Abstract
Since it was first detected in 1996, the Goose/Guangdong/1/1996 (Gs/GD) H5N1 influenza virus and its reassortants have spread to over 60 countries, with over 20 distinct genetic reassortants previously recognized. However, systematic analysis of their interrelationship and the development of genetic diversity have not been explored. As each of those reassortants was first detected in China, here 318 full- length H5N1 virus genomes isolated from 1996–2006 in this region were phylogenetically analyzed. Our findings revealed two major group reassortment events in 2001 and 2002 that were responsible for the generation of the majority of the 44 distinct Gs/GD genotypes identified, excepting those 1997 variants. Genotype replacement and emergence occurred continually, with 34 transient genotypes detected while only 10 variants were persistent. Two major replacements of predominant genotypes were also observed: genotype B replaced by Z in 2002 and then genotype Z replaced by the now predominant genotype V in 2005.
Keywords: genotype, reassortant, avian influenza
Introduction
Highly pathogenic avian influenza (HPAI) H5N1 viruses derived from Goose/Guangdong/1/96 (Gs/GD) lineage, have affected more than 60 countries across Eurasia and Africa (FAO, 2007). The viruses have become endemic and panzootic in some of these countries and have caused repeated poultry outbreaks and human infection, posing an unprecedented global pandemic threat (Webster and Gorokova, 2007). Over 20 H5 and H7 subtype HPAI outbreaks have been recorded in the last 100 years (Alexander, 2007), but only the Gs/GD lineage H5N1 HPAI has become so geographically widespread and persisted for such an extended period of time. While the lineages and sublineages of HPAI H5N1 have been discussed in detail, there is less systematic analysis of the virus “genotype”, as defined to mean all eight genes of the virus. All recorded HPAI H5 and H7 viruses, with the exception of the Gs/GD lineage, contained a single genotype, or reassortant, in each outbreak event (Donatelli et al., 2001, Fouchier et al., 2004, Horimoto et al., 1995). In contrast, more than twenty genetic reassortants of the Gs/GD lineage H5N1 viruses have so far been recognized (Chen et al., 2006, Guan et al., 2002, Li et al., 2004). The reasons underlying the emergence and maintenance of those Gs/GD lineage reassortants remain an enigma in influenza ecology.
The Hong Kong/156/97 (HK/97)-like virus that was responsible for the Hong Kong ‘bird flu’ incident was a reassortant that derived its hemagglutinin (HA) gene from Gs/GD-like viruses and the other genes from H9N2 or H6N1 viruses found in quail and other game birds sold in live bird markets (Guan et al., 1999, Hoffman et al., 2000, Subbarao et al., 1998, Xu et al., 1999). It has been suggested by a World Health Organization (WHO) task force in early 1998 that the HK/97-like reassortant was generated locally in Hong Kong rather than being introduced from another geographic region. The depopulation of poultry in Hong Kong in December 1997 aborted that H5N1 influenza outbreak and no further human cases were detected (Shortridge et al., 1998). Since then, a virus with the HK/97-like gene constellation has not been isolated in our surveillance program in Hong Kong or southern China and it appeared that the virus was completely eradicated by these control measures. However, a recent report that the HK/97-like H5N1 virus was isolated in China from shell washes of duck and goose eggs confiscated in Guangzhou from travelers entering from Vietnam in 2005 remains an exception to be explained (Li et al., 2006).
Since 2000, reassortment of H5N1 Gs/GD lineage viruses was found to have generated multiple novel genotypes, or reassortants (Guan et al., 2002). The genetic evolution was initially described when those reassortants were first recognized in the outbreaks of 2001 and 2002 in live-poultry markets of Hong Kong Special Administration Region (Hong Kong SAR) (Guan et al., 2002, 2004). Previous studies revealed that most of those novel genotypes isolated since 2000 in Hong Kong were all recognized in our surveillance network in mainland China (Chen et al., 2006, Li al., 2004). In addition to those genotypes, recent available information suggested that some H5N1 viruses from central China appear to have undergone reassortment as early as 1997. Phylogenetic analysis revealed that some of those earlier H5N1 reassortants contained viral gene segments from influenza viruses isolated in the 1970’s (Duan et al., 2007).
Phylogenetic analyses of H5 subtype influenza viruses from Eurasia isolated from 1976 to 2005 suggested that the Gs/GD-like virus was most likely derived from a low pathogenic H5 subtype virus carried by migratory waterfowl along the western Pacific coastal region (Duan et al., 2007). Since 2003 the HPAI H5N1 viruses spread to other countries and gradually developed into a global issue by three major 5 transmission waves, i.e., 2003, 2005 and 2006 (Chen et al., 2005, Li et al., 2004, Smith et al., 2006). Our recent studies have showed that the direct precursors of those wave 1 H5N1 influenza viruses now endemic in Vietnam and Indonesia were present in Yunnan and Hunan provinces, respectively, prior to the 2003 outbreaks in Southeast Asia (Wang et al., 2008). However, there remains a gap in the understanding of the genesis and development of H5N1 influenza virus within China from 1996 to 2003. Recently, sequence data of H5N1 influenza viruses isolated from provinces in China outside our surveillance areas from 1997–2005 have become publicly available. This data, together with data accumulated through our systematic influenza surveillance, enables us to now explore the evolution and development of the genetic diversity of the Gs/GD lineage.
A critical component in describing the genetic diversity and development of the H5N1 lineage has been a genotyping system that was largely defined using viruses isolated from Hong Kong and Guangdong between 2001 and 2002 (Guan et al., 2002, Li et al 2004). However, definition of those genotypes relied on partial-length sequence data, particularly of the polymerase genes. Some studies have also designated genotypes without reference to all the genomic information already available (Chen et al., 2004, Kou et al., 2005, Wan et al., 2005). It was therefore necessary to re-assess and update the H5N1 genotype nomenclature using full-length genomes. Furthermore, there has been a large increase in available H5N1 sequence data in public databases, especially of viruses from mainland China, providing an opportunity to obtain a better understanding of the reassortment events in the evolutionary history of H5N1 virus.
Phylogenetic analysis of 318 H5N1 viruses isolated in different regions of China from 1996 to 2006 revealed that Gs/GD virus lineage viruses underwent extensive reassortment. A total of 44 different reassortants or genotypes were recognized. Even though most of those H5N1 reassortants were generated from 2000 to 2002, novel genotypes continued emerging in China throughout the study period. Our findings also suggest that almost all of those genotypes resulted from two major multiple reassortment events; the first generating genotype B, Z and W viruses, and the other generating genotype X series viruses. Furthermore, these analyses also show that all major contemporary genotypes resulted from the further reassortment of earlier viruses (e.g. genotypes G and V are derived from genotype Z). Two major genotype replacement events were observed in China, from genotype B to Z in 2002, and from genotype Z to V in 2005. Both genotype replacements were associated with the enlargement of H5N1 activity in this region. With the availability now, of full-length genome sequence information, the Clade 2.3.4 (Fujian-like variants) viruses that were previously described as genotype Z (Smith et al., 2006) now are reclassified as genotype V. The distribution of genotypes in different provinces at the same time period was similar, suggesting that major H5N1 reassortants affected many different geographic regions. The current study, once more, highlights the necessity of continued systematic influenza surveillance in poultry to track the emergence and reemergence of novel viruses with pandemic potential.
