Abstract
High-throughput screening (HTS) is increasingly being adopted in academic institutions, where the decoupling of screening and drug development has led to unique challenges, as well as novel uses of instrumentation, assay formulations, and software tools. Advances in technology have made automated unattended screening in the 1,536-well plate format broadly accessible and have further facilitated the exploration of new technologies and approaches to screening. A case in point is our recently developed quantitative HTS (qHTS) paradigm, which tests each library compound at multiple concentrations to construct concentration-response curves (CRCs) generating a comprehensive data set for each assay. The practical implementation of qHTS for cell-based and biochemical assays across libraries of > 100,000 compounds (e.g., between 700,000 and 2,000,000 sample wells tested) requires maximal efficiency and miniaturization and the ability to easily accommodate many different assay formats and screening protocols. Here, we describe the design and utilization of a fully integrated and automated screening system for qHTS at the National Institutes of Health's Chemical Genomics Center. We report system productivity, reliability, and flexibility, as well as modifications made to increase throughput, add additional capabilities, and address limitations. The combination of this system and qHTS has led to the generation of over 6 million CRCs from > 120 assays in the last 3 years and is a technology that can be widely implemented to increase efficiency of screening and lead generation.
Introduction
In recent years, continuous advances in HTS technologies have combined with the increasingly challenging drug development environment1 to produce two seismic shifts in the use of HTS. First, the current generation of screening instrumentation (liquid dispensers, microplate readers, and control and analysis software), having especially evolved during the past decade,2,3 is so robust, user-friendly, and application-diverse that screening is being utilized to investigate entirely new areas of biology and chemistry.4 Second, with large pharmaceutical companies trying to manage the risk inherent in new targets and fragmented markets, early stages of drug development previously performed solely in biopharmaceutical companies are being carried out increasingly in academic institutions.5,6 HTS operations in the academic sector within the United States have benefited from a major investment by the National Institutes of Health (NIH) Roadmap in screening and chemistry center infrastructure, a small molecule repository that performs centralized procurement, quality control, and storage/distribution of the compound collection to the centers, and support for assay development, diverse compound library synthesis, and cheminformatics.7 The NIH Molecular Libraries Screening Centers Network of 10 HTS facilities was established in 2004 with the intramural NIH Chemical Genomics Center (NCGC) (www.ncgc.nih.gov); nine extramural centers were added in 2005.7 The network collaborates with individual academic investigators to develop chemical probes of biology and starting points for drug development; its scope, diversity, and policy of public release of screening data are unique (see http://mli.nih.gov).
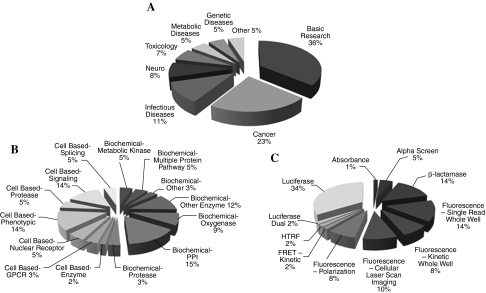
The NCGC's mission is complementary to that of the biopharmaceutical sector: it produces chemical probes for biology and target evaluation rather than drugs from validated targets; it focuses on the chemical biology of novel targets, rare or neglected diseases, and paradigms to increase the efficiency of the probe development process from assay development through screening, informatics, and medicinal chemistry; and it publishes its results and data with the explicit intention of enabling the chemical biology and drug development research communities to utilize it in their own research. This mission—particularly the focus on the > 90% of targets and diseases that are currently “undrugged” (i.e., no chemical modulator exists) and the need for screening data of sufficient quality to be of utility to the research community—dictated the screening technologies and paradigms implemented at the NCGC. The targets screened at the NCGC during the last 3 years are indeed quite distinct from the conventional “druggable genome” target classes that are the focus of the majority of drug development activity in the private sector (Fig. 1). Examples include rare genetic disorders, such as Gaucher's disease8 and β-thalassemia (PubChem BioAssay identifier [AID] 910), unconventional anti-infective targets such as anthrax toxin internalization (PubChem AID 912), pathway interrogations such as CRE signaling (PubChem AID 903, 168), protein-protein interactions like the BRCA1:pBACH protein carboxy-terminus–phosphopeptide interaction (PubChem AID 892),9 epigenetic gene regulation (PubChem AID 597),10 and various proteins of unknown function (PubChem AID 605, 886, 893). The NCGC developed its quantitative HTS (qHTS) technology to increase the efficiency of the probe development process,7 as well as to allow the population of a usable “chemical genomics” database given the well-known high false-positive and false-negative rates of conventional single-concentration screening.11 In this approach, compounds are screened in seven or more concentrations across an approximately four-log range of concentrations, mitigating these issues. As the dilution series is present on different plates,12 the loss of a single plate due to equipment problems rarely requires the rescheduling and screening of library plates because the remaining test concentrations, present on separate plates, are usually adequate to construct a reliable concentration–response curve (CRC) and thus assign activity. Conversely, inactive compounds are reliably assigned even in the presence of artifactual activity at a single concentration. Lastly, complex biological responses are readily apparent from the curve shape and are automatically recorded. qHTS shifts the burden of reliable chemical activity identification from labor-intensive post-HTS confirmatory assays to automated primary HTS and is therefore more efficient, but it requires more screening throughput. Thus the NCGC's screening infrastructure needed to be both unprecedently high-throughput and unusually flexible. These factors made the design of the NCGC's principal screening system particularly challenging.
FIG. 1.
NCGC assay portfolio during the 2005–2007 period. Breakdown charts include (A) disease areas represented, (B) target types screened, and (C) the detection methods utilized. GPCR, G-protein coupled receptor; PPI, protein-protein interaction.
In implementing a screening system, emphasis was placed on minimizing three factors that limit efficiency (i.e., productivity per unit cost and unit time): reagent use, system reliability, and requirement for human operator involvement. Stated differently, emphasis was placed on miniaturization and precision, reliability, and human-independent operation. To address the first, we chose to establish 1,536-well-based sample handling and testing as our standard; other plate formats should be usable but only in unusual circumstances.12 This, in turn, required high precision in liquid (reagent and compound) dispensing. We wished to eliminate the labor and reagent use associated with just-in-time compound library preparation for each screen, obviate the need for duplicate screening systems by utilizing a system with < 5% downtime (i.e., percentage of time system is unusable), and reduce the need for personnel by implementing an integrated, walk-away platform.
The system we implemented has allowed us to achieve all of these objectives. The technologies used in the system were developed at the Genomics Institute of the Novartis Research Foundation (GNF) (La Jolla, CA) and commercialized by Kalypsys, Inc. (San Diego, CA). Key design features include random-access on-line compound library storage carousels, extremely reliable plate handling, an innovative lidding system, multifunctional reagent dispensers employing solenoid valve technology, aspirators, a 1,536-pin array for rapid compound transfer, scheduling software, and fail-safe anthropomorphic arms for plate transport and delidding. A forerunner of this system has been described in a U.S. patent assigned to Aurora Biosciences.2 Though screening systems based on GNF/Kalypsys technologies have been utilized by several biopharmaceutical companies,13–15 the system at the NCGC was the first to be installed in a non-commercial organization. Over the past three years the NCGC has used this system to run a wide variety of assays8,9,16–23 (Fig. 1 and Table 1); this experience serves as the basis for this report.
Table 1.
Examples of Assay Target Categories and Detection Platforms Screened on the System
| Target type | Examples | Measurement type | Detection signal | Detector | PubChem AIDs |
|---|---|---|---|---|---|
| Profiling | Fluorescence, aggregation, luciferase inhibition | End-point, kinetic read | Fluorescence, absorbance, luminescence | ViewLux | 411, 587, 588, 590, 591, 592, 593, 594, 584, 585 |
| Biochemical | Enzyme reactions, protein-protein interactions, protein-ligand interactions | End-point, kinetic read, multiwavelength ratiometric | Fluorescence, absorbance, luminescence, fluorescence polarization, time-resolved FRET, FRET, Alphascreen | ViewLux EnVision | 360, 448, 603, 605, 875, 892, 893, 886, 879, 880, 887, 888 |
| Cell-based | Luciferase reporter gene, GFP induction, cell death | End-point, multireader, multichannel | Fluorescence, luminescence, object enumeration/scoring | ViewLux, EnVision, Acumen | 168, 357, 444, 445, 450, 451, 530, 540, 542, 543, 597, 662, 910, 912 |
In the present review, we describe the integrated NCGC screening system and our utilization of it to perform both routine and unusual assay and screening paradigms. Performance, reliability, and selection of peripheral units, including detectors, are described, as well as system performance, reliability, and examples of special assay situations. Generally applicable as well as system-specific lessons learned are detailed, including reasons for system limitations and failures and modifications/improvements introduced and planned to address them.
System Description
Overview and main components
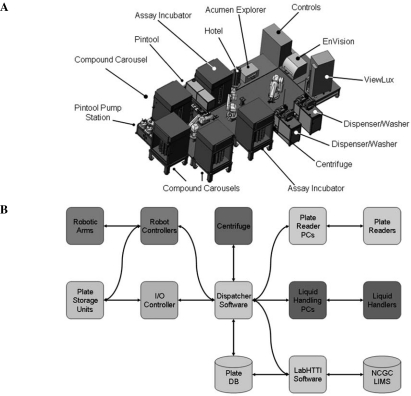
The NCGC robotic screening system is capable of storing compound collections, performing assay steps, and measuring various assay outputs in a fully integrated manner.24–28 It consists of peripheral units including assay and compound plate carousels, liquid dispensers, plate centrifuge, and plate readers, all of which are serviced by three high-precision Stäubli (Duncan, SC) robotic arms to execute hands-free biochemical and cell-based screening protocols (Fig. 2). The major system components and their functions are described below.
FIG. 2.
