Abstract
Background
The pancreatic regenerating (reg I) gene and its protein product are derived from acinar cells, and are mitogenic to β- and ductal cells. We studied the mechanism of this mitogenic response.
Methods
ARIP (rat ductal) and RIN 1046–38 (rat β-) cell lines were exposed to exogenous reg I in culture or transfected with a reg I expression vector. Mitogenesis was assessed by MTS assay, and cellular mRNA was subjected to gene microarray analysis to determine signal transduction pathways. Yeast two-hybrid technology was then used to determine intracellular binding of reg I protein.
Results
Cells exposed to exogenous reg I showed a mitogenic response; cells transfected with reg I expression vector showed inhibited growth. Microarray analysis of the former showed induction of cyclin pathways and MKP-1, cyclins were inhibited in the latter. Northern analysis confirmed gene induction of cyclin D1 and MKP-1; JNK was phosphorylated prior to expression of both. Yeast two-hybrid analysis confirmed a protein-protein interaction with MKP-1; this was confirmed by immunoprecipitation.
Conclusions
Pancreatic derived cells exposed to reg I grow by activation of signal transduction pathways involving the mitogen-activated protein (MAP) kinase phosphatases and cyclins, with concomitant induction of MKP-1. But, high intracellular levels of reg I lead to decreased growth, likely via a binding to and inactivation of MKP-1. Inhibition of cell growth, and possible induction of apoptosis, may lead to differentiation of these cells to other cell types.
Keywords: Pancreatic reg I, pancreatic ductal cells, pancreatic β-cells, cyclin D1, MKP-1, EXTL3, mitogenesis
INTRODUCTION
The regenerating (reg) family of genes is predominantly expressed in cells of the gastrointestinal tract (1–3). Reg I is an acinar cell-derived product of the pancreas, is constitutively expressed and secreted from the acinar cells, and can be ectopically induced within regenerating islets. Reg I protein is an established mitogen to pancreatic β-, ductal and mucosal cells of the GI tract including the stomach and colon (2, 4–5). We have shown that purified recombinant reg I is bioactive as well (6), and is mitogenic to primary cultures of ductal cells (6).
The mechanism of reg I mitogenesis is likely paracrine. A transmembrane receptor has been identified which is homologous to the multiple exostoses-like gene family (EXTL3) (7). EXT genes are mutated in bone disease, and the function of the EXT and EXTL proteins are in assembly of peptide-glycans, specifically heparan sulfate (8–9). Specifically, they catalyze the polymerization of glycosaminoglycans into heparan sulfate; EXTL3 actually initializes the event (10). However, EXT proteins have not been linked to cell growth, or cell cycle pathways, so precisely how reg I exerts its mitogenic effect is still unknown. While reg I does bind to the cell surface transmembrane EXTL3 protein, EXT gene products localize within the cell - specifically in the Golgi apparatus and endoplasmic reticulum (11). Interestingly, Simmons et al showed that EXT1 and EXT2 directly interacts with intracellular proteins (12). This raises the possibility that reg I might have a direct intracellular effect, by either trafficking into the cell by means of the receptor or after induction of its gene within the proliferating cells.
To determine which intracellular signaling cascades are induced during mitogenesis by reg I, we employed microarray analysis and found it to act through mitogen-activated protein (MAP) kinase pathways in both ductal and β-cells. We also explored which proteins reg I associates with inside the cell using yeast two-hybrid technology and have determined that it actually binds to the MAP kinase phosphatase (MKP-1).
MATERIALS & METHODS
Cell proliferation and gene transcription
ARIP (rat ductal, American Type Culture Collection (Rockville, MD)) or RIN 1046–38 (rat insulinoma) cells (6) were used. We defined 10nM as the optimal dose for mitogenesis studies, since previous studies in our laboratory (6,13) showed that reg I protein was mitogenic at doses ranging in concentration from 0.1 pM to 10 nM to both ARIP and RIN cells, but growth began to show inhibition at 100 nM.
To measure the effect of endogenously expressed reg I on these cell lines, a 540 bp cDNA sequence of the coding region of reg I (6) was cloned into the pcDNA3 vector (Invitrogen, Carlsbad, CA) between BamHI and Not1 sites, under the control of the CMV promoter. Controls were cells transfected with pcDNA3 vector alone. Transfected cells were selected with 500ug/ml G418 selection medium (Sigma, St. Louis, MO). Reg I protein expression was verified through Western blotting with a monoclonal anti-reg I antibody 2B3-F12 developed in our laboratory. The antibody was created using reg I purified by serial ammonium sulfate precipitations from human pancreatic juice as previously described (13). Monoclonal antibodies to reg I were raised according to established protocols. Briefly, female BALB/c mice were immunized with reg I in Freund’s adjuvant (GIBCO Life Technologies, Rockville, MD). Hybridomas were produced using mouse myeloma cell line Sp2/0 (ATCC, Rockville, MD) and fused to mouse splenocytes using Hybrid MAX PEG Solution (Sigma, St Louis). Supernatants of the clones were screened against purified rat and human reg I by ELISA, and the positive clones confirmed by Western blotting, using anti-mouse Ig –HRP (Amersham Biosciences, New Jersey) as a second antibody. Clone 2B3-F12 was chosen since it cross reacted with both rat and human. Injection of the hybridoma into mice and harvesting of resultant ascites typically yielded a titer of 1:12,8000 by ELISA.
