Abstract
The E3 ubiquitin ligase Pellino can be activated by phosphorylation in vitro, catalyzed by IL-1 receptor-associated kinase 1 (IRAK1) or IRAK4. Here, we show that phosphorylation enhances the E3 ligase activity of Pellino 1 similarly with any of several E2-conjugating enzymes (Ubc13-Uev1a, UbcH4, or UbcH5a/5b) and identify 7 amino acid residues in Pellino 1 whose phosphorylation is critical for activation. Five of these sites are clustered between residues 76 and 86 (Ser-76, Ser-78, Thr-80, Ser-82, and Thr-86) and decorate a region of antiparallel β-sheet, termed the “wing,” which is an appendage of the forkhead-associated domain that is thought to interact with IRAK1. The other 2 sites are located at Thr-288 and Ser-293, just N-terminal to the RING-like domain that carries the E3 ligase activity. Unusually, the full activation of Pellino 1 can be achieved by phosphorylating any one of several different sites (Ser-76, Thr-86, Thr-288, or Ser-293) or a combination of other sites (Ser-78, Thr-80, and Ser-82). These observations imply that dephosphorylation of multiple sites is required to inactivate Pellino 1, which could be a device for prolonging Pellino's E3 ubiquitin ligase activity in vivo.
Keywords: Toll-like receptor, innate immunity, Lysine63-linked polyubiquitination
During infection by bacteria, bacterial products engage Toll-like receptors (TLRs) in immune cells, triggering the activation of signaling pathways that lead to the production of proinflammatory cytokines, chemokines, and interferons. Signaling via all TLRs (except TLR3) or the receptor for the proinflammatory cytokine IL-1 leads to the recruitment of proteins, such as myeloid differentation factor 88 (MyD88) (1, 2) and the serine/threonine-specific protein kinases IL-1 receptor-associated protein kinase 1 (IRAK1) and IRAK4 (3, 4). IRAK4 activates IRAK1, which is followed by the autophosphorylation of IRAK1 at several sites (5), its release from MyD88 (6), and its association with an E3 ubiquitin ligase, termed TNF receptor-associated factor 6 (TRAF6) (7) with which it propagates the signal.
Signaling by the IRAK1 (8, 9) and TRAF6 (10, 11) involves the Lys-63-linked polyubiquitination of both proteins. The IL-1-stimulated polyubiquitination of IRAK1 was first described >10 years ago (12) and initially thought to be a Lys-48-linked polyubiquitination event that led to the proteasomal degradation of IRAK1, explaining the rapid disappearance of IRAK1 under these conditions. However, more recently, we (8) and others (9, 13) established, through the use of ubiquitin mutants, Lys-63-linked polyubiquitin-binding proteins, and antibodies that recognize Lys-63-linked polyubiquitin chains specifically, that IL-1 stimulates the rapid Lys-63-linked polyubiquitination of IRAK1. Moreover, we showed that incubation with proteasomal inhibitors did not enhance the levels of endogenous polyubiquitinated IRAK1 nor did it prevent the IL-1-stimulated disappearance of unmodified IRAK1 (8). Indeed, the IL-1 induced “disappearance” of IRAK1 was reported not to be caused by proteolytic destruction at all, but simply to result from conversion to a variety of phosphorylated and polyubiquitinated species, as demonstrated by the regeneration of unmodified IRAK1 upon treatment of cell extracts with a protein phosphatase and a deubiquitinase (9).
The Lys-63-linked polyubiquitination of IRAK1 was found to be critical for the activation of the transcription factor NF-κB, probably by recruiting the NEMO-IKKα/β complex (14, 15) and inducing conformational changes (16) that facilitate its activation by TRAF6-associated TAK1 (10, 11, 17). However, a major unresolved question is how IL-1 or TLR agonists stimulate the formation of K63-linked TRAF6 and K63–pUb–IRAK1. Isoforms of Pellino, which were shown some years ago to interact with IRAK1 and IRAK4 (18–22), were recently reported to be E3 ubiquitin ligases (23–25). Moreover, we showed that Pellino isoforms only became competent to form K63–pUb chains after phosphorylation by IRAK1 or IRAK4 in vitro, an effect reversed by dephosphorylation (24). The cotransfection of vectors encoding wild-type IRAK1 and wild-type Pellino 2 induced the phosphorylation of Pellino 2 and the formation of K63–pUb–IRAK1 in cells. In contrast, transfection of wild-type IRAK1 and an E3 ligase-deficient mutant of Pellino induced the phosphorylation of Pellino 2 but not the formation of K63–pUb–IRAK1, whereas cotransfection of a catalytically-inactive mutant of IRAK1 with wild-type Pellino 2 did not induce the phosphorylation of Pellino 2 or the formation of K63–pUb–IRAK1 (24). These observations were consistent with the notion that, at least in cotransfection experiments, IRAK1 catalyzes the phosphorylation and activation of Pellino 2, which then polyubiquitinates IRAK1. This mechanism can potentially explain how IL-1 and TLR agonists stimulate the formation of K63–pUb–IRAK1 in cells. However, when incubated in vitro with the E2-conjugating enzyme Ubc13–Uev1a, IRAK1-activated Pellino 1 mediates the formation of free Lys-63-linked polyubiquitin chains that are not linked to either IRAK1 or Pellino 1 (24). This result suggests that the polyubiquitination of IRAK1 may be a 2-step process. For example, Pellino may first combine with an E2 distinct from Ubc13–Uev1a to monoubiqutinate IRAK1 and then with Ubc13–Uev1a to produce Lys-63-linked polyubiquitinated IRAK1.
To investigate whether IL-1 induces the phosphorylation of Pellino isoforms in vivo, it is first necessary to identify the amino acid residues phosphorylated by IRAK1 and IRAK4 in vitro and determine which phosphorylation sites are critical for activation of Pellino's E3 ligase activity. Here, we show that Pellino is phosphorylated at multiple sites by IRAK1 and IRAK4 and that activation can be achieved by phosphorylating any one of several sites or a combination of other sites. These observations imply that dephosphorylation of multiple sites is required to inactivate Pellino 1, which could be a device for prolonging Pellino's E3 ubiquitin ligase activity in vivo.