(A) System components and (B) controls. I/O, input/output; DB, database; LIMS, Library Information Management System.
Assay and compound plate storage
In its present configuration, the system has a total capacity of 2,565 plates, with 1,458 positions dedicated to compound storage and the remaining 1,107 positions dedicated to assay plate storage. Every storage point on the system is random access, thus allowing complete access to any individual plate at any given time. The total amount of compound samples that can be stored on the system is over 2.2 million, which represents approximately 300,000 compounds prepared as a seven-point concentration series.12 There is also area on the system to allow for the future expansion of compound storage capacity, as needed (see Conclusions and Future Directions). Three 486-position plate incubators capable of controlling temperature, humidity, and CO2 are present on the system. These multiple incubators allow for a variety of assay types to be run simultaneously, as each incubator can be individually controlled.27,28
Each incubator is built as a rotating carousel containing 18 columns, each having 27 plate positions. To access these positions, the gripper (discussed later) of the robotic arm pushes through VCR doors (designed in a manner similar to the spring-actuated front doors that have been in place in household videocassette players/recorders for decades) that help maintain the environmental control of the incubator. Each VCR door uses a simple spring-and-bearing mechanism to swing open when pressed, at which point a drawbar is actuated to hold the door down. Once a plate has been either deposited or retrieved, the drawbar is released, and the VCR door closes. Each incubator is controlled by programs running on the Adept Robotic Controller (Adept Robotics, Livermore, CA), with digital input and output lines used to set the carousel position for plate access and to return various status signals back to the controller (Fig. 2B). Five 27-position fixed hotels at ambient temperature can be used for additional plate storage or as intermediate locations during the course of a screen to minimize plate movements and optimize scheduling (one is shown in Fig. 2A).
Design and use of plates and lids
Because of the proprietary design of the dovetail joint fit that needs to be accomplished when the gripper handles the plates, the system can only be loaded with manufacturer-certified microtiter plates, all of which are produced based on an extrusion mold originally developed by GNF and Greiner Bio-One (Monroe, NC). These plates also require a featureless top to accept the rubber gasket-sealed plate lids (see below). Currently, these plates are supplied by Greiner Bio-One, Corning (Corning, NY), and Aurora Biotechnologies (San Diego). All common plate densities (384 and 1,536 well) and types are accepted and have been utilized on the system, including solid- and clear-bottom black and white assay plates, sterilized and tissue culture-treated plates, and polypropylene library storage plates. The plate's orientation is crucial for recognizing the compound and control locations; therefore, all plates handled by the Kalypsys system are barcoded with unique pairs of numbers, odd and immediate-increment even, on the two narrow sides, respectively.
Two types of plate lids are utilized in the system for covering compound library plates and assay plates (Fig. 3A) (described in detail by Mainquist et al.26). The assay lids differ from the compound lids primarily by an array of pin holes on their tops, with the latter designed for allowing air exchange while minimizing evaporation. The plate lids are machined out of a solid block of stainless steel and fitted with an autoclavable sealing rubber gasket. The rubber gasket is installed into a machined groove within the lid's periphery, and its shape and size are such that it can bend and partly spread over the plate edge under the weight of the metal lid, thereby increasing its plate contact area, ensuring isolation of the well samples from the outside environment, and minimizing the shared headspace over the wells. The lid weight of approximately 1 pound, combined with the gasket's features, provides an efficient seal. As there is no adhesive surface or locking mechanism involved, there is no limit on the number of times a plate lid can be removed and replaced.
FIG. 3.
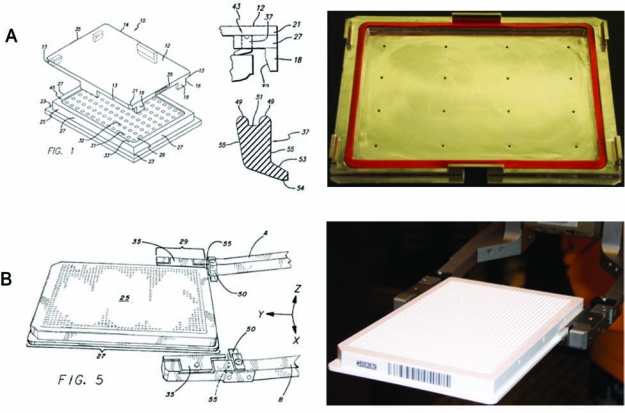
(Right panels) Plate (A) lid and (B) gripper. (Left panels) Highlighted from U.S. Patents 6,534,01426 and 6,592,324,25 respectively, are the plate lid and its flexible rubber seal and the plate gripper with its special groove designed for secure plate grasping and transport.
Plate gripping and transportation
Plates are transported to different stations on the system through the use of three industrial-grade six-axis Stäubli robotic arms, hereinafter referred to as cells, which have integrated grippers and barcode readers providing a high degree of accuracy and precision to the plate handling. The plate gripper (Fig. 3B) (described in detail by Downs and Weselak25 and Downs et al.29) contains grooves along the sides and back tabs to create a dovetail joint that prevents plates from being dropped by the robot when gripping a plate along the ridge on the bottom edges. The unique grooved pivot members (number 35 in Fig. 3B) are attached in a semiflexible fashion, and in combination with the two stop tabs (number 50 in Fig. 3B), the gripper can grasp the plate edges, thus aligning itself in all three dimensions before securely gripping the plate in preparation for movement. Since first becoming operational in August 2005 until the present, no plate has ever been dropped by this system. A barcode reader, connected to each gripper, scans every plate immediately prior to pickup, and the data are returned to the Adept Robot Controller. The plate gripper is connected to an air release wrist that will disengage in the event of a collision and trigger an emergency stop of the system.
Each Robot Controller runs a real-time operating system capable of executing multiple programs simultaneously. The Robot Controller communicates with the Dispatcher screening control software via Transmission Control Protocol (TCP)/Internet Protocol (IP) (see System control and monitoring and Fig. 2B). Movement programs are downloaded to the controller from the Dispatcher PC via a file sharing protocol. Each Robot Controller has multiple digital inputs and outputs to control various electrical components on the system, in addition to RS 232 ports to control external devices such as the barcode reader on each robot gripper. Plate handoff and de-lidding stations are used to pass plates between cells on the system. De-lidding stations are present at various points of the system for the removal of plate lids for operations such as a liquid dispense, compound transfer, or a plate reading on a detector.
Compound transfer
A Pin Transfer Station performs direct 1,536-compound to 1,536-assay plate compound transfer, with each slotted pin transferring approximately 23 nl of compound from a source plate into a destination plate.30 Respectively, each compound plate contains up to 1,408 compound samples located in columns 5–48. Typically, assay-specific controls are placed in multiple wells within the left four columns of the assay plate and sourced out of a dedicated assay-specific control plate. The controls-only plate is prepared in multiple copies (each plate has enough material to transfer into approximately 250 plates) to sustain a full collection screen of currently over 1,000 plates. To wash the pins, there are three consecutive solvent baths and a drying station incorporated into the deck. The first, second, and third baths contain dimethyl sulfoxide, 1:1 (vol/vol) isopropanol:water, and methanol, respectively. The washing protocol combines multiple dips, as well as soaking periods, into the baths, the parameters of which can be modified by the user. After the pins are soaked in each bath for a controlled period of time they are finally moved into the drying station for a prescribed time.
Liquid dispensers
Two solenoid “bottle valve” dispensers are used for reagent addition and liquid removal via aspiration. Each dispenser has two heads, each with eight tips capable of a dispense volume range of 200 nl to 20 μl (coefficient of variance [CV] < 10%). One head dispenses vertically onto the bottom of the well, and the other at an angle of approximately 45° into the back wall of the well. Most often the vertical-dispense head is used unless there is a special circumstance, for example, if a reagent contains detergent and bubble formation is a concern, or if fragile cell layers could be disrupted by a 90° direct-dispense of a reagent stream (see below for further discussion). Additional variables that can be modified to accommodate different reagent types or specific needs include bottle valve pressure, number of tips used (one, two, four, or eight), predispense volume, and motion control (serpentine or carriage-return schemes). Volume or gravimetric scaling can be a tedious process when multiple tips are used in a protocol, but this ensures each tip dispenses the same volume into the wells. If an exceptionally high degree of uniformity is sought (better than 10%) or if time permits, a single-tip dispense can be used to gain well-to-well dispense precision. The dispense time is ∼1–3 min per plate, depending upon the specific protocol. Having two dispensers on the system is very useful in splitting up the supply of the assay's reagents to increase productivity of the screen by reducing resource competition.
Each dispenser has a unique aspiration head with 32 thin-walled stainless-steel tubes to enable column-wise liquid removal out of a 1,536-well plate. Optimizing aspiration involves adjusting several factors: vacuum flow level, aspiration-tip well depth, and dwell time. The vacuum flow level for the aspiration head is controlled by manually adjusting a needle valve, and dwell time and aspiration well depth are software-controlled. This provides precise control over the volume of liquid removed from each well (CV < 10%). The combination of high-precision positioning (within 0.1 mm) and the high speed of the aspirator head and plate stage allow the liquid to be removed from the entire 1,536-well plate within only 90 s. This aspiration head has been used for cell washes and/or fixing.15,16
Detectors
The system currently integrates three different types of detectors, enabling a wide variety of assay formats. The EnVision™ and ViewLux™ detectors (PerkinElmer, Waltham, MA) together cover almost the entire spectrum of fluorescence, absorbance, and luminescence measurement techniques used in HTS. The ViewLux is a multimodal charge-coupled device (CCD)-based imager capable of ultrafast detection of luminescence, fluorescence intensity, absorbance, time-resolved fluorescence resonance energy transfer (FRET), and fluorescence polarization signals. Read times are very short, sometimes lasting a total of 30 s (including plate transport into and out of the reader), which makes this instrument convenient for HTS. The EnVision multilabel photomultiplier tube (PMT)-based plate reader is highly customizable and capable of detecting many wavelength regions, but unlike the ViewLux, the EnVision can be used for measuring AlphaScreen® assay outputs, increasingly utilized in HTS,31–39 and allows bottom plate reading, a requirement for cell-based β-lactamase reporter assays, which is not straightforwardly accommodated by the ViewLux once integrated onto a screening platform. The third detector, the Acumen® Explorer (TTP LabTech, Royston, UK), is a PMT-based microplate cytometer that uses laser line scanning to image and enumerate fluorescent characteristics from individual cells,40 complementing the aggregate-type (e.g., total well signal) outputs of the other two detectors.