Normal and transfected ARIP (100 × 103 cells/well) and RIN (100 × 103 cells/well) were plated in 96-well plates, in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum, penicillin, and streptomycin (GIBCO). After an overnight incubation, each well was washed three times with 1 ml of phosphate-buffered saline (PBS) (Sigma). Normal cells were inoculated with 10 nM recombinant reg I protein with 1% serum replacement medium (Sigma); pcDNA3-reg I transfected cells were compared to cells transfected with blank vector. Cells were incubated for 48 hrs and mitogenesis was assayed by the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) -tetrazoleum assay (CellTiter 96, Promega, Inc., Madison, WI) following the manufacturer’s protocol. Absorbance was measured at 490nm directly from 96-well assay plates with a microplate reader (Bio-Rad, Inc., Hercules, CA). Each experiment was done in triplicate and repeated at least twice.
For assessment of mitogen-activated kinase, ARIP ductal cells were inoculated with 10 nM recombinant reg I protein. A 24 hr time course (0.5, 1, 4, 8, 24 hour) of phosphorylation of the mitogen-activated kinases (MAPK), P44-42 (ERK1/2), P38 and SAPK/JNK was assessed on 10ug of cellular protein by Western analysis using antibodies directed against the phosphorylated forms of the MAPKs (New England Biolabs, Ipswich, MA).
Microarray analysis of reg I gene activation
100 ug RNA was extracted from control and experimental ARIP and RIN cells with the RNeasy kit (Qiagen, Inc., Valencia, CA) and resuspended in diethylpyrocarbonate (DEPC) treated water. Probes were prepared by annealing 17ul/probe of resuspended RNA with oligo dT at 65°C and then labeled with either CY-3 (control) dUTP (green color) or CY-5 (experimental) dUTP (red color) fluorescent nucleotides with the Superscript-RT kit (GIBCO) during reverse transcription into cDNA. After RNase treatment, cDNA was concentrated (Amicon, Inc. Beverly, MA), and hybridized with mouse 9M gene chips obtained from, and following the protocol of, the Albert Einstein College of Medicine (AECOM) Microarray Core Facility. The gene chips were then scanned and analyzed with ScanAnalyze2 software (M.B. Eisen, Stanford University). The cutoff ratios for up-regulation and down-regulation were defined as 1.5 and 0.67, respectively. Each experiment was repeated three times.
Isolation of rat reg I binding proteins by the Yeast Two-Hybrid System
To identify genes encoding proteins that associate with reg I protein, we employed the Hybrid ZAP-2.1 two-hybrid vector system (Stratagene, La Jolla, CA). The rat reg I coding sequence was cloned into pBD-GAL4 Cam (bait) phagemid vector, and a cDNA library of the target cell, ARIP, which was stimulated with recombinant reg I protein (10nM for 2 days) was cloned into the pAD-GAL4-2.1 (target) phagemid vector using the manufacturer’s guidelines.
The rat reg I coding sequence was established using PCR primers to directionally clone it into the pBD-GAL4 Cam (bait) vector using Sal I and Pst I at each end (reg I-Sal I primer was: 5′ ACG CGT CGA CTC ATG ACT CGC AAC AAA TAT TTC 3′ (Sal I in italics), the sequence of reg I - Pst I primer was: 5′ GGC A CTG CA G TCA GGC TTT GAA CTT GCA GAC 3′ (Pst I in italics). The recombinant pBD-GAL4-reg I plasmid was verified by DNA sequencing.
pBD-GAL4-reg I plasmid was transformed into freshly prepared yeast (YRG-2) competent cells, plated on SD agar plates without tryptophan, and incubated at 30°C for 2–4 days. Total RNA was extracted from the yeast colonies, and reg I mRNA was verified by RT-PCR. Reg I protein was confirmed by Western Blotting with the anti-reg I monoclonal antibody described above.
Plasmid DNA from the pAD-GAL4-2.1 target library was transformed into the YRG-2 yeast containing pBD-GAL4-reg I. Selection was done on agar plates lacking histidine, leucine and tryptophan. Colonies were transferred to nitrocellulose paper, permeabilized in liquid nitrogen, and assayed for expression of the LacZ reporter gene by the detection of β-galactosidase.
Plasmid DNA was isolated from the His+LacZ+ yeast colonies, and transformed into XL1-Blue MRF’ competent cells. The target plasmid (pAD-GAL4-2.1) was selected by plating the transformant mixture on LB-Ampicillin agar plates. The target plasmid DNA was isolated using the B101 RPM Rapid Pure Miniprep Kit (BIO 101, La Jolla, CA). The size of the inserts in the target plasmids was analyzed using the Expand Long Template PCR System (Roche, Mannheim, Germany). The sequence of the 5′AD primer was: 5′ AGG GAT GTT TAA TAC CAC TAC 3′, the sequence of the 3′AD primer was: 5′ GCA CAG TTG AAG TGA ACT TGC 3′. The PCR conditions were as follows: initial denaturation at 94°C for 3 min, then 30 cycles of denaturation at 94°C for 30sec, annealing at 56°C for 30sec, and elongation at 68°C for 6 min, followed by 1 cycle of final elongation at 68°C for 7 min. Resultant plasmid DNA was subjected to DNA sequencing, and the sequences were analyzed in GenBank using BLAST software.