Results
IRAK-Catalyzed Activation of Pellino 1 Requires Phosphorylation of Pellino 1.
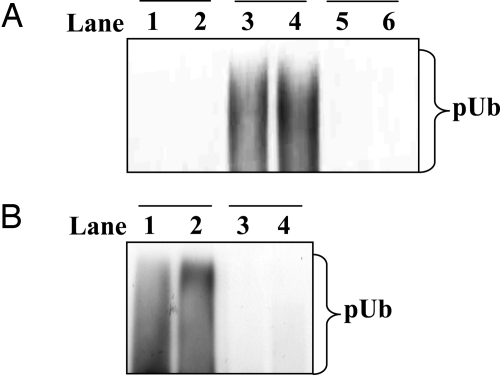
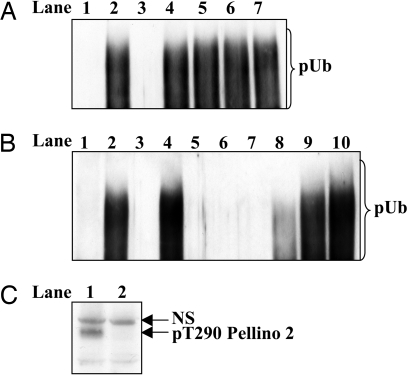
The E3 ligase activity of Pellino isoforms is greatly enhanced by phosphorylation catalyzed by IRAK1 or IRAK4 in vitro, an effect reversed by treatment with a protein phosphatase (24). These findings suggested but did not prove that activation had resulted from the phosphorylation of Pellino 1. In particular, it remained possible that activation was triggered by the autophosphorylation of IRAK1 and/or IRAK4, which then bound to Pellino, inducing a conformational change that activated its latent E3 ligase activity. To distinguish between these possibilities, we compared the activity of a phosphorylated IRAK1–unphosphorylated Pellino 1 complex with a phosphorylated IRAK1–phosphorylated Pellino 1 complex. These experiments demonstrated that the phosphorylated IRAK1–unphosphorylated Pellino 1 complex had the same activity as unphosphorylated Pellino 1 when assayed with Ubc13–Uev1a as the E2-conjugating enzyme (Fig. 1A), demonstrating that the autophosphorylation of IRAK1 per se was not sufficient to induce the activation of unphosphorylated Pellino 1 via a protein–protein interaction. We also found that phosphorylated Pellino 1 remained fully active when freed from its activator IRAK4, again showing that phosphorylation of Pellino 1 is required for activation (Fig. 1B).
Fig. 1.
The IRAK-catalyzed activation of Pellino 1 requires the phosphorylation of Pellino 1. (A) The E3 ubiquitin ligase activities of three different Pellino complexes were compared by using Ubc13–Uev1a as the E2-conjugating enzyme. Lanes 1 and 2 show the activity of a phosphorylated IRAK1–unphosphorylated Pellino 1 complex, lanes 3 and 4 show a phosphorylated IRAK1–phosphorylated Pellino 1 complex, and lanes 5 and 6 show unphosphorylated Pellino 1 in the absence of IRAK1. The preparation of each complex and their assay are detailed in Experimental Procedures. pUb, polyubiquitin. (B) GST–Pellino 1 was phosphorylated with IRAK4 (lanes 1 and 2) or left unphosphorylated by incubation without IRAK4 (lanes 3 and 4). GST–Pellino 1 was then freed from IRAK4 and assayed for E3 ligase activity as described in Experimental Procedures.
Identification of in Vitro Phosphorylation Sites on Pellino 1.
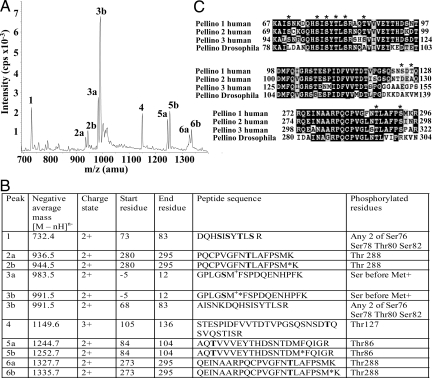
To identify the sites of Pellino 1 phosphorylation, Pellino 1 was phosphorylated with IRAK1 (24), digested with trypsin, and analyzed by MS (Fig. 2A) (26, 27). Diphosphorylated forms of phospho-peptides comprising residues 73–83 and 68–83 were identified, and the MS data were consistent with phosphorylation of any 2 of the 4 residues, Ser-76, Ser-78, Thr-80, and Ser-82 (Fig. 2B). Three other phosphorylation sites were identified as Thr-86, Thr-127, and Thr-288 (Fig. 2B). We compared the sequences of the 3 human Pellino isoforms, which show >80% amino acid similarity, and Drosophila Pellino, and observed that Ser-76, Ser-78, Thr-80, Ser-82, Thr-86 and Thr-288 were conserved or phosphorylatable residues in all 4 species of Pellino, but Thr-127 was not (Fig. 2C).
Fig. 2.
Identification of phosphorylation sites in Pellino 1 by LC/MS. (A) Pellino 1 was phosphorylated by IRAK1 in vitro and subjected to SDS/PAGE, and the band corresponding to phosphorylated Pellino 1 was excised, digested with trypsin, and analyzed by LC/MS with precursor of 79 scanning (27). (B) Identification of the major peaks in A. Phosphorylated residues are shown in bold, and methionine sulfoxide is indicated by M*. The first 5 residues of phosphopeptides 3a and 3b, precede the initiating methionine (M+) of Pellino 1 for reasons explained in Results. (C) Comparison of the amino acid sequences surrounding the identified phosphorylation sites in human and Drosophila Pellinos. The major sites of IRAK1/4-catalyzed phosphorylation identified in this study are denoted by *. The white letters on a black background indicate identities between the sequences and black letters on a gray background indicate conservative replacements.
The removal of the GST tag from Pellino 1 by cleavage with PreScission protease leaves a pentapeptide sequence GPLGS, preceding the initiating methionine residue of Pellino 1. We found that the serine in this sequence was also phosphorylated by IRAK1 (Fig. 2), but its mutation to Ala had no effect on the ability of IRAK1 to activate Pellino 1 (Fig. S1). All subsequent experiments were therefore carried out with a mutant in which this residue was changed to Ala.