System control and monitoring
The system is instructed to run a screen via two files: Method and Assay. The Method File contains all protocol steps of the assay, such as dispensers utilized, dispense protocols, incubations, plate centrifugation, and detector reads. Methods are created using an application developed by the vendor to control each step of the assay, stored as a Method File. The Assay File is a “.csv” file that lists the barcodes of all assay plates for the screen, in addition to all library plates of compounds to be pin-transferred to an assay plate. During the screen, those assay plates are run in the precise order specified. Assays are controlled and monitored using vendor-supplied software called the Dispatcher.
Screens are started from the Dispatcher by loading the Method File describing the process and the Assay File listing the plates designated for the screen. Multiple assays can be conducted simultaneously, limited by the shared resources required for each assay scheduled (see Examples of screening strategies). Data files generated from the detectors are saved locally to the Dispatcher PC. We have developed an application to control the Dispatcher as well as adding more functionality, primarily dealing with generated data and trace files (discussed later). Errors that occur while the system is running are displayed on the monitor, and staff can be paged via an automatic dialing system. Internal software allows for error messages to be reported via e-mail, using the Outlook Scheduler account so that there is a record of any runtime errors.
Backup power supply
Power requirements for robotic rooms are derived from a complex mixture of computers and motors that create an adverse combination of sensitive electronics with noise-producing devices. The NCGC system requires a 100-kVA uninterrupted power supply (UPS) (480 V at 100 A, three-phase) and we have installed two of these at our site. It is expected that the robotic system will operate at 30–40% power utilization, but the 100 kVA will ensure full support. The system has to be on a backup generator as the UPS will only act in a “ride-through” capacity with a typical 5–15-min runtime being sufficient. In addition, the large robotic arms can produce sizable but short-lived (<100 ms) in-rush currents at system power-up; therefore, the UPS should be able to handle start-up in-rush currents of several times the system's rated capacity or be equipped so that the UPS can be put into “bypass” mode for system start-up. Additionally, we ensured that all other utilities required by the Kalypsys system have separate back-up support (e.g., compressed air, vacuum, CO2, and N2). Further, most power outlets inside the robotics lab are on UPS to ensure that peripheral instruments and the computers that control them are covered.
System Utilization for qhts
Assay technologies and detector selection
The automated screening system at NCGC is particularly flexible with respect to integration of peripheral detectors. However, once the instruments have been software-integrated and physically bolted to the robot platform, they cannot be removed or exchanged freely. Thus, a careful selection of plate readers is necessary to maximally utilize the limited floor space while being capable of addressing the numerous and changing needs for signal detection. Our selection of the ViewLux as the main reader was driven primarily by the fact that it is the fastest such detector available and has been established as an industry standard during the past 6 years. The EnVision fills an important niche in enabling AlphaScreen assays, the easy selection of bottom versus top read, the wider choice of detection wavelengths, and simultaneous two-channel detection, while also serving as a backup to the ViewLux. Together, these two readers account for approximately three-quarters of the data generated on the system.
For bridging the gap between the plate readers mentioned above where the entire well signal is quantified (i.e., population averaged) and the high level of image complexity and resolution collected by CCD-based wide-field and confocal-based high-content screening systems, we have pioneered the application of laser scanning cytometry to automated robotic systems using the Acumen Explorer and the next-generation three-laser eX3 (TTP LabTech). The eX3 is equipped with three lasers (405, 488, and 633 nm) and four PMTs, allowing for multi-parametric population distribution analysis of fluorescence events on a cell-by-cell basis (the whole well area is measured) with 1,536-well plate read times as low as 7 min. Scan speed and data acquisition are effected by the x and y resolution. Because only one section of the plate is read at a time, as opposed to the ViewLux where the entire plate is imaged at once, scan times are typically 10 min per plate. This microplate cytometer can monitor up to four fluorescent signals from single objects using one laser for excitation; however, if all three lasers are used theoretically 12 different signals could be collected from a single well. We have found the Acumen and eX3 to be useful in the measurement of fluorescent protein expression, cell shape, or simple cellular redistribution events such as cytoplasmic-to-nuclear translocation.16 Examples of the utilization of this unique microplate cytometer are provided in later sections.
Optimization to robotic screen
Initially the screening scientist completes assay miniaturization and optimization using stand-alone equipment similar or identical to the stations on the robotic platform. Two assay validations are typically performed; the first using stand-alone equipment (“off-line”) and the second using the integrated robotic system (“on-line”). Both validations are a qHTS of the Library of Pharmacologically Active Compounds (LOPAC)1280 collection of annotated bioactives at seven or eight fivefold dilutions typically beginning at > 40 μM. Once the off-line validation has been completed successfully a robotic validation is scheduled.
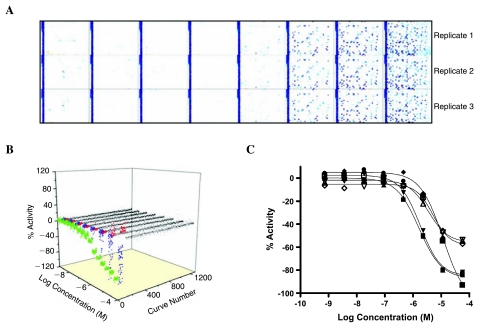
Use of the LOPAC or a similar collection for validation offers several advantages: known modulators of the assay target or pathway present in the library can determine the biological validity of the assay, compounds of higher potency and efficacy can be identified for use as assay controls, and the assay's sensitivity can be assessed by the number and potency of actives identified. While many assays are not significantly modulated by LOPAC library members, often sufficient perturbation of an assay response is observed at the high compound concentrations tested in the qHTS to aid in the evaluation of the process's precision (see below). After successful off-line validation, the project team biologist consults with the HTS Operations Core to discuss the assay protocol for implementation of a triplicate LOPAC validation on the robotic screening system. The on-line validation tests the assay protocol that will be used for the full screen and will determine the appropriate configuration of the robot to ensure high throughput. The triplicate LOPAC qHTS allows assessment of assay repeatability by comparing the potencies of actives from each LOPAC run as well as measure of assay performance (such as signal:background [S:B] ratio and Z′ of control wells and 50% inhibitory concentration and minimum significant ratio [MSR] of control titrations41) across the entire qHTS validation (Fig. 4). Successful validation requires sufficient and stable S:B ratio, CV, and Z′ over the entire run. Furthermore, for triplicate LOPAC validations, the potencies of actives must correlate well between each of the runs (i.e., fall within the 95% confidence interval on correlation plots). If the on-line validation fails, assay conditions are modified, and the validation is repeated.
FIG. 4.
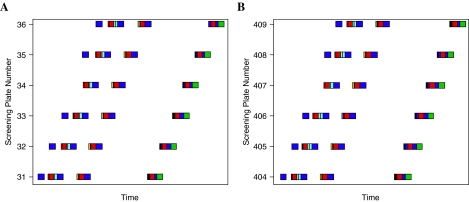
Examples of validation data. (A) Plate activity heatmaps of an eight-concentration LOPAC screen repeated three times. Concentrations are shown from lowest to highest. Each plate contains an intra-plate control titration. (B) Activity of validation run shows samples identified as inhibitors (blue), activators (red), or inactives (gray); control titration (green) presents as near-overlapping curves indicating excellent assay stability and reproducibility (control 50% inhibitory concentration = 2 μM, control MSR = 1.9). (C) An example of three different compounds showing excellent triplicate CRC reproducibility: samples A(1) (▪), A(2) (▴), A(3) (▾), B(1) (♦), B(2) (•), B(3) (□), C(1) (▵), C(2) (▿), and C(3) (◊).
The 1,536-well plate format offers a wealth of possibilities for inclusion of information-rich controls, above and beyond those typically utilized in 96- and 384-well plates. In lower plate densities, such as the 96-well plate, the eight or 16 wells allocated to low and high normalization controls are frequently insufficient to provide useful statistics during large-scale screening. By comparison, one column of eight wells in a 96-well plate is equivalent to four columns totaling 128 wells in a 1,536-well plate (16 96-well source plates can populate one 1,536-well final plate).12 This 16-fold increase in available wells makes it possible to add information content to each assay plate by further partitioning the control area. The 128 available wells can be apportioned between the most commonly used control pairs (basal/stimulated, with or without enzyme, and free/bound tracer), various levels of control stimulus (10%, 50%, or 90% effective concentration), calibration curves of an appropriate fluorophore to better assess for example enzymatic product formation, or a titration series of a control antagonist or agonist. Control titrations, while not required for signal normalization, are useful for measuring “the pulse” of the assay. For example, the stability of assay reagents and sensitivity to a control compound can be monitored by the MSR of the control titration for each assay plate throughout the screen progression.9,20,41
As the time required for running an on-line validation is much less than that required to complete a full-library screen, numerous on-line validations can be run quickly within a given period, thus creating a queue of ready-to-go full-library screens and minimizing idle time of the screening systems. Assays that pass the on-line validation are presented internally for review and approval for screening of the entire collection.
