Abstract
RS1, also known as retinoschisin, is an extracellular discoidin domain containing protein that has been implicated in maintaining the cellular organization and synaptic structure of the vertebrate retina. Mutations in the gene encoding RS1 are responsible for X-linked retinoschisis, a retinal degenerative disease characterized by the splitting of the retinal cell layers and visual impairment. To better understand the role of RS1 in retinal cell biology and X-linked retinoschisis, we have studied the interaction of wild-type and mutant RS1 with various carbohydrates coupled to agarose supports. RS1 bound efficiently to galactose-agarose and to a lesser extent lactose-agarose, but not agarose, N-acetylgalactosamine-agarose, N-acetylglucosamine-agarose, mannose-agarose or heparin-agarose. RS1 cysteine mutants (C59S/C223S and C59S/C223S/C40S) which prevent disulfide-linked octamer formation showed little if any binding to galactose-agarose. The disease-causing R141H mutant bound galactose-agarose at levels similar to wild-type RS1, whereas the R141S mutant resulted in a marked reduction in galactose-agarose binding. RS1 bound to galactose-agarose could be effectively displaced by incubation with isopropyl β-D-1-thiogalactopyranoside (IPTG). This property was used as a basis to develop an efficient purification procedure. Anion exchange and galactose affinity chromatography was used to purify RS1 from the culture media of stably transformed Sf21 insect cells that express and secrete RS1. This cell expression and protein purification method should prove useful in the isolation of RS1 for detailed structure-function studies.
Keywords: Discoidin domain, retinoschisin, RS1, affinity chromatography
RS1, also known as retinoschisin, is a discoidin domain containing extracellular protein that plays a crucial role in maintaining the cellular organization and synaptic structure of the vertebrate retina. Mutations in the gene encoding RS1 are responsible for X-linked juvenile retinoschisis (XLRS), an early onset retinal degenerative disease characterized by a splitting of the retinal layers and a loss in vision (1, 2). Mice deficient in RS1 exhibit a marked disorganization of the retinal cell layers, gaps between bipolar cells of the inner retina, disruption of the photoreceptor-bipolar synaptic structure and signal transmission, and photoreceptor degeneration (3, 4).
The RS1 gene encodes a 24 kDa protein which is expressed and secreted from retinal photoreceptor and bipolar cells as a homo-oligomeric complex (2, 5–7). RS1 consists of an N-terminal 23 amino acid signal peptide followed by a 39 amino acid unique Rs1 domain, a 157 amino acid discoidin domain, and a 5 amino acid C-terminal segment (8). The signal peptide is removed in the lumen of the endoplasmic reticulum to generate a mature polypeptide that is assembled and secreted from cells as a disulfide-linked homo-octameric complex (6). Intermolecular disulfide bonds between cysteine residues in the Rs1 domain and C-terminal segment are responsible for octamer formation (6, 9).
The discoidin domain is the principal structural and functional unit of RS1. This domain was first identified in discoidin proteins of Dictyostelium discoideum and subsequently found in a variety of extracellular and membrane proteins including the blood coagulation Factor V and Factor VIII, milk fat globule protein, neuropilins, neurexin IV, and discoidin domain receptor proteins (10–12). The high resolution structures of a number of discoidin domains have been determined (12–14). In all cases these domains consist of a 5-strand antiparallel β-sheet that packs against a 3-strand antiparallel β-sheet to form a core barrel-like structure. An intramolecular disulfide bond between conserved cysteine residues at the beginning and end of the discoidin domain stabilizes the three dimensional structure. At the opposite end spikes or loops protrude from the core β-barrel structure to define a groove or cleft that serves as the ligand binding site. Homology modeling suggests that the discoidin domain of RS1 has a similar structure (9, 15).
In vitro expression studies indicate that disease-associated missense mutations in RS1 result in a nonfunctional protein (9, 16–18). Mutations in the signal sequence cause mislocalization of the polypeptide to the cytoplasm of the cell and rapid proteasomal degradation. Mutations in the C59 residue of the Rs1 domain and C223 residue in the C-terminus prevent octamer formation, while most mutations in the discoidin domain cause protein misfolding, aggregation, and retention inside the cell by the quality control system of the ER.
Discoidin domain containing proteins interact with a diverse set of ligands to initiate any of a variety of biological processes including cell adhesion and aggregation, migration, signaling, and development (10, 11). Discoidin proteins from Dictyostelium discoideum function as lectins with a high affinity for galactose residues to promote cell aggregation (19, 20). Factors V and VIII bind phosphatidylserine on the surface of platelets and endothelial cells through their discoidin domains as a crucial step in initiating the blood coagulation cascade (21, 22). Discoidin domain receptors interact with collagen to regulate cell proliferation and extracellular matrix modeling via activation of their tyrosine kinase activities (23). Finally, the discoidin domains of SED1 mediate the interaction of sperm and egg as part of the fertilization process (24). More recently, the interaction of RS1 with the surface of retinal cells has been investigated. Co-immunoprecipitation and co-localization studies suggest that RS1 is anchored to the surface of photoreceptor and bipolar cells through its interaction with a Na/K ATPase-SARM1 complex (25).
Although significant progress has been made toward defining the role of RS1 in maintaining the cellular organization and structure of the retina (3, 25) and developing gene therapy as a possible treatment for XLRS (4, 26, 27), we still know relatively little about the structure-function relationships of this unique discoidin domain containing protein. This is due in part to the lack of both a cell system capable of producing milligram quantities of RS1 and an efficient method to purify RS1 from cell extracts. In this study, we have examined the interaction of RS1 with carbohydrates. We found that RS1 specifically and reversibly binds to modified galactose residues. This property has been used as a basis to develop an efficient affinity based procedure to purify RS1 from cell media. We also describe the development of a stably transformed insect cell line that is capable of expressing and secreting sufficient quantities of RS1 for structure-function studies.
EXPERIMENTAL PROCEDURES
Materials
Galactose-agarose was purchased from Pierce (Rockford, IL); lactose-agarose, N-acetylglucosamine-agarose, N-acetylgalactosamine-agarose, mannose-agarose, and heparin-agarose were obtained from Sigma (St. Louis, MO); and 6% beaded agarose was obtained from Amersham (Buckinghamshire, UK). The RS1 3R10 monoclonal antibody has been previously described (3, 9). Isopropyl β-D-1-thiogalactopyranoside (IPTG) was obtained from Gold Biotechnologies (St Louis, MO). Human Factor Va and an anti-human factor V antibody were obtained from Haematologic Technologies Inc, (Essex Junction, VT).