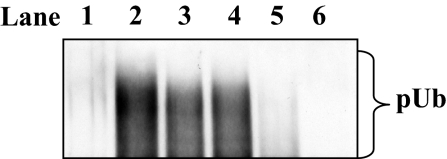
To identify phosphorylated residues that were critical for activation of Pellino's E3 ligase activity, the 7 sites identified in Fig. 2 were mutated to Ala both separately and in combination, and the ability of Pellino 1 to mediate the formation of free Lys-63-linked polyubiquitin chains in the presence of Ubc13–Uev1a was studied. Surprisingly, even when all 7 sites (Ser-76, Ser-78, Thr-80, Ser-82, Thr-86, Thr-127, and Thr-288) were mutated to Ala, IRAK4 (Fig. 3, compare lanes 2 and 3) was able to activate this form of Pellino 1 (termed Pellino 1[7A]) similarly to wild-type Pellino 1, suggesting that an additional site(s) of phosphorylation had been missed that was critical for activation. Pellino 1[7A] was therefore phosphorylated by incubation with IRAK4 and Mg-[γ-32P]ATP and found to incorporate 25% of the 32P-radioactivity introduced into wild-type Pellino 1. MS analysis of the tryptic digest as in Fig. 2 identified Ser-70 as an additional site of phosphorylation, but Pellino 1[8A], in which Ser-70 and the other 7 identified sites were mutated to Ala, could also be activated by IRAK4 (Fig. 3, lane 4) similarly to wild-type Pellino 1.
Fig. 3.
Activation of wild-type and mutant forms of Pellino 1 by IRAK4 in vitro. Pellino 1 was phosphorylated with (lanes 2–6) or without (lane 1) IRAK4, and its E3 ligase activity was measured with Ubc13-Uevla for 3 min as described in Experimental Procedures. Lanes 1 and 2, wild-type Pellino 1; lane 3, Pellino 1[7A]; lane 4, Pellino 1[8A]; lane 5, Pellino 1[10A]; lane 6, Pellino 1[10A] in which Ala-125 and Ala-127 were back-mutated to Ser and Thr, respectively. In Pellino 1[7A] residues 76, 78, 80, 82, 86, 127, and 288 all were mutated to Ala. Pellino 1[8A] contained the same mutations as Pellino 1[7A] except for the further mutation of Ser-70 to Ala. Pellino 1[10A] contained the same mutations as Pellino 1[8A] except for the further mutation of Ser-125 and Ser-293 to Ala (see Fig. 2C for the amino acid sequences of these regions).
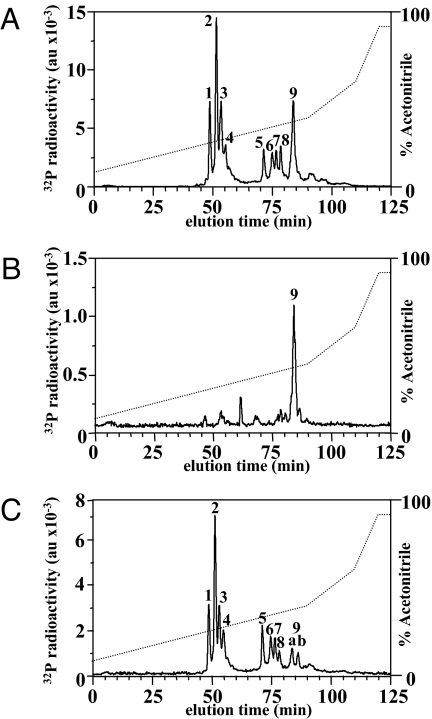
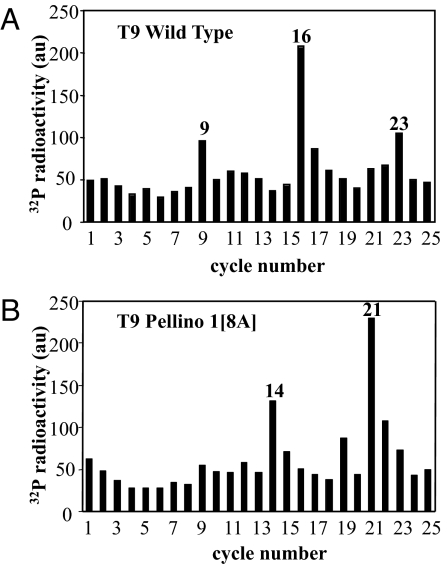
We next phosphorylated wild-type Pellino 1 and Pellino 1[8A] with IRAK4 and Mg-[γ32P]ATP but, after tryptic digestion, we used RP-HPLC to separate the 32P peptides. These experiments revealed that the major peptide (T9) in the digest of Pellino 1[8A] (Fig. 4B) was the latest eluting of the many 32P peptides present in the digest of wild-type Pellino 1 (Fig. 4A). MS analysis of this fraction from both digests revealed the presence of the same 3 phosphopeptides corresponding to amino acid residues 280–295, 273–295 and 105–136, the first of these arising from cleavage of the Arg–Pro bond between residues 279 and 280, which did not occur in the second peptide. Solid-phase Edman sequencing of this fraction from wild-type Pellino 1 showed that 32P radioactivity was released from the peptides after the ninth, 16th, and 23rd cycles of Edman degradation (Fig. 5A), indicating that phosphorylation had taken place at Thr-288 (the ninth and 16th residues of the peptides 280–295 and 273–295, respectively) and Thr-127 (the 23rd residue in the peptide 105–136), 2 of the sites detected earlier in this study by a different method (Fig. 2). In contrast, solid-phase sequencing of the same fraction from the tryptic digest of Pellino 1[8A] released 32P radioactivity after the 14th and 21st cycles of Edman degradation (Fig. 5B). These results demonstrated that Ser-293 (the 14th and 21st residue of the peptides 280–295 and 273–295, respectively) had become phosphorylated now that Thr-288 was no longer available for phosphorylation, and that Ser-125 (the 21st residue of the peptide 105–136) had become phosphorylated now that Thr-127 was unavailable for phosphorylation. Ser-125 is replaced by an amino acid residue that cannot be phosphorylated in Pellino 2, Pellino 3, and Drosophila Pellino (Fig. 2C). Ser-293 is conserved in the 3 human Pellino isoforms, although not in Drosophila Pellino (Fig. 2C).