Prescreen checklist
The standard operating procedure developed at the NCGC is guided by a “checklist” containing steps that must be completed by both the biologist and the HTS Operations group prior to initiation of a screen. The biologist ensures that all of the required plates and reagents to run the screen are present and available and that the on-line detection instrumentation needed for the screen has the necessary protocols and optics configured. Members of the HTS Operations group make certain that the selected screening system is in proper working order, with a specific checklist for every piece of instrumentation and any specific chemical library modifications. Once both the biologist and the HTS Operations group are satisfied with the prescreen preparations, the actual screen can start.
Screen data processing
The LabHTTI software (discussed later; see Controller hardware and software enhancements) is integrated with the plate database used by the Dispatcher software, so the complete inventory of the compound library can be tracked by the NCGC Laboratory Information Management System. During a screen, instrument data files are streamed into a central server, and key assay parameters are made available to the staff to judge daily performance of ongoing screens. After the screen, raw data are loaded in our system, and the plate log is processed to link assay plates with compound plates. A predefined analysis template is used in conjunction with a flexible built-in formula engine to create calculated data layers that synthesize readouts together (for example, computing ratios of two optical channels) to yield a single activity. This is particularly useful for multichannel data as it allows different phenotypes or readouts to be combined dynamically to create summarized values for each well. Next, the layers undergo automated correction of systematic errors that arise during screening. Insertion of vehicle-only control plates uniformly throughout the course of a screen allows systematic artifacts such as signal drift and dispense variability to be corrected. Once data layers have been corrected, plate-well-based data are converted to sample-oriented titration–response data. ActivityBase is used for the compound and plate registrations, while an in-house database and software tools are used for compound concentration mappings and building sample titration relationships. Automated fitting and characterization of titration curves are performed using an algorithm developed in-house (http://ncgc.nih.gov/pub/openhts/).
Examples of screening strategies
Time course-based screens
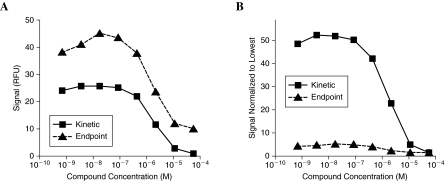
The high speed of plate measurement afforded by the ViewLux has made it possible to acquire time interval or “kinetic” data for a number of screens. In the past, semiautomated screening has involved processing assay plates in batches, typically loaded into readers by plate stackers, limiting data collection to primarily end-point measurements. For fully automated screening systems, collection of time course data is conceptually possible but is performed infrequently because of slow plate reader speeds and the complexity of processing multiple time points. With our system, we utilize time course assay data collection whenever possible for several reasons. First, time course measurements of reaction progress are inherently more robust and provide a more accurate quantification of the reaction rate. Second, computation of reaction rate based on signal change results in markedly improved S:B ratios and thus enables screening of traditionally difficult systems such as chromogenic assays associated with small increase in absorbance42 and well-to-well volume and meniscus variation, or assays involving weak natural fluorophores like NAD(P)H (Fig. 5).20 Third, for a number of fluorogenic assays operating in the blue-shifted region of the ultraviolet–visible spectrum, collecting kinetic data minimizes the interfering effect of compound autofluorescence.20,21
FIG. 5.
Implementation of kinetic assay (▪). In enzyme assays associated with minimal or noisy signal changes (due to dim fluorophore or otherwise unfavorable assay chemistry or enzymology), the end-point method (▴) may simply preclude the use of these assays in HTS (concentration–response plot using raw signals in [A]). The boost in S:B afforded by performing kinetic measurement and computing the activity using the change in signal (concentration–response plot using normalized signals in [B]) transforms many of these weak signal systems into screenable assays. RFU, relative fluorescent units.
While the benefits of time course reads may be intuitively obvious, the practical implementation of kinetic-read assays requires a balance between the total time for multiple reads of each plate and the need to maintain high throughput. Currently, pintool transfer (of both compound and control plates) or single-tip dispenses are the slowest steps on our system, taking between 2.5 and 3.5 min, including arm movements. For kinetic assays for online screening, we restrict plate residence time inside the ViewLux to 5 min or less, including plate entry and exit. This requirement leads to two screening strategies. Enzyme assays associated with a relatively fast signal change are run as one continuous time course measurement, for example, by acquiring eight data points every 30 s.20 Slower reactions are handled in a discontinuous manner by measuring the signal immediately after reaction initiation, incubating the assay plate at a designated location (assay incubator or auxiliary hotel; see Assay and compound plate storage), and returning the plate to the reader for collection of the second data point. In the slower assay case, only two data points are typically collected per reaction, but these are sufficient to calculate a signal change (PubChem AID 893).
Time course data, especially combined with concentration–response screening, have allowed us to rapidly and reliably distinguish enzyme inhibitors from false-positives or mixed-effect compounds acting as assay signal attenuators. We have developed data storage and processing algorithms that allow the combined analysis and viewing of both activity, as calculated from the slope or difference of the signal change and the starting datum value (y-intercept) associated with each compound, at each concentration. In this manner, compounds that exhibit high fluorescence or absorbance are identified by the anomalous shift of the assay progress curves.
The contentious screen: multiple timed dispense and read steps
In certain screening assays, only end-point data collection is possible because of the requisite addition of detection reagent added at the end of a reaction. The combination of multiple reagent dispenses, such as substrate and stop/developing solution, the timing of these steps, and the collection of several separate reads per plate can create an unusually high level of contention for the robotic arm that services both the dispensers and readers.
We encountered such a “busy screen” with an endpoint assay for DNA polymerase holoenzyme in which the double-stranded DNA product was detected by PicoGreen® (Invitrogen, Carlsbad, CA) staining (PubChem AID 603). In order to produce a more informative data set, we measured fluorescence after compound addition (to record fluorescent library members) and after the end of the enzymatic reaction but before PicoGreen addition (to identify profluorescent DNA intercalators). The convergence of two timed reagent dispenses (substrate dispense following a 15-min incubation of compounds with the enzyme and PicoGreen addition taking place after 35-min enzyme reaction) and three coupled plate reads (first before substrate addition, a second read before, and the last read after PicoGreen addition) required a high degree of optimization in order to maintain the throughput of the screen. This was achieved by the careful calibration and utilization of all 16 available tips across both heads of one of the liquid dispensers to deliver the substrate and stop solutions at the highest speed possible. Additionally, device dependencies were configured such that highest priority was given to the stop solution dispense precisely 35 min after the substrate was delivered to ensure constant reaction time for all microtiter plates. As is evident in Fig. 6, the screen of over 400 plates proceeded successfully, with the timing and plate sequence maintained throughout the operation.
FIG. 6.
“Busy screen.” The multiple dispense, incubation, and read steps are indicated on the Spotfire (TIBCO Software, Somerville, MA) plots: Dispense ( ), Incubate (
), Incubate ( ), Read (
), Read ( ), Read2 (
), Read2 ( ), Read3 (
), Read3 ( ), and Transfer (
), and Transfer ( ). The order and timing of all protocol steps remained stable throughout the screen, as evidenced from the near-identical snapshots obtained from six (A) early and (B) late screening plates.
). The order and timing of all protocol steps remained stable throughout the screen, as evidenced from the near-identical snapshots obtained from six (A) early and (B) late screening plates.
Interleaved screens
On limited occasions, two screens sharing the same overall protocol but utilizing different reagents have been processed concurrently in an interleaved fashion so that each compound library plate is tested against the two assays in rapid succession. A recent screen involved a probe displacement fluorescence polarization assay for inhibitors of a protein–phosphopeptide interaction. To assess the effect of library compound fluorescence on the assay performance, we screened the library against separate fluorescein- and rhodamine-based fluorescence polarization assays with each compound being tested in the two assays at immediately adjacent time points (PubChem AID 875 and 892).9 This requirement presented a challenge because the robotic software did not contain a function allowing explicit protocol-to-protocol dependencies. That is, the operator could not specify to the robot to process every compound plate pairwise against the fluorescein and immediately afterward against the rhodamine assay protocol. Further complications arose because the two assay protocols utilized shared resources, such as the pintool transfer station, the ViewLux reader, and the robotic arms, and as such could not be run in parallel.
To circumvent this software limitation, we interleaved the two screens by adding a preincubation time to the second assay in order to shift its starting point enough to allow uninterrupted passage of alternating-assay plates through the system. The preincubation time was determined by the pintool-mediated compound transfer, the rate-limiting step utilizing a shared device. While this spacing resolved the contention problem with the first shared resource, additional steps were taken to ensure that contention for the shared robotic arm and the ViewLux reader at the end of the assay protocol would not lead to “throwing off” the interleaving and accumulation of “lagging-assay” plates. We thus implemented a “zero pad” by appending a series of extra plates to the end of each screen. Those assay plates received dimethyl sulfoxide in place of library compounds and served as ending sequences to both screens to absorb any lagging plates that appeared because of occasional shifts in the interleaving pattern. This approach is based on the zero padding applied in digital signal processing where a string of zeros is added to the end of a time domain signal sequence to increase the resolution of the frequency domain sampling.
Role of dispense head angle
As stated earlier, the integrated nanoliter dispensers offer a selection of vertical-and angled-tip arrays. The rational choice for dispensing reagents into wells containing sensitive cell monolayers or suspensions is the angled head due to its ability to send the incoming reagent stream in a manner that is least disruptive to the cells. A mismatch of experimental conditions therefore occurs when prescreen assay development and validations are performed on different dispenser types. Traditionally, our off-line assay validation protocols often rely heavily on the BioRAPTR™ Flying Reagent Dispenser™ (FRD™; Aurora Discovery, presently Beckman Coulter, Fullerton, CA) to deliver low volumes of reagents. Reagents are dispensed at a 90° angle directly above the well surface, at a velocity meant to induce mixing of well components and prevent air bubbles from forming. While this delivery mechanism is ideal for many assays that require thorough mixing upon reagent addition, some assays may benefit from less vigorous mixing of reagents within the well. An example of one such assay is a β-lactamase reporter gene assay where suspension cell lines were employed. The assay used Jurkat cells expressing the Ml receptor with a nuclear factor of activated T-cells–β-lactamase reporter (GeneBLAzer™, Invitrogen, Carlsbad, CA), and we used the next-generation Acumen, the eX3 laser-scanning microplate cytometer, as the detector. One of the main advantages of using the eX3 for fluorescent cell-based assays is that it allows the user to define fluorescent cell populations within a well, while disregarding background fluorescence and autofluorescence of objects in the well other than cells (e.g., lint contamination). We observed that delivery of reagents (assay buffer or loading dye) at a 90° angle directly above wells with cells in suspension caused cells to move to the outside of the wells and clump together (Fig. 7, top right panel). This occurred even with delivery of as little as 1 μl of reagent. Upon reading the plate using the eX3, we found that only about 20% of the 750 cells per well seeded were counted as individual cells and thus included in the data analysis. The majority of the cells were clumped together and could not be distinguished as part of the cell population.