Generation of cDNA Constructs
The cloning of human WT-RS1 cDNA and the generation of the RS1 mutants C59S/C223S, C40S/C59S/223S, R141G, R141A, R141V, R141S, and R141H in pCEP4 have been described previously (6, 28). For expression in insect cells, WT-RS1 was amplified by PCR and cloned in p2Zop2F vector (29) via the HindIII and EcoRI restriction sites using the following primers: P1: CCCAAGCTTATGTCACGCAAGATAGAAGGC; P2: GGGGAATTCTCAGGCACACTTGCTGACGC. All constructs were sequenced to verify the absence of random mutations.
Cell Culture and RS1 Expression
HEK 293T cells (American Type Culture Collection) were grown in Dulbecco’s Modified Eagle Medium (DMEM) with L-glutamine and 10% fetal calf serum. Transfections were carried out in 10 cm dishes with 20 μg of DNA per dish using a calcium phosphate transfection procedure. Briefly, 500 μl of BES-buffered saline (50 mM N,N-bis(2-hydroxyethyl)-2-aminoethane, 280 mM NaCl, 1.4 mM Na2PO4, pH 6.96) was added dropwise to a DNA solution containing 250 mM calcium chloride and incubated for 20 min at room temperature. DNA was then added to exponentially growing HEK 293 T cells at 37°C under 5% CO2. The transfection medium was replaced with regular medium the next day, and the cells were harvested two days later.
Sf21 Cell Transfection and Stable Cell Line Selection
Prior to transfection, Sf21 cells were grown at 26°C to mid-log phase in ESF-AF serum free, protein free medium (Expression Systems, Woodland, CA). For transient transfections, 1–2 ×106 cells were seeded in a 6 well tissue culture plate with 1 ml of Grace’s minimal medium (Invitrogen) and allowed to attach. CellFectin™ (Invitrogen) liposome reagent (5 or 10μl) was mixed with 0.5, 1.0 or 2.0 μg of plasmid DNA in 1 ml of Grace’s medium, incubated for 30 min and added to the seeded Sf21 cells. Following a 4 h incubation, 2 ml of ESF-AF medium were added to the transfection mix and the cultures incubated at 26°C for 48 h. For the selection of stable cell lines, the cells from an entire well were transferred to T25 tissue culture flasks and allowed to grow for 3 days on 750 μg/ml Zeocin and then to confluence in the presence of 1 mg/ml Zeocin™. Selection on 1 mg/ml Zeocin™ was maintained for a total of three passages to obtain a stable polyclonal cell line. Selected lines were tested for RS1 expression levels and the highest expressing line was scaled up to 50 ml in 250 ml Erlenmeyer flasks and incubated at 26 °C at 100 rpm. When required, the culture was further scaled up to several 250 ml cultures using 1 liter Erlenmeyer flasks and grown as noted above.
Carbohydrate affinity chromatography
Approximately, 250 μl of culture fluid from HEK 293 cells expressing WT or mutant RS1 or 5 μg of purified Factor Va was incubated with 50 μl of agarose conjugated with various carbohydrates (1:1 packed beads to TBS buffer) for 1 h at 4°C. The unbound fraction was retained for analysis and the beads were washed three times with TBS buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.5) in an Ultrafree filter unit (Millipore, Billerica, MA). The bound protein was dissociated from the affinity matrix with 75 μl of 4% SDS or TBS containing 1M of specified sugar for 30 min at 23°C or the indicated temperature. The samples were collected by low speed centrifugation.
Extraction of RS1 from retinal membranes
Bovine retinal membranes were isolated on sucrose gradients as previously reported (25). Membranes in 20 mM Tris-HCl, 150 mM NaCl, pH 7.5 were incubated with 1 M IPTG for 1 h. The supernatant fraction was separated from the membrane fraction by centrifugation in a Beckman T55 rotor at 100,000g and the fractions were analyzed by SDS gel electrophoresis and Western blotting.
Purification of RS1
RS1 secreted from cultured insect cells was purified by a two-step procedure involving ion exchange and affinity chromatography. Typically, 100 ml of insect cell culture was centrifuged at 6000 rpm for 20 min to remove cells and residual cell debris. The supernatant was directly loaded onto a column containing 5 ml of DEAE Sephacryl (Amersham) prewashed in Tris buffer (20 mM Tris-HCl, pH 7.5). The column was washed with 2 volumes of Tris buffer and the bound protein was subsequently eluted with 2 column volumes of Tris buffer containing increasing concentrations of sodium chloride from 0.1 to 1 M. A sample from each step was run on a 10% SDS-polyacrylamide gel to assess the presence of RS1 by western blotting and the purity of the sample by Coomassie blue staining. The fraction containing the RS1 (typically the fraction that eluted with 0.3M NaCl) was further purified on a galactose-agarose affinity column. The sample (typically 10 ml) was applied to a column containing 500 μl of galactose-agarose beads. The column was then washed with 5 column volumes of TBS (20 mM Tris, 150 mM NaCl, pH 7.5). The bound RS1 was subsequently eluted with TBS containing 1M IPTG.
Binding affinity of IPTG for RS1
The affinity of RS1 for IPTG was determined by incubating 500μl of insect cell supernatant containing RS1 (or affinity purified RS1) with 20μl galactose-agarose in the presence of varying concentrations of IPTG for 10 min at 23°C. The beads were washed three times at 4°C to remove unbound protein and eluted twice with 50μl TBS containing 4% SDS. The elutions were combine and 30 μl of each sample was run on a 10% SDS gel for analysis by western blotting. The intensity of the bands was quantified with the Licor Odyssey Imaging System. Three independent measurements were made. The data was fit to a simple one site model (single set of binding sites) using Grafit 6.0.5 (Erithacus Software).
SDS-polyacrylamide gel electrophoresis and western blotting
Samples were denatured in SDS-denaturing buffer (10 mM Tris-HCl pH 6.8, 4% SDS, and 20% sucrose with or without 4% β-mercaptoethanol) and separated by electrophoresis on a 10% polyacrylamide gel. For western blotting, proteins were electroblotted onto Immobilon-FL membranes (Millipore, Bedford, MA). The blots were blocked for 30 min at room temperature in 0.5% skimmed milk in PBS and subsequently incubated with RS1 3R10 hybridoma supernatant diluted 1:10 in PBS containing 0.05% Tween 20 (PBS-T) for 1 h at 22 °C. The blots were rinsed in PBS-T and labeled for 1 h at 22 °C with anti-mouse IgG conjugated with Alexa Fluor® 680 or LI-COR IRDye™ 800 diluted 1:15,000 in PBS-T containing 0.5% skimmed milk and 0.02% SDS. Bands were visualized and quantified using a LI-COR Odyssey® system (Lincoln, NE).