Fig. 4.
Separation of tryptic phosphopeptides from Pellino 1. 32P-labeled Pellino 1 obtained by incubation with Mg-[γ32P]ATP and either IRAK4 (A and B) or IRAK1 (C) was subjected to SDS/PAGE, the gel was stained with Coomassie blue, and Pellino 1 was excised, digested with trypsin, and subjected to RP-HPLC on a Vydac C18 column equilibrated in 0.1% (vol/vol) trifluoroacetic acid. The column was developed with an acetonitrile gradient in 0.1% (vol/vol) trifluoroacetic acid. 32P radioactivity in arbitrary units (au) is shown by the full line, and the acetonitrile gradient is indicated by the diagonal broken lines. (A and C) Wild-type Pellino 1. (B) Pellino 1[8A].
Fig. 5.
Solid-phase sequencing of peptide T9 from Fig. 4. Peak T9 from wild-type Pellino 1 (Fig. 4A) and Pellino 1[8A] (Fig. 4B) were subjected to solid-phase sequencing (36) to identify the cycles of Edman degradation at which 32P radioactivity (filled bars) was released from the phosphopeptides present in these fractions.
The results described above led us to generate a further mutant, termed Pellino 1[10A], in which Ser-125 and Ser-293, and the other 8 sites in Pellino 1[8A], were mutated to Ala. This mutant could hardly be activated by IRAK4 (Fig. 3, lane 5) compared with wild-type Pellino 1 (Fig. 3, lane 2) or Pellino 1[8A] (Fig. 3, lane 4), suggesting that the key activating sites were located within the 10 Ser/Thr residues that had been mutated. Like the other Pellino mutants, Pellino 1[10A] retained the trace basal activity of unphosphorylated wild-type Pellino 1, indicating that the mutation of multiple Ser/Thr residues to Ala had not affected the ability of Pellino 1 to function as an E3 ligase.
We also analyzed the 32P-tryptic peptides T1–T8 in the digest of wild-type Pellino 1 (Fig. 4A). Peptides T1-T4 were found to contain several phosphopeptides spanning the region between amino acids 68 and 83. T1 and T2 contained triply and doubly phosphorylated versions, respectively, of a peptide comprising residues 73–83. Like peptide T2, peptide T3 also contained the doubly phosphorylated derivative of the peptide comprising residues 73–83 plus a peptide comprising residues 68–83 with 3 and 4 phosphorylated residues. Peptide T4 contained doubly and singly phosphorylated forms of the peptide 73–83 and a doubly phosphorylated version of the peptide comprising residues 68–83. Solid-phase sequencing of phosphopeptides T1–T4 was consistent with phosphorylation taking place at Ser-76, Ser-78, Thr-80, and Ser-82 (Fig. S2A). Peptide T5 contained the peptide 84–104 phosphorylated at Thr-86 and the peptide 280–295 phosphorylated at both Thr-288 and Ser-293. T6 contained a diphosphorylated version of the peptide 273–295, phosphorylated at both Thr-288 and Ser-293, and a monophosphorylated version of the peptide 280–295. Peptide T7 also contained the peptide 273–295 but phosphorylated at Thr-288 alone, plus the peptide comprising residues 280–295 phosphorylated at Thr-288 and the peptide comprising 105–136. Peptide T8 also contained a monophosphorylated form of the peptide 273–295, which was phosphorylated at Thr-288, and the peptide 280–295, which was phosphorylated at both Thr-288 and Ser-293. Thus, the major phosphopeptides detected in this analysis were consistent with the results obtained in Fig. 2, but identified the precise sites of phosphorylation more definitively (Table S1A).
Comparison of the Sites Phosphorylated by IRAK1 and IRAK4.
The pattern of phosphopeptides detected after RP-HPLC of tryptic digests of Pellino 1 phosphorylated by IRAK1 (Fig. 4C) was similar to that observed after phosphorylation by IRAK4 (Fig. 4A) and was further confirmed by solid-phase sequencing of phosphopeptides T1 to T4 (Fig. S2B). IRAK1 did, however, appear to phosphorylate Thr80 preferentially to Ser82 and the region comprising amino acid residues 105–136 (containing Ser-125 and Thr-127) much more weakly than IRAK4 (Table S1B).
Identification of Amino Acid Residues Whose Phosphorylation Activates Pellino 1.
To identify which phosphorylation sites were critical for activation, we back-mutated particular Ala residues in Pellino 1[10A] to the Ser or Thr residues present in wild-type Pellino 1. Like Pellino 1, Pellino 2 and Pellino 3 can be activated by IRAK1 and IRAK4 in vitro (23, 24), but Ser-125 and Thr-127 are not present in these isoforms (Fig. 2C). For this reason, it seemed unlikely that they would be the key activating sites and so Ala-125 and Ala-127 were initially reconverted to Ser and Thr, respectively, while keeping all of the other phosphorylation sites as Ala residues. This mutant could not be activated by IRAK4 in vitro (Fig. 3, lane 6), demonstrating that activation could not be achieved by the phosphorylation of Ser-125 and/or Thr-127 alone.
We next restored amino acid residues 288 and/or 293 to Thr and Ser, respectively. These experiments revealed that the reconversion of residue 288 to Thr and/or residue 293 to Ser was sufficient to restore activation to a level similar to that observed with wild-type Pellino 1, even when all of the other 8 sites were unavailable for phosphorylation (Fig. 6A, compare lanes 4–6 with lanes 2 and 3). However, Pellino 1 in which both Thr-288 and Ser-293, but no other residue, had been mutated to Ala could also be activated by IRAK4 (Fig. 6A, lane 7). This result indicated that phosphorylation of one or more of the Ser and Thr residues between Ser-70 and Thr-86 was also sufficient for activation if neither Thr-288 nor Ser-293 was available for phosphorylation.
Fig. 6.