FIG. 7.
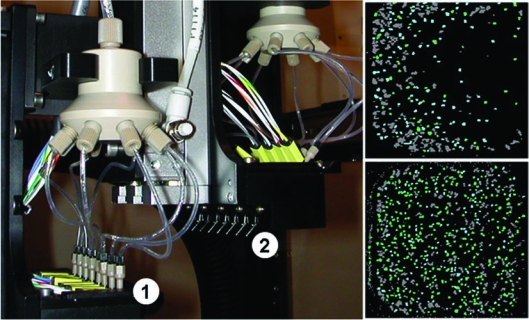
Use of the Kalypsys angled-head dispenser. (Left panel) The array of eight tip dispensers for the straight head (1) and the angled head (2). (Right panels) Images from the whole-well scan of a 1,536-well plate obtained on the Acumen Explorer using GeneBLAzer M1 receptor/nuclear factor of activated T-cells-β-lactamase expressed in Jurkat cells, a cell line grown in suspension. Reagents were dispensed with either (top panel) a straight head or (bottom panel) the angled head. Cells were plated at a density of 750 cells per well. Cell clumps are shown in dark gray, while individual cells included in the cell population are light blue or green depending on the level of β-lactamase expression. (Top panel) Reagent addition to wells at 90° by the Bio-RAPTR FRD causes suspension cells to move to the sides of the well and clump together. High content data analysis that relies on defining individual cells thus becomes difficult, as fewer individual cells are counted. (Bottom panel) Addition of reagents with the Kalypsys angled-head dispenser does not cause significant movement of suspension cells within the well, as reagent is dispensed at an angle at which fluid hits the side of the well and runs down to the bottom. Cells, therefore, remain dispersed throughout the well volume and show significantly less clumping, providing a significantly higher number of individual cells to be analyzed and counted.
In an effort to include a greater percentage of the cells plated in the data analyzed, the Kalypsys angled-head dispenser (Fig. 7, left panel) was employed to deliver the reagents at a 45° angle. In this case, the reagents are dispensed such that the volume contacts the side of the well. We found that addition of reagent to the wells in this manner helped to prevent cells clumping at the well perimeter and allowed the cells to remain dispersed across the well (Fig. 7, bottom right panel). Therefore, a significantly greater percentage of cells plated were counted as part of the cell population—approximately 70% of the 750 cells per well.
Asymmetric concentration-response screens
The online storage of our entire collection in the form of inter-plate dilution series and the random-access option with respect to compound plates have allowed us to not only realize concentration-response screening, but to also utilize the library in new ways to maximize the information output from a screen while minimizing its cost. An example is the customization of the titration curve based on the underlying biology/biochemistry of the assay. Enzyme assays, where the target is often employed at low nanomolar concentration, typically allow the detection of a broad range of compound potencies, from tens of micromolar to low nanomolar. The same logic applies to cell-based assays where, depending on the exact biology and detection format, compound potencies in the low nanomolar range are often observable. In those cases, performing a qHTS on all available concentration points is highly relevant and necessary to enable a high-confidence structure-activity relationship analysis. Conversely, low-affinity binding assays where target and labeled ligand/tracer are present at high concentration (e.g., ≥ 100 nM) do not require testing of the library at the lowest concentrations but rather call for a highly customized concentration-response testing, ideally shifting the titration to higher compound concentrations.
The random-access feature of the present screening system makes the realization of the above scenario as simple as selecting a series of barcodes out of an inventory list. We recently performed a qHTS of a weak binding interaction involving a complex between fluorescently labeled 2 μM ligand and protein used at 11 μM (PubChem AID 605). Since no compound activity was expected below the single-digit micromolar level, we omitted three of the low-concentration library plates and instead introduced a new, twofold higher top concentration point by performing a double pin-transfer of compound solution.12 By selectively biasing the concentrations tested, we conserved protein reagent and generated appropriately right-shifted titration data set to allow better curve fitting through the compound responses.
Use of laser cytometry
Incorporation of the Acumen Explorer (and later eX3) on our system provides access to assays requiring high content or population distribution analysis. When deciding on a high content screening system for the robotic platform we considered several factors. The reader must support data acquisition from typical 1,536-well plates. The system should provide rapid throughput to support screening of approximately 200,000 samples per day, and data file sizes and analysis time should be minimized. The Acumen Explorer and eX3 come equipped with an F-theta scan lens that has a depth of focus sufficiently large (25–30 μm) to compensate for variations in the flatness of plastic microtiter plates that allows for rapid data acquisition times as on-the-fly focusing is not required. The x resolution can be set at predetermined intervals, and the y resolution is set by the user; we have found that a 1- × 8-μm resolution is sufficient for most assays40 (Fig. 8). At this resolution, 200 whole well scans per min can be achieved that gives a plate throughput of approximately six plates per hour or 200,000 samples per day. The amount of image data saved can also be determined by the user. In one mode, a 1- × 8-μm whole well scan of a 1,536-well plate generates a 50 Mb file as the scanned images are saved. However, the desired end-point measurements can be selected so that only the relevant data are saved in a file size < 200 kB per plate. This eliminates both the need to store large image files and subsequent imaging processing time.
FIG. 8.
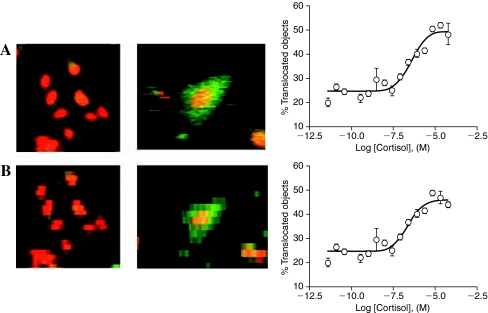
Effect of cytometer scan resolution on CRC determination. A 1,536-well plate assay for glucocorticoid receptor nuclear translocation was performed by fixing U2OS cells and staining nuclei with propidium iodide. The amount of GFP signal within the nucleus was then measured on the Acumen laser cytometer. Representative images of untreated U2OS cells stained for nuclei (left panels) (red is the propidium iodide channel) or cells (middle panels) (green is the GFP channel; red is the propidium iodide channel) are shown with CRC data (right panels) obtained when scanned at either (A) 1 × 0.5 μm or (B) 1 × 8 μm resolution.
The Acumen Explorer and eX3 are ideally suited for fluorescent protein-based assays, and the majority of assays performed on this instrument (10% of the total assays) used either green fluorescent protein (GFP) or GFP/red fluorescent protein (RFP) reporter systems. We have measured both GFP- and RFP-expressing cell lines, including dual reporter systems where both GFP and RFP are expressed in the same cell line to provide a ratio-metric assay signal. In this case, one reporter serves to monitor cell numbers and nonspecific effects on the reporter system, while the other reporter provides a target-specific signal. Such a ratiometric assay was performed on the Acumen where the specific signal occurred in approximately 50% of the cells under the assay conditions. The optimized protocol used a 5-μl assay volume and a 30-h incubation time, and the robotic screen against approximately 1 million wells showed a Z-factor of 0.66 with an average S:B ratio of 2.2. We have also utilized the washing feature of the Kalypsys dispenser to fix and stain adherent cells for the purpose of Acumen-based detection. For example, we have assayed nuclear translocation of the glucocorticoid receptor in U2OS cells using an optimized 1,536-well plate washing protocol followed by staining of nuclei with propidium iodide (Fig. 8).16
System Device Reliability
Overview
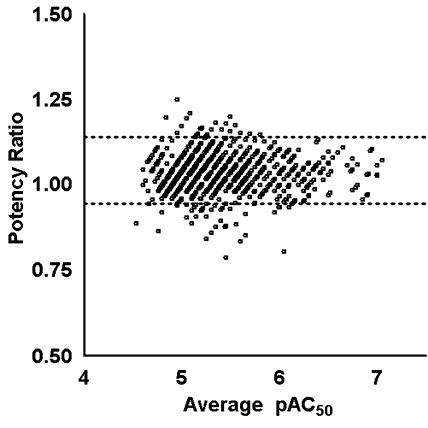
During the three years of operation, the system has been used in over 200 screens ranging from small-size validations of 10–30 plates to full-collection screens utilizing over 1,300 plates (Table 2). The reliability and reproducibility of our qHTS processes are indicated by the excellent correlation of 50% active concentration (AC50) data derived for a pyruvate kinase-luciferase-coupled biochemical assay performed in September 2005 and again in March 2007 (Fig. 9). Separate copies of the library were obtained, diluted, and stored on the robotic system in each screen. Both the 2005 and 2007 qHTS runs were performed with fresh copies of the library (less than 2 weeks after plating). The AC50 values derived from 3,260 active samples exhibited excellent reproducibility across the entire potency range (MSR = 1.24).41
Table 2.