Immunoprecipitation/Western blots
Whole cell lysates were prepared from ARIP and RIN cells which were either normal or transfected with pcDNA3-reg I expression vector. 500 ug of lysate was immunoprecipitated (14–15) with mouse anti-human reg I monoclonal antibody (1:50) using Protein G-Sepharose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Immunoprecipitates were subjected to 12% Sodium Dodecyl Sulfate – Polyacrylamide Gel Electrophoresis (SDS-PAGE) under reducing conditions, and proteins were electrophoretically transferred to a nitrocellulose membrane. For Western blotting, the membrane was blocked with Tris Buffered Saline – Tween 20 (TBS-T) containing 5% non-fat dry milk for 1 hour, washed several times with TBS-T, and incubated with a 1:200 dilution of the primary rabbit anti-mouse MKP-1 polyclonal antibody (Santa Cruz). After 1 hour, the membrane was washed several times again with TBS-T, incubated with a 1:5000 dilution of horse radish peroxidase conjugated bovine anti-rabbit IgG as the secondary antibody for 1 hour, washed again several times with TBS-T, and subjected to detection with an ECL-plus chemiluminescence autoradiography kit (Amersham, Inc. Piscataway, NJ).
RNAse Protection Analysis
Cyclin D1 was assayed by RNase Protection Assay with the Ribo-Quant Multi-Probe Template Set mCYC-1 (BD Biosciences, San Diego, CA) as previously described (16). Briefly, total RNA was extracted from ARIP cells treated with 10 pM of reg I at 0, 2, 4 & 24 hours post-treatment. 15 μg of RNA from each timepoint, hybridized to an antisense 32P labeled probe cocktail containing the templates for cyclins A2, B1, C, D1, D2, D3, A1, & B2, was subjected to 5% SDS-PAGE on a vertical apparatus (IBI, Standard Thermoplate Sequencer, New Haven, CT), along with the appropriate controls and probes, and run as per the manufacturer’s instructions. The gel was then adsorbed to filter paper, dried under vacuum, and placed in a PhosphorStorage screen. Quantification of mRNA was performed (Storm 860 PhosphorImager, Molecular Dynamics, Sunnyvale, CA) and analysis of radioactive bands was undertaken (ImageQuant software, Molecular Dynamics) according to the manufacturer’s recommendations. Data, represented as phosphoimager units, are from three independent experiments. To adjust for differences in sample processing, hybridization signals in each sample were divided by the signal for the housekeeping ribosomal protein mRNA (L32).
RESULTS
Studies of reg I treated and pcDNA3-regI transfected cells
While exogenous reg I treatment increased RIN and ARIP cell growth rates by at least 20%, cells transfected with the pcDNA3-regI plasmid showed reduced cell growth rates compared to controls (Figure 1). Similar depression in cell growth has been observed by our laboratory in the AR42J acinar cell line transfected with a similar reg I expression vector (not shown, ref 17).
Figure 1.
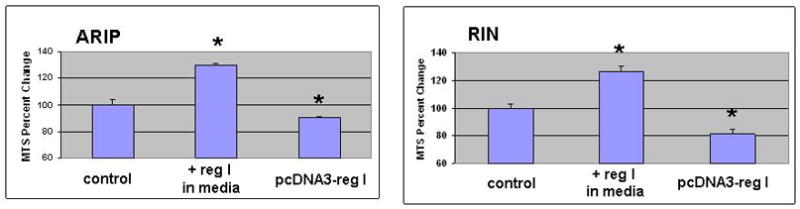
MTS assay demonstrating proliferation rates of ARIP ductal or RIN β-cells exposed to exogenous reg I protein (10nM), or cells transfected with pcDNA3-reg I plasmid which induced reg I protein within cells. Both lines express the reg I receptor, and grow in response to exogenous reg I; growth is significantly inhibited when reg I is expressed within the cell (p<0.05 compared to controls). There was no difference between normal control cells and cells expressing blank pcDNA3 vector (not shown).
Transfection of the pcDNA3-reg I expression vector into RIN or ARIP cells resulted in reg I gene and protein expression, and even secretion of reg I protein into the media (not shown). Interestingly, when compared to non-transfected RIN cells, insulin responses were lower in the culture medium of RIN pcDNA3-reg I cells at increasing glucose concentrations (not shown).
Microarray studies
cDNA microarray analysis of ARIP and RIN cells exogenously treated with reg I showed induced regions of cell cycle and downstream signal transduction genes, including MAP kinase phosphatase 1 (MKP-1) and the cyclin pathways; in cells which overexpressed reg I, these genes were inhibited. For example, when compared to normal controls, exogenously treated ARIP cells increased cyclin D2, cyclin B1 (EST) and cyclin kinase (EST) levels 3.5, 2.1 and 2.0 fold, respectively (p<0.05 compared to untreated controls). By comparison, ARIP cells expressing endogenous reg I showed depression to 0.8, 0.6 and 1.2 fold, respectively (p<0.05; controls were cells with blank expression vector). Microarray analysis also showed that MKP-1 was induced by exogenous reg I (below).
cDNA microarray analysis of RIN cells treated with exogenous reg I showed 2-fold increase in expression of MKP-1 (p<0.05). Figure 2 confirms, using Northern analysis, that MKP-1 mRNA increased in RIN cells exposed to exogenous reg I; similar data was shown for ARIP cells. Another depressed gene seen in RIN cells was protein tyrosine phosphatase, receptor type A (PTPra) to 0.42 fold of controls (p<0.05).