Activation of different mutants of Pellino 1. Wild-type and mutated forms of Pellino 1 were phosphorylated with (lanes 2–7) or without (lane 1) IRAK4, and their E3 ligase activity was measured for 3 min as described in Experimental Procedures with Ubc13-Uevla. Pellino 1[10A] is a form of Pellino 1 in which the 10 phosphorylation sites at residues 70, 76, 78, 80, 82, 86, 125, 127, 288, and 293 are all mutated to Ala. (A) Lanes 1 and 2, wild-type Pellino 1; lane 3, Pellino 1[10A] in which Ala-125 and Ala-127 had been back-mutated to Ser and Thr, respectively; lane 4, same as lane 3, except for the additional back mutation of Ala-288 to Thr; lane 5, same as lane 3 except for the additional back mutation of Ala-293 to Ser; lane 6, same as lane 3, except that both Ala-288 and Ala-293 were back-mutated to Thr and Ser, respectively; lane 7, Pellino 1 in which Thr-288 and Ser-293 (but no other residue) was mutated to Ala. (B) Lanes 1 and 2, wild-type Pellino 1; lane 3, Pellino 1[10A] in which Ala-125 and Ala-127 were back-mutated to Ser and Thr, respectively; lanes 4, 5, 6, 7, and 8 show the activities of Pellino 1[10A] in which Ala-76 alone, Ala-78 alone, Ala-80 alone, Ala-82 alone, or Ala-86 alone, respectively were back-mutated to the Ser or Thr present in wild-type Pellino 1; lane 9, Pellino 1[10A] in which Ala-78, Ala-80, and Ala-82 were backmutated to Ser, Thr, and Ser, respectively; lane 10, Pellino 1[10A] in which Ala-78, Ala-80, Ala-82, and Ala-86 were back-mutated to Ser, Thr, Ser, and Thr, respectively. (C) DNA vectors encoding HA-Pellino 2 and either wild-type flag–IRAK1 (lane 1) or the catalytically-inactive flag–IRAK1[K239A] mutant (lane 2) were cotransfected into IRAK1-null IL-1R cells. Twenty-four hours later the cells were lysed and 25 μg of extract protein was subjected to SDS/PAGE and immunoblotted with a phospho-specific antibody that recognizes Thr-288 of Pellino 1 and the equivalent residue in Pellino 2 (Thr-290). NS, nonspecific band recognized by the antibody.
To identify other residues whose phosphorylation was capable of activating Pellino 1 we back-mutated Pellino 1[10A] to restore residue 76 to Ser, or residue 78 to Ser, or residue 80 to Thr, or residue 82 to Ser, or residue 86 to Thr. These experiments revealed that the back-mutation of residue 76 alone to Ser was sufficient to restore activation by IRAK4 (Fig. 6B, lane 4) to a level similar to that observed with wild-type Pellino 1. The back mutation of residue 86 to Thr alone allowed partial activation by IRAK4 (Fig. 6B, lane 8), but the individual back mutation of residue 78 or residue 80 or residue 82 did not by themselves permit any activation by IRAK4 (Fig. 6B, lanes 5–7). However, when residues 78, 80, and 82 or residues 78, 80, 82, and 86 were back-mutated to Ser or Thr in combination, while keeping residues 76, 125, 127, 288, and 293 as Ala, these species could also be activated by IRAK4 (Fig. 6B, lanes 9 and 10) similarly to wild-type Pellino 1.
Thr-290 of Pellino 2 Is Phosphorylated in Cells.
To investigate whether Pellino was phosphorylated in cells at a residue critical for activation, we raised a phospho-specific antibody that recognizes Pellino 1 phosphorylated at Thr-288 and also recognizes Pellino 2 phosphorylated at the equivalent residue (Thr-290). The cotransfection of Pellino 2 with wild-type IRAK1 in IRAK1-null IL-1R cells led to the phosphorylation of Pellino 2 at Thr-290, but this did not occur if wild-type IRAK1 was replaced with a catalytically-inactive mutant (Fig. 6C). Pellino 2 was used in this experiment instead of Pellino 1, because Pellino 1 was difficult to express in these cells.
Effect of Phosphorylation on the E3 Ligase Activity of Pellino 1 with Different E2-Conjugating Complexes.
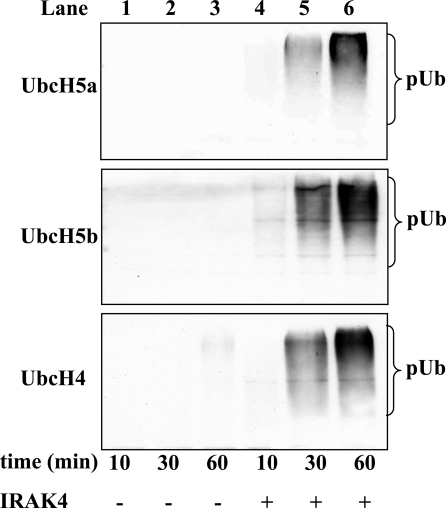
In the experiments described above, the activity of Pellino 1 was studied in vitro by using the E2-conjugating enzyme Ubc13–Uev1a with which it forms free Lys-63-linked polyubiquitin chains not attached to any other protein. However, we have reported that Pellino 1 can form polyubiquitin chains of different topology when incubated with other E2-conjugating complexes. For example, when incubated with UbcH4 or UbcH5a/5b the polyubiquitin chains formed were mainly linked via Lys-11 and Lys-48 of ubiquitin (24). Moreover, in contrast to the free Lys-63-linked polyubiquitin chains formed with Ubc13–Uev1a, the polyubiquitin chains formed with UbcH4/5a/5b are conjugated to IRAK1 and Pellino (24). It was therefore important to know whether the phosphorylation of Pellino 1 also increased its E3 ubiquitin ligase activity when assayed with E2s other than Ubc13–Uev1a. These experiments revealed that the phosphorylation of Pellino 1 also greatly enhanced its activity with UbcH5a, UbcH5b, and UbcH4 (Fig. 7). After tryptic digestion and analysis by MS, we identified Lys-355 and Lys-397 in IRAK1 and Lys-169, Lys-202, and Lys-266 of Pellino 1 as amino acid residues that became conjugated to ubiquitin in the presence of UbcH4 in vitro (Fig. S3).