Examples of Screening Assays and Performance
| Index | Assay name (PubChem AID) | Detection (number of data layers) | Assay target type | Detector | 1536-Well plates screened | Total wells screened | CRCs generated | Average Z′ | Rate-limiting step | Total screening time (h) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Prx2 (448) | Fluorescence: kinetic whole well (16) | Biochemical: multiple protein | ViewLux | 453 | 671,232 | 71,028 | 0.76 | Dispense and read linked | 75 |
| 2 | BRCA1-Green (875) BRCA1–Red (892) | Fluorescence polarization (3) | Biochemical: protein-protein interaction | ViewLux | 943 | 1,443,840 | 151,104 | 0.87 | Compound transfer | 54 |
| 3 | Luciferase (411) | Luciferase (1) | Biochemical: other enzyme | ViewLux | 450 | 665,624 | 72,365 | 0.92 | Compound transfer | 24 |
| 4 | IκBα (445) | Luciferase-dual (2) | Cell-based signaling | ViewLux | 748 | 1,926,344 | 118,049 | 0.52 | Plate read | 62 |
| 5 | CRE (662) | β-Lactamase (2) | Cell-based signaling | EnVision | 492 | 755,712 | 74,000 | 0.60 | Plate read | 150 |
| 6 | AmpC + detergent (584) AmpC − detergent (585) | Absorbance: kinetic (12) | Biochemical: other enzyme | ViewLux | 870 | 1,336,320 | 141,126 | 0.80 | Dispense and read linked | 116 |
| 7 | Inositol monosphatase (901) | HTRF (2) | Cell-based signaling | ViewLux | 883 | 2,165,760 | 123,824 | 0.61 | Compound transfer | 54 |
| 8 | β-Thalassemia (925) | Fluorescence: protein laser scan imaging (9) | Cell-based splicing | Acumen | 497 | 763,392 | 114,484 | 0.73 | Plate read | 142 |
| 9 | Glucocerebrosidase (360) | Fluorescence: single read whole well (1) | Biochemical: other enzyme | ViewLux | 365 | 611,318 | 59,815 | 0.67 | Plate read | 37 |
| 10 | Heat shock protein 90 (595) | AlphaScreen (1) | Biochemical: protein-protein interaction | EnVision | 477 | 732,672 | 71,974 | 0.65 | Plate Read | 83 |
HTRF®, homogeneous time-resolved fluorescence (Cisbio International, Bagnols/Cèze, France).
FIG. 9.
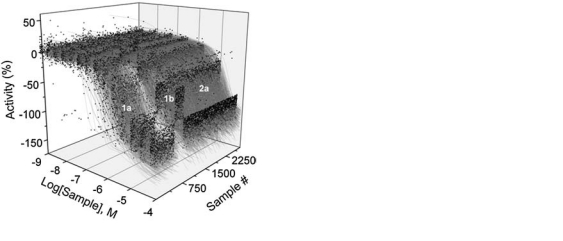
Reproducibility of the qHTS process on the Kalypsys robotic system. (Left panel) 3D-scatter plot with CRC fits shown for samples assayed in the pyruvate kinase-luciferase coupled assay. CRCs are annotated by curve fit quality as described in Inglese et al.11 (Right panel) Bland-Altman plot comparing the pAC50 values from a pyruvate kinase-luciferase-coupled biochemical assay performed in September 2005 and again in March 2007. Separate copies of the chemical library were obtained, titrated, and stored on the robotic system in each screen. Both the 2005 and 2007 qHTS runs were performed with freshly titrated copies of the library (less than 2 weeks). The pAC50 values derived from the high-quality CRCs from each dataset are plotted (MSR = 1.24, n = 3,260 samples).41 The 95% limits of agreement are shown as dotted lines.
The system and its components have proven to be unusually reliable, allowing the high degree of productivity described. In the last full year of operation, for example, the system was down only 11 days, 6 of those for scheduled maintenance. At the same time, no highly complex automated system can operate without failure, particularly when used in different modes and run every week for up to 7 full days of uninterrupted screening. Even the most well-designed systems can fail for unforeseen reasons, often as the result of a combination or series of otherwise inconsequential or minor errors, referred to as “system fracture.”43 During the 36 months of system utilization, the primary mode of failure and system downtime has been integration or failure of plate readers rather than the robotics themselves. These have included, e.g., Acumen Explorer software freezes and motherboard burnout, ViewLux camera and shutter failures, and EnVision first-row signal biases when in AlphaScreen mode. In contrast, the robotic components, such as grippers, controllers, plate latching mechanisms, and power supplies, have run largely failure-free during this period. The reliability of the robotic arms on the NCGC system can be attributed to their evolution during over a decade of use in automobile assembly factories, where reliability is a paramount consideration. However, combining such robust automation with the relatively delicate processes and systems required for biological application creates unique challenges. We describe here three non-user-attributable failures that illustrate this principle and the unanticipated screening system design issues they raise.
Premature failure of VCR doors
The requirement of keeping microtiter plates in environmentally controlled conditions during HTS requires the integration of large incubators with microplate carousels in a manner that provides easy access. We have found that the most common form of mechanical failure of the system occurs when one of the VCR doors of such plate incubators becomes jammed and causes a crash when a robotic arm tries to deposit or retrieve a plate from the stuck VCR door. This happens when the springs in the VCR mechanism become stiff enough so that even when the pressing force is removed, the door remains in the downward position after the drawbar is released. Therefore, the stuck open door acts as a mechanical barrier preventing the door beneath it from opening, and this leads to a major crash when attempting to access the position below the jammed VCR door. Such crashes can be very damaging to the gripper, and on two occasions the gripper was sent back to the manufacturer for repair and recalibration.
Currently, these VCR doors are viewed as the mechanical liability for the system, as there is no method to prevent this error aside from manually exercising every VCR door before each screen takes place to test for ease of retractability. Therefore we check to determine if a door appears difficult to open, and if so the springs associated with it are replaced, although we have found that even recently replaced springs can jam. Lowering of the incubator temperature and humidity can cause the VCR door springs to stiffen, so when this process is performed, we exercise the doors before the screen is run.
Lack of device timeout
A type of system failure that can be very costly is one not recognized by the controlling software as an error and an operator is not notified. This can happen when there is a communication failure between the automation Dispatcher and the Bridge, an application that acts as an interface between the Dispatcher and vendor-provided software for a particular peripheral device, most commonly a plate reader. For example, the software used to control the Acumen Explorer provided by the manufacturer acts as a COM server, allowing external software applications to control the instrument remotely. During a screen, when a plate is to be read, the Dispatcher sends a message to the Bridge application requesting that the read take place. The Bridge application then sends the appropriate commands to the Acumen Explorer software to initiate the read. Any errors that may occur during the course of a plate read are immediately reported to the Acumen software, which passes the error message to the Bridge application. The Bridge then sends the error message to the Dispatcher, which has the capability of automatically paging the operator and stops the screen from running until the error is acknowledged. If the plate is read without errors, the Acumen software sends a message to the Bridge application that the read is complete, with this message being relayed from the Bridge to the Dispatcher. Of note is that all of these messages and responses are event-driven, meaning no action is taken by any of the software applications used unless a message has been received. Although an event-driven software architecture has many advantages, the lack of any sort of timeout warning can lead to problems in the event that a message is not received for any reason as was the case in the failure described below.
A failure that resulted from lack of timeout between the Dispatcher and Bridge software applications illustrates this issue. Halfway through an automated overnight screen, the Acumen Explorer cytometer software experienced a hard crash during a plate read and terminated abruptly without sending any message back to the Bridge. The Bridge application remained in a permanent waiting mode, expecting a response from the cytometer application that would never come. Because this failure was not recognized as an error, the Dispatcher continued to execute the other assay protocol steps for all subsequent plates in the screen normally without notifying the operator of any errors. As this assay used cultured cells with a short time window for response measurement, subsequent plates that had received all necessary reagents expired because the failure occurred late in the evening with no call from the system that an error had transpired. Therefore, the problem was not discovered until the operator arrived early the next morning. Several hundred plates containing cells and detection reagent were lost because of this failure, which stresses the need for timeout warnings in the event of unforeseen software failures. We also note that all the microplate reader peripherals underwent an extensive site acceptance testing without failure; however, given the thinner margin for error when processing hundreds of thousands of samples per day, it would be prudent to force microplate reading failures during the site acceptance testing to diagnose and prevent these types of errors.
Detector failures
The most common type of system failures have been the result of errors with the integrated detectors, which as mentioned above can be attributed to the detectors' integration with optimized automation resulting in a high degree of use.
The most heavily used detector to date has been the ViewLux, with more than 100,000 plate reads generated over a 30-month period. Several failures were directly due to core components failing after extended usage, such as stepper motors burning out and causing mechanical failures, or various parts such as the light sources, optical components, or the CCD camera itself failing over time. These breakdowns were relatively easy to diagnose because of the immediate error reporting from the reader.
One malfunction difficult to diagnose and correct occurred when the ViewLux computer intermittently rebooted without warning and then failed to boot properly, instead only displaying a message that the temperature of the mother board was too high. Given the reported error message, troubleshooting of the cooling components of the motherboard was attempted first, with both the heat sink and fans being determined to work properly. Further investigation revealed that the manufacturer of the computer had a known problem with capacitors on the motherboard leaking, leading to intermittent and then eventual total failure unless the motherboard was replaced. This illustrates the complexity involved with peripheral device integration as the components of the plate reader were not at fault but the computer system that controlled the plate reader failed, leading to a failure of the entire system.
The Acumen Explorer cytometer40 was a first-time integration on the Kalypsys system at the time of our system construction. The examples given below serve to illustrate the factors that need to be considered when integrating any new reader into an automated process. The primary source of cytometer failures has been mechanical associated with the anvil, an electromechanical component that holds the microtiter plate within the instrument. The anvil rests underneath the plate and is a hollow square tube whose function is to press against the bottom of the plate to assist in the xy positioning of each well relative to the laser beam. The plate is lifted off of the in/out carrier when the anvil is raised, and then the carrier is moved out of the way so that the plate can be realigned once the region of interest being scanned is complete and the plate is to be moved to the next region. The anvil is controlled by a DC motor, which can either raise or lower it. The height at which the anvil can be raised is fixed, and during installation shims are added at the base of it to adjust the height it can raise according to the plate types being used.