Figure 2.
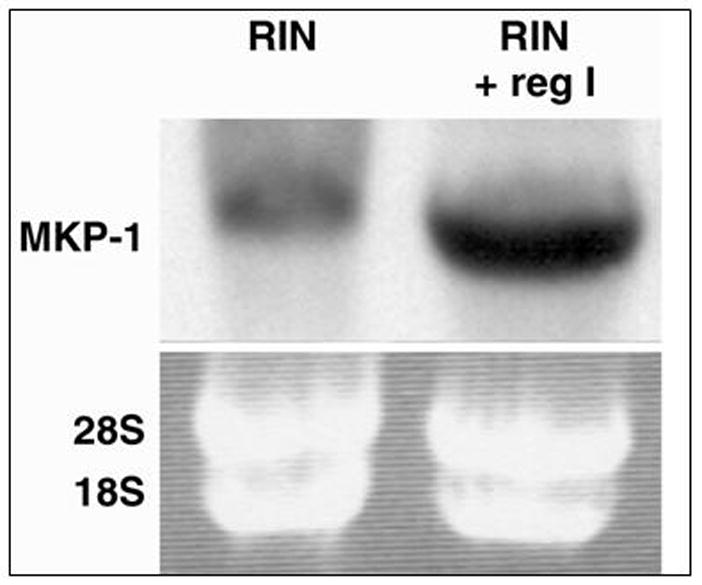
Northern blot analysis verified upregulation of MKP-1 mRNA gene expression by exogenous reg I treatment of RIN cells. Bottom panel is ethidium bromide staining of gel prior to transfer, confirming integrity of mRNA, and concentration of RNA loaded.
Yeast Two-hybrid analysis of Reg I Binding Proteins (Rbp)
In order to identify protein-protein interactions with reg I protein, reg I was cloned into a yeast two-hybrid system, and probed with a cDNA library of genes isolated from ARIP cells stimulated by reg I.
Fourteen positive colonies were identified and sequenced. Interestingly, after a homology search using NCBI BLAST software, none of these clones had any homology to the EXTL3-reg receptor gene (7) or other EXT genes, and 3 full sequence clones were found of particular interest. Clone 53, named Reg-binding protein-2 (Rbp-2), was shown to be identical to rat MKP-1, a protein important in controlling cell growth. Clone 13 was named Rbp-1, and we submitted the cDNA sequence to GenBank (18). It has subsequently been identified as pyridoxal (vitamin B6) phosphate phosphatase. Clone 145 was named Rbp-III and is 99% identical to ribosomal protein L37, another protein important in cell division (19).
Reg I and kinase pathways
To investigate whether reg I influences cyclin gene expression as part of cell proliferation, we determined the expression of cyclins in ARIP cells treated with reg I in a 24 hour period. The most dramatic change noted was an increase in cyclin D1 expression of approximately 1.6 fold after 2 hours, 3.1 fold at 4 hours and 5.3 fold at 24 hours (Figure 3); this was not observed in cells transfected with the pcDNA3-reg I expression vector.
Figure 3.
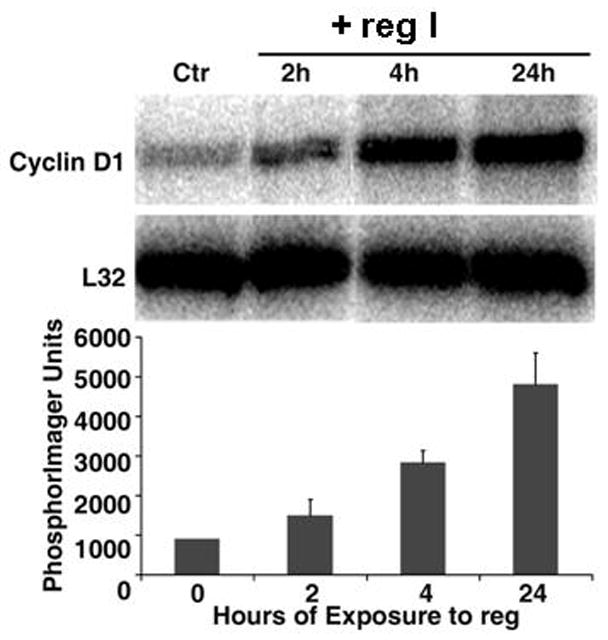
Time course of reg I effect on cyclin levels in ARIP cells. Upper: RNase protection analysis of ARIP mRNA is shown. Total RNA was extracted from ARIP cells treated with 10 pM of reg I at indicated times; Each lane contained 15 μg of total RNA hybridized to an antisense RNA probe cocktail which contained the templates for cyclin genes, protected fragments of which were separated on a 5% DNA sequencing gel, and are indicated. Lower: The graph represents the quantification results from three independent experiments. Cyclin D1 signals were normalized to those of L32 (mRNA for ribosomal protein subunit).
Since cyclins are modulated by MAP kinase pathways, the effect of reg I on p38 MAPK, ERK1/2 and JNK pathways was explored. Western analysis of cellular protein of cells exposed to reg I showed increased levels of phosphorylated SAPK/JNK within 1 hour, P44-42 (ERK 1/2) at 4 hr, and progressive activation of MAPK p38 at 24 hours. The induction of JNK preceded the expression of cyclin D1 (Figure 4).