RESULTS
Binding of RS1 and Factor Va to carbohydrates
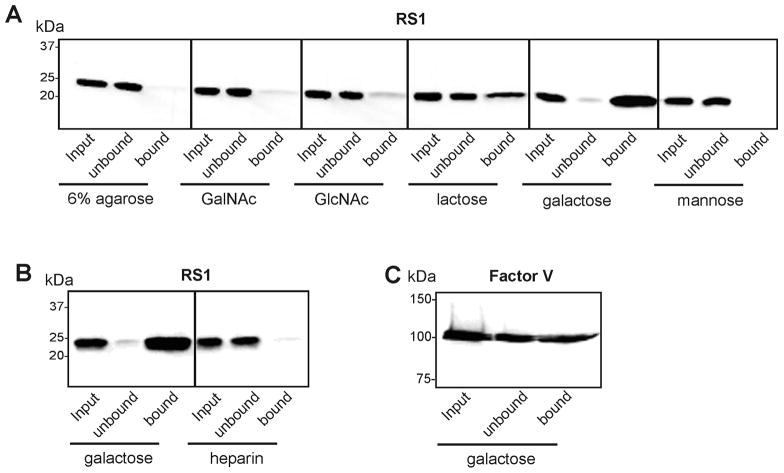
The binding of RS1 to various mono- and disaccharides was examined by carbohydrate-agarose affinity chromatography. RS1 secreted from Weri-Rb1 cells (30) was incubated with agarose or agarose conjugated with N-acetyl-D-galactosamine, N-acetyl-D-glucosamine, D-(+) lactose, D-(+) galactose, or mannose for 1 hour. After removal of unbound protein, the bound RS1 was eluted with SDS for analysis on western blots labeled with the RS1 R310 anti-RS1 monoclonal antibody. Fig 1A shows that most RS1 efficiently bound to galactose-agarose as indicated by the intensely stained band in the bound fraction and the weakly stained band in the unbound fraction. In contrast, only moderate binding of RS1 was observed for lactose-agarose and little or no binding was seen for agarose, N-acetylgalactosamine-agarose, N-acetylglucosamine-agarose, or mannose-agarose. Since the discoidin domain of neuropilin has been reported to bind heparin (31), we also investigated the binding of RS1 to heparin-agarose. Fig 1B shows that RS1 does not bind to heparin-agarose.
Figure 1.
Binding of RS1 and Factor Va to carbohydrates immobilized on agarose. Culture medium from HEK 293 cells expressing recombinant RS1 or purified Factor Va was incubated with agarose or agarose conjugated to carbohydrates. The unbound protein was collected and after washing the agarose matrix, the bound protein was eluted with 4% SDS. The input, unbound and bound fractions were analyzed on Western blots labelled with an antibody to either RS1 or Factor Va. A. Binding of RS1 to agarose conjugated with N-acetylgalactosamine – GalNAc, N-acetylglucosamine – GlcNAc, lactose, galactose, or mannose, or heparin. B. Binding of RS1 to agarose conjugated to either galactose or heparin. C. Binding of Factor Va to agarose conjugated with galactose.
Since RS1 as well as discoidin I and II proteins bind galactose, we reasoned that galactose binding may extend to other discoidin domain containing proteins. This was investigated by examining the interaction of activated blood coagulation Factor Va containing C1 and C2 discoidin domains to galactose-agarose. Fig 1C shows that Factor Va binds to the galactose-agarose, but with a lower efficiency than RS1. No binding of Factor Va to galactose-agarose was observed in the presence of phosphatidylserine (data not shown) indicating that binding of Factor Va to galactose-agarose occurred through its discoidin domains.
Binding of oligomeric forms of RS1 to galactose-agarose
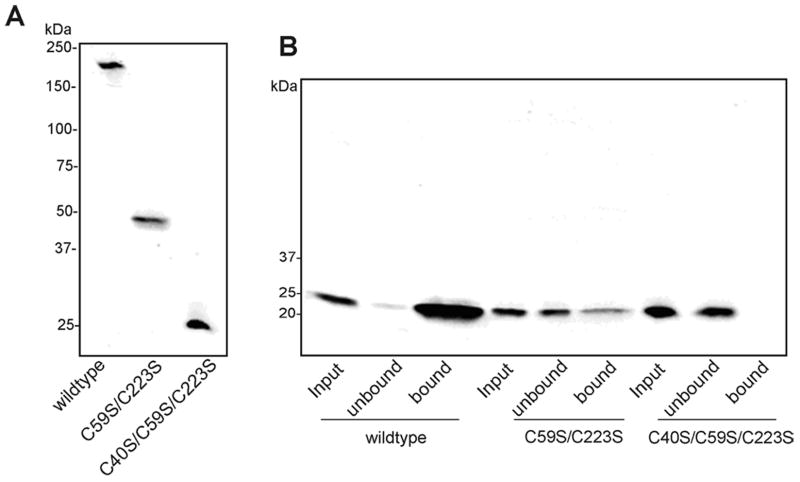
As shown in Fig 2A, WT RS1 is assembled and secreted from cells as a disulfide-linked octamer, while the double (C59S/C223S) and triple (C40S/C59S/C223S) cysteine mutants are secreted as a dimer and monomer, respectively. To determine the effect of the oligomeric state of RS1 on its interaction with galactose, the binding of the C59S/C223 double mutant and C40S/C59S/C223S triple mutant to galactose-agarose was compared with WT octameric RS1. The C59S/C223S mutant showed only limited binding to galactose-agarose, while no binding was detected for the C40S/C59S/C223S mutant (Fig 2B). This indicates that the octameric structure of RS1 is essential for efficient binding of RS1 to galactose-agarose.
Figure 2.
Binding of oligomeric forms of RS1 to galactose-agarose. Culture medium from HEK 293 cells expressing wild-type, C59S/C223S double mutant or C40S/C59S/C223S triple mutant were incubated with galactose agarose at 4°C for 1 h. After the matrix was washed to remove unbound protein, the bound RS1 was eluted with 4% SDS for analysis by SDS gel electrophoresis and Western blotting. A. Western blots of wild-type (octamer), C59S/C223S (dimer) and C40S/C59S/C223S (monomer) RS1 analyzed on non-reducing SDS gels. B. Binding of wild-type and mutant RS1 to galactose-agarose as analyzed on reducing SDS gels.
Effect of carbohydrates and temperature on the elution of RS1 from galactose-agarose
Next we determined the extent to which various mono- and di-saccharides were able to displace RS1 from galactose-agarose by treating the matrix with 1M saccharide followed by 4% SDS. Fig 3A shows that very little RS1 was eluted with glucose, lactose, or even galactose relative to SDS. In contrast over half the RS1 was eluted with IPTG (Fig 3A).
Figure 3.
The effect of various carbohydrates and temperature on the dissociation of RS1 from galactose-agarose. A. RS1 bound to galactose-agarose was eluted with 1 M saccharide at 25°C for 1 h followed by elution with 4% SDS for analysis on western blots labelled for RS1. The lane at the right side of the panel represents the direct elution with 4% SDS. B. RS1 bound to galactose-agarose was eluted sequentially with IPTG and SDS at the indicated temperature. Top panel shows the effect of IPTG and SDS on the dissociation of RS1 from galactose-agarose. Bottom panel shows the % dissociation of RS1 relative to that obtained by SDS (left lane). Data is for 3 independent experiments +/− SD.