After repeated instrument use, the plastic shims compressed under the force of the anvil and made it impossible to completely separate the plate from its carrier. Thus, when the carrier moved out of the way during scanning, it rotated the plate. This rotation changed the scan region, and in many cases only partial wells were scanned, leading to inconsistent data, which dramatically affected the assay performance without triggering an actual error. This failure, which occurred on both the system-integrated and standalone readers, can be attributed to a design weakness and was very difficult to diagnose. Ultimately, at our suggestion, the manufacturer changed the shim material from plastic to metal.
A serious cytometer failure involved the motion control of the anvil. The DC motor used to control the anvil positioning is driven by a motion control board within the instrument. The board drives the DC motor to the requested position until it triggers a limit switch which cuts current to the motor, at which point movement is stopped. In one instance, the motion control board never received the limit switch response from the motor, and the motor continued to draw current in an attempt to move past its mechanical limit. As a result, components on the motion control board eventually overheated, further causing several on-board integrated circuit chips to melt. This caused the entire instrument to fail and could have led to combustion of the reader if the problem had not been detected. The reasons for this failure could not be determined but prompted the manufacturer to develop a safety circuit to prevent failure in the event a limit switch signal is never sent.
When choosing microplate readers for an automated system two considerations are important. First, it is prudent, when possible, to procure from a single vendor to reduce the number of sources of peripheral devices that can aid diagnosis of system failures. Second, one should choose microplate readers that have been tested on such systems whenever possible. However, as in our case with the cytometer, one is often required to become the “early-adopter” of a new technology, and in this case there will be inherent risks, and troubleshooting must be accepted as part of this choice. Increased system monitoring should be employed during the first year of use for new microplate readers.
System Modifications and Enhancements
Based on our experience, a variety of modifications were made over the last 3 years that enhance the functionality of the system and provide directions for further technology development.
Controller hardware and software enhancements
Automatic data file redirecting
The Dispatcher software can only store data files to one location as specified by the user when creating a method file. This location remains constant regardless of the number of screens running, which can make it difficult to process trace files for multiple screens. To address this deficiency, we have modified the High Throughput Target Identification (HTTI) software component of the robot controller. The modified application, dubbed LabHTTI, was developed using LabView to control the Dispatcher Application (Fig. 10). All GNF/Kalypsys screening systems come with an ActiveX control that provides a limited set of control and status reporting of the Dispatcher through an automation server. Once the LabHTTI Application has been launched within the Dispatcher environment, the “Connect” button is pressed to create a reference to the Dispatcher automation server ActiveX object, which will be used to control and monitor the properties, methods, and events associated with the object. Next, all of the events associated with the object are registered, which allows the LabHTTI Application to capture these events as they are generated by the Dispatcher. Finally, the “Connect” method of the ActiveX object is invoked to establish a connection between the LabHTTI Application and the Dispatcher automation server.
FIG. 10.
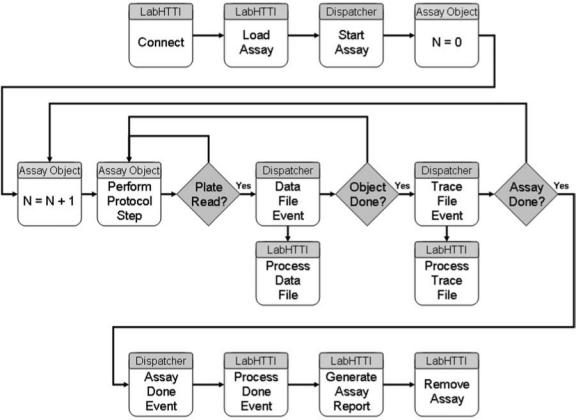
Enhancement of the Dispatcher by the LabHTTI Application. LabHTTI communicates with the Dispatcher automation server via an ActiveX component. The assay objects are defined as the mother plates and their associated daughter plates, with “N” representing a single object. Multiple assay objects can be running in parallel as needed, with the Dispatcher handling the scheduling aspects. LabHTTI connects to the Dispatcher and then launches an assay. As the Dispatcher performs various functions, LabHTTI captures all events generated by the Dispatcher and handles them as required.
With the connection established, LabHTTI controls the Dispatcher, in addition to capturing events sent from Dispatcher and processing these events as necessary. Within this environment, Assay and Method Files are loaded in order to execute a screen as described above (see System control and monitoring). A key new feature developed within LabHTTI is the Report Destination listbox of file paths entered by the user that will act as the destination for all output plate read files generated during the course of the assay. We utilized the “PlateReadComplete” event generated by the Dispatcher at the end of a plate read to make a copy of or move the file to one or more locations as specified in the “Report Destination” field. Thus, if multiple reads are collected from the same assay plate, LabHTTI can create multiple subdirectories automatically based on the assay logic. Importantly, because of the continuous nature of the file copying/redirecting, on-the-fly data analysis can be performed for screen quality control purposes. Analogously, we have developed a Trace Destination listbox to assist with the analysis of log files generated by multiple screens that are run simultaneously.
Pintool protocol improvements
The pintool wash cycle is a major component of the compound transfer process, and as a result this step in HTS has frequently been rate limiting. The initially established pin transfer protocol (not including plate movement) was 1 min 40 s per plate, of which 1 min 15 s was spent on pin washing and drying. Over time and through experimentation we have optimized the washing cycle to take a significantly shorter time while continuing to effectively clean the pins. The optimized version of this protocol was obtained after adjusting dwell time in the baths and drying station and number of immersions in the baths. The full cycle time of one pin transfer, wash, and dry process was reduced by 31 s, which has led to an increase of the screening speed by up to 50% in the cases of assays where the pin transfer steps was rate determining.
System monitoring via webcams
In order to improve the way errors and crashes are investigated, we installed three web-enabled video cameras to monitor the system. Each camera acts as a Web Server, allowing complete control from an external location. These cameras are programmed by the operator to view specific locations. At the start of any assay, LabHTTI connects to these cameras to point each one to specific cell locations. Each camera can store a buffer of images for up to 30 s. In the event of an error, a message is sent to retrieve those buffered images and to then store them to a network location using FTP. These images are then combined to create a video that a user can watch to investigate the exact motions and steps directly leading to the error.
Further reader and system upgrades
The most frequent modification performed on a screening system is the upgrade or complete changeover of a plate reader as optical detection technologies continue to evolve. Thus far, we have exchanged the Analyst® GT reader (Molecular Devices, Sunnyvale, CA), initially integrated on the system, with an EnVision unit in order to address our growing utilization of the AlphaScreen assay technology. Additionally, the Acumen Explorer has been upgraded from a single- to three-laser light source eX3 model to permit further cell assay multiplexing.
With the expansion of our compound collection and the introduction of sophisticated multistep and multireader assays, the need for increased system capacity has become evident. Therefore, we added second EnVision and eX3 readers that will allow incorporation of various pre- and post-reads into screening protocols, provide an overall increase in throughput, and introduce a level of redundancy should a reader fail during a screen. Further, a major system upgrade associated with replacement of the two RX-90 arms with RX-120 units and the attachment of additional compound storage carousels, plate de-lidding and handoff stations, and an additional pintool and dispenser will increase the system storage and plate handling capacity while further improving the efficiency of screening.
Conclusions and Future Directions
The NCGC screening system has provided a uniquely robust, reliable, and comprehensive solution to the demanding screening requirements of academic HTS. Its unique approach to on-line compound plate storage and rapid access enabled the rapid operationalization of the qHTS concept and thus led to the generation and public dissemination of an unprecedented volume and quality of small molecule bioactivity data over the last 3 years. Several lessons may drawn from this experience. First, a properly designed, fabricated, and operated integrated screening system allows extremely efficient screening, which then allows new paradigms to be considered that can drive improvements in efficiency in subsequent informatics and medicinal chemistry steps of the drug development process. Second, the potential of powerful and sophisticated screening systems such as this is only realized with highly skilled, experienced, and creative operation scientists; technology is necessary but not sufficient. Third, such a screening system should not be viewed as static; rather, tracking of performance metrics and causes of failures should guide continual modifications, driving evolution of the system to address shifting weak points and thus allowing continual improvement in efficiency, throughput, and flexibility. Innovations such as these promise in the short term to improve chemical probe and lead generation productivity and in the long term to transform HTS from a specialized and narrowly directed search for small numbers of statistical actives to a commonly performed enumerator of biological activity profiles of large chemical libraries. This transformation will allow enumeration of generalizable relationships between chemical and biological space and ultimately drive truly “rational” drug development based on understanding of chemical genomic principles.
Abbreviations
- AC50
50% active concentration
- AID
PubChem BioAssay identifier
- CCD
charge-coupled device
- CRC
concentration-response curve
- CV
coefficient of variance
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- GNF
Genomics Institute of the Novartis Research Foundation
- HTTI
High Throughput Target Identification
- LOPAC
Library of Pharmacologically Active Compounds
- MSR
minimum significance ratio
- NCGC
National Institutes of Health Chemical Genomics Center
- NIH
National Institutes of Health
- PMT
photomultiplier tube
- qHTS
quantitative high-throughput screening
- RFP
red fluorescent protein
- S:B
signal:background
- UPS
uninterrupted power supply.