Figure 4.
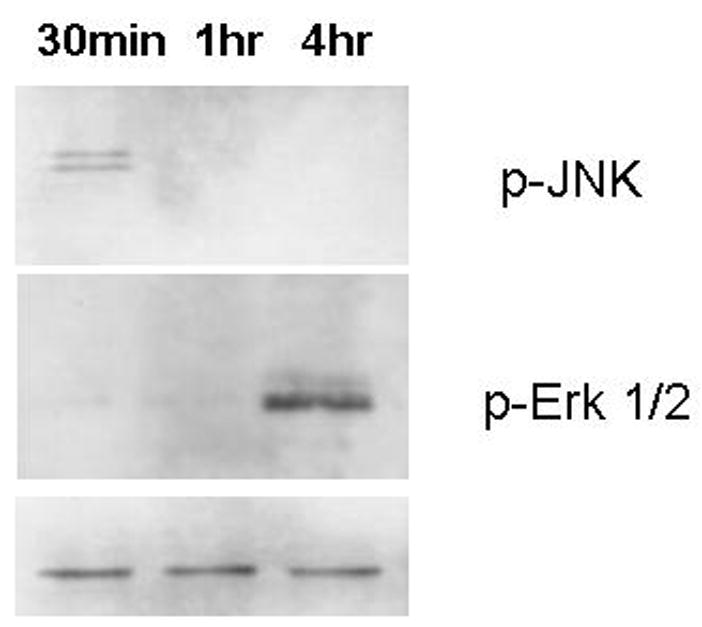
JNK and ERK mitogen kinase activation in ARIP cells after reg treatment. A 24-hour time course was assessed on 10 μg of cellular protein by western blotting analysis using antibodies to the phosphorylated forms of both proteins. P38 showed no change until 24 hours, where an increase was noted (not shown). Bottom panel is non-phosphorylated JNK protein, approximately 50Kd.
Reg I and MKP-1
As both our yeast two-hybrid and microarray experiments identified MKP-1 as a gene of interest in cells exposed to reg I, we proposed that reg I acts via a MAP kinase pathway, and that MKP-1 was induced to control cell growth. As noted above, Northern analysis confirmed that MKP-1 mRNA increased in RIN cells exposed to exogenous reg I (Figure 2).
Microarray, Northern and yeast two-hybrid data suggested a physical interaction between reg I and MKP-1. MKP-1 activity is an established modulator of mitogen kinase phosphorylation (20). To explore a potential protein-protein interaction of MKP-1 and reg I, we employed immunoprecipitation techniques. Figure 5 shows that in cells overexpressing reg I protein, MKP-1 co-precipitated with reg I. Co-precipitation was not observed in control cells transfected with blank vector or in cells exposed to exogenous reg I. This indicates that while exogenously administered reg I induces MKP-1 gene expression, endogenously expressed reg I binds directly to MKP-1 in the cell, and might lead to inhibition of cell division (20).
Figure 5.
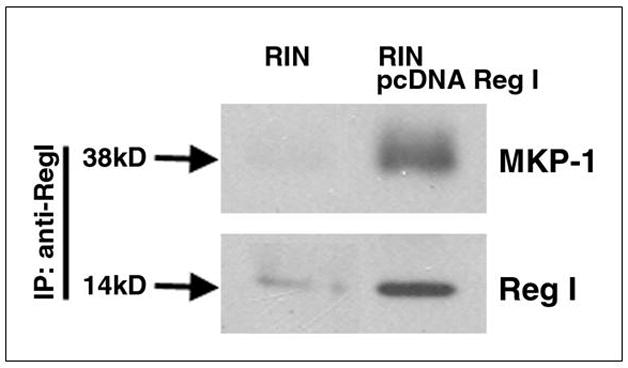
Reg I and MKP-1 co-precipitated in RIN cells overexpressing reg I protein. Whole cell lysates were generated from confluent RIN or RIN transfected pcDNA3-regI. Cell lysates were immunoprecipitated with mouse anti-reg monoclonal antibody overnight at 4°C and immunoprecipitates were resolved by SDS-12% PAGE; precipitated proteins were detected by immunoblotting with a bovine anti-MKP-1 and by a mouse anti-reg antibody.
DISCUSSION
In this study, we have shown that when pancreatic ductal and β-cells are exposed to exogenous reg I protein, proliferation occurs, in conjunction with induction of genes involved in the cell cycle. Mitogenesis by reg I is well established (13,21); it likely acts via the recently characterized reg I receptor, a transmembrane protein which is homologous to the EXTL3 gene (7). While the reg I receptor is found on cells which grow in response to reg I, and not on cells which do not express the gene (22), there is presently no data which shows that these receptors are involved in signal transduction pathways of cell cycle genes or in cellular mitogenesis. The only study which showed any possibility of an intracellular interaction was by Simmons et al, who found that EXT1 and EXT2 bind intracellular proteins (12).
We therefore explored the cellular pathways of reg-mediated mitogenesis. Using cDNA microarray technology we characterized the effect of reg I on cell proliferation and gene transcription in RIN cells (a pancreatic β-cell line) and ARIP cells (a rat ductal cell line) under two conditions: cells cultured in medium containing reg I protein and over-expression of reg I in cells which normally express the gene.