The effect of temperature on the elution of RS1 from galactose-agarose by IPTG was examined (Fig 3B). The amount of RS1 eluted with 1M IPTG increased with rising temperature. Over 75% of the bound RS1 was eluted with IPTG at 37°C compared to about 40% at 4°C.
Extraction of RS1 from retinal membranes with IPTG
The finding that IPTG can release RS1 from galactose-agarose prompted us to determine if IPTG could also release RS1 from retinal membranes. A retinal membrane extract was treated with 1 M IPTG for 1 h at 4°C and subsequently separated into a membrane pellet and soluble fraction by high speed centrifugation. Figure 4 shows that IPTG treatment of retinal membranes resulted in the release of about 50% of the RS1 as determined by SDS gel electrophoresis and western blotting. A number of additional retinal proteins were also present in the supernatant faction as revealed by Coomassie Blue staining. In control studies treatment of retinal membrane extracts with buffer in the absence of IPTG (5) or in the presence of glucose failed to release RS1 from membrane extracts (data not shown).
Figure 4.
IPTG extraction of RS1 from retina membranes. Membranes from bovine retina were treated with 1 M IPTG for 1 hour at 25°C. The supernatant fraction was then separated from the membrane fraction by high-speed centrifugation and analyzed on SDS gels stained with Coomassie Blue (CB) or on Western blots labelled for RS1 with the RS1 3R10 antibody. lane 1: Retina membranes; lane 2: Supernatant fraction.
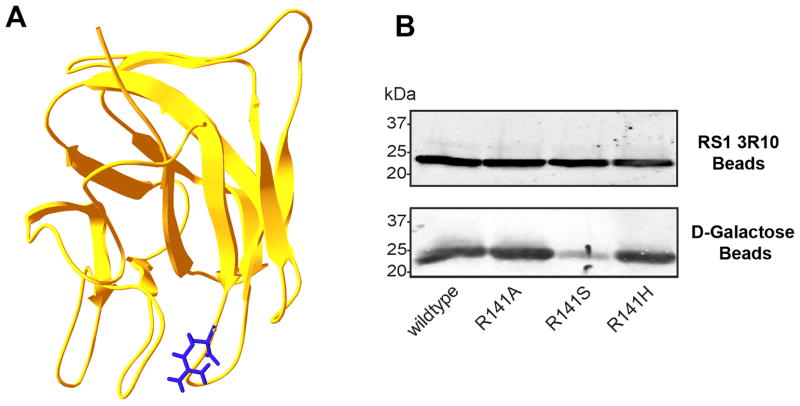
Binding of R141 mutants to galactose-agarose
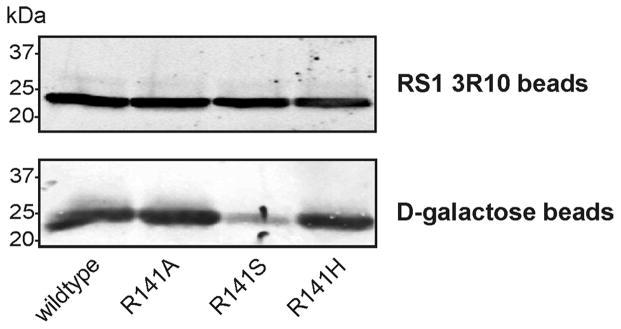
Previous heterologous cell expression studies have shown that most disease-linked missense mutations in the discoidin domain of RS1 results in misfolded, aggregated protein that is retained in the ER of the cell (9, 16). The R141H mutation is an exception, in that this disease-associated mutant is secreted from cells as an octameric complex at levels similar to WT RS1 (17, 28). Mixed results were obtained when arginine at position 141 located on loop or spike 3 of the discoidin domain (Fig 5A) was replaced with other amino acids. The R141G and R141V mutants were largely retained in HEK 293 cells as misfolded, aggregated protein, while R141A and R141S mutants, like the R141H mutant, were secreted from HEK 293 cells as a homo-octameric complex (28). As part of this study, we examined the binding of various secreted R141 mutants to galactose-agarose. Fig 5B shows that the R141A, R141S and R141H mutants were secreted from cells and captured on a RS1 3R10 antibody matrix in similar amounts as the WT RS1. However, while the R141A and R141H mutants bound to galactose-agarose at levels similar to WT RS1, the R141S bound at a significantly reduced level.
Figure 5.
Binding of R141 mutants to galactose-agarose. A. Model showing the location of the R141 residue on spike 3 of the RS1 discoidin domain (model adapted from (9)). B. The secreted fraction of HEK 293 cells expressing wild-type RS1 or R141A, R141S, and R141H mutants were applied to either an anti-RS1 3R10 antibody-Sepharose beads (top panel) or a galactose-agarose beads (bottom panel). After washing to remove unbound protein, bound RS1 was eluted with 4% SDS for analysis by western blotting. The R141A and R141H mutants bound to galactose-agarose at similar levels to wild-type RS1, whereas the R141S mutant showed reduced binding.
Expression and secretion of RS1 from stably transformed Sf21 insect cell line
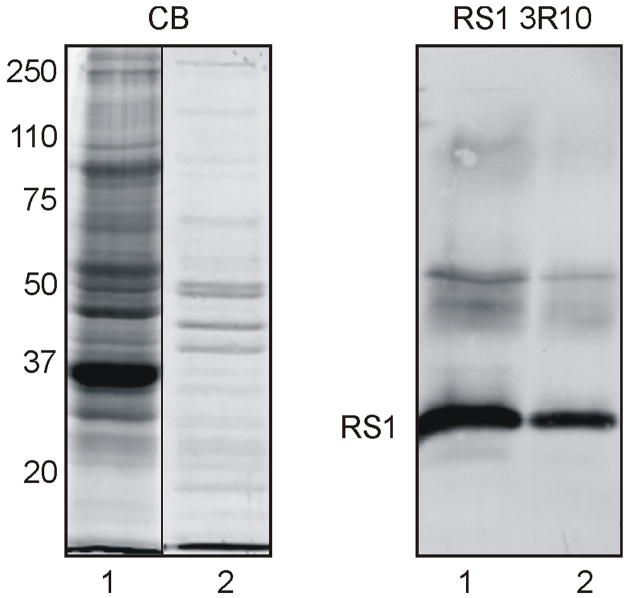
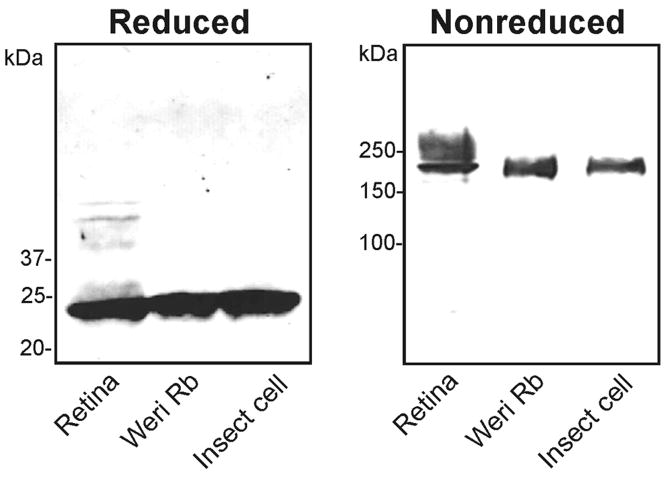
RS1 is expressed and secreted from a number of cell lines including Weri-retinoblastoma cells and transiently transfected COS7 and HEK293 cells (9, 16, 30). The amount of protein that can be recovered in the medium of these cells, however, is limiting for structure-function analysis. To overcome this problem, we have generated a stably transformed Sf21 insect cell line which expresses and secretes RS1 at a level 4–6 times higher than Weri-retinoblastoma cells. Fig 6 compares RS1 secreted from insect cells with RS1 secreted from Weri-retinoblastoma cells and RS1 from retinal membrane extracts. RS1 secreted from insect cells, like RS1 from Weri-retinoblastoma cells and retinal extracts, migrated as a characteristic 24 kDa monomer under disulfide reducing conditions and a 185 kDa octameric complex under nonreducing conditions. As previously reported, RS1 from retinal extracts migrated as a diffuse band under nonreducing conditions, possibly due to posttranslational modifications (6, 32).
