Abstract
In this study we investigate the ability of Salmonella enterica serovar Typhi (S. Typhi) surface structures to influence invasion and adhesion in epithelial cell assay systems. In general, S. Typhi was found to be less adherent, invasive and cytotoxic than S. enterica serovar Typhimurium (S. Typhimurium). Culture conditions had little effect on adhesion of S. Typhi to cultured cells but had a marked influence on invasion. In contrast, bacterial growth conditions did not influence S. Typhi apical invasion of polarized cells. The levels of S. Typhi, but not S. Typhimurium, invasion were increased by application of bacteria to the basolateral surface of polarized cells. Expression of virulence (Vi) capsule by S. Typhi resulted in a modest reduction in adhesion, but profoundly reduced levels of invasion of non-polarized cells. However, Vi capsule expression had no affect on invasion of the apical or basolateral surfaces of polarized cells. Mutation of the staA, tcfA or pilS genes did not affect invasion or adhesion in either the presence or the absence of Vi capsule.
INTRODUCTION
Salmonella enterica subspecies I serovar Typhi (S. Typhi) is the causative agent of the human systemic infectious disease typhoid (or enteric) fever (Parry et al., 2002). S. Typhi infections are human-restricted, although some higher primates, such as the chimpanzee, can be infected experimentally (Edsall et al., 1960). In contrast, some other serovars of S. enterica such as Typhimurium are commonly associated with a wide range of hosts, and cause both gastroenteritis and invasive disease in humans. Although little is known about the mechanisms responsible for the differences in pathogenicity and host range exhibited by S. Typhi and S. Typhimurium, genome sequencing of these pathogens has revealed differences in gene content and coding capacity. Although 90% of the S. Typhi and S. Typhimurium genomes share a common ancestry, the remaining genes are unrelated (Parkhill et al., 2001; McClelland et al., 2001; Deng et al., 2003). The S. Typhi-specific gene content includes a large pathogenicity island, termed Salmonella pathogenicity island 7 (SPI-7) that encodes the viaB locus that directs the biosynthesis and export of a polysaccharide termed the Vi (virulence) capsule (Liu & Sanderson, 1995; Parkhill et al., 2001; Pickard et al., 2003). A further difference in coding capacity arises from the presence of over 200 putative pseudogenes in the S. Typhi genome, most of which are apparently functional in S. Typhimurium (Parkhill et al., 2001).
DNA sequencing of two different S. Typhi genomes, strains CT18 and Ty2, has shown that this serovar encodes 13 fimbrial operons, including a single member of the nucleating (csg) and 12 members of the chaperone-usher fimbrial families. However, at least five of the chaperone-usher-family encoding loci (sef, bcf, ste, stg and sth) harbour potentially inactivating pseudogenes in S. Typhi, but not in S. Typhimurium (McClelland et al., 2001; Townsend et al., 2001). Additionally, the fim chaperoneusher fimbrial operon contains a disruption in the minor fimbrial subunit gene (fimI) in S. Typhi strain CT18 but not in Ty2 (A. Bishop, unpublished observation). Little is known of the role of these fimbriae in host–pathogen interactions. S. Typhi also harbours two intact fimbrial operons (tcf and sta), which are not encoded by the S. Typhimurium genome, and a type IVb fimbrial operon (pil) (Zhang et al., 2000) encoded on SPI-7 (Pickard et al., 2003), which can target the human cystic fibrosis transmembrane conductance regulator (Tsui et al., 2003). The distinct repertoire of fimbriae present in S. Typhi may contribute, along with the Vi capsule, to the specific virulence traits of this serovar, perhaps by influencing adhesion to and invasion of host cells. The contributions of a number of S. Typhi loci to adhesion and invasion have been characterized previously using in vitro models. The SPI-1-encoded type III secretion system (TTSS) facilitates S. Typhi and S. Typhimurium invasion of human epithelial cell lines (Galan & Curtiss, 1991), whereas the expression of Vi capsule has been reported to decrease invasion efficiency (Miyake et al., 1998). Interestingly, the expression of both SPI-1 and Vi capsule is influenced by environmental osmolarity, mediated through the actions of the RcsB–RcsC and OmpR–EnvZ regulatory systems in a manner that indicates an inverse relationship between their expression levels (Arricau et al., 1998; Pickard et al., 1994; Virlogeux et al., 1996; Zhao et al., 2001).
Here we report an investigation of the influence of the sta, tcf, pil and Vi capsule biosynthesis genes, encoding S. Typhi surface structures that are not present in S. Typhimurium, upon the ability of S. Typhi to adhere to, and subsequently invade, human epithelial cells.