Our data indicate that in pancreatic ductal and β-cells exposed to reg I protein, signal transduction pathways involving the mitogen-activated protein (MAP) kinase phosphatases and cyclins are induced. Northern analysis confirmed that MKP-1 gene expression and cyclin D1 are induced by reg I. Yeast two-hybrid analysis identified that MKP-1 protein actually binds reg I.
The cyclin D1 pathway has previously been identified as an important pathway in the mitogenic response of β-cells to reg I protein. Takasawa et al, like us, showed that cyclin D1 is activated within 60 minutes of exposure to reg I by phosphorylation of ATF-2 by a phosphoinositide-3 kinase (PI(3)K) dependent pathway (23). This in turn inhibits retinoblastoma protein Rb and induces the cell cycle. Also, Kodawaki et al (24) have shown that reg I is mitogenic via the classical MAPK-ERK1/2 pathway. This is different from our observation that the JNK pathway was stimulated first, but their measurements were within 15 minutes of exposure, earlier than ours.
MKP-1 activity can modulate JNK phosphorylation. Even though many believe that MKP-1 works to inhibit JNK-stimulated cell growth, Wu et al (20) clearly showed that inhibition of MKP-1 can result in decreased cell growth, increased apoptosis, and enhanced cell death. These experiments, performed in fibroblasts harvested from MKP-1 knockout mice, predicted that MKP-1 can have an anti-apoptotic function. This concept can be important if the MKP-1 is bound by reg I protein and the anti-apoptotic effect is inhibited and cell division stops. We have yet to do the studies on reg I and apoptosis, but preliminary work in our laboratory shows that high doses of exogenous reg I can induce caspase-3 activity.
This hypothesis is supported by our current observation that when pancreatic-derived cells were transformed with an expression vector to overexpress reg I, cell growth was retarded. In fact, we and other laboratories have previously observed that high extracellular levels of over 100nM reg I can inhibit cell growth (6,13,24) in culture. These data indicate that reg I may have a growth-inhibiting effect at high levels, and maybe it is modulated by intracellular effects.
Supporting this concept is our yeast two-hybrid experiment which showed that reg I physically associates with MKP-1 (as reg-binding protein-2, Rbp-2), and inhibition of MKP-1 abrogates its anti-apoptotic effect (20). The physical interaction between the proteins was confirmed by immunoprecipitation studies. It is possible that the reg I protein has an inhibitory effect on the MKP-1 molecule, the same gene it induces.
We postulate that when cells are exposed to exogenous reg I protein, the protein binds to the EXTL3 protein, and stimulates mitogenesis via activation of an unknown signal transduction pathway related to the EXTL3 receptor. This most likely occurs via a MAP kinase pathway leading through JNK to cyclin D1 to induce β- or ductal cellular growth (Figure 6). The MKP-1 induction by exogenous reg I is likely a secondary response to stimulation of MAP kinases, as part of the normal feedback control (20,25).
Figure 6.
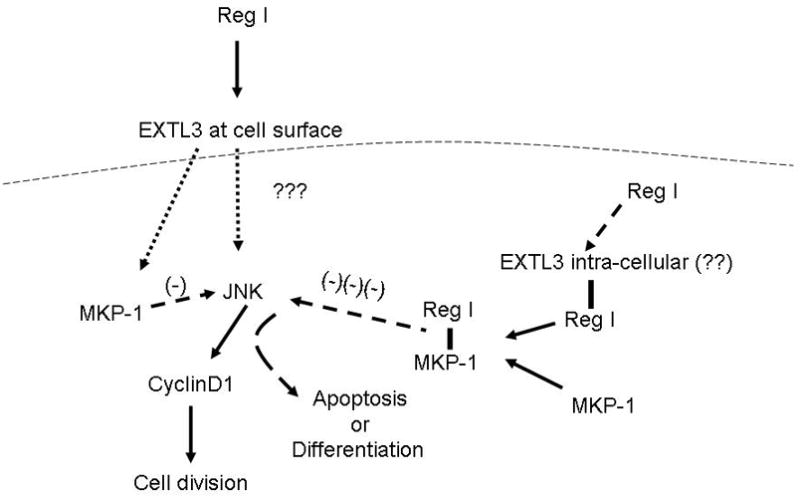
Proposed mechanism of action of pancreatic reg I on ductal and β-cells. Exogenous reg I binds the EXTL3 receptor and induces cell growth via signal transduction through JNK and cyclin D1. Intracellular reg I, however, can inhibit cell growth by direct interaction with MKP-1, inducing a pathway other than mitogenesis (dotted line represents cell membrane).
But, very high levels of exogenous reg I can result in elevation of intracellular levels of reg which adversely affect cell growth. Whether or not reg enters the cell via binding to intracellular EXTL proteins (9) is unknown, but the intracellular effect of reg I is clearly mediated by binding MKP-1, inhibiting cellular division.
Others have shown that high intracellular levels of reg I can retard growth. Yamaoka et al showed in vivo that in transgenic mice, reg I overexpression within islet cells resulted in β-cell apoptosis, compensatory islet hypertrophy and an overall depressed insulin content (26). In a similar set of experiments, we observed depressed mitogenesis in the acinar cell line AR42J after transfection with a reg I expression vector (17), along with differentiation toward an acinar cell lineage, a more differentiated state, and away from the ductal phenotype.