Figure 6.
Analysis of RS1 secreted from stably transformed Sf21 insect cells. The medium from Sf21 cells (insect cell) or Weri-retinoblastoma cells (Weri Rb) were immunoprecipitated on a RS1 3R10 antibody-Sepharose matrix. These samples together with RS1 from retinal extracts were separated on reducing and nonreducing SDS gels. Similar amounts of RS1 were applied to each lane. RS1 was detected on western blots labelled with the RS1 3R10 antibody. RS1 migrated as a 24 kDa monomer under disulfide reducing conditions and a 185 kDa octamer under nonreducing conditions. RS1 from retina showed a broad band above 185 kDa as previously reported (6)
Purification of recombinant RS1 from insect cells
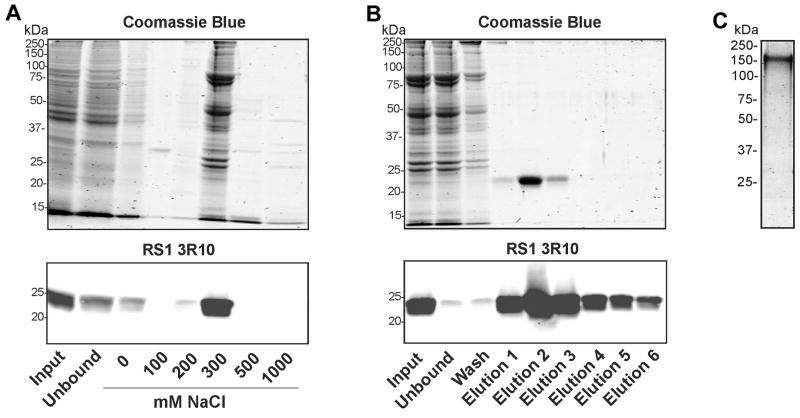
A combination of anion exchange and galactose affinity chromatography was used to purify RS1 from the secreted fraction of stably transformed Sf21 insect cell cultures. The secreted fraction was first applied to a DEAE Sephacryl column and eluted with increasing NaCl concentrations. Total protein from the column fractions was monitored on SDS gels stained with Coomassie Blue and RS1 was detected by Western blotting. Fig 7A shows the isolation of RS1 by anion chromatography. RS1 that bound to the DEAE-Sephacryl column eluted with 300 mM NaCl as detected by Western blotting. Numerous other proteins were also present in this fraction as visualized by Coomassie blue staining.
Figure 7.
Purification of recombinant RS1 from Sf21 insect cell medium. A. Cell medium from Sf21 insect cells expressing RS1 was applied to a DEAE-Sephacryl column and bound protein was eluted with 20 mM TrisCl buffer, pH 7.5 containing increasing concentrations of NaCl. B. The fraction that eluted from the anion exchange column with 300 mM NaCl was applied to a galactose-agarose column. After removing the unbound protein, bound RS1 was eluted 6 times with 1 M IPTG with the majority of RS1 present in elutions 1–3. Samples from the anion and affinity columns were analyzed on SDS gel stained with Coomassie Blue to detect total protein and Western blots labelled with the RS1 3R10 antibody to detect RS1. C. Purified RS1 migrates as a 186 kDa octameric complex as revealed on SDS gels stained with Coomassie Blue.
The RS1 enriched fraction from the DEAE column was applied to a galactose-agarose column. Essentially all proteins except RS1 were recovered in the unbound and wash fractions (Fig 7B). Bound RS1 was selectively eluted from the galactose-agarose column with 1 M IPTG. No other Coomassie blue stained proteins were observed in this IPTG eluted fraction. The purified RS1 ran as a 185 kDa protein on nonreducing SDS gels confirming the retention of the disulfide-linked octameric stoichiometry of RS1 after purification (Fig 7C).
The purification of RS1 from the secreted fraction of 100 ml of insect cell medium is summarized in Table I. A 19-fold purification of RS1 was obtained after anion exchange chromatography with a yield of about 50%. An additional 17-fold purification was achieved following galactose-agarose affinity chromatography. A small discrepancy between total protein (117.8 μg) and RS1 (86.1 μg) in the final fraction may be due either to the presence of low quantities of contaminating protein or proteolysis not detected by Coomassie blue staining or more likely the inherent limitation in determining actual protein concentrations by the Bradford procedure. At any rate approximately 86 μg of highly pure RS1 was obtained from 100 ml of insect cell medium under the conditions employed here.