METHODS
Bacterial strains
S. Typhi BRD948 (Ty2 ΔaroC aroD htrA) and its mutant derivatives were routinely grown in Luria–Bertani (LB) broth containing (where stated) different salt concentrations (standard, 0.17 M NaCl; low-salt, 0.09 M NaCl; high-salt, 0.3 M NaCl) supplemented with a mixture of aromatic amino acids (aro mix; 0.04 g phenylalanine l−1, 0.04 g tryptophan l−1, 0.01 g para-aminobenzoic acid l−1 and 0.01 g dihydrobenzoic acid l−1) and 0.04 g tyrosine l−1. Static or shaken (at 200 r.p.m.) cultures (5 ml) were routinely incubated overnight at 37 °C, unless otherwise stated. Mutant S. Typhi were grown with 30 mg kanamycin l−1 (Roche) or 15 mg chloramphenicol l−1 (Sigma-Aldrich) selection where appropriate. S. Typhi BRD948 (Ty2 ΔaroC aroD htrA) was used instead of S. Typhi Ty2 for reasons of biosafety. When S. Typhi BRD948 (Ty2 ΔaroC aroD htrA) was compared with S. Typhimurium, strain BRD807 (SL1344 ΔaroA htrA) was employed (Chatfield et al., 1992), or for data shown in Fig. 3(a, b), SL3261 (SL1344 ΔaroA) and its isogenic invA::aph mutant (Avogadri et al., 2005). These strains of S. Typhi and S. Typhimurium contain well-characterized attenuating mutations in the aro locus making them auxotrophic for aromatic compounds. These mutations affect the ability to grow in the intracellular compartment due to limitation of exogenous aromatic metabolites at this site. htrA mutations are also known to compromise intracellular survival, possibly due to increased susceptibility to damage by agents such as H2O2 (Lowe et al., 1999). Importantly, these mutations do not affect the ability of these bacteria (cultured in aromatic-replete media) to adhere to or invade cultured cells (Lowe et al., 1999; Chatfield et al., 1992).
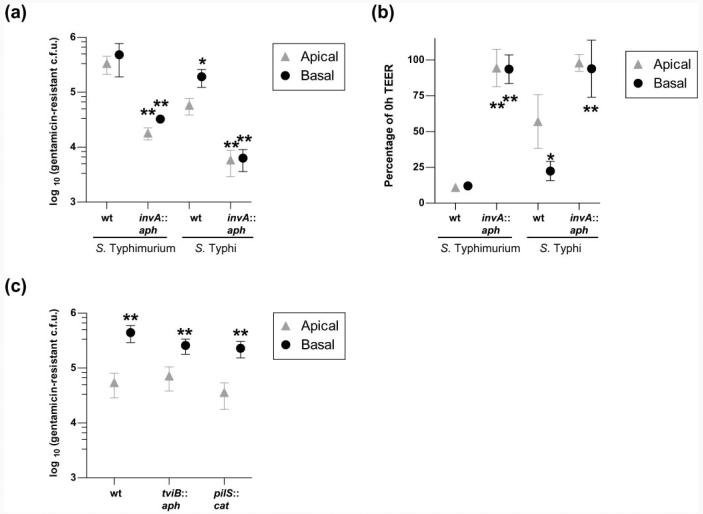
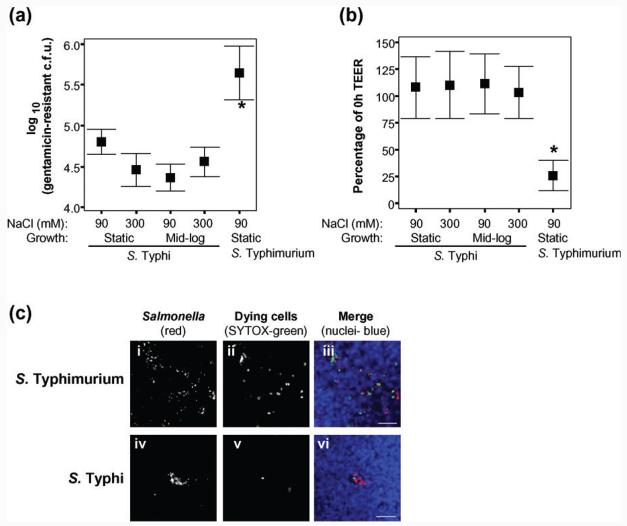
Fig. 3.