There is other evidence that cellular overexpression of reg I may lead to transdifferentiation to other cell types. The transgenic mice above, which ectopically expressed reg I, showed pancreatic and other non-islet tumors - cervical, hepatocellular, uterine, and ovarian. Transgenic reg I overexpression in gastric mucosa results in differentiation of progenitor cells into chief and parietal cells (27). Finally, ectopic expression of reg I in the colon is associated with cancer formation (5). But, this concept is controversial - Okamoto’s group did show that ectopic expression of reg I in islets can result in β-cell growth and islet hypertrophy (28), and, reg I knockout mice have smaller β-cell mass and poor regenerative capacity (29).
We postulate that reg I protein exhibits a dual action on cells. As depicted in Figure 6, when applied exogenously in media to cells which express the EXTL3/reg receptor, it induces growth via a MAP-kinase pathway, then activates cyclin D1, and is modulated by MKP-1. When over-expressed within cells or in very high concentration, it inhibits growth by binding with MKP-1; this inhibition directs cells to transdifferentiate to other types.
Acknowledgments
The authors recognize the technical support of Drs. Haiyan Wu, Sameer Patel, Zuoheng Fan, and P. Rengabashyam; supported by NIH 1 RO1 DK54511.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Okamoto H. The Reg gene family and Reg proteins: with special attention to the regeneration of pancreatic beta-cells. J Hepatobiliary Pancreat Surg. 1999;6:254–62. doi: 10.1007/s005340050115. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Zheng Q, Rengabashyam P, Zenilman ME. A brief history of pancreatic reg: Implications regarding its clinical importance. Einstein Quarterly. 2001;17:178–189. [PMC free article] [PubMed] [Google Scholar]
- 3.Graf R, Schiesser M, Reding T, Appenzeller P, Sun LK, Fortunato F, Perren A, Bimmler D. Exocrine meets endocrine: pancreatic stone protein and regenerating protein--two sides of the same coin. J Surg Res. 2006;133:113–20. doi: 10.1016/j.jss.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita Y, Ishihara S, Kadowaki Y, Fukui H, Chiba T. Reg protein is a unique growth factor of gastric mucosal cells. J Gastroenterol. 2004;39:507–13. doi: 10.1007/s00535-004-1354-5. [DOI] [PubMed] [Google Scholar]
- 5.Zenilman ME, Kim S, Levine BA, Lee C, Steinberg JJ. Ectopic expression of reg protein: A marker of colorectal mucosa at risk for neoplasia. J Gastrointest Surg. 1997;1:194–201. doi: 10.1016/s1091-255x(97)80109-6. discussion 201–2. [DOI] [PubMed] [Google Scholar]
- 6.Levine JL, Patel KJ, Zheng Q, Shuldiner AR, Zenilman ME. A recombinant rat regenerating protein is mitogenic to pancreatic derived cells. J Surg Res. 2000;89:60–5. doi: 10.1006/jsre.1999.5800. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi S, Akiyama T, Nata K, Abe M, Tajima M, Shervani NJ, Unno M, Matsuno S, Sasaki H, Takasawa S, Okamoto H. Identification of a receptor for reg (regenerating gene) protein, a pancreatic beta-cell regeneration factor. J Biol Chem. 2000;275:10723–6. doi: 10.1074/jbc.275.15.10723. [DOI] [PubMed] [Google Scholar]
- 8.Lind T, Tufaro F, McCormick C, Lindahl U, Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J Biol Chem. 1998;273:26265–8. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi S, Morimoto K, Shimizu T, Takahashi M, Kurosawa H, Shirasawa T. Association of EXT1 and EXT2, hereditary multiple exostoses gene products, in Golgi apparatus. Biochem Biophys Res Commun. 2000;268:860–7. doi: 10.1006/bbrc.2000.2219. [DOI] [PubMed] [Google Scholar]
- 10.Busse M, Feta A, Presto J, Wilén M, Grønning M, Kjellén L, Kusche-Gullberg M. Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J Biol Chem. 2007;282:32802–10. doi: 10.1074/jbc.M703560200. [DOI] [PubMed] [Google Scholar]
- 11.Duncan G, McCormick C, Tufaro F. The link between heparan sulfate and hereditary bone disease: finding a function for the EXT family of putative tumor suppressor proteins. J Clin Invest. 2001;108:511–6. doi: 10.1172/JCI13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons AD, Musy MM, Lopes CS, Hwang LY, Yang YP, Lovett M. A direct interaction between EXT proteins and glycosyltransferases is defective in hereditary multiple exostoses. Hum Mol Genet. 1999;8:2155–64. doi: 10.1093/hmg/8.12.2155. [DOI] [PubMed] [Google Scholar]