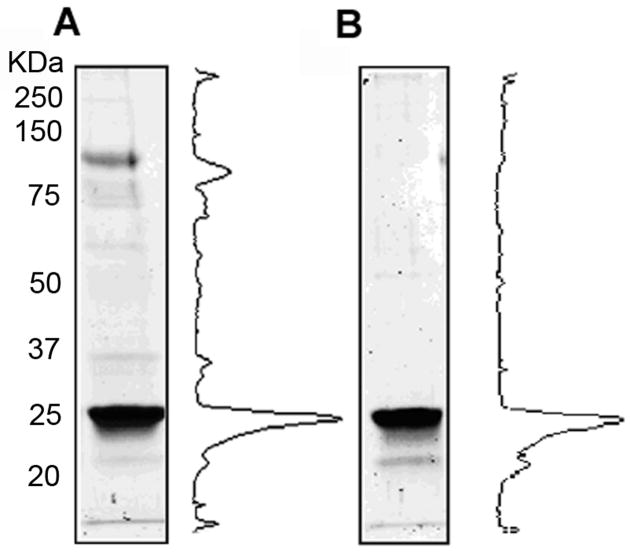
Finally, we compared the purification of RS1 by the two-step anion exchange/galactose affinity chromatography procedure with a single step galactose affinity procedure (Figure 8). The one-step procedure shows a prominent protein band at ~ 120 kDa and numerous minor protein bands between 25 kDa and 120 kDa, whereas the two-step purification procedure shows a predominant 24 kDa RS1 band and in some, but not all, preparations an additional lower molecular weight protein band which most likely represents a proteolytic product of RS1 (compare figure 8B with figure 7B).
Figure 8.
Comparison of a two-step and one-step procedure for RS1 purification. Cell media from Sf21 insect cells expressing RS1 was applied either directly to a galactose-agarose column (A) or subjected to anion exchange chromatography followed by galactose-agarose chromatography (B). Coomassie blue stained gels and densitometry profiles for each procedure are shown.
Affinity of RS1 for IPTG
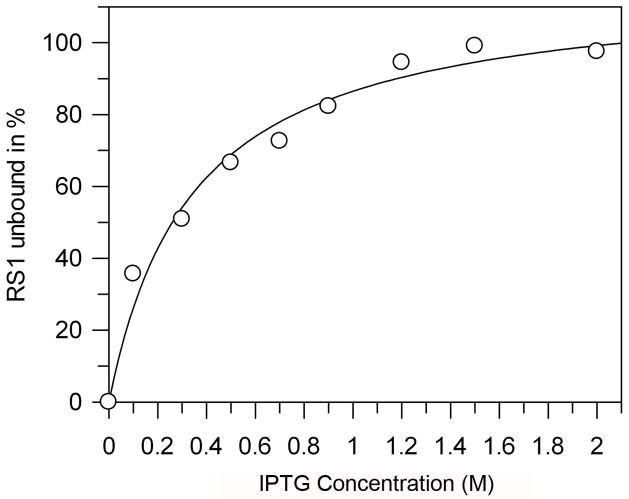
The high concentration of IPTG required to release RS1 from galactose-agarose suggested that the affinity of RS1 for IPTG is low. This was confirmed by determining the effective concentration of IPTG that inhibits RS1 binding to galactose agarose (Fig 9). The data fitted with a curve for a single set of binding sites gave an apparent dissociation constant of 0.35 M ± 0.06 at 23°C.
Figure 9.
Inhibition of RS1 binding to galactose-agarose by IPTG. Binding of RS1 to galactose-agarose was measured in the presence of increasing concentrations of IPTG. Bound RS1 was quantified by western blotting. The data is fitted with a curve for a single set of binding sites with a Kd of 0.35 M ± 0.06.
DISCUSSION
The discoidin domain is a conserved protein fold found in over 100 eukaryotic and 300 prokaryotic proteins (10, 11). It has a characteristic 8 β-strand core barrel structure maintained primarily by hydrophobic interactions between conserved amino acids (22). Loops or spikes protruding from the core barrel structure are composed of nonconserved amino acids and contribute to the diversity of ligand binding properties of these domains. The binding of RS1 to phospholipids and membrane proteins has been recently examined. In one study the recombinant discoidin domain of RS1 fused to GST was reported to bind anionic lysophospholipids immobilized on nitrocellulose strips (33). However, recently it has been shown that native oligomeric RS1 fails to bind synthetic phospholipids including phosphatidylserine and retinal lipids reconstituted into liposomes or immobilized onto microtiter plates (25). Instead, RS1 was shown to be anchored to photoreceptor and bipolar cell surfaces through its interaction with Na/K ATPase (25).
In this study we have investigated the interaction of RS1 with carbohydrates using a variety of mono- and di-saccharides conjugated to an agarose matrix. Our studies indicate that RS1 selectively binds to galactose-agarose and to a lesser extent lactose-agarose. Galactose binding appears to occur through the spike or loop regions of the discoidin domains since replacement of arginine on spike 3 (Fig 5A) with serine at position 141 in spike 3 reduces galactose binding without significantly altering the folding or oligomeric assembly of the RS1 subunits. The actual three dimensional structure of the RS1 discoidin domain and its binding site for galactose remains to be determined. Therefore, it is not possible to explain why the R141S mutant exhibits a lower affinity to galactose-agarose than either WT or the R141A and R141H mutants. However, one can speculate that the presence of a polar serine side chain may limit hydrophobic interaction with galactose thereby lowering its effective affinity. Multiple interactions between the RS1 discoidin domains and the galactose-agarose matrix are required to promote strong binding. This is evident in the finding that the octameric RS1 complex efficiently binds to galactose-agarose, while dimeric and monomeric RS1 show little or no binding. Importantly, the binding of native RS1 to galactose is reversible. High concentrations of IPTG, a derivative of galactose, can effectively displace RS1 from the galactose-agarose column under nondenaturing condition. Free galactose is less effective than IPTG in displacing RS1 from galactose-agarose than IPTG suggesting that derivatization of galactose at the C1 position enhances binding to RS1. The low affinity of RS1 for IPTG is consistent with the low affinity that proteins typically have for carbohydrates. This affinity, however, is increased significantly as a result of the multivalent nature of carbohydrate binding proteins. This is particularly evident for the binding of octameric RS1 to galactose-agarose.
The physiological relevance of RS1 binding to galactose is not known. RS1 has been shown to form a complex with Na/K ATPase on photoreceptors and bipolar cells (25). This abundant membrane protein consists of a large α3 catalytic subunit and a smaller β2 accessory subunit (34). The β2 subunit has a single membrane spanning segment and a relatively large extracellular domain with 9 oligosaccharide chains. It is possible that RS1 binds to one or more of these glycosylation sites on the β2 subunit of Na/K ATPase. Peanut agglutinin (PNA), a lectin with an affinity for galactose sugars, has been used to examine the distribution of galactose containing proteins in the retina. PNA labeling has been observed in the interphotoreceptor matrix surrounding inner and outer segments as well as the synaptic region of cone photoreceptor cells (35). RS1 exhibits partial co-localization with PNA binding proteins to cone photoreceptors of the outer retina as visualized by immunofluorescence microscopy (5, 36). However, it remains to be determined if there is a direct interaction between RS1 and PNA binding proteins in the retina. Alternatively, the interaction of RS1 with galactose may represent an evolutionarily conserved property of discoidin domains. Sequence and three dimensional structural analyses have revealed that the D1 domain of galactose oxidase that coordinates carbohydrate binding is a distant member of the discoidin domain family (11). A number of other prokaryotic and eukaryotic proteins also contain discoidin domains which are known to interact with carbohydrates including discoidin I and II, hemocytin and sialidases (11, 20, 37, 38). In the present study, we have shown that Factor Va can bind galactose-agarose although less efficiently than RS1. This interaction appears to be mediated by the discoidin domains of Factor V since no binding was observed in the presence of phosphatidylserine, the ligand for Factor V discoidin domains. The inefficient binding of Factor V to galactose agarose may be due in part to the fact that Factor Va has only 2 discoidin domains, whereas RS1 has 8 discoidin domains.
