial electro-genetic input device by a factor of 15 000 (electrolyte volume: 16.7 µl versus 250 ml). To achieve this reduction factor, we have cast a 17% (v/v) ethanol-containing hydrogel into a PVDF-based hollow fiber membrane which was powered via two platinum electrodes (Figure 4a). Application of an electric input between 1 µA and 4 µA (5–15 V) produced a dose-dependent output signal when linked to a miniaturized CPU (mCPU) (Figure 4b). As with electronic devices, miniaturization of the electro-genetic control device reduced power consumption to achieve maximum transgene expression levels by 30 000-fold (60 µW, compared to 1.8 W of a standard CPU).
Figure 4.
Design and characterization of the mCPU. (a) Miniaturized input device. The input device consists of a PVDF hollow fiber membrane filled with an ethanol (17%, v/v)-containing hydrogel (16.7 µl) connected to platinum electrodes at both ends. (b) mCPU validation. The mCPU consisting of the miniaturized input device, a CPU (Figure 1a) and an enzymatic–optical output device (Figure 1a) was exposed to increasing DC for 1 h and the resulting electric output signal was scored after 24 h.
Although microelectronic devices are dominating our daily life and control most of the analytical instruments advancing life science research, our molecular understanding of how electricity impacts biological function remains largely limited to specialized electrogenic cells such as nerve and muscle cells and appropriate applications such as the pacemaker. The electro-genetic device described here provides a first example of modulating transgene expression in response to an electric current by coupling an electricity-triggered electrochemical reaction to a synthetic gene network engineered into mammalian cells. Such electro-genetic devices may define novel interfaces between microelectronic and biological transcription circuits and such electro-genetic information crosstalk may one day control therapeutic transgene expression or process disease signals of prosthetic implant devices thereby harnessing the full potential of progress in electronics sectors for human therapy.
FUNDING
Swiss National Science Foundation (grant no. 3100A0-112549); and the EC Framework 6 (COBIOS, 043379). Funding for open access charge: ETH Zurich.
ACKNOWLEDGEMENTS
We thank Max Wohlwend for construction of electronic devices; Christian Schirm for scientific advice; Evelyne Perriard, Anna Bogdanova and Nikolai Bogdanov for providing neonatal rat cardiomyocytes; and Francesca Faraci for supplying HD-MEAs and analysing HD-MEA-based data.
REFERENCES
- 1.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J. Biotechnol. 2007;130:329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006–0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 5.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 6.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 7.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 8.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 9.May T, Eccleston L, Herrmann S, Hauser H, Goncalves J, Wirth D. Bimodal and hysteretic expression in mammalian cells from a synthetic gene circuit. PLoS ONE. 2008;3:e2372. doi: 10.1371/journal.pone.0002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 11.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 12.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc. Natl Acad. Sci. USA. 2007;104:10435–10440. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson ML, Sayler GS, Fleming JT, Applegate B. Whole-cell biocomputing. Trends Biotechnol. 2001;19:317–323. doi: 10.1016/s0167-7799(01)01691-2. [DOI] [PubMed] [Google Scholar]
- 15.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. Synthetic biology: engineering Escherichia coli to see light. Nature. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat. Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 18.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 19.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 21.Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli F, De Francesco R, Cortese R. A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep. 2003;4:159–165. doi: 10.1038/sj.embor.embor734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber W, Schoenmakers R, Spielmann M, El-Baba MD, Folcher M, Keller B, Weber CC, Link N, van de Wetering P, Heinzen C, et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 2003;31:e71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malphettes L, Weber CC, El-Baba MD, Schoenmakers RG, Aubel D, Weber W, Fussenegger M. A novel mammalian expression system derived from components coordinating nicotine degradation in arthrobacter nicotinovorans pAO1. Nucleic Acids Res. 2005;33:e107. doi: 10.1093/nar/gni107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullick A, Xu Y, Warren R, Koutroumanis M, Guilbault C, Broussau S, Malenfant F, Bourget L, Lamoureux L, Lo R, et al. The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnol. 2006;6:43. doi: 10.1186/1472-6750-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber W, Link N, Fussenegger M. A genetic redox sensor for mammalian cells. Metab. Eng. 2006;8:273–280. doi: 10.1016/j.ymben.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 2004;22:1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 27.Boorsma M, Nieba L, Koller D, Bachmann MF, Bailey JE, Renner WA. A temperature-regulated replicon-based DNA expression system. Nat. Biotechnol. 2000;18:429–432. doi: 10.1038/74493. [DOI] [PubMed] [Google Scholar]
- 28.Weber W, Marty RR, Link N, Ehrbar M, Keller B, Weber CC, Zisch AH, Heinzen C, Djonov V, Fussenegger M. Conditional human VEGF-mediated vascularization in chicken embryos using a novel temperature-inducible gene regulation (TIGR) system. Nucleic Acids Res. 2003;31:e69. doi: 10.1093/nar/gng069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl Acad. Sci. USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer BP, Weber W, Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- 31.Auerbach D, Bantle S, Keller S, Hinderling V, Leu M, Ehler E, Perriard JC. Different domains of the M-band protein myomesin are involved in myosin binding and M-band targeting. Mol. Biol. Cell. 1999;10:1297–1308. doi: 10.1091/mbc.10.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Bustamante CD, Frey U, Kelm JM, Hierlemann A, Fussenegger M. Modulation of cardiomyocyte electrical properties using regulated bone morphogenetic protein-2 expression. Tissue Eng. Part A. 2008;14:1969–1988. doi: 10.1089/ten.tea.2007.0302. [DOI] [PubMed] [Google Scholar]
- 33.Mitta B, Rimann M, Fussenegger M. Detailed design and comparative analysis of protocols for optimized production of high-performance HIV-1-derived lentiviral particles. Metab. Eng. 2005;7:426–436. doi: 10.1016/j.ymben.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Schlatter S, Rimann M, Kelm J, Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- 35.Frey U, Heer F, Pedron R, Hafizovic S, Greve F, Sedivy J, Kirstein KU, Hierlemann A. An 11k-electrode 126-channel highdensity microelectrode array to interact with electrogenic cells. IEEE International Solid-State Circuits Conference 2007. 2007:158–159. [Google Scholar]
- 36.Tremiliosi-Filho G, Gonzalez ER, Motheo AJ, Belgsir EM, Léger J.-M, Lamy C. Electro-oxidation of ethanol on gold: analysis of the reaction products and mechanism. J. Electroanal. Chem. 1998;444:31–39. [Google Scholar]
- 37.Vigier F, Coutanceau C, Hahn F, Belgsir EM, Lamy C. On the mechanism of ethanol electro-oxidation on Pt and PtSn catalysts: electrochemical and in situ IR reflectance spectroscopy studies. J. Electroanal. Chem. 2004;563:81–89. [Google Scholar]
- 38.Ghosh-Choudhury N, Abboud SL, Chandrasekar B, Ghosh Choudhury G. BMP-2 regulates cardiomyocyte contractility in a phosphatidylinositol 3 kinase-dependent manner. FEBS Lett. 2003;544:181–184. doi: 10.1016/s0014-5793(03)00507-6. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh-Choudhury N, Abboud SL, Mahimainathan L, Chandrasekar B, Choudhury GG. Phosphatidylinositol 3-kinase regulates bone morphogenetic protein-2 (BMP-2)-induced myocyte enhancer factor 2A-dependent transcription of BMP-2 gene in cardiomyocyte precursor cells. J. Biol. Chem. 2003;278:21998–22005. doi: 10.1074/jbc.M302277200. [DOI] [PubMed] [Google Scholar]