Fig. 7.
Effect of phosphorylation of Pellino 1 on its E3 ligase activity in the presence of different E2-conjugating enzymes. Pellino 1 was phosphorylated by incubation for 30 min at 30 °C without (lanes 1–3) or with (lanes 4–6) IRAK4, and its E3 ligase activity then measured for the times indicated by using UbcH5a (Top), UbcH5b (Middle), or UbcH4 (Bottom) as the E2-conjugating enzyme (see Experimental Procedures). pUb, polyubiquitin.
The failure to detect Lys-134 and Lys-180 of IRAK1 was initially surprising as their polyubiquitination was reported to be critical for signaling “downstream” of IRAK1 (9). However, inspection of the sequence of IRAK1 revealed that Lys-134 and Lys-180 should both be present in a single 75-residue tryptic peptide. The large size of this peptide probably explains the failure to detect it in MS analysis.
Discussion
In this article we demonstrate that Pellino 1 is phosphorylated at multiple sites by IRAK1 or IRAK4 in vitro. The key residues involved in activation are located between residues 76 and 86 (Ser-76, Ser-78, Thr-80, Ser-82, and Thr-86) and at Thr-288 and Ser-293, just N-terminal to the RING-like domain that carries the E3 ligase activity. Unusually, we found that the phosphorylation of Ser-76 or Thr-288 or Ser-293 alone was sufficient for maximal activation, under the assay conditions that we studied, whereas phosphorylation of Thr-86 alone caused partial activation, even when all of the other phosphorylation sites were mutated to Ala. Moreover, if Ser-76, Thr-86, Thr-288, and Ser-293 all were mutated to Ala, full activation by IRAK1 or IRAK4 could still occur if Ser-78, Thr-80, and Ser-82 were available for phosphorylation (Fig. 6). All of these sites are conserved in Pellino 2 and 3 and in Drosophila Pellino, apart from Thr-86, which is Ser in Pellino 3 and Ala in Drosophila Pellino (Fig. 2C). Moreover, because Pellino 2 and Pellino 3 can also be activated by IRAK1/IRAK4, these observations imply that all forms of Pellino are likely to be activated by a common mechanism. A key outstanding question is whether IRAK1, IRAK4, or both protein kinases mediate the activation of Pellino isoforms in vivo.
The finding that Pellino 1 (and presumably other Pellino isoforms) can be activated by the phosphorylation of several different sites, or sets of sites, implies that the dephosphorylation of multiple residues is needed before any decrease in the E3 ligase activity of Pellino could occur. This may be a device for maintaining the activation of Pellino isoforms and prolonging the Lys-63-linked polyubiquitination of IRAK1. This situation is unusual, although it is reminiscent of the phosphorylation of mitogen-activated protein kinase kinase 1 (MKK1) by c-Raf, where the phosphorylaton of either Ser-218 or Ser-222 is sufficient for activation, and dephosphorylation of both sites is needed for inactivation (28). Similarly, the activity of glycogen synthase can be decreased by phosphorylating different sets of serine residues, although in this case distinct protein kinases phosphorylate the different sites (29).
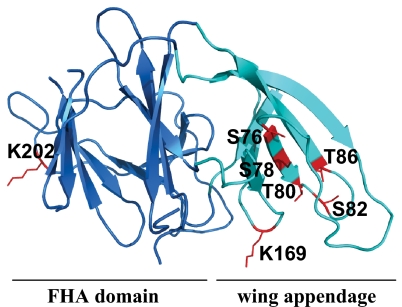
While this study was in progress, the 3D structure of Pellino 2 (but lacking the C-terminal RING-like domain) was solved by X-ray crystallography (30). These studies unexpectedly revealed the presence of a “cryptic” Forkhead-associated (FHA) domain formed by several disparate regions of Pellino 2. The cluster of phosphorylation sites between amino acid residues 76 and 86 decorate a region of antiparallel β-sheet that forms an appendage or wing of the FHA domain (Fig. 8). The presented structure terminates at residue 258 of Pellino 2, so that residues 288 and 293 of Pellino 1 (equivalent to residues 290 and 295 of Pellino 2), which are located just before the RING domain, are not present, but are clearly not situated within the FHA domain. FHA domains are known to bind to phosphothreonine-containing peptides, and one role of this domain in Pellino could be to bind the autophosphorylated form of IRAK1. Such an interaction might facilitate the IRAK1-catalyzed phosphorylation Pellino or position IRAK1 in such a way that it can be polyubiquitinated efficiently by Pellino–E2 complexes. Interestingly, 2 of the lysine residues in Pellino 1 that underwent autopolyubiquitination in vitro upon incubation with UbcH4, namely Lys-169 and Lys-202 (equivalent to Lys-171 and Lys-204 of Pellino 2), are located in insertions that loop out from the wing and the FHA domain, respectively (Fig. 8). The third lysine (Lys-266 of Pellino 1/Lys-268 of Pellino 2) is located C-terminal to the fragment of Pellino 2 whose structure was presented (30).
Fig. 8.
Location of the N-terminal phosphorylation sites and autoubiquitination sites in the structure of Pellino 1. A homology model of Pellino 1 was generated based on the recently published structure of Pellino 2 (30) using the program Modeller (37). Ser-76, Ser-78, Thr-80, Ser-82, and Thr-86 of Pellino 1 (shown in red) are located in a region of antiparallel β-sheet, termed the wing (turquoise), which is an appendage of the cryptic FHA domain (blue) thought to interact with phosphorylated IRAK1. Lys-169 and Lys-202 of Pellino 1, which were detected as sites of autoubiquitination, are also shown in red.