Regardless of the physiological relevance of the interaction of RS1 with galactose, this property can be used for the purification of RS1. In this study, we have developed an efficient two-step procedure to isolate RS1 from the supernatant fraction of cells expressing and secreting RS1. In the first step anion exchange chromatography was employed to separate RS1 from the bulk of the protein in the cell medium. In the second step galactose-agarose chromatography was used to obtain a highly pure preparation of RS1. The final RS1 product retains its characteristic native disulfide-linked octameric structure and is devoid of contaminating proteins as revealed by SDS gel electrophoresis. Although this procedure has been developed to isolate RS1 from culture cell media, it can also be used to isolate RS1 from retinal extracts.
In addition to an effective purification procedure, it is important to have a cell based system capable of expressing high levels of protein for structure-function studies. Previous studies have shown that stably transformed insect cell lines can be employed to express and secrete significant amounts of recombinant protein (39). As part of this study we have developed a stable, transformed Sf21 cell line capable of secreting RS1. This insect cell line secreted over 6 mg of RS1 per liter of cells, significantly higher quantities than can be obtained from mammalian cell lines expressing RS1. The secreted recombinant protein exists in its characteristic native disulfide-linked octameric state and retains the biochemical properties exhibited by native RS1 from retina extracts and RS1 expressed and secreted from mammalian cells. By further scaling up the production and purification of RS1 from this insect cell line, it should be possible to obtain sufficient quantities of RS1 for detailed structure-function studies.
Table 1.
Purification of RS1 from Insect Cell Culture Fluid
| Purification step | Total protein | Total RS1 | Total RS1/Total protein | Purification |
|---|---|---|---|---|
| Insect cell supernatant | 296 mg | 646 μg | 0.0022 | --- |
| DEAE sephacel elution | 7.8 mg | 336 μg | 0.043 | 19.5 |
| D-galactose column | 117.8 μg | 86 μg | 0.73 | 332 |
Total protein concentration was determined by the method of Bradford. RS1 concentration was determined by western blot against a calibration standard curve of different amounts of highly purified RS1 protein.
Footnotes
Supported by Grants from the Canadian Institutes for Health Research (MT 5822) and National Institutes of Health NEI (EY 02422). F.M.D was supported on a Foundation Fighting Blindness –Canada Postdoctoral Fellowship and an Arthur and June Willms Postdoctoral Fellowship. R.S.M. holds a Canada Research Chair in Vision and Macular Degeneration.
Abbreviations: WT, wild-type; IPTG, isopropyl β-D-1-thiogalactopyranoside; XLRS, X-linked retinoschisis; PBS, phosphate-buffered saline; ER, endoplasmic reticulum; PNA, peanut agglutinin
References
- 1.George ND, Yates JR, Moore AT. X linked retinoschisis. Br J Ophthalmol. 1995;79:697–702. doi: 10.1136/bjo.79.7.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer CG, Gehrig A, Warneke-Wittstock R, Marquardt A, Ewing CC, Gibson A, Lorenz B, Jurklies B, Weber BH. Positional cloning of the gene associated with X-linked juvenile retinoschisis. Nat Genet. 1997;17:164–170. doi: 10.1038/ng1097-164. [DOI] [PubMed] [Google Scholar]
- 3.Weber BH, Schrewe H, Molday LL, Gehrig A, White KL, Seeliger MW, Jaissle GB, Friedburg C, Tamm E, Molday RS. Inactivation of the murine X-linked juvenile retinoschisis gene, Rs1h, suggests a role of retinoschisin in retinal cell layer organization and synaptic structure. Proc Natl Acad Sci U S A. 2002;99:6222–6227. doi: 10.1073/pnas.092528599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Y, Takada Y, Kjellstrom S, Hiriyanna K, Tanikawa A, Wawrousek E, Smaoui N, Caruso R, Bush RA, Sieving PA. RS-1 Gene Delivery to an Adult Rs1h Knockout Mouse Model Restores ERG b-Wave with Reversal of the Electronegative Waveform of X-Linked Retinoschisis. Invest Ophthalmol Vis Sci. 2004;45:3279–3285. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- 5.Molday LL, Hicks D, Sauer CG, Weber BH, Molday RS. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Invest Ophthalmol Vis Sci. 2001;42:816–825. [PubMed] [Google Scholar]
- 6.Wu WW, Wong JP, Kast J, Molday RS. RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer. J Biol Chem. 2005;280:10721–10730. doi: 10.1074/jbc.M413117200. [DOI] [PubMed] [Google Scholar]
- 7.Takada Y, Fariss RN, Tanikawa A, Zeng Y, Carper D, Bush R, Sieving PA. A retinal neuronal developmental wave of retinoschisin expression begins in ganglion cells during layer formation. Invest Ophthalmol Vis Sci. 2004;45:3302–3312. doi: 10.1167/iovs.04-0156. [DOI] [PubMed] [Google Scholar]
- 8.Molday RS. Focus on molecules: retinoschisin (RS1) Exp Eye Res. 2007;84:227–228. doi: 10.1016/j.exer.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Wu WW, Molday RS. Defective discoidin domain structure, subunit assembly, and endoplasmic reticulum processing of retinoschisin are primary mechanisms responsible for X-linked retinoschisis. J Biol Chem. 2003;278:28139–28146. doi: 10.1074/jbc.M302464200. [DOI] [PubMed] [Google Scholar]
- 10.Kiedzierska A, Smietana K, Czepczynska H, Otlewski J. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim Biophys Acta. 2007;1774:1069–1078. doi: 10.1016/j.bbapap.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–1631. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL. Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature. 1999;402:439–442. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 13.Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 1999;402:434–439. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- 14.Lee CC, Kreusch A, McMullan D, Ng K, Spraggon G. Crystal structure of the human neuropilin-1 b1 domain. Structure. 2003;11:99–108. doi: 10.1016/s0969-2126(02)00941-3. [DOI] [PubMed] [Google Scholar]