Acknowledgments
This research was supported by the NIH Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute, NIH. We thank Paul Queeney, Rose Hughes, and Ben Baumann (Kalypsys) for expert help with system set-up and initial training and Daniel Sipes (GNF Systems) for helpful discussions.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hughes B. 2007 FDA drug approvals: a year of flux. Nat Rev Drug Discov. 2008;7:107–109. doi: 10.1038/nrd2514. [DOI] [PubMed] [Google Scholar]
- 2.Stylli C. Beckey SS. Shumate CB. Coassin PJ inventors; Aurora Biosciences Corporation, assignee. Systems and methods for rapidly identifying useful chemicals in liquid samples. Nov 16, 1999. US Patent 5,985,214.
- 3.Kornienko O. Lacson R. Kunapuli P. Schneeweis J. Hoffman I. Smith T, et al. Miniaturization of whole live cell-based GPCR assays using microdispensing and detection systems. J Biomol Screen. 2004;9:186–195. doi: 10.1177/1087057103260070. [DOI] [PubMed] [Google Scholar]
- 4.Inglese J. Auld DS. Application of high throughput screening techniques to chemical biology. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons; Hoboken, NJ: 2008. [DOI] [Google Scholar]
- 5.Gray NS. Drug discovery through industry-academic partnerships. Nat Chem Biol. 2006;2:649–653. doi: 10.1038/nchembio1206-649. [DOI] [PubMed] [Google Scholar]
- 6.Verkman AS. Drug discovery in academia. Am J Physiol Cell Physiol. 2004;286:C465–C474. doi: 10.1152/ajpcell.00397.2003. [DOI] [PubMed] [Google Scholar]
- 7.Austin CP. Brady LS. Insel TR. Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 8.Zheng W. Padia J. Urban DJ. Jadhav A. Goker-Alpan O. Simeonov A, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease. Proc Natl Acad Sci U S A. 2007;104:13192–13197. doi: 10.1073/pnas.0705637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simeonov A. Yasgar A. Jadhav A. Lokesh GL. Klumpp C. Michael S, et al. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem. 2008;375:60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson RL. Huang W. Jadhav A. Austin CP. Inglese J. Martinez ED. A quantitative high-throughput screen identifies potential epigenetic modulators of gene expression. Anal Biochem. 2008;375:237–248. doi: 10.1016/j.ab.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inglese J. Auld DS. Jadhav A. Johnson RL. Simeonov A. Yasgar A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasgar A. Shinn P. Jadhav A. Auld D. Michael S. Zheng W, et al. Compound management for quantitative high-throughput screening. J Assoc Lab Automat. 2008;13:79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blow N. Lab automation: tales along the road to automation. Nat Methods. 2008;5:109. doi: 10.1038/nmeth0108-109. [DOI] [PubMed] [Google Scholar]
- 14.Melnick JS. Janes J. Kim S. Chang JY. Sipes DG. Gunderson D, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc Natl Acad Sci U S A. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassaday J. Shah T. Murray J. O'Donnell GT. Kornienko O. Strulovici B, et al. Miniaturization and automation of an ubiquitin ligase cascade enzyme-linked immunosorbent assay in 1,536-well format. Assay Drug Dev Technol. 2007;5:493–500. doi: 10.1089/adt.2007.076. [DOI] [PubMed] [Google Scholar]
- 16.Auld DS. Johnson RL. Zhang YQ. Veith H. Jadhav A. Yasgar A, et al. Fluorescent protein-based cellular assays analyzed by laser-scanning microplate cytometry in 1536-well plate format. Methods Enzymol. 2006;414:566–589. doi: 10.1016/S0076-6879(06)14029-X. [DOI] [PubMed] [Google Scholar]
- 17.Auld DS. Southall NT. Jadhav A. Johnson RL. Diller DJ. Simeonov A, et al. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 18.Davis RE. Zhang YQ. Southall N. Staudt LM. Austin CP. Inglese J, et al. A cell-based assay for IκBα stabilization using a two-color dual luciferase-based sensor. Assay Drug Dev Technol. 2007;5:85–103. doi: 10.1089/adt.2006.048. [DOI] [PubMed] [Google Scholar]
- 19.Honson NS. Johnson RL. Huang W. Inglese J. Austin CP. Kuret J. Differentiating Alzheimer disease-associated aggregates with small molecules. Neurobiol Dis. 2007;28:251–260. doi: 10.1016/j.nbd.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simeonov A. Jadhav A. Sayed AA. Wang Y. Nelson ME. Thomas CJ, et al. Quantitative high-throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade. PLoS Negl Trop Dis. 2008;2(1):e127. doi: 10.1371/journal.pntd.0000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simeonov A. Jadhav A. Thomas CJ. Wang Y. Huang R. Southall NT, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 22.Zhu PJ. Zheng W. Auld DS. Jadhav A. Macarthur R. Olson KR, et al. A miniaturized glucocorticoid receptor translocation assay using enzymatic fragment complementation evaluated with qHTS. Comb Chem High Throughput Screen. 2008;11:545–559. doi: 10.2174/138620708785204045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Titus S. Neumann S. Zheng W. Southall N. Michael S. Klumpp C, et al. Quantitative high-throughput screening using a live-cell cAMP assay identifies small-molecule agonists of the TSH receptor. J Biomol Screen. 2008;13:120–127. doi: 10.1177/1087057107313786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burow KM. Caldwell JS. Downs RC. Lesley SA. Mainquist JK. Meyer AJ, et al. inventors; IRM LLC, assignee. High throughput processing system and method of using. Jun 24, 2008. US Patent 7,390,458.
- 25.Downs RC. Weselak MR inventors; IRM LLC, assignee. Gripper mechanism. Jul 15, 2003. US Patent 6,592,324.
- 26.Mainquist JK. Downs RC. Weselak MR. Meyer AJ. Burow KM. Sipes DG, et al. inventors; IRM LLC, assignee. Specimen plate lid, method of using. Mar 18, 2003. US Patent 6,534,014.
- 27.Weselak M. Burow K inventors; IRM LLC, assignee. Compound storage system. Nov 25, 2004. US Patent Application 20040 236463.
- 28.Weselak MR. Downs RC inventors; IRM LLC, assignee. High throughput incubation devices. Feb 12, 2008. US Patent 7,329,394.
- 29.Downs RC. Weselak MR. Mainquist JK inventors; IRM LLC, assignee. Gripping mechanisms, apparatus, methods. Aug 23, 2005. US Patent 6,932,557.
- 30.Cleveland PH. Koutz PJ. Nanoliter dispensing for uHTS using pin tools. Assay Drug Dev Technol. 2005;3:213–225. doi: 10.1089/adt.2005.3.213. [DOI] [PubMed] [Google Scholar]
- 31.Binder C. Lafayette A. Archibeque I. Sun Y. Plewa C. Sinclair A, et al. Optimization and utilization of the SureFire phospho-STAT5 assay for a cell-based screening campaign. Assay Drug Dev Technol. 2008;6:27–37. doi: 10.1089/adt.2007.111. [DOI] [PubMed] [Google Scholar]
- 32.Gabriel D. Vernier M. Pfeifer MJ. Dasen B. Tenaillon L. Bouhelal R. High throughput screening technologies for direct cyclic AMP measurement. Assay Drug Dev Technol. 2003;1:291–303. doi: 10.1089/15406580360545107. [DOI] [PubMed] [Google Scholar]
- 33.Glickman JF. Wu X. Mercuri R. Illy C. Bowen BR. He Y, et al. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J Biomol Screen. 2002;7:3–10. doi: 10.1177/108705710200700102. [DOI] [PubMed] [Google Scholar]
- 34.Gray A. Olsson H. Batty IH. Priganica L. Peter Downes C. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 35.Li Y. Cummings RT. Cunningham BR. Chen Y. Zhou G. Homogeneous assays for adenosine 5′-monophosphate-activated protein kinase. Anal Biochem. 2003;321:151–156. doi: 10.1016/s0003-2697(03)00397-x. [DOI] [PubMed] [Google Scholar]
- 36.Rouleau N. Turcotte S. Mondou MH. Roby P. Bosse R. Development of a versatile platform for nuclear receptor screening using AlphaScreen. J Biomol Screen. 2003;8:191–197. doi: 10.1177/1087057103252605. [DOI] [PubMed] [Google Scholar]
- 37.Sehr P. Pawlita M. Lewis J. Evaluation of different glutathione S-transferase-tagged protein captures for screening E6/E6AP interaction inhibitors using AlphaScreen. J Biomol Screen. 2007;12:560–567. doi: 10.1177/1087057107301246. [DOI] [PubMed] [Google Scholar]
- 38.Von Leoprechting A. Kumpf R. Menzel S. Reulle D. Griebel R. Valler MJ, et al. Miniaturization and validation of a high-throughput serine kinase assay using the AlphaScreen platform. J Biomol Screen. 2004;9:719–725. doi: 10.1177/1087057104268805. [DOI] [PubMed] [Google Scholar]
- 39.Warner G. Illy C. Pedro L. Roby P. Bosse R. AlphaScreen kinase HTS platforms. Curr Med Chem. 2004;11:721–730. doi: 10.2174/0929867043455693. [DOI] [PubMed] [Google Scholar]
- 40.Bowen WP. Wylie PG. Application of laser-scanning fluorescence microplate cytometry in high content screening. Assay Drug Dev Technol. 2006;4:209–221. doi: 10.1089/adt.2006.4.209. [DOI] [PubMed] [Google Scholar]
- 41.Eastwood BJ. Farmen MW. Iversen PW. Craft TJ. Smallwood JK. Garbison KE, et al. The minimum significant ratio: a statistical parameter to characterize the reproducibility of potency estimates from concentration-response assays and estimation by replicate-experiment studies. J Biomol Screen. 2006;11:253–261. doi: 10.1177/1087057105285611. [DOI] [PubMed] [Google Scholar]
- 42.Feng BY. Simeonov A. Jadhav A. Babaoglu K. Inglese J. Shoichet BK, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 43.Chiles JR. Inviting Disaster: Lessons from the Edge of Technology. HarperCollins; New York: 2001. [Google Scholar]