Results
Phylogenetic analysis of the surface genes
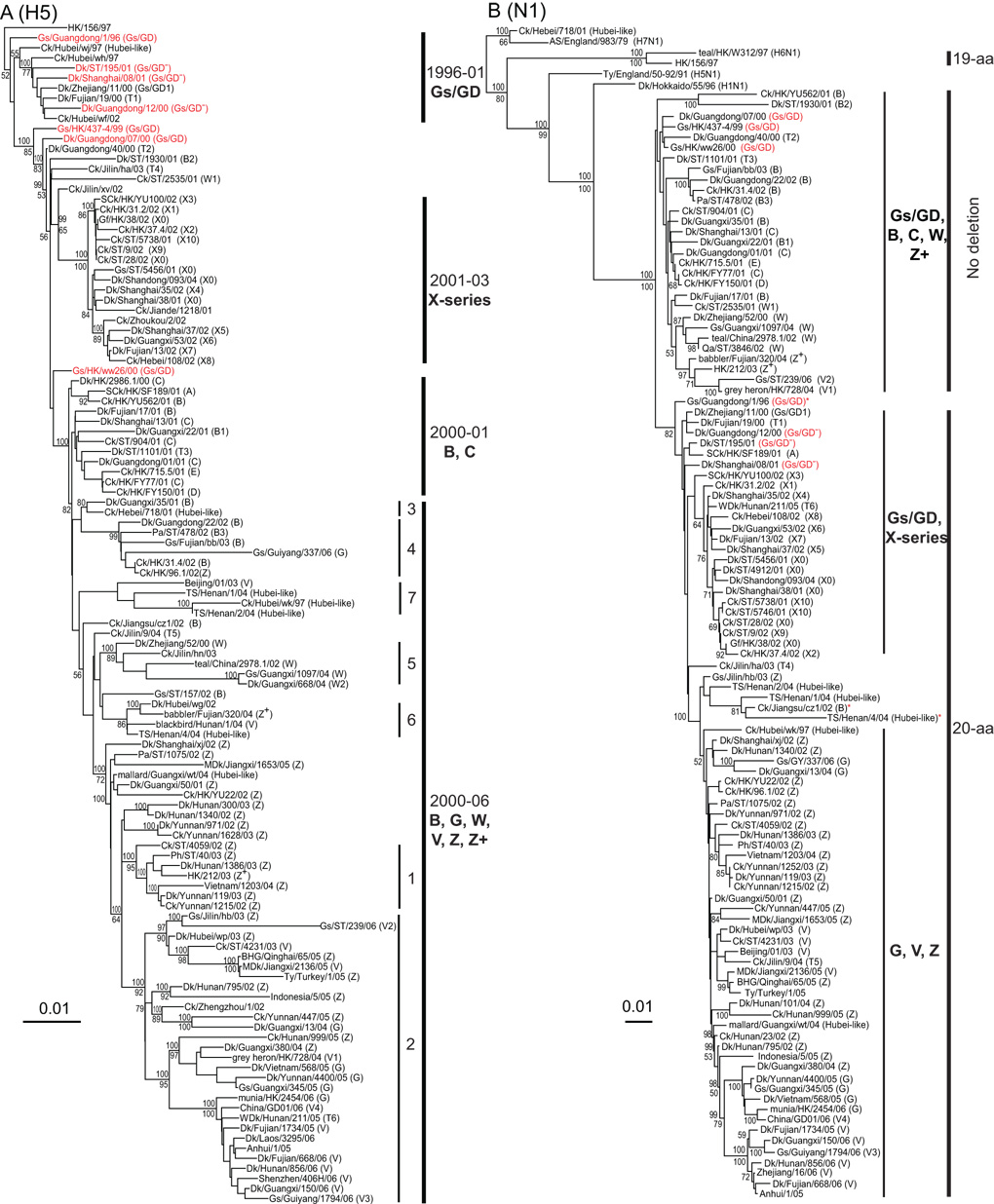
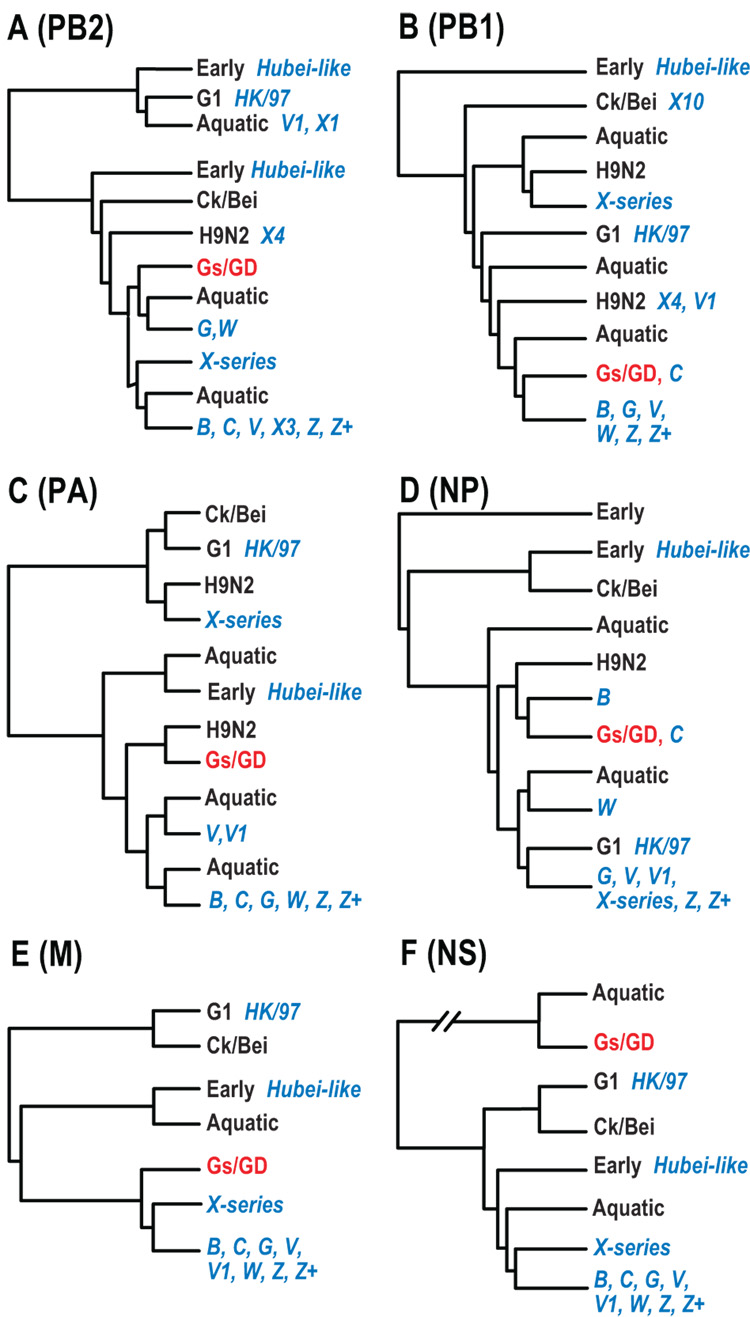
Phylogenetic analysis of H5-HA genes of Gs/GD lineage H5N1 viruses isolated from China during 1996–2006 revealed that, except for HK/97-like viruses, the Gs/GD lineage could be distinguished into four major groups (Fig. 1A). This first group contained viruses mainly isolated from 1996 to 2001 that resulted from the early establishment of Gs/GD-like viruses following early interspecies transmission and reassortment events. This group included the Hubei-like variants that contained many internal genes related to viruses isolated in the 1970’s (Duan et al., 2007). The second group included only genotype X series viruses, which were detected from 2001 to 2004, and these contained 11 distinct internal gene constellations (Fig. 1A; genotype descriptions are presented below). The third group contained viruses mostly from genotypes A–E isolated from 2000–01, that resulted from a series of reassortment events, and caused a major H5N1 outbreak in Hong Kong in 2001 (Guan et al., 2002). Viruses from this group were also detected in Fujian, Guangdong, Guangxi and Shanghai (Fig. 1A). The fourth group consisted of H5N1 virus genotypes isolated from 2001 onwards, and could be further divided into a number of subgroups that correspond to WHO designated H5N1 clades 1 to 7 (http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en). Clades 3 and 4 contain mostly genotype B, and also genotypes G and Z viruses isolated from 2001 to 2006. Clade 7 consisted mostly of transient Hubei-like viruses, along with the earliest detected genotype V virus (Beijing/01/03). Interestingly, Clade 7 viruses were all isolated after 2002 except for a single 1997 isolate (Ck/Hubei/wk/97) with an unusually long terminal branch (Fig. 1A). Clade 5 viruses belonged to genotype W and were isolated from 2000 to 2004, while Clade 6 included genotypes V, Z+ and Hubei-like transient reassortants. Clade 1 was composed of genotype Z and Z+ viruses isolated from 2002 to 2003, including the viruses from Vietnam and Thailand and their progenitors. Clade 2 contained contemporary H5N1 viruses isolated from 2003 onwards, belonging to genotypes Z, V and G. This clade could be further separated into several subclades, including Clade 2.1 (Indonesia-like, genotype Z), 2.2 (Qinghai-like, predominantly genotype Z) and 2.3.4 (Fujian-like, genotype V) (Fig 1A). It is interesting to note that the emergence of the genotype X series, some Hubei- like and Clade 2 variants (e.g. Qinghai-like and Fujian-like) was associated with unusually long branches in the phylogenetic tree, indicating the accumulation of a high number of mutations (Worobey, 2008). This was particularly apparent for individual strains such as Ck/Hubei/wk/97, Gs/GY/337/06, Gs/GX/1097/04 and Gs/ST/239/06.
Fig. 1.
Phylogenetic relationships of the HA (A) and NA (B) genes of representative influenza A viruses. Analyses were based on nucleotides 1–1,696 of the HA gene and nucleotides 1–1,355 of the NA gene. The HA and NA gene trees were rooted to duck/Hokkaido/51/96 and chicken/Scotland/59, respectively. The numbers above and below the branch nodes indicate Bayesian posterior probabilities of ≥95 and neighbor-joining bootstrap values of ≥50%, respectively. H5N1 prototype virus names are in red. Virus genotypes are given in parenthesis following the virus names. Major genotypes are indicated with bold text. Numbers labeled on the HA tree indicate WHO H5N1 clade designations (http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en). In the NA tree, 19-aa and 20-aa indicate the presence of amino acid deletions in the stalk region. *Indicates viruses without the 20-aa NA deletion. Scale bar, 0.01 nucleotide substitutions per site. Abbreviations: AS, African starling; BHG, bar-headed goose; Ck, chicken; Dk, duck; Gf, Guinea fowl; Gs, goose; HK, Hong Kong; MDk, migratory duck; Pa, partridge; Ph, pheasant; SCk, silky chicken; ST, Shantou; TS, tree sparrow; Ty, Turkey; WDk, wild duck.