- 15.Fraternali F, Cavallo L, Musco G. Effects of pathological mutations on the stability of a conserved amino acid triad in retinoschisin. FEBS Lett. 2003;544:21–26. doi: 10.1016/s0014-5793(03)00433-2. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Waters CT, Rothman AM, Jakins TJ, Romisch K, Trump D. Intracellular retention of mutant retinoschisin is the pathological mechanism underlying X-linked retinoschisis. Hum Mol Genet. 2002;11:3097–3105. doi: 10.1093/hmg/11.24.3097. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, Zhou A, Waters CT, O’Connor E, Read RJ, Trump D. Molecular pathology of X linked retinoschisis: mutations interfere with retinoschisin secretion and oligomerisation. Br J Ophthalmol. 2006;90:81–86. doi: 10.1136/bjo.2005.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iannaccone A, Mura M, Dyka FM, Ciccarelli ML, Yashar BM, Ayyagari R, Jablonski MM, Molday RS. An unusual X-linked retinoschisis phenotype and biochemical characterization of the W112C RS1 mutation. Vision Res. 2006;46:3845–3852. doi: 10.1016/j.visres.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Poole S, Firtel RA, Lamar E, Rowekamp W. Sequence and expression of the discoidin I gene family in Dictyostelium discoideum. J Mol Biol. 1981;153:273–289. doi: 10.1016/0022-2836(81)90278-3. [DOI] [PubMed] [Google Scholar]
- 20.Valencia A, Pestana A, Cano A. Spectroscopical studies on the structural organization of the lectin discoidin I: analysis of sugar- and calcium-binding activities. Biochim Biophys Acta. 1989;990:93–97. doi: 10.1016/s0304-4165(89)80017-0. [DOI] [PubMed] [Google Scholar]
- 21.Ortel TL, Devore-Carter D, Quinn-Allen M, Kane WH. Deletion analysis of recombinant human factor V. Evidence for a phosphatidylserine binding site in the second C-type domain. J Biol Chem. 1992;267:4189–4198. [PubMed] [Google Scholar]
- 22.Fuentes-Prior P, Fujikawa K, Pratt KP. New insights into binding interfaces of coagulation factors V and VIII and their homologues lessons from high resolution crystal structures. Curr Protein Pept Sci. 2002;3:313–339. doi: 10.2174/1389203023380639. [DOI] [PubMed] [Google Scholar]
- 23.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal. 2006;18:1108–1116. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Ensslin MA, Shur BD. Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell. 2003;114:405–417. doi: 10.1016/s0092-8674(03)00643-3. [DOI] [PubMed] [Google Scholar]
- 25.Molday LL, Wu WW, Molday RS. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J Biol Chem. 2007;282:32792–32801. doi: 10.1074/jbc.M706321200. [DOI] [PubMed] [Google Scholar]
- 26.Janssen A, Min SH, Molday LL, Tanimoto N, Seeliger MW, Hauswirth WW, Molday RS, Weber BH. Effect of Late-stage Therapy on Disease Progression in AAV-mediated Rescue of Photoreceptor Cells in the Retinoschisin-deficient Mouse. Mol Ther. 2008;16:1010–1017. doi: 10.1038/mt.2008.57. [DOI] [PubMed] [Google Scholar]
- 27.Min SH, Molday LL, Seeliger MW, Dinculescu A, Timmers AM, Janssen A, Tonagel F, Tanimoto N, Weber BH, Molday RS, Hauswirth WW. Prolonged recovery of retinal structure/function after gene therapy in an Rs1h-deficient mouse model of x-linked juvenile retinoschisis. Mol Ther. 2005;12:644–651. doi: 10.1016/j.ymthe.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Dyka FM, Molday RS. Coexpression and interaction of wild-type and missense RS1 mutants associated with X-linked retinoschisis: its relevance to gene therapy. Invest Ophthalmol Vis Sci. 2007;48:2491–2497. doi: 10.1167/iovs.06-1465. [DOI] [PubMed] [Google Scholar]
- 29.Hegedus DD, Pfeifer TA, Hendry J, Theilmann DA, Grigliatti TA. A series of broad host range shuttle vectors for constitutive and inducible expression of heterologous proteins in insect cell lines. Gene. 1998;207:241–249. doi: 10.1016/s0378-1119(97)00636-7. [DOI] [PubMed] [Google Scholar]
- 30.Grayson C, Reid SN, Ellis JA, Rutherford A, Sowden JC, Yates JR, Farber DB, Trump D. Retinoschisin, the X-linked retinoschisis protein, is a secreted photoreceptor protein, and is expressed and released by Weri-Rb1 cells. Hum Mol Genet. 2000;9:1873–1879. doi: 10.1093/hmg/9.12.1873. [DOI] [PubMed] [Google Scholar]
- 31.Mamluk R, Gechtman Z, Kutcher ME, Gasiunas N, Gallagher J, Klagsbrun M. Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J Biol Chem. 2002;277:24818–24825. doi: 10.1074/jbc.M200730200. [DOI] [PubMed] [Google Scholar]
- 32.Reid SN, Yamashita C, Farber DB. Retinoschisin, a photoreceptor-secreted protein, and its interaction with bipolar and muller cells. J Neurosci. 2003;23:6030–6040. doi: 10.1523/JNEUROSCI.23-14-06030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vijayasarathy C, Takada Y, Zeng Y, Bush RA, Sieving PA. Retinoschisin is a peripheral membrane protein with affinity for anionic phospholipids and affected by divalent cations. Invest Ophthalmol Vis Sci. 2007;48:991–1000. doi: 10.1167/iovs.06-0915. [DOI] [PubMed] [Google Scholar]
- 34.Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am J Physiol. 1998;275:F633–650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 35.Blanks JC, Johnson LV. Specific binding of peanut lectin to a class of retinal photoreceptor cells. A species comparison. Invest Ophthalmol Vis Sci. 1984;25:546–557. [PubMed] [Google Scholar]
- 36.Steiner-Champliaud MF, Sahel J, Hicks D. Retinoschisin forms a multi-molecular complex with extracellular matrix and cytoplasmic proteins: interactions with beta2 laminin and alphaB-crystallin. Mol Vis. 2006;12:892–901. [PubMed] [Google Scholar]
- 37.Barondes SH, Cooper DN, Haywood-Reid PL. Discoidin I and discoidin II are localized differently in developing Dictyostelium discoideum. J Cell Biol. 1983;96:291–296. doi: 10.1083/jcb.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotani E, Yamakawa M, Iwamoto S, Tashiro M, Mori H, Sumida M, Matsubara F, Taniai K, Kadono-Okuda K, Kato Y, et al. Cloning and expression of the gene of hemocytin, an insect humoral lectin which is homologous with the mammalian von Willebrand factor. Biochim Biophys Acta. 1995;1260:245–258. doi: 10.1016/0167-4781(94)00202-e. [DOI] [PubMed] [Google Scholar]
- 39.Pfeifer TA, Guarna MM, Kwan EM, Lesnicki G, Theilmann DA, Grigliatti TA, Kilburn DG. Expression analysis of a modified factor X in stably transformed insect cell lines. Protein Expr Purif. 2001;23:233–241. doi: 10.1006/prep.2001.1503. [DOI] [PubMed] [Google Scholar]