The E2-conjugating enzyme Ubc13–Uev1a appears to play a key role in directing the formation of Lys-63-linked polyubiquitin chains in vivo. However, the observation that Ubc13–Uev1a only catalyzes the formation of free Lys-63-linked polyubiquitin chains when incubated in vitro with E3 ligases, such as TRAF6, CHIP (31), or Pellino 1 (24, 25), raises the question of how Lys-63-linked polyubiquitin chains actually become conjugated to proteins in vivo, because the cotransfection of Pellino with IRAK1 leads to the formation of Lys-63-linked polyubiquitinated IRAK1 in cells (24). The most likely explanation for this discrepancy is that a second E2-conjugating enzyme, distinct from Ubc13–Uev1a, is required to attach the first ubiquitin moiety to IRAK1, whereas Pellino–Ubc13–Uev1a complexes promote the elongation of the monoubiquitinated species. Indeed, in budding yeast, there is strong genetic evidence that proliferating cell nuclear antigen (PCNA), a key protein in the DNA repair pathway, is monoubiquitinated by the E2-conjugating enzyme RAD6 and the E3 ligase RAD18 and then Lys-63-polyubiquitinated by the E2-conjugating enzyme Ubc13–Mms2 and the E3 ligase RAD5 (32). Similarly, the Lys-63-linked polyubiquitination of MHC class 1 molecules in HeLa cells mediated by the K3 gene product of Karposi's sarcoma herpes virus involves UbcH5b/c mediated monoubiquitination followed by Ubc13-dependent Lys-63-linked polyubiquitination (33). A 2-step model for Lys-48-linked polyubiquitination has also been proposed for the anaphase-promoting complex in yeast, with UbcH4 promoting monoubiquitination and Ubc1, the yeast homologue of the human E2 25K protein, promoting chain extension (34). It therefore seems likely that a similar situation occurs in the innate immune system, with the monoubiquitnation of IRAK1 being catalyzed by an E2 distinct from Ubc13–Uev1a.
It was recently reported that IL-1 also stimulated the Lys-48-linked polyubiquitination of IRAK1 in IL-1R cells. However, in contrast to the rapid Lys-63-linked polyubiquitination of IRAK1, which was detected at 5 min and was maximal between 30 and 60 min (13), Lys-48-linked polyubiquitination of IRAK1 was much slower, peaking between 90 and 150 min. Whether a Pellino isoform mediates the slow Lys-48-linked polyubiquitination of IRAK1 in conjunction with an E2-conjugating enzyme distinct from Ubc13–Uev1a, is unknown. We have reported that Pellino participates in the formation of Lys-48-linked polyubiquitin chains when incubated with UbcH3 in vitro (24), but additional experiments have shown that this results from the UbcH3-catalyzed formation of free Lys-48-linked polyubiquitin chains, without any contribution from Pellino 1.
In summary, the results presented here, together with the work of other laboratories, suggest the following model by which IL-1 may stimulate the Lys-63-linked polyubiquitination of IRAK1. In this model, the interaction of IL-1 with its receptor first recruits MyD88, IRAK4, and IRAK1. IRAK4 then activates IRAK1 followed by the autophosphorylation of IRAK1 at multiple sites. This causes the dissociation of IRAK1 and IRAK4 from MyD88 and phosphorylated IRAK1 to bind to the FHA domain of Pellino. IRAK1 and/or IRAK4 then catalyze the phosphorylation of Pellino, followed by a 2-step process in which activated Pellino first combines with an E2, distinct from Ubc13–Uev1a to monoubiquitinate IRAK1 and then with Ubc13–Uev1a to produce Lys-63-linked polyubiquitinated IRAK1. However, the involvement of other E3 ligases, such as TRAF6 (9), in the attachment of the first or subsequent ubiquitins to IRAK1 is not excluded. Further work is therefore needed to identify all of the E3 ligases and E2-conjugating enzymes involved in the polyubiquitination of IRAK1.
Experimental Procedures
DNA Constructs.
DNA and expression vectors have been described (24). Mutations were made following the QuikChange site-directed mutagenesis method (Stratagene), but using KOD Hot Start Polymerase.
Protein Expression and Purification.
Pellino 1 was expressed as a GST-fusion protein in Escherichia coli with a PreScission protease cleavage site between the GST and the Pellino 1. The GST tag was removed by cleavage with PreScission protease as described (24), which leaves a sequence of 5 amino acids before the normal initiating methionine residue of Pellino 1. Pellino 1 that had been freed from GST in this way was used for all experiments unless specified otherwise. IRAK1 and IRAK4 were expressed as active GST-tagged proteins in insect Sf21 cells and purified on glutathione-Sepharose, whereas IRAK4 was also expressed with a His6 tag at its N terminus instead of GST and purified on nickel-nitrilo-triacetate agarose. The E2-conjugating enzymes UbcH4, UbcH5a, and UbcH5b were produced as His6-tagged proteins in E. coli and purified on nickel nitrilotriacetate agarose (24), and the E2-conjugating complex Ubc13–Uev1a and E1 enzyme were produced as described (35). Ubiquitin was purchased from Sigma.
Antibodies.
A synthetic phosphopeptide CPVGFNT*LAFPS (corresponding to residues 282–293 of Pellino 1, where T* is phosphothreonine) was conjugated to both keyhole limpet hemocyanin and BSA and injected into a sheep (sheep S393C). The serum from the third bleed was affinity-purified against the phosphopeptide antigen that had been immobilized on agarose, and the purified antibody was used for immunoblotting at 1 μg/mL in the presence of 10 μg/mL of the unphosphorylated form of the immunogen. Antiubiquitin was purchased from DakoCytomation, and anti-rabbit IgG was from Pierce.
Preparation of Different Pellino-1–IRAK1 Complexes.
A phosphorylated IRAK1–phosphorylated Pellino 1 complex was prepared by incubating IRAK1 (1 μM) with Pellino 1 (1 μM) for 30 min at 30 °C in 50 mM Tris·HCl (pH 7.5), 5 mM MgCl2, and 2 mM ATP. After 30 min IRAK1 was inhibited by adding staurosporine to a final concentration of 75 μM. A phosphorylated IRAK1–unphosphorylated Pellino 1 complex was prepared by incubating IRAK1 with MgATP as described above and only adding Pellino 1 after the IRAK1 had been inactivated with staurosporine. Unphosphorylated Pellino 1 was prepared by taking it through the same procedure in the absence of IRAK1.
Removal of IRAK4 from Pellino.