In the NA tree all H5N1 viruses analyzed, with two exceptions, had NA genes derived from Gs/GD-like viruses (Fig. 1B). The HK/97-like NA genes, which possess a 19-amino acid (aa) deletion in the stalk region, were most closely related to H6N1 viruses, while the NA of a single virus (Ck/Hebei/718/01) was closely related to an early H7N1 reference strain (African starling/England-Q/983/79) (Liu et al., 2006). Within the Gs/GD-like NA genes there were two major groups, although statistical support was generally lacking throughout the tree (Fig. 1B). The first is composed entirely of viruses isolated from 1999 to 2006 that do not have any deletion in the NA stalk, and which includes some non-reassortant Gs/GD-like viruses along with genotypes B, C, W, Z+ and transient variants (Fig 1B). The second group contains viruses with and without the 20-aa amino acid deletion in the NA stalk and can be further divided into two subgroups (Fig. 1B). The first subgroup contains mostly genotype X viruses isolated from 2000 to 2006 that have a 20-aa deletion in their stalk region, except for the prototype virus, Gs/GD/1/96, without the deletion. Viruses in the second subgroup are almost all contemporary genotype G, V and Z viruses isolated from 2001 onwards that also have the 20-aa stalk deletion (Fig. 1B). However, this subgroup also contains Ck/Hubei/wk/97, which is the earliest known virus to possess the 20-aa stalk deletion. Interestingly, there was also an intermediate group of three viruses that incorporated isolates with (TS/Henan/1/04) and without (Ck/Jiangsu/cz1/02 and TS/Henan/4/04) the 20-aa NA deletion (Fig 1B).
Phylogenetic analysis of the internal genes
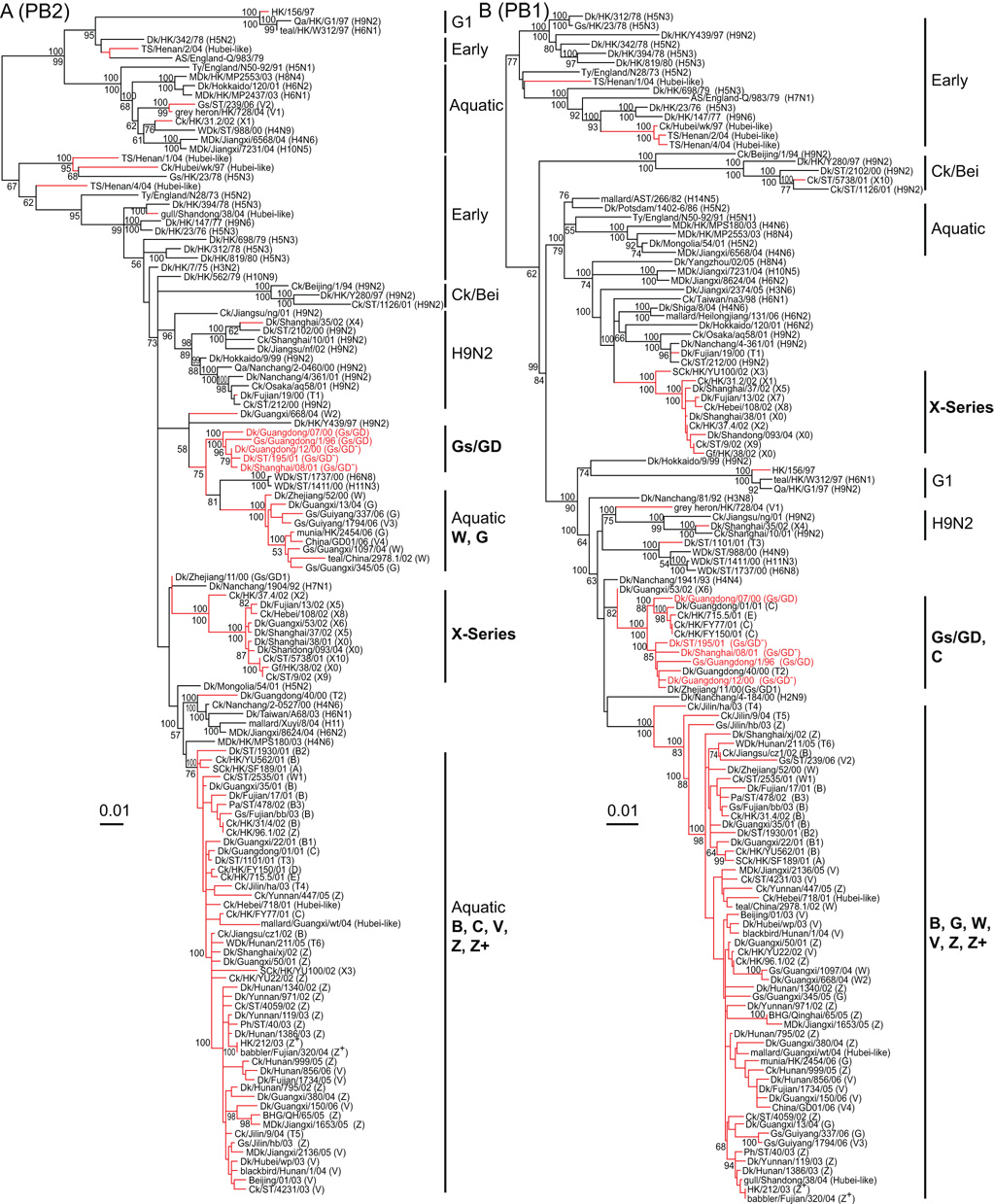
Phylogenetic analysis of the Gs/GD lineage PB2 genes revealed that all of the viruses clustered into the Eurasian gene pool but with different origins, suggesting their incorporation through multiple reassortment events (Fig. 2A). The PB2 of HK/97 viruses were derived from Qa/HK/G1/97-like virus (H9N2) while Hubei-like variants were related to viruses isolated in the 1970’s. In addition to the Gs/GD-like PB2, three major lineages of PB2 gene in H5N1 viruses were recognized (Fig. 2A). The first group mainly contained genotype G and W viruses that had a sister-group relationship with two viruses isolated from aquatic birds (WDk/ST/1737/00, H6N8; WDk/ST/1411/00, H11N3). The source of this group was previously defined as Gs/GD-like (Chen et al., 2006). The second group contained genotype X series viruses that were associated with the PB2 gene from a single H7N1 isolate (Dk/NC1904/92). The third group included PB2 genes from genotypes B, C, V, Z, Z+ and a few transient variants, which may be derived directly from the influenza gene pool in aquatic birds, e.g. MDk/HK/MPS180/03 (H4N6) (Fig. 2A).
Fig. 2.
Phylogenetic relationships of the polymerase genes PB2 (A) and PB1 (B) of representative influenza A viruses isolated in Eurasia. Analysis was based on nucleotides 1–2,277 of the PB2 gene and 1–2,271 of the PB1 gene. The PB2 and PB1 trees are rooted to equine/Prague/1/56 (H7N7), pintail duck/Alberta/628/79, respectively. The numbers above and below the branch nodes indicate Bayesian posterior probabilities of ≥95 and neighbor-joining bootstrap values of ≥50%, respectively. Red branches indicate H5N1 viruses with H5N1 prototype virus names in red. Virus genotypes are given in parenthesis following the virus names. Major genotypes are indicated with bold text. Lineage names are indicated in normal text: Aquatic, contemporary Eurasia aquatic virus; Ck/Bei, Ck/Beijing/1/94-like (H9N2) virus; Early, 1970s Eurasia aquatic virus; G1, Qa/HK/G1/97-like (H9N2) virus; H9N2, contemporary reassortant H9N2 virus. Scale bar, 0.01 substitutions per site. Abbreviation: AS, African starling; BHG, bar-headed goose; Ck, chicken; Dk, duck; Gf, Guinea fowl; Gs, goose; HK, Hong Kong; MDk, migratory duck; Pa, partridge; Ph, pheasant; Qa, Quail; SCk, silky chicken; ST, Shantou; TS, tree sparrow; Ty, Turkey; WDk, wild duck.
Except for a few transient genotype viruses, the PB1 genes of the perpetuating H5N1 variants clustered into three different groups, including Gs/GD-like, the genotype X series, and a dominant clade of genotype B, G, V, W, Z and Z+ viruses (Fig. 2B). The genotype X series clade appears to be derived from viruses in aquatic birds as it clustered with many different subtype viruses from aquatic birds in this region. The Gs/GD-like group contained viruses isolated from 1996 to 2001 including genotype C. The third group held most contemporary H5N1 variants (genotypes B, G, V, W, Z and Z+) that first emerged in 2000, e.g. Dk/Zhejiang/52/00 (Fig. 2B). It was noted that three viruses isolated in northern China (Ck/Jilin/ha/03, Ck/Jilin/hb/03 and Ck/Jilin/9/04) had an ancestral relationship with the PB2 gene all currently circulating H5N1 genotype viruses. As observed in other genes transient H5N1 variants incorporated PB1 genes from multiple sources, including 1970’s viruses (Hubei-like), H9N2 variants as well as directly from other subtype viruses in the current gene pool (Fig. 2B).