S. Typhi basolateral invasion of polarized T84 cells is higher than apical invasion. Invasion of S. Typhi or S. Typhimurium at the basolateral or apical surfaces of polarized T84 cells, grown on 3 μm-pore supports, measured as gentamicin-resistant viable counts (a), and the associated effect of invasion upon TEER (b). For S. Typhi, but not for S. Typhimurium, both invasion levels (a) and the associated drop in TEER (b) were greater after infection at the basolateral surface than at the apical surface (one-way ANOVA and post-hoc t tests with Tukey's correction, S. Typhi apical versus basal; *P<0.05). Invasion (a) and the drop in TEER (b) were largely invA-dependent [one-way ANOVA and post-hoc t tests with Tukey's correction, wild-type BRD948 (wt) versus isogenic invA mutant; **P<0.005]. S. Typhimurium SL3261 (ΔaroA), rather than BRD807, and its isogenic invA mutant were used for this experiment. (c) S. Typhi BRD948 and isogenic tviB : : Km and pilS : : Cm mutants all behave similarly, in terms of viable gentamicin-resistant invasion counts, during apical and basolateral invasion of polarized T84 cells (t tests apical versus basal for each strain; **P<0.005). Mid-exponential high-salt bacterial growth conditions were used for assays shown in (c). In each case data are shown from three independent experiments in which each condition was tested in triplicate. Means±sd are shown.
Generation of S. Typhi mutants
Mutants of S. Typhi BRD948 were made using the Red-recombinase system, based on a method described elsewhere (Datsenko & Wanner, 2000). S. Typhi was transformed with pKD46 plasmid carrying the Red-recombinase genes. PCR of cassettes was carried out using either pKD3, encoding the chloramphenicol acetyl transferase (cat) gene, or pKD13 aminoglycoside phosphotransferase (aph) template DNA at 0.1 ng μl−1, with appropriate primers at 0.1 pmol μl−1 and Red Taq polymerase according to the manufacturer's instructions (Sigma-Aldrich). PCR conditions were as follows: 94 °C for 30 s; 30 cycles of 94 °C for 30 s, 57 °C for 30 s and 72 °C for 2 min; final elongation 72 °C for 5 min. Primers were designed using the Ty2 genome sequence as a reference (Deng et al., 2003) and were as follows (the 38–50 bp regions that facilitate homologous recombination with each gene are underlined): invA::aph, ΔinvAF GCTATCTGCTATCTCACCGAA AGATAAAACCTCCAGATCGTGTAGGCTGGAGCTGCTTC and ΔinvAR GCATTATCGATC AGTACCAGCCGTCTTATCTTGATTGAATTCCGGGGATCCGTCGACC; tviB::aph, ΔtviBF GGGTGGATGTCAATCTGGAAACCACTGAAGAAGAATTACGGTGTAGGCTGGAGCTGCTTC and ΔtviBR TTAAAGGTAAAGCCGAGAATCAGCACGCTGGAACCCTCATTCCGGG GATCCGTCGACC; staA::cat, ΔstaAF AATGGTTATGGCTATGGGTTCTACTTCCGCA ATGGCAGGTGTAGGCTGGAGCTGCTTC and ΔstaAR AGTCACTTCTTTAGAAGCATCGGC ACGAACGTAAGACGCATATGAATATCCTCCTTAG; tcfA::cat, ΔtcfAF CGGGGTGTTT CTCTGTGTCGCTATGTTTGCATGTGGTCGTGTAGGCTGGAGCTGCTTC and ΔtcfAR CAGC AACACCCCCCAGACAAGATTCACACTTACGGATGCATATGAATATCCTCCTTAG; pilS::cat, ΔpilSF ATGAAACAGAGGGAAAGATGATGAATGAGGTATCAACATTAAATCCATG CGTGTAGGCTGGAGCTGCTTC and ΔpilSR GCCGTTACTTTCGCATCGGTGTGGTTATT GCCGTTTATTTTAATACCGGACATATGAATATCCTCCTTAG. PCR product (3–5 μg) was transformed into BRD948-pKD46 that had been either grown to OD600 0.3 and then induced to express Red-recombinase with 1 mM arabinose for 1 h or grown to OD600 0.5 in the presence of 0.1 mM arabinose. Mutant clones that were able to grow on the appropriate selection media were then cultured overnight at 43 °C to destabilize pKD46. Clones that had lost the ampicillin resistance from the pKD46 plasmid, but were still positive by selection for the inserted chloramphenicol- or kanamycin-resistance cassette, were screened by colony PCR and tested for the presence of a single cassette by Southern blotting (Amersham ECL kit) (data not shown). The growth of each mutant in LB broth (with 10 g NaCl l−1) was not significantly different to that of BRD948, which itself showed no growth defects, with aro mix and tyrosine supplementation, compared with wild-type Ty2 (data not shown).
Mammalian cell culture
INT-407 cells (European Collection of Cell Cultures #85051004) were grown for up to 20 passages. INT-407 cells were routinely cultured in low-glucose Dulbecco's Modified Eagle's Medium (DMEM; containing 1000 mg glucose l−1) supplemented with 10 % fetal calf serum (FCS) and 2 mM l-glutamine (Sigma-Aldrich). T84 cells (American Type Culture Collection #CCL-248) are human colonic carcinoma cells. T84 cells were used for up to 10 passages and were maintained in medium composed of a 1 : 1 mixture of high-glucose DMEM (4500 mg glucose l−1) supplemented with 4 mM l-glutamine and Hams F12 medium, to which was added 5 % FCS (Sigma-Aldrich).