GST–Pellino 1 (1 μM) was incubated for 30 min at 30 °C in 50 mM Tris·HCl (pH 7.5), 5 mM MgCl2, and 2 mM ATP in the presence or absence of His6-IRAK4 (1 μM). After 30 min at 30 °C, glutathione-Sepharose (5 μL of packed volume) was added and the reactions kept on ice for 45 min with occasionally stirring. After brief centrifugation to pellet the glutathione Sepharose to which the GST–Pellino was now bound, the pellets were washed 3 times with 1 mL of 50 mM Tris·HCl (pH 7.5), 0.5 M NaCl and 3 times with 50 mM Tris·HCl (pH 7.5).
Assay of the E3 Ligase Activity of Pellino 1.
The assays of Pellino 1 (1 μM) were carried out in 20-μL incubations with E1 enzyme (0.1 μM), E2-conjugating enzyme (1 μM), ubiquitin (0.1 mM), magnesium chloride (5 mM), ATP (2 mM), and IRAK1/4 where indicated (1 μM) in 50 mM Tris·HCl (pH 7.5) at the final concentrations indicated in parentheses. The reactions were terminated by denaturation in SDS, subjected to SDS/PAGE, transferred to nitrocellulose, and immunoblotted with antiubiquitin.
Analysis of Phosphorylation and Ubiquitination by MS.
Phosphorylated and ubiquitinated proteins were separated by SDS/PAGE, stained with Colloidal Coomassie blue and digested with trypsin as described (26). When the sites of phosphorylation could not be assigned from the MS/MS spectra acquired on the 4000 Q-Trap, a second aliquot of the digest was analyzed by LC-MS with multistage activation on a ThermoElectron LTQ-orbitrap system coupled to a Dionex 3000 HPLC system. The resultant MS/MS spectra were searched by using Mascot (Matrixscience) run on a local server, allowing for phosphoserine/phosphothreonine, phosphotyrosine, and Gly–Gly attached to a ubiquitinated lysine residue as revealed by an enhanced mass of 114.1 Da. The mass tolerances used were +/− 1Da for 4000 Q-Trap experiments and +/− 20 ppm for orbitrap experiments.
Supplementary Material
Acknowledgments.
We thank Dr. Kathryn Ferguson for providing the coordinates of the structure of Pellino 2 (pdb file 3egb) and Dr. Xiaoxia Li for the IRAK1-null IL-1R cells. We thank Dr. Edmond Wong for producing a homology model of the structure of Pellino 1 and personnel in the Division of Signal Transduction Therapy and DNA Sequencing Service, Medical Research Council Protein Phosphorylation Unit, Dundee, for protein and antibody production and DNA sequencing. This work was supported by the U.K. Medical Research Council, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck–Serono, and Pfizer (P.C.) and a Radical Solutions for Researching the Proteome grant from the Biotechnology and Biological Sciences Research Council and Medical Research Council (to Nicolas Morrice). P.C. is a Royal Society Research Professor. H.S. is the recipient of a Wellcome Trust Prize Studentship, and E.C. is the recipient of an Engineering and Physical Sciences Research Council training grant.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2008.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900774106/DCSupplemental.
References
- 1.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 2.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: An adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: A novel member of the IRAK family with the properties of an IRAK kinase. Proc Natl Acad Sci USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin M, Bol GF, Eriksson A, Resch K, Brigelius-Flohe R. Interleukin-1-induced activation of a protein kinase coprecipitating with the type I interleukin-1 receptor in T cells. Eur J Immunol. 1994;24:1566–1571. doi: 10.1002/eji.1830240717. [DOI] [PubMed] [Google Scholar]
- 5.Kollewe C, et al. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem. 2004;279:5227–5236. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 8.Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008;28:1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 11.Deng L, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 12.Yamin TT, Miller DK. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- 13.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 14.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 15.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 16.Bloor S, et al. Signal processing by its coil zipper domain activates IKKγ. Proc Natl Acad Sci USA. 2008;105:1279–1284. doi: 10.1073/pnas.0706552105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanayama A, et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Grobhans J, Schnorrer F, Nusslein-Volhard C. Oligomerization of Tube and Pelle leads to nuclear localisation of Dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 19.Jensen LE, Whitehead S. A. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J Immunol. 2003;171:1500–1506. doi: 10.4049/jimmunol.171.3.1500. [DOI] [PubMed] [Google Scholar]
- 20.Resch K, Jockusch H, Schmitt-John T. Assignment of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet Cell Genet. 2001;92:172–174. doi: 10.1159/000056895. [DOI] [PubMed] [Google Scholar]
- 21.Rich T, Allen RL, Lucas AM, Stewart A, Trowsdale J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics. 2000;52:145–149. doi: 10.1007/s002510000249. [DOI] [PubMed] [Google Scholar]
- 22.Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–161. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- 23.Butler MP, Hanly JA, Moynagh PN. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: Direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J Biol Chem. 2007;282:29729–29737. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- 24.Ordureau A, et al. The IRAK-catalyzed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLP/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Pozuelo Rubio M, et al. 14–3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson BL, Marchese J, Morrice NA. Automated identification and quantification of protein phosphorylation sites by LC/MS on a hybrid triple quadrupole linear ion trap mass spectrometer. Mol Cell Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Alessi DR, et al. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. EMBO J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakielny S, Campbell DG, Cohen P. The molecular mechanism by which adrenalin inhibits glycogen synthesis. Eur J Biochem. 1991;199:713–722. doi: 10.1111/j.1432-1033.1991.tb16175.x. [DOI] [PubMed] [Google Scholar]
- 30.Lin C-C, Huoh Y-S, Schmitz KR, Jensen LE, Ferguson K. Pellino proteins contain a cryptic FHA domain that mediates interaction with phosphorylated IRAK1. Cell Structure (London) 2008;16:1806–1816. doi: 10.1016/j.str.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–729. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- 32.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 33.Duncan LM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, et al. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 36.Morton S, et al. Phosphorylation of the ARE-binding protein DAZAP1 by ERK2 induces its dissociation from DAZ. Biochem J. 2006;399:265–273. doi: 10.1042/BJ20060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eswar N, et al. Current Protocols in Bioinformatics. New York: Wiley; 2006. Comparative protein structure modeling using Modeller. Chapter 5: Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.