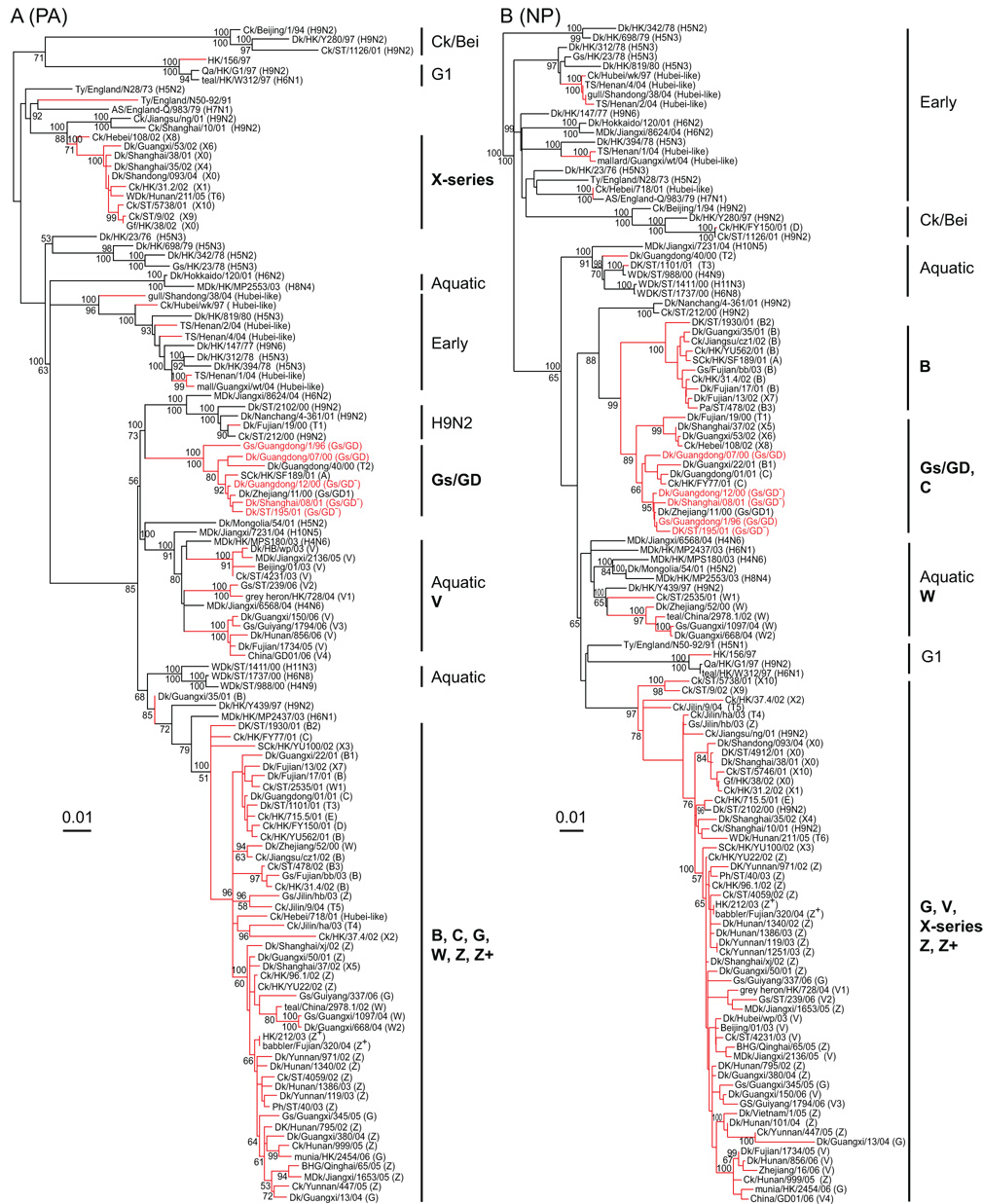
In the PA gene tree, except for those Gs/GD-like and transient variants, H5N1 viruses incorporated PA genes from three different origins (Fig. 3A). The first group contained genotype X series viruses that have a sister-group relation to H9N2 viruses from southern China. The second group consisted of genotype V viruses that appear to be directly derived from the aquatic gene pool (Fig. 3A). It is noteworthy that the PA gene of most of Fujian-like (Clade 2.3.4) viruses clustered with this group. The third group contained contemporary H5N1 variants, including genotypes B, C, G, W, Z and Z+ that were likely derived directly from viruses isolated from aquatic birds although the “gene donor” remains unidentified at present (Fig. 3A).
Fig. 3.
Phylogenetic relationships of polymerase acidic (PA) (A) and nucleoprotein (NP) (B) genes of representative influenza A viruses isolated in Eurasia. Analysis was based on nucleotides 1–2,148 of the PA gene and 1–1,482 of the NP gene. The PA and NP trees are rooted to equine/Prague/1/56 (H7N7). The numbers above and below the branch nodes indicate Bayesian posterior probabilities of ≥95 and neighbor-joining bootstrap values of ≥50%, respectively. Red branches indicate H5N1 viruses with H5N1 prototype virus names in red. Virus genotypes are given in parenthesis following the virus names. Major genotypes are indicated with bold text. Lineage names are provided in the Figure 2 legend. Scale bar, 0.01 substitutions per site. Abbreviation: AS, African starling; BHG, bar-headed goose; Ck, chicken; Dk, duck; Gf, Guinea fowl; Gs, goose; HK, Hong Kong; MDk, migratory duck; Pa, partridge; Ph, pheasant; Qa, Quail; SCk, silky chicken; ST, Shantou; TS, tree sparrow; Ty, Turkey; WDk, wild duck.
Phylogenetic analysis of the NP gene also revealed extensive reassortment events, with four major groups recognized (Fig. 3B). The first was predominantly genotype B viruses, while the second group consisted of a subgroup dominated by Gs/GD-like and genotype C viruses. The third group appeared to be derived directly from the aquatic gene pool and consisted of viruses from genotype W (Fig. 3B). The final major group was comprised of viruses that have been perpetuated in poultry since 2001, including some genotype X viruses and the currently circulating genotypes G, V, Z and Z+.
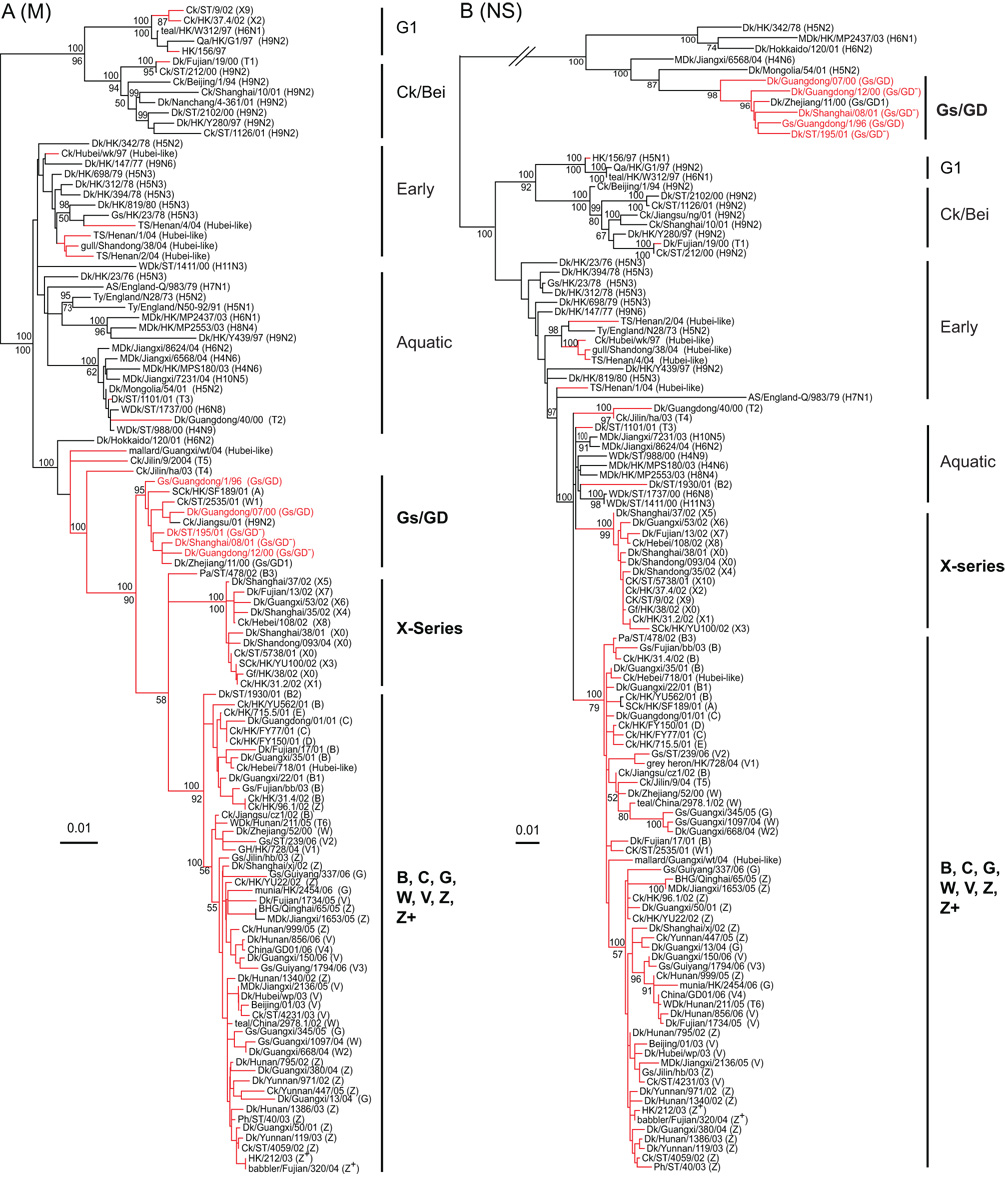
Phylogenetic analysis indicated that three major M gene groups could be distinguished (Fig. 4A). The first group was predominantly Gs/GD-like viruses, and the second group was composed exclusively of genotype X series viruses. The third group contained most of the current H5N1 variants, including viruses from genotypes B, C, G, W, V, Z and Z+ (Fig. 4A). These major groups formed a monophyletic clade but each of them had significant bootstrap support. This phylogeny may have been generated by incorporating M genes from different unknown sources, or divergence from a single source, e.g. Gs/GD-like.