Gentamicin-protection invasion assay
Gentamicin-protection assays were carried out as described elsewhere (Weinstein et al., 1998) with specific minor modifications as follows. At 48 h prior to addition of bacteria, 2×105 INT-407 cells were added to each well of a 24-well plate. Where cells were to be visualized by immunofluorescence microscopy coverslips were added prior to cells. At the time of infection, 80–100% confluent monolayers had been formed with an average of 4×105 cells per well. For T84 cell infections, 2.5×105cells were seeded onto 24 mm (six-well) transwells containing 0.4 μm-pore polyester supports (Corning Life Sciences). For comparison of apical and basolateral invasion, 3 μm-pore supports were utilized. Medium was changed every day during the polarization process. The progression to polarization was monitored using an epithelial Volt Ohm meter (World Precision Instruments) and cells were infected after 8–10 days, when a minimum trans-epithelial electrical resistance (TEER) of 700 Ω cm−2 was achieved. Fresh medium was placed onto the cells at least 1 h prior to infection. For T84 cells, the TEER was read just before addition of bacteria (0 h) and at the end of the experiment, after gentamicin treatment.
Where alternative bacterial growth conditions were tested, bacterial cultures were grown with shaking for 6 h at 200 r.p.m., subcultured by 1:100 dilution in fresh medium, and grown overnight at 37 °C with or without shaking at 200 r.p.m., or in altered-NaCl-concentration LB broth. For mid-exponential growth, overnight shaken cultures were diluted 1:50 into fresh LB broth with high or low NaCl and grown to OD600 0.5 (∼3×108 bacteria ml−1). S. Typhi had a longer lag phase than S. Typhimurium, but grew at a similar growth rate in exponential phase (data not shown). Approximately 1 ml of bacterial culture (volumes corrected to give an OD600 of 0.5) was centrifuged at 14 000 g for 2 min, and bacterial pellets were resuspended in 1 ml cell culture medium. Infections were routinely carried out in 0.5 ml cell culture medium, or in 1 ml of medium for basolateral infections in order to maintain submersion of the transwell insert. When S. Typhi was compared with S. Typhimurium, 0.025 ml bacterial suspension was added to each well, giving an m.o.i. of ∼50 c.f.u. per cell. Where BRD948 was compared with mutant strains, 0.1 ml of this bacterial suspension was used, giving an m.o.i. of ∼200 c.f.u. per cell. Inoculum counts were checked by enumeration after dilution in PBS and plating on LB agar. Infections were carried out for 2 h at 37 °C in 5% CO2. Bacteria were then removed and replaced with 1 ml of the appropriate cell growth medium containing 0.1 mg gentamicin ml−1 and incubated at 37 °C in 5% CO2 for 1 h. Cells were washed three times in Dulbecco's PBS (Sigma-Aldrich) to wash away gentamicin and remove dead bacteria. For polarized cells, both the upper and lower compartments were treated with gentamicin and washed with PBS. For immunofluorescence staining, cells were fixed for 15 min in 4 % formaldehyde solution in PBS at room temperature, and coverslips (or transwell inserts) were placed into fresh PBS prior to staining. Gentamicin-resistant intracellular bacterial counts were enumerated, after lysis of cells for 30 min at 37 °C with 1 ml 0.1% Triton X-100 in H2O, by serial 10-fold dilution in PBS and plating on LB agar.
SYTOX-green (Invitrogen Molecular Probes) staining was carried out in order to visualize cell death. Cells become permeable to this nucleic acid stain only when their membranes are compromised. After gentamicin treatment, cells were incubated for a further 20 min with the appropriate cell culture medium, freshly prepared and containing 0.5 μM SYTOX-green. Cells were washed in PBS, fixed in formaldehyde and stained.
Adhesion assays
For the 4 °C adhesion assay, INT-407 cells prepared in 24-well plates, as described above, were pre-incubated in 0.5 ml 20 mM HEPES-buffered cell growth medium at 4 °C for 20 min prior to infection. Bacteria were resuspended in 20 mM HEPES-buffered cell growth medium at 4 °C. Infections were carried out at 4 °C for 1 h, and cells were washed five times with 1 ml Dulbecco's PBS at 4 °C and processed for enumeration of c.f.u. or immunofluorescence staining as described above. The ability of bacteria that had adhered at 4 °C subsequently to invade was determined by warming plates to 37 °C in fresh culture medium for 1 h, after the standard adhesion procedure.
For the cytochalasin D (CD)-dependent adhesion assay, INT-407 cells prepared in 24-well plates, as described above, were pre-treated with 10 μg CD ml−1 (Sigma-Aldrich) for 1 h. CD treatment was maintained during the 2 h infection period. Cells were washed five times in Dulbecco's PBS and processed for immunofluorescence or c.f.u. determination by serial dilution.