- 13.Zenilman ME, Magnuson TH, Swinson K, Egan J, Perfetti R, Shuldiner AR. Pancreatic thread protein is mitogenic to pancreatic-derived cells in culture. Gastroenterology. 1996;110:1208–14. doi: 10.1053/gast.1996.v110.pm8613011. [DOI] [PubMed] [Google Scholar]
- 14.Jung JJ, Kim CW. Interaction between chicken protein tyrosine phosphatase 1 (CPTP1)-like rat protein phosphatase 1 (PTP1) and p60v-src in v-src-transformed Rat-1 fibroblasts. Exp Mol Med. 2002;34:476–480. doi: 10.1038/emm.2002.66. [DOI] [PubMed] [Google Scholar]
- 15.Barnes H, Larsen B, Tyers M, van der Geer P. Tyrosine-phoshorylated low density lipoprotein receptor-related protein 1 (LRP1) associates with the adaptor protein SHC in SRC-transformed cells. J Biol Chem. 2001;276:19119–19125. doi: 10.1074/jbc.M011437200. [DOI] [PubMed] [Google Scholar]
- 16.Bluth MH, Kandil E, Mueller CM, Shah V, Lin Y-Y, Zhang H, Dresner L, Lempert L, Nowakowski M, Gross R, Schulze R, Zenilman ME. Sophorolipids block lethal effects of septic shock in rats in a cecal ligation and puncture model of experimental sepsis. Crit Care Med. 2006;34:E188–195. doi: 10.1097/01.ccm.0000196212.56885.50. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez D, Mueller CM, Zenilman ME. Pancreatic Reg I and Acinar Cell Differentiation: Influence On Cellular Lineage. Surgery. doi: 10.1097/mpa.0b013e3181a1d9f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NLM accession #NM_020097 http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=9910529
- 19.Su S, Bird RC. Cell cycle, differentiation and tissue-independent expression of ribosomal protein L37. Eur J Biochem. 1995;232:789–97. [PubMed] [Google Scholar]
- 20.Wu JJ, Bennett AM. Essential Role for Mitogen-activated Protein (MAP) Kinase Phosphatase-1 in Stress-responsive MAP Kinase and Cell Survival Signaling. J Biol Chem. 2005;280:16461–16466. doi: 10.1074/jbc.M501762200. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe T, Yonemura Y, Yonekura H, Suzuki Y, Miyashita H, Sugiyama K, Moriizumi S, Unno M, Tanaka O, Kondo H, et al. Pancreatic beta-cell replication and amelioration of surgical diabetes by Reg protein. Proc Natl Acad Sci U S A. 1994;91:3589–92. doi: 10.1073/pnas.91.9.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Z, Wu H, Patel S, Zenilman M. Differential growth effect of regenerating (reg) protein on a rat β-cell line. Gastroenterology. 2001;120:A338–339. [Google Scholar]
- 23.Takasawa S, Ikeda T, Akiyama T, Nata K, Nakagawa K, Shervani NJ, Noguchi N, Murakami-Kawaguchi S, Yamauchi A, Takahashi I, Tomioka-Kumagai T, Okamoto H. Cyclin D1 activation through ATF-2 in Reg-induced pancreatic beta-cell regeneration. FEBS Lett. 2006;580:585–91. doi: 10.1016/j.febslet.2005.12.070. [DOI] [PubMed] [Google Scholar]
- 24.Kadowaki Y, Ishihara S, Miyaoka Y, Rumi MA, Sato H, Kazumori H, Adachi K, Takasawa S, Okamoto H, Chiba T, Kinoshita Y. Reg protein is overexpressed in gastric cancer cells, where it activates a signal transduction pathway that converges on ERK1/2 to stimulate growth. FEBS Lett. 2002;530:59–64. doi: 10.1016/s0014-5793(02)03398-7. [DOI] [PubMed] [Google Scholar]
- 25.Schliess F, Kurz AK, Haussinger D. Glucagon-induced expression of the MAP kinase phosphatase MKP-1 in rat hepatocytes. Gastroenterology. 2000;118:929–936. doi: 10.1016/s0016-5085(00)70179-x. [DOI] [PubMed] [Google Scholar]
- 26.Yamaoka T, Yoshino K, Yamada T, Idehara C, Hoque MO, Moritani M, Yoshimoto K, Hata J, Itakura M. Diabetes and tumor formation in transgenic mice expressing Reg I. Biochem Biophys Res Comm. 2000;278:368–376. doi: 10.1006/bbrc.2000.3813. [DOI] [PubMed] [Google Scholar]
- 27.Miyaoka Y, Kadowaki Y, Ishihara S, Ose T, Fukuhara H, Kazumori H, Takasawa S, Okamoto H, Chiba T, Kinoshita Y. Transgenic overexpression of Reg protein caused gastric cell proliferation and differentiation along parietal cell and chief cell lineages. Oncogene. 2004;23:3572–9. doi: 10.1038/sj.onc.1207333. [DOI] [PubMed] [Google Scholar]
- 28.Terazono K, Uchiyama Y, Ide M, Watanabe T, Yonekura H, Yamamoto H, Okamoto H. Expression of reg protein in rat regenerating islets and its co-localization with insulin in the beta cell secretory granules. Diabetologia. 1990;33:250–2. doi: 10.1007/BF00404804. [DOI] [PubMed] [Google Scholar]
- 29.Unno M, Nata K, Noguchi N, Narushima Y, Akiyama T, Ikeda T, Nakagawa K, Takasawa S, Okamoto H. Production and characterization of Reg knockout mice: reduced proliferation of pancreatic beta-cells in Reg knockout mice. Diabetes. 2002;51(Suppl 3):S478–83. doi: 10.2337/diabetes.51.2007.s478. [DOI] [PubMed] [Google Scholar]