Fig. 4.
Phylogenetic relationships of matrix (M) (A) and non-structural (NS) (B) genes of representative influenza A viruses isolated in Eurasia. Analysis was based on nucleotides 1–1,002 of the M gene and 1–863 of the NS gene. The M and NS trees are rooted to equine/Prague/1/56 (H7N7). The numbers above and below the branch nodes indicate Bayesian posterior probabilities of ≥95 and neighbor-joining bootstrap values of ≥50%, respectively. Red branches indicate H5N1 viruses with H5N1 prototype virus names in red. Virus genotypes are given in parenthesis following the virus names. Major genotypes are indicated with bold text. Lineage names are provided in the Figure 2 legend. Scale bar, 0.01 substitutions per site. Abbreviation: AS, African starling; BHG, bar-headed goose; Ck, chicken; Dk, duck; Gf, Guinea fowl; Gs, goose; HK, Hong Kong; MDk, migratory duck; Pa, partridge; Ph, pheasant; Qa, Quail; SCk, silky chicken; ST, Shantou; TS, tree sparrow; Ty, Turkey; WDk, wild duck.
Phylogenetic analysis of the NS gene revealed three distinct groups that were incorporated into H5N1 variants (Fig. 4B). The first was Gs/GD-like viruses that belonged to NS allele A, while the remaining groups belonged to allele B. The second group contained viruses from the genotype X series for which there was no clear source (Fig. 4B). The third group had most of the current H5N1 variants prevailing in the field, including genotypes B, C, G, W, V, Z and Z+. The NS gene of all viruses in this group had a 5-aa deletion at position 80–84 of NS1 protein that was first acquired as early as 2000 (Fig. 4B).
Taken together, results of the phylogenetic analyses show that the Gs/GD H5N1 virus lineage continued evolving primarily through a series of reassortment events with other viruses. These analyses revealed that each of those internal genes had two to three origins in addition to the prototype Gs/GD-like virus genes. There were also over 20 transient variants that incorporated viral genes from 1970’s and contemporary viruses from aquatic birds, H9N2 variants or some unknown sources. It is noteworthy that in six out of eight genes, a few intermediate strains (e.g. Ck/Jinlin/9/04, Ck/Jinlin/ha/03 and Gs/Jilin/hb/03) consistently maintained an ancestral relationship with major H5N1 variants such as genotypes B, G, V and Z, suggesting that these Jilin strains may be the progenitors for those viruses prevailing in the field.
Genotyping and chronology
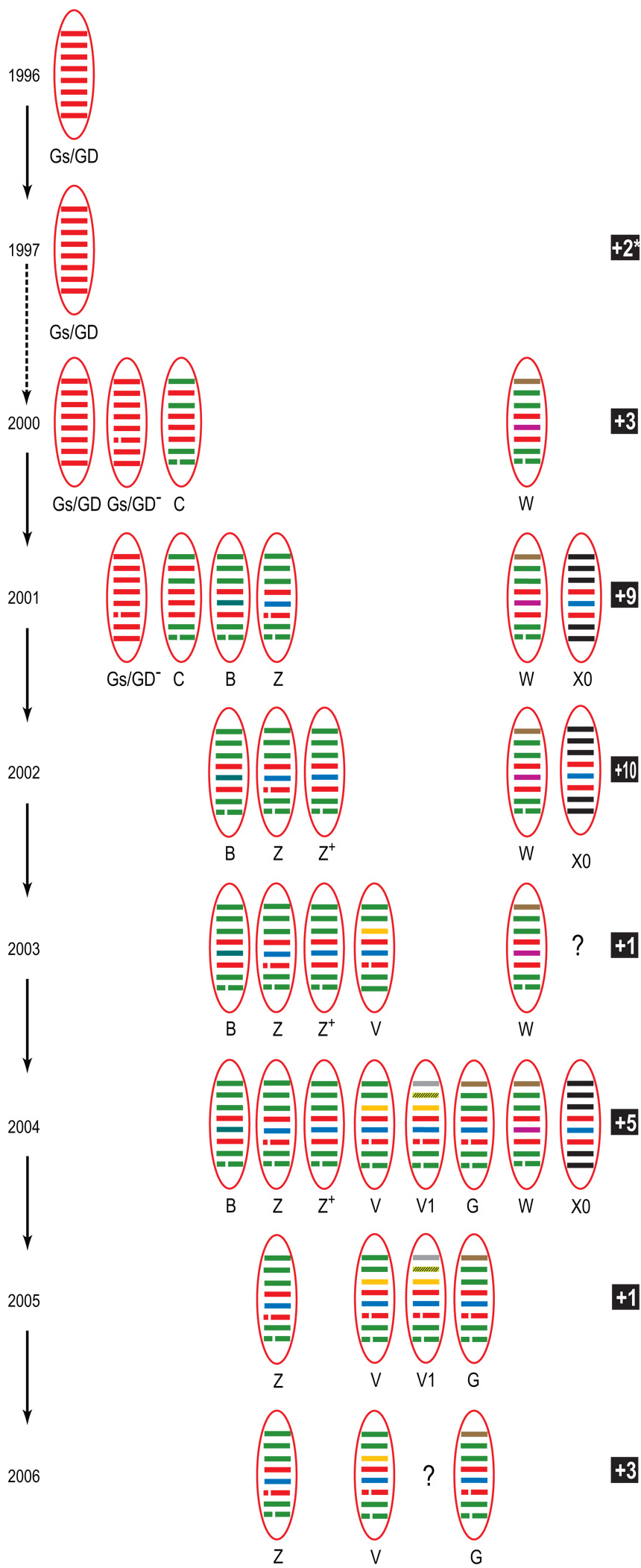
Based on these phylogenetic analyses we could identify 44 H5N1 reassortants or genotypes from the 318 viruses genotyped (Fig 5, Fig 6 and Fig S1; Table S1). With the exception of the HK/97-like and Hubei-like viruses the earliest reassortants, detected in 2000, were genotypes C and W along with three transient genotypes and a non-reassortant Gs/GD-like virus with the 20-aa deletion in the NA gene, designated as Gs/GD−(Fig 5, Fig 6 and Fig S1). In 2001, there was dramatic increase in genetic diversity with 15 genotypes detected, including genotypes Gs/GD−, C, B, Z, W, X0 and nine transients. This is also the earliest detection of a genotype Z virus (Dk/Guangxi/50/01) that was previously first recognized in 2002 in Hong Kong (Li et al., 2004). In 2002, an additional 10 transient genotypes were detected including nine genotype X series viruses (X1–X9), with five genotypes (B, Z, Z+, V, W and X0) persistent from 2001 (Fig 5, Fig 6 and Fig S1). In 2003 there was a reduction in the number of genotypes detected, mainly due to the disappearance of the entire genotype X series, with only six genotypes detected. Another new genotype (V) was first recognized and four genotypes (B, Z, Z+ and W) persisted from the previous year (Fig 5, Fig 6 and Fig S1). In 2004 eight major genotypes were detected, including the first record of genotype G and V1 viruses and the reappearance of genotype X0, along with five transient genotypes. In 2005, genotype number was reduced again as only four major genotypes persisted and only one transient was detected (Fig 5, Fig 6 and Fig S1). While in 2006, only genotypes Z, V and G were detected from the field, along with three transient genotypes.