Immunofluorescence staining
Infected cells were washed, fixed in formaldehyde and placed into PBS as described above. Transwell inserts were carefully cut from their supports with a scalpel prior to staining. Free aldehyde groups were quenched with 10 mM NH4Cl in PBS for 10 min, followed by PBS washes. Differential staining of extracellular (before permeabilization, red or red/green populations) and intracellular (after permeabilization, green population) bacteria was carried out in a similar way to that described elsewhere (Heesemann & Laufs, 1985). Extracellular bacteria were stained with goat anti-Salmonella common surface antigen serum (CSA-1, Insight Biotechnology) in PBS at 0.0025 mg ml−1 for 1 h, followed by PBS washes and incubation with rhodamine-conjugated donkey anti-goat IgG at 2 μg ml−1 (Stratech-Jackson ImmunoResearch) in PBS for 30 min and extensive PBS washes. Cells were permeabilized with 0.2 % saponin in PBS for 10 min. Subsequent antibody incubations were carried out in 0.2 % saponin in PBS. For intracellular antibody staining, cells were then incubated with directly FITC-conjugated CSA-1 antibody at 0.005 mg ml−1 for 1 h, followed by PBS washes. In order to visualize the epithelial cells, a further 30 min incubation was carried out with Alexa 633 phalloidin at 0.0002 mg ml−1 (Invitrogen Molecular Probes). After PBS washes, and a deionized H2O rinse, cells were mounted in ProLong Gold Antifade Reagent (Invitrogen Molecular Probes). It should be noted that using this protocol the majority of extracellular bacteria appear red, rather than both red and green, due to the use of a directly FITC-conjugated anti-Salmonella antibody for the staining of intracellular bacteria. Since the extracellular bacterial staining was saturating, this approach resulted in few red/green co-stained extracellular bacteria. For SYTOX-green-stained cells, saponin permeabilization was carried out prior to staining with CSA-1, followed by a rhodamine-conjugated secondary antibody and far-red DNA stain at 0.0002 mg ml−1 (TO-PRO-3, Invitrogen Molecular Probes), to reveal all epithelial cells and cell-associated Salmonella. Samples were visualized using confocal microscopy with a Zeiss Axioplan2 microscope and LSM 510 Laser Scanning Confocal System (Carl Zeiss). Z-stacks were collected to encompass all bacteria within the frame and were recombined into 3D projections using LSM 510 Image software (Carl Zeiss).
RT-PCR
RNA was stabilized by adding RNAProtect reagent (Qiagen) directly to cultures before extraction. RNA was extracted using the Qiagen RNeasy kit, and DNA was removed using the Ambion DNA-free kit according to the manufacturer's instructions. For each RNA preparation, DNA contamination was detected by using 0.5 μg RNA as a template in a 25 μl PCR reaction using RedTaq mix (Sigma-Aldrich), according to the manufacturer's instructions, and 16S forward and reverse primers (16SF GTGAAATGCGTAGAGATCTGGAGG and 16SR CGAGCTGACGACAGCCATGC) (0.40 pmol μl−1). cDNA was synthesized using 1 μg RNA template (assessed by A260) using Qiagen Omniscript reverse transcriptase (Qiagen), random hexamer primers (Invitrogen) and RNase inhibitor (Invitrogen) according to the manufacturer's instructions. BRD948 mutants lacking the appropriate gene were tested as negative controls for each primer set (except for 16S). PCR was carried out using 2.3 μl cDNA template in a 25 μl reaction, with 0.40 pmol μl−1 of each primer set and Red Taq polymerase mix (Sigma-Aldrich) according to the manufacturer's instructions. PCR cycle parameters were as follows: 94 °C for 30 s; 94 °C for 30 s, 54 °C for 30 s and 72 °C for 1 min for 30 cycles; 72 °C for 2 min. For semi-quantification, reactions were held at 24 °C after 10, 15, 20, 25, 30 and 35 cycles for removal of reactions, which were analysed for detectable products by separation on 2% agarose ethidium bromide-stained gels. The number of amplification cycles after which bands were semi-quantified was chosen so that amplification products were clearly visible on agarose gels, but an increase could still be detected with increased cycle number, suggesting that linear amplification was in progress. Cycle numbers and primer sequences for each gene analysed were as follows: 16S, 10 cycles for RT-PCR and 20 cycles for genomic DNA control PCR (primer sequences above); invA, 30 cycles (CTACCTTGCTGATGGATTGTTGG and TGGTTGTTACGGCTATTTTGACC); tviB, 20 cycles (CTGCCTCAGCAACTTTGATGC and GATATGTTGGGCTTCCTCTGG); pilS, 30 cycles (CAGAGGGAAAGATGATGAATGA and GTGTGGTTATTGCCGTTTATTT); staA, 30 cycles (CTTTAGAAGCATCGGCACGAAC and CGCAATGGTTATGGCTATGGG); tcfA, 30 cycles (GCAGTTGCCAGCCCGAATAAG and CGGGGTGTTTCTCTGTGTCGC). Band densities were analysed from digital images using ImageJ software (National Institute of Mental Health), background density levels were subtracted, and measurements for each sample were expressed relative to 16S rRNA.