Fig. 5.
Genotypes of H5N1 influenza viruses in southern China, 1996–2006. The eight gene segments, represented by horizontal bars are, from top to bottom, PB2, PB1, PA, HA, NP, NA, M and NS. Each different color represents a distinct phylogenetic lineage. Genotype definitions are described in the results section. Only persistent genotypes that were detected for more than two years are represented. Numbers of transient genotypes detected in only one year is given in boxed text. All detected genotypes are given in Figure S1 and a representative virus for each genotype in each year is listed in Table S1.
Fig. 6.
Simplified cladograms representing major lineages of the PB2 (A), PB1 (B), PA (C), NP (D), M (E) and NS (F). The cladograms are based on the phylogenetic trees shown in Figure 1–Figure 4. Major H5N1 genotypes are shown in blue text and prototype virus lineages are shown in red text. Lineage names are provided in the Figure 2 legend.
In March 2005 a new H5N1 variant (Clade 2.3.4 or Fujian-like) was detected in southern China that rapidly replaced other H5N1 HA clades. These Fujian-like viruses were initially described as belonging to either genotypes G or Z (Smith et al., 2006). However, the present study shows that all those Fujian-like (Clade 2.3.4) viruses that were originally described as genotype Z, using partial length sequences, are actually genotype V viruses when full-length genomes were analyzed (Fig 3B, Fig 5 and Fig 6). Genotypes V and Z have seven genes from a common source while their PA genes have different origins (Chen et al., 2006). The single exception is Gs/Guiyang/1794/2006, which is the single representative of the transient genotype V3 (Fig 1 and Fig S1).
In summary, within the Gs/GD lineage H5N1 variants two major series of multiple reassortment events could be identified. The first series was associated with the genesis of genotype B and a few transient reassortants (B1, B2, D and E) that shared a majority of their internal genes with genotype B (Fig. S1). Comparison of phylogenies suggests that genotype B virus was probably the progenitor of genotype Z viruses, and that genotypes G and V are further reassortants of genotype Z (Fig 5 and Fig 6). The second series of reassortment events resulted in the genesis of the genotypes X series. These viruses were first detected during an H5N1 outbreak in Hong Kong in 2002. While their surface genes and the majority of their internal genes (PB2, PB1, M and NS) form distinct lineages, these viruses also incorporated gene segments from a variety of unidentified sources (Fig. 6).
Geographical expansion
The distribution and occurrence of H5N1 activity in China is summarized in Table 1. Based on available sequence data, after the Gs/GD-like virus outbreak in 1996 the prototype virus was mainly prevalent and maintained in geese in Guangdong from 1996 to 1999 (Webster et al., 2002). In 1997 H5N1 viruses were detected in Hong Kong and Hubei, however, each of these two outbreaks resulted from two independent reassortment events (Table 1). In 2000, the Gs/GD lineage viruses were detected in five provinces of eastern and southern China with novel reassortants or genotypes beginning to be detected (Table 1 and Fig. S1). In 2001 eight provinces were affected and almost all isolates were novel reassortants. Genotype B viruses were identified in three provinces for which sequence data were available. There was a further expansion of H5N1 activity in 2002 with the virus detected in four additional provinces (Jilin, Hubei, Hunan and Yunnan) while the virus persisted for all provinces detected in the previous year (Table 1). From 2003 to 2004, H5N1 virus activity was continually observed in 11 and 12 provinces, respectively. It is noteworthy that genotype V virus has gradually replaced genotypes Z since the emergence of the Fujian-like (Clade 2.3.4) H5N1 variant in early 2005 (Smith et al., 2006). As such, in 2006 genotype V was detected in eight of 10 provinces where genotypes were defined, while genotypes Z and G were only detected in three provinces each (Table 1).
Table 1.
H5N1 influenza virus activity and genotype distribution in China, 1996–2006
| Year | Province (genotype) |
|---|---|
| 1996 | Guangdong#(Gs/GD) |
| 1997 | Guangdong (Gs/GD, HK/97-like), Hubei (Hubei-like) |
| 1999 | Guangdong (Gs/GD) |
| 2000 | Fujian (F), Guangdong (Gs/GD, Gs/GD−, C, T1), Guangxi (ND*), Jiangsu (ND),b Zhejiang (Gs/GD1, W) |
| 2001 | Fujian (B, W), Guangdong (Gs/GD−, A–E, X0, X10, W1, B2, T3), Guangxi (B, B1,Z), Hebei (Hubei-like), Henan (ND), Jiangsu (ND), Shanghai (Gs/GD−, C, X0), Zhejiang (ND). |
| 2002 | Fujian (X7), Guangdong (X0-X3, X9, B, B3, Z, Z+,W), Guangxi (X6), Hebei (X8), Henan (ND), Hubei (ND), Hunan (Z), Jiangsu (B), Jilin (ND), Shanghai (X4, X5, Z), Yunnan (Z), Zhejiang (ND). |
| 2003 | Beijing (V), Fujian (B, W), Guangdong (Z, V), Guangxi (ND), Henan (ND), Hubei (V), Hunan (Z), Jiangsu (ND), Jilin (T4, Z), Shangdong (ND), Yunnan (Z) |
| 2004 | Anhui (ND), Fujian (Z+), Guangdong (Z, V, V1, Hubei-like), Guangxi (Z, W, G, W2, Hubei-like), Henan (Z, Hubei-like), Hubei (Z), Hunan (Z, V), Jiangsu (ND), Jilin (T5), Shandong (B, X0, Hubei-like), Shanghai (Z), Yunnan (Z) |
| 2005 | Anhui (ND), Fujian (Z, V), Guangdong (Z, V, V1), Guangxi (Z, V, G), Guizhou (Z, G, V), Hebei (V), Hubei (ND), Hunan (Z, V, G, T6), Jiangsu (ND), Jiangxi (Z, V), Qinghai (Z), Yunnan (Z, V, G), Zhejiang (ND). |
| 2006 | Fujian (V), Guangdong (Z, V, G, V2, V4), Guangxi (V, G), Guizhou (G, V, V3), Hunan (Z, V), Jiangsu (ND), Qinghai (Z), Shanxi (V), Yunnan (V), Zhejiang (V) |
Includes Hong Kong S.A.R and Shantou
ND, denotes genotype could not be determined as full genome is not available
Discussion
In the present study, comprehensive phylogenetic analyses of Gs/GD H5N1 virus lineage full genomes identified 44 different reassortants or genotypes from China during 1996–2006. Two major series of multiple reassortment events were recognized in 2001 and 2002 that were responsible for the genesis of most of those reassortants; genotype B, Z, W, V and G viruses, and genotype X series viruses, respectively. The counterparts of each of the major genotypes detected outside of mainland China were also detected in China at an earlier time.
The earliest reassortants of the Gs/GD virus lineage were detected in 1997 (Guan et al., 1999, Hoffman et al., 2000, Xu et al., 1999), and subsequently, a diversity of H5N1 genotypes were recognized in the field in 2001 and 2002. This suggests that the multiple reassortment events likely occurred from 2000 to 2001, as genotypes C and W were first detected in 2000 while the earliest detection of genotype X viruses was in November 2001 (Guan et al., 2004). Therefore, the reassortment events resulting in genotypes B, C, Z and W viruses very likely occurred in 2000; preceding the genesis of genotype X series viruses. As both of these genotype series share the same NP gene sources, it is likely that they both arose from the same primary reassortment event. However, as most genotype X viruses had different internal genes sources to the other major genotypes (e.g. B, Z and W), these reassortant groups subsequently had distinct evolutionary pathways.
This study also demonstrates that novel reassortants, including transients, were detected in each year from 2000 onwards. We were also able to recognize several genotypes earlier than previously known: genotypes B and Z in 2001, V in 2003, V1 and G in 2004. Genotype B was the major reassortant detected in 2001; however, genotype Z gradually became predominant from 2002 to mid-2005. In late 2005, genotype Z was gradually replaced by genotype V (Clade 2.3.4, i.e. the Fujian-like variant) in southern China and adjacent countries. Each of these genotype replacements were associated with observed poultry outbreaks (Guan et al., 2002, 2004, Li et al., 2004). Currently, genotype Z viruses are still dominant in Southeast Asia (Clades 1 and 2.1) and Europe and Africa (Clade 2.2), while genotype V viruses (Clade 2.3.4) predominate in southern China and have been detected in Vietnam and Laos in 2006–2007 (Al-Azemi et al., 2008; Boltz et al., 2006, Nguyen et al., 2008; Smith et al., 2006). The mechanism underlying these genotype replacements is unknown; however, as the Gs/GD H5N1 lineage remains endemic in poultry in this region such replacements are likely to occur again. Since many of these genotypes have infected humans with lethal consequences, it is possible that these successive genotype replacements may generate and/or select a pandemic influenza strain (Peiris et al., 2007). It is interesting to note that, to date, almost all H5N1 reassortant viruses were first detected in China. Why no new genotypes appear to have been generated in Vietnam, Thailand, Indonesia and other H5N1 endemic regions, despite similar farm and market structures with mixed bird populations, remains to be answered.