Statistics
The data were normally distributed; therefore, mean values are displayed. Differences between two independent groups were analysed using two-tailed t tests. For multiple comparisons, one-way ANOVA was used with post-hoc t tests and Tukey's correction for multiple comparisons. A P value <0.05 was taken as significant in all cases. All tests were performed using SPSS statistical software.
RESULTS
S. Typhi adhesion to and invasion of non-polarized epithelial cells
Invasion of tissue culture cells by S. Typhi has been reported to be more efficient following bacterial culture in growth medium containing high salt concentrations (Mills & Finlay, 1994; Tartera & Metcalf, 1993). Since the two-component regulators RcsB–RcsC and OmpR–EnvZ influence Vi capsule and SPI-1 expression, this phenomenon may be explained, at least in part, by the effect of osmolarity on the expression of key surface-associated structures. Consequently, the ability of S. Typhi to mediate adhesion to and invasion of INT-407 human epithelial cells was investigated following culture under a number of different growth conditions using S. Typhimurium as a control.
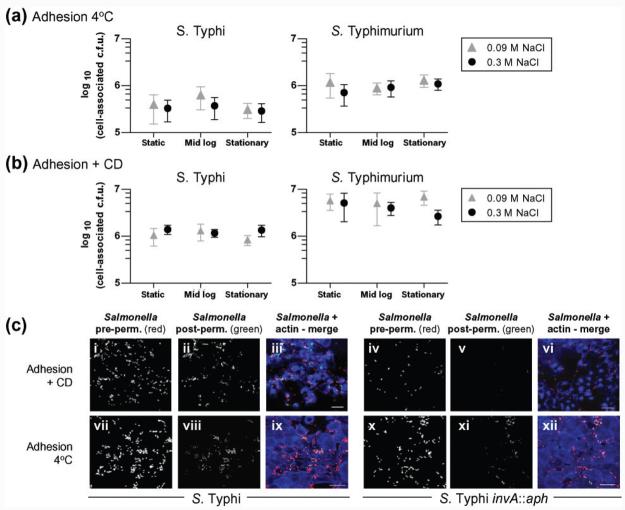
Initially, the ability of S. Typhi BRD948 to adhere to INT-407 epithelial cells was determined under conditions in which invasion was inhibited by reduced temperature or by the presence of the actin polymerization inhibitor CD. Bacterial adhesion to host cells was determined at 4 °C or in the presence of CD at 37 °C. The levels of cell-associated S. Typhi (measured as viable counts) that were protected from gentamicin were about 0.003% after incubation at 4 °C for 1 h followed by gentamicin treatment for 1 h at 37 °C, and about 1% at 37 °C for 2 h in the presence of CD followed by gentamicin treatment for 1 h at 37 °C. Differential staining of intracellular (green) and extracellular (red, red/ green) bacteria confirmed the presence of few intracellular bacteria after incubation at 4 °C or CD treatment (Fig. 1c), further indicating that invasion levels were low and that bacteria were largely cell surface-associated under these conditions. Interestingly, the distribution of bacteria was different in the two adhesion assays. After incubation at 4 °C bacteria were mainly associated with the periphery of cells, in contrast to their localization on the top surface of the cells in CD-treated adhesion assays (Fig. 1c). Adhesion of S. Typhimurium was consistently greater than that of S. Typhi (P<0.005) in both assays (Fig. 1a, b). However, no differences due to bacterial culture conditions in adhesion of S. Typhi to epithelial cells were detected using either assay (Fig. 1a, b).
Fig. 1.