All H5N1 variants currently circulating in Eurasia and Africa were first detected in the field in mainland China. The findings of the present study revealed that as early as 2000, Gs/GD lineage reassortants were detected in five provinces and in eight provinces in 2001. From 2002 onwards, H5N1 virus activity was recorded in more than ten provinces every year. Also, a similar pattern of genotype variation or replacement was observed in multiple provinces during the same periods of time (Table 1) indicating that H5N1 activity was much broader and more severe than previously understood. Unfortunately, the lack of systematic influenza surveillance in poultry in the wider region has made it impossible to trace the H5N1 virus transmission pathway, just as it is difficult to directly identify the source of many current human infections.
Historically, more than 20 highly pathogenic H5 and H7 influenza outbreaks have been recorded since the 20th century (Alexander, 2007), but only the Gs/GD H5N1 lineage has undergone such extensive reassortment. It is possible that these reassortment events and subsequent dominance of certain genotypes reflects the emergence of viruses with a fitness advantage. Although many of these viruses were isolated from apparently healthy birds in live-poultry markets, it is unclear whether the birds were infected before transport or within the market, and whether they may have eventually succumbed to the infection. The detection and maintenance of these viruses in poultry raises the question of whether these strains are evading the immunity afforded either by vaccination or hetero-subtypic immunity induced by other viruses endemic in poultry (e.g. H9N2).
It was interesting to note that some Hubei-like and Clade 2 variants (e.g. Qinghai-like and Fujian-like) were associated with unusually long branches in the phylogenetic tree, particularly for the HA gene, indicating the accumulation of a high number of mutations (Worobey, 2008). This was particularly apparent for a number of individual strains (e.g. Ck/Hubei/wk/97). Whether poultry vaccination would accelerate the evolutionary rate of the Gs/GD lineage has not been investigated, but a previous study suggested that the Mexican H5N2 lineage viruses underwent increased genetic and antigenic drift that was most likely due to vaccination pressure (Lee et al., 2004). Therefore, it is possible that the emergence and re-emergence of those multiple Gs/GD variants may also be due to the large-scale vaccination program in China. However, extraordinarily long branches in the phylogenetic trees are also expected for laboratory-adapted viruses that have experienced many rounds of replication while grown in cell culture or chicken eggs (Worobey, 2008).
The prototype strain of Ck/Bei/1/94-like (H9N2) virus could cause significant mortality for chickens under laboratory conditions (Guo et al., 2000). It was reported that a H9N2 subtype vaccine had been also used in this region (Xu et al., 2004). The HK/97-like virus is thought to have resulted from double or triple reassortment between Gs/GD-like, H9N2 and H6N1viruses (Guan et al., 1999, Hoffman et al., 2000, Xu et al., 1999). Recent reports also revealed that many reassortant H9N2 and H6N1 viruses share many internal gene segments with contemporary Gs/GD H5N1 reassortants (Cheung et al., 2007, Xu et al., 2007). In this regard, H5N1 and H9N2 subtype viruses were not only co-circulating in the field, but vaccines for these subtypes were also co-manufactured and applied during the same period (http://www.fao.org/AG/AGAINFO/Programmes/en/empres/vaccine_producers.htm). The unusual field isolate, Ck/Hebei/718/01 has NA and NP genes almost identical to African starling/England-Q/983/79 (H7N1), a virus that was investigated as a vaccination candidate in China (Liu et al., 2006) and raises the possibility that such an isolate may have arisen from vaccine viruses. The gene constellations of Hubei-like variants, initially detected in 1997 and thereafter sporadically isolated, contained at least some internal genes that are closely related to genes from the 1970’s aquatic influenza gene pool, providing further support for this hypothesis.
Given the lack of systematic surveillance in most regions of China, it may not be possible to profile the detailed genesis and evolution of Gs/GD lineage in more precise detail. Here we have collated available data to address these questions. As the pandemic threat is still persists, it is essential to conduct systematic influenza surveillance in both poultry and humans to track the continued emergence and reemergence of viruses with pandemic potential so as to provide an early warning system for global public health.
Materials and methods
Virus and viral sequence data
To evaluate the evolutionary pathway of each gene segment and to establish a thorough H5N1 genotyping system, we sequenced 46 representative viruses available in our repository to provide full-length genomes. Viral genome sequencing was conducted as previously described (Guan et al., 2002, Li et al., 2003, Smith et al., 2006).
Phylogenetic analysis
All sequence data of Gs/GD lineage H5N1 viruses isolated from China during 1996–2006 available in GenBank were aligned together with sequences from this study using the NCBI Influenza Virus Resource (Bao et al., 2008). Only sequences that were full-length or almost full-length were included in subsequent analyses, resulting in alignments of 318 virus full genomes. The program MrModeltest 2.2 (Nylander, 2004) was used to determine the appropriate DNA substitution model and γ-rate heterogeneity, and the generated models were used in all subsequent analyses. Neighbor-joining (NJ) trees were constructed by using PAUP* 4.0 (Swofford, 2001). Bayesian analysis was conducted with MrBayes 3.1 (Huelsenbeck and Ronquist, 2001) using two replicates of 2 to 4 million generations with six chains, sampled every 200 generations. Convergence of the Bayesian replicates were assessed visually using Tracer v 1.3 (http://beast.bio.ed.ac.uk/Tracer). Phylogenetic supports were calculated by performing 1,000 NJ and Bayesian posterior probabilities were calculated from the consensus of no less than 15,000 trees after excluding the first 10 to 20% of trees as burnin, except for a burnin of 50% for the PB1 gene.
Genotyping
Consistent with previous studies, those H5N1 viruses with a Gs/GD/1/96 lineage hemagglutinin (HA) gene were classified into genotypes that were defined by the phylogenies of each of the six internal gene segments (Guan et al., 2002, Li et al., 2004). A distinct phylogenetic lineage was identified based on NJ bootstrap support of ≥70% or Bayesian posterior probability of ≥95% as indicating a common origin. A genotype was described when the phylogenies of the internal genes resulted in a unique gene constellation for one or more isolates. However, genotypes were also named in cases where viruses had the same gene constellation but differed by some molecular marker such as an amino-acid deletion, e.g. genotypes Z and Z+ differ only in the presence or absence of a 20-aa deletion in the neuraminidase (NA) protein (Guan et al., 2004). Genotypes were also further distinguished into two groups, 1) those major genotypes that detected in the field for two years or more and 2) transient genotypes that were only detected within a single year and that were often represented by just a single virus isolate. In cases wherein a transient genotype differed from a major genotype in the source of just a single gene segment, we named those transients as a variant of the major genotype, e.g. genotypes B1 and V1.
Chronology and geographical development
As there has been a lack of information on H5N1 activity in China from 1996 to 2003, we described the development of H5N1 virus based on the geographic distribution and timeline of emergence of the H5N1 variants from the virus names, which provided both place and year of isolation.
Definition of terminology
In the present study, all H5N1 influenza viruses with a hemagglutinin gene derived from Gs/Guangdong/1/96 were designated as belonging to the “Gs/GD lineage”. Non-reassortant viruses with all eight gene segments closely related to Gs/GD/1/96 were referred to as “Gs/GD-like”. However, in describing the source of gene segments of reassortant viruses, we referred to genes derived from the Gs/GD- like virus as being “Gs/GD-like”. Viruses isolated during the Hong Kong ‘bird flu’ incident in 1997, represented by HK/156/97, were named as “HK/97-like”. The viruses that were first isolated from Hubei province in 1997, and which incorporated internal genes closely related to the viruses isolated from aquatic birds in the 1970’s, were referred to as “Hubei-like”. The remaining H5N1 reassortants from the Gs/GD lineage were referred to by their genotype designation. The previously agreed nomenclature on H5 hemagglutinin clades and sublcades has been adhered to (http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en).
Nucleotide sequence accession numbers
The 129 new nucleotide sequences reported in this paper have been deposited in the GenBank database (accession numbers CY030878–CY031006). A further 114 partial gene sequences from 29 viruses have also been updated to provide full-length genomes. These viruses and the GenBank accession numbers are provided in Table S2.
Supplementary Material
Genotypes of H5N1 viruses in southern China, 1996–2006. The eight gene segments, represented by horizontal bars are, from top to bottom, PB2, PB1, PA, HA, NP, NA, M and NS. Each color represents a distinct phylogenetic lineage. Genotype definitions are described in the results section. ‘Persistent’ represents genotypes that were detected for more than two years, while the ‘transients’ are those that were detected in only one year. A representative virus for each genotype in each year is listed in Table S1.
Acknowledgements
This study was supported by the Li Ka Shing Foundation, the National Institutes of Health (NIAID contract HHSN266200700005C), and the Area of Excellence Scheme of the University Grants Committee (Grant AoE/M-12/06) of the Hong Kong SAR Government.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Azemi A, Bahl J, Al-Zenki S, Al-Shayji Y, Al-Ahmad S, Chen H, Guan Y, Peiris JSM, Smith GJD. Avian influenza A virus (H5N1) outbreaks, Kuwait, 2007. Emerg. Infect. Dis. 2008;14:958–961. doi: 10.3201/eid1406.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- Bao Y, Dernovoy PBD, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The Influenza Virus Resource at the National Center for Biotechnology Information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz DA, Douangngeun B, Sinthasak S, Phommachanh P, Rolston S, Chen H, Guan Y, Peiris JSM, Smith GJD, Webster RG. H5N1 influenza viruses in Lao People's Democratic Republic. Emerging Infect. Dis. 2006;12:1593–1595. doi: 10.3201/eid1210.060658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Deng G, Li Z, Tian G, Li Y, Jiao P, Zhang L, Liu Z, Webster RG, Yu K. The evolution of H5N1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, Cheung CL, Xu KM, Duan L, Huang K, Qin K, Leung YHC, Wu WL, Lu HR, Chen Y, Xia NS, Naipospos TSP, Yuen KY, Hassan SS, Bahri S, Nguyen TD, Webster RG, Peiris JSM, Guan Y. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJD, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JSM, Guan Y. H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Vijaykrishna D, Smith GJD, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, Poon LLM, Peiris JSM, Chen H, Guan Y. Establishment of influenza a virus (H6N1) in minor poultry species in southern China. J. Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donatelli I, Campitelli L, Di Trani L, Puzelli S, Selli L, Fioretti A, Alexander DJ, Tollis M, Krauss S, Webster RG. Characterization of H5N2 influenza viruses from Italian poultry. J. Gen. Virol. 2001;82:623–630. doi: 10.1099/0022-1317-82-3-623. [DOI] [PubMed] [Google Scholar]
- Duan L, Campitelli L, Fan XH, Leung YHC, Vijaykrishna D, Zhang JX, Donatelli I, Delogu M, Li KS, Foni E, Chiapponi C, Wu WL, Kai H, Webster RG, Shortridge KF, Peiris JSM, Smith GJD, Chen H, Guan Y. Characterization of low-pathogenic H5 subtype influenza viruses from Eurasia: Implications for the origin of highly pathogenic H5N1 viruses. J. Virol. 2007;81:7529–7539. doi: 10.1128/JVI.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agricultural Organization of the United Nations. Avian Influenza Disease Emergence Bulletin, issue 48. 2007 http://www.fao.org/avianflu/en/AIDEnews.html.
- Fouchier RAM, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SAG, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, van Doornum GJJ, Koch G, Bosman A, Koopmans M, Osterhaus ADME. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Peiris JSM, Lipatov AS, Ellis TM, Dyrting KC, Krauss S, Zhang LJ, Webster RG, Shortridge KF. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8950–8955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Poon LLM, Cheung CY, Ellis TM, Lim W, Lipatov AS, Chan KH, Sturm-Ramirez KM, Cheung CL, Leung YHC, Yuen KY, Webster RG, Peiris JSM. H5N1 influenza: A protean pandemic threat. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8156–8161. doi: 10.1073/pnas.0402443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: Were they the donors of the "internal" genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. U.S.A. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YJ, Krauss S, Senne DA, Mo IP, Lo KS, Xiong XP, Norwood M, Shortridge KF, Webster RG, Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin PS, Peiris M, Shortridge KF, Webster RG. Characterization of the influenza A virus gene pool in avian species in southern China: Was H6N1 a derivative or a precursor of H5N1? J. Virol. 2000;74:6309–6315. doi: 10.1128/jvi.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster RG. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza-virus in Mexico. Virology. 1995;213:223–230. doi: 10.1006/viro.1995.1562. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hulse DJ, Perez DR, Webster RG. Molecular determinants of the pathogenicity of H5N1 influenza viruses in chickens. Int. Congr. Ser. 2004;1263:114–117. [Google Scholar]
- Kou Z, Lei FM, Yu J, Fan ZJ, Yin ZH, Jia CX, Xiong KJ, Sun YH, Zhang XW, Wu XM, Gao XB, Li TX. New genotype of avian influenza H5N1 viruses isolated from tree sparrows in China. J. Virol. 2005;79:15460. doi: 10.1128/JVI.79.24.15460-15466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J. Virol. 2004;78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KS, Guan Y, Wang J, Smith GJD, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie ATS, Chaisingh A, Auewarakul P, Long HT, Hanh NTH, Webby RJ, Poon LLM, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JSM. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- Li KS, Xu KM, Peiris JSM, Poon LLM, Yu KZ, Yuen KY, Shortridge KF, Webster RG, Guan Y. Characterization of H9 subtype influenza viruses from the ducks of southern China: A candidate for the next influenza pandemic in humans? J. Virol. 2003;77:6988–6994. doi: 10.1128/JVI.77.12.6988-6994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin Z, Shi J, Qi Q, Deng G, Li Z, Wang X, Tian G, Chen H. Detection of Hong Kong 97-like H5N1 influenza viruses from eggs of Vietnamese waterfowl. Arch. Virol. 2006;151:1615–1624. doi: 10.1007/s00705-006-0738-7. [DOI] [PubMed] [Google Scholar]
- Liu LL, Wang XR, Chen HL, Tian GB, Sun K, Wei P. The growth characteristics of different subtypes of avian influenza virus in the MDCK cell line. Chinese Journal of Veterinary Medicine (Zhongguo Shou Yi Za Zhi) 2006;42:18–20. [Google Scholar]
- Nguyen TD, Nguyen TV, Vijaykrishna D, Guan Y, Peiris JSM, Smith GJD. Multiple lineages of influenza A (H5N1) viruses in Vietnam (2006–2007) Emerg. Infecti. Dis. 2008;14:632–636. doi: 10.3201/eid1404.071343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. MrModelTest version 2. Evolutionary Biology Centre, Uppsala University. Program distributed by the author. 2004 [Google Scholar]
- Peiris JSM, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti KG, Norwood M, Senne D, Sims L, Takada A, Webster RG. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- Smith GJD, Fan XH, Wang J, Li KS, Qin K, Zhang JX, Vijaykrishna D, Cheung CL, Huang K, Rayner JM, Peiris JSM, Chen H, Webster RG, Guan Y. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16936–16941. doi: 10.1073/pnas.0608157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJD, Naipospos TSP, Nguyen TD, de Jong MD, Vijaykrishna D, Usman TB, Hassan SS, Nguyen TV, Dao TV, Bui NA, Leung YHC, Cheung CL, Rayner JM, Zhang JX, Zhang LJ, Poon LLM, Li KS, Nguyen VC, Hien TT, Farrar J, Webster RG, Chen H, Peiris JSM, Guan Y. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology. 2006;350:258–268. doi: 10.1016/j.virol.2006.03.048. [DOI] [PubMed] [Google Scholar]
- Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–396. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 beta. Sunderland, MA: Sinauer Associates; [Google Scholar]
- Wan XF, Ren T, Luo KJ, Liao M, Zhang GH, Chen JD, Cao WS, Li Y, Jin NY, Xu D, Xin CA. Genetic characterization of H5N1 avian influenza viruses isolated in southern China during the 2003-04 avian influenza outbreaks. Arch. Virol. 2005;150:1257–1266. doi: 10.1007/s00705-004-0474-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Vijaykrishna D, Duan L, Bahl J, Zhang JX, Webster RG, Peiris JSM, Chen H, Smith GJD, Guan Y. Identification of the progenitors of Indonesia and Vietnam avian influenza A (H5N1) viruses from southern China. J. Virol. 2008;82:3405–3414. doi: 10.1128/JVI.02468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Govorkova EA. H5N1 Influenza - Continuing evolution and spread. N. Engl. J. Med. 2007;355:2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- Webster RG, Guan Y, Peiris M, Walker D, Krauss S, Zhou NN, Govorkova EA, Ellis TM, Dyrting KC, Sit T, Perez DR, Shortridge KF. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J. Virol. 2002;76:118–126. doi: 10.1128/JVI.76.1.118-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M. Phylogenetic evidence against evolutionary stasis and natural abiotic reservoirs of influenza A virus. J. Virol. 2008;82:3769–3774. doi: 10.1128/JVI.02207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan W, Wei R, Zhao H. Isolation and identification of swine influenza recombinant A/Swine/Shangdong/1/2003 (H9N2) virus. Microbes Infect. 2004;6:919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Xu KM, Smith GJD, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, Webster RG, Chen H, Peiris JSM, Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotypes of H5N1 viruses in southern China, 1996–2006. The eight gene segments, represented by horizontal bars are, from top to bottom, PB2, PB1, PA, HA, NP, NA, M and NS. Each color represents a distinct phylogenetic lineage. Genotype definitions are described in the results section. ‘Persistent’ represents genotypes that were detected for more than two years, while the ‘transients’ are those that were detected in only one year. A representative virus for each genotype in each year is listed in Table S1.