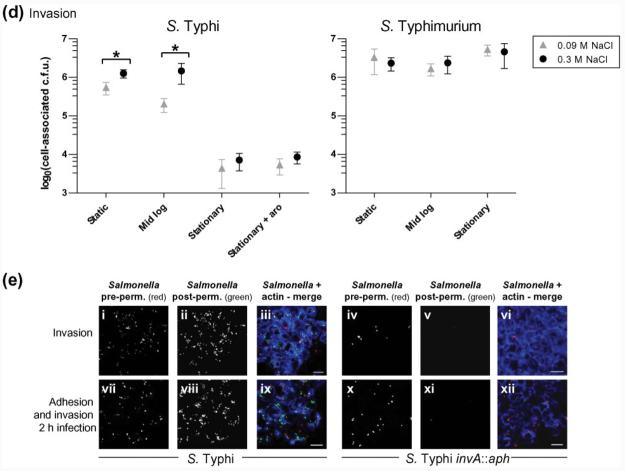
S. Typhi and S. Typhimurium adhesion or invasion after growth under various conditions. (a) Adhesion at 4 °C. The mean cell-associated adherent c.f.u. (±1sd) are shown from five experiments in which samples were tested in triplicate and each condition was tested in at least two independent experiments. (b) Adhesion in the presence of CD. The mean cell-associated adherent c.f.u. (±1sd) are shown from two experiments in which samples were tested in triplicate. (c, e) Z-stack 3D projected images showing extracellular (red or red/green) and intracellular (green) S. Typhi BRD948 or isogenic invA : : aph mutant (as indicated) and INT-407 cells (visualized with actin staining, blue). Examples are shown from adhesion assays with CD or after 4 °C incubation (c), and from invasion assays or combined invasion and adhesion assays (e), as indicated. pre-perm., pre-permeabilization; post-perm., post-permeabilization. Bars, 20 μm. (d) Invasion of cultured INT-407 cells by S. Typhi or S. Typhimurium cultured under various conditions. Mean cell-associated c.f.u. (±1sd) are shown following gentamicin treatment from six experiments in which samples were tested in triplicate and each condition was tested in at least three independent experiments.
Invasion of INT-407 cells by S. Typhi was ∼10-fold lower than for S. Typhimurium, although invasion was increased following culture at elevated salt concentrations (Fig. 1d). Similar invasion results were seen with wild-type S. Typhi strain Ty2, from which BRD948 was derived, after growth under different conditions (data not shown). In contrast, no significant effects of growth phase or salt concentration were observed for S. Typhimurium invasion using this assay system (Fig. 1d). Stationary shaken cultures of S. Typhi exhibited dramatically decreased levels of invasion, while this was not the case for S. Typhimurium (Fig. 1d). S. Typhi culture to late-exponential phase in high-salt media has been reported to result in maximal invasion (Tartera & Metcalf, 1993; Mills & Finlay, 1994). Here, although we use mid-exponential and not late-exponential cultures, it should be noted that the increase in invasion found by Tartera & Metcalf (1993) from mid- (OD600 0.5) to late- (OD600 1.5) exponential phase was approximately twofold, while an ∼10-fold increase in invasion was observed under low- compared with high-salt conditions at either growth stage.
Following our observation that S. Typhi invasion of, but not adhesion to, INT-407 epithelial cells was dependent on growth conditions, the involvement of the SPI-1-encoded TTSS and Vi capsule was investigated. Strains with an inactivated SPI-1-encoded TTSS (invA : : aph) or Vi capsule (tviB : : aph) were employed. The parental S. Typhi exhibited 1000-fold greater invasion than S. Typhi invA : : aph under all growth conditions tested, highlighting the importance of the SPI-1 locus (Figs 1e and 5a, c). In contrast, S. Typhi invA : : aph remained adherent at 4 °C or after CD treatment of cells (Figs 1c and 5b, d, e). This adherent, but poorly invasive, S. Typhi invA : : aph mutant was also tested for adhesion at 37 °C after mid-exponential or static growth in high- or low-NaCl media. No significant differences in adhesion were observed for this non-invasive mutant grown under these four conditions (data not shown).
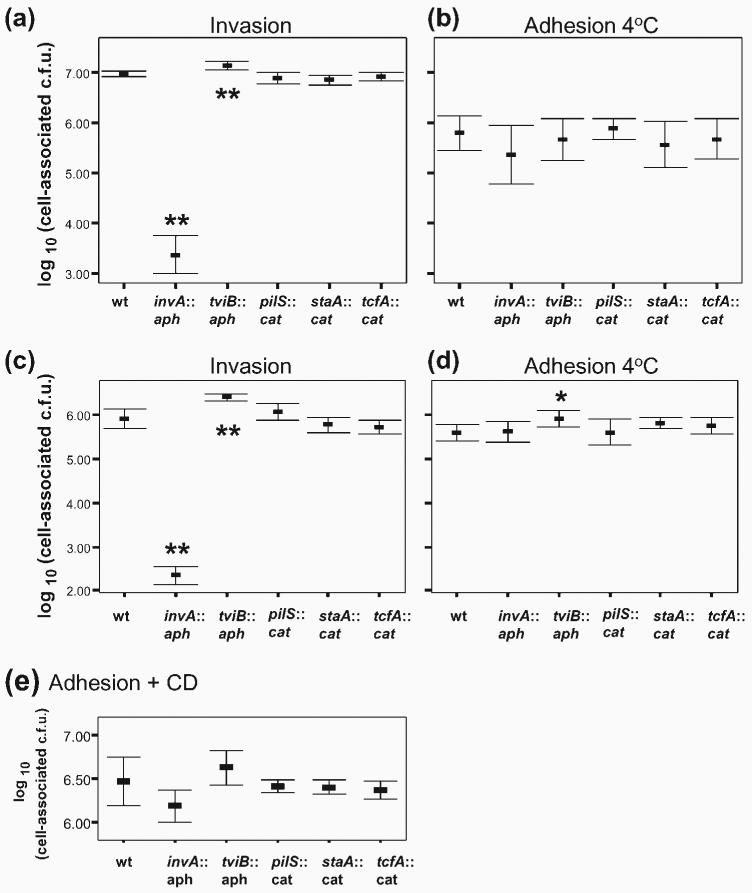
Fig. 5.
Invasion and adhesion for S. Typhi and its isogenic mutants lacking various surface structures. (a, b) Bacteria grown to mid-exponential phase under high-salt conditions, which are maximal conditions for BRD948 invasion (Fig. 1d), or (c, d) grown statically in high-salt LB broth, which are maximal conditions for expression of staA and tcfA fimbrial genes and pilS (Fig. 4), were assayed for invasion (a, c) or adhesion after incubation at 4 °C (b, d). S. Typhi BRD948 (wild-type, wt) and its isogenic invA, tviB, pilS, staA or tcfA mutants (as indicated) were tested. Previously reported severe invasion attenuations (invA mutant) or enhancements (tviB mutant) were evident (one-way ANOVA and post-hoc t tests with Tukey's correction, mutant compared with BRD948 wild-type; **P<0.01). No effects upon invasion due to mutations in pilS or major fimbrial subunits (staA or tcfA) were detected (one-way ANOVA, P>0.05), nor were there any detectable adhesion defects for any of the BRD948 mutants (one-way ANOVA, P>0.05). Under static high-salt growth conditions (d), a small but significant increase in adhesion was seen for the tviB mutant compared with BRD948 under the same conditions (one-way ANOVA and post-hoc t tests with Tukey's correction; *P<0.05). Data are shown from three, or two (c), independent experiments. (e) Adhesion in the presence of CD for S. Typhi BRD948 and its isogenic mutants grown to mid-exponential phase with high salt. There were no significant differences between each mutant and BRD948 for adhesion in the presence of CD (one-way ANOVA, P>0.05). Data are shown from four independent experiments, each in triplicate, in which each strain was tested in at least three experiments. In all cases, means±sd are shown.
To investigate the relationship between host cell invasion and Vi capsule expression, invasion of INT-407 cells by S. Typhi-tviB : : aph was compared with that of the parent strain cultured under different growth conditions. S. Typhi-tviB exhibited a higher level of invasion compared with the parental S. Typhi under all conditions, although the difference was least pronounced after culture in high-salt media to mid-exponential phase, where Vi capsule expression was low (Fig. 5a for mid-exponential and Fig. 5c for static high-salt growth conditions; data not shown for low-salt growth conditions).
S. Typhi invasion of polarized epithelial cells
The ability of S. Typhi cultured under different conditions to invade polarized T84 cells on transwell inserts was determined. Both S. Typhi and S. Typhimurium invaded polarized cells ∼10-fold less efficiently than non-polarized cells. Furthermore, S. Typhi invasion efficiency was lower than that of S. Typhimurium (Fig. 2a) as observed with non-polarized cells. Greater invasion by S. Typhimurium was reflected in a dramatic decrease in TEER (Fig. 2b). Similar levels of invasion to that of S. Typhi BRD948 were obtained with the parental wild-type strain Ty2 (data not shown). Fixed transwell membranes permeabilized and stained with anti-Salmonella antibody showed small patches of intracellular S. Typhi, whereas S. Typhimurium was more evenly distributed across the monolayer but was still associated with a small percentage of cells (Fig. 2c). As might be expected due to the bacterial burden, SYTOX-green staining (a marker for non-viable cells) was greater in the T84 monolayer infected with S. Typhimurium compared with those infected with S. Typhi (Fig. 2c). Indeed, the proportion of SYTOX-green-positive cells following the addition of S. Typhi was similar to that for uninfected monolayers (data not shown). The conditions of S. Typhi culture had no measurable effect on the levels of invasion of polarized T84 cells (Fig. 2a) or TEER during the assay (Fig. 2b).
Fig. 2.
Apical invasion of polarized T84 cells by S. Typhi and S. Typhimurium. (a) Apical invasion of polarized T84 cells (grown on 0.4 μm-pore supports), determined by viable counts, for S. Typhi BRD948 grown under different conditions and S. Typhimurium SL3261 grown only under static conditions with low salt. S. Typhi invasion of polarized T84 cells was lower than that of S. Typhimurium (t test for static low-salt growth conditions; *P<0.05). (b) TEER after 2 h infection and 1 h gentamicin treatment, expressed as a percentage of TEER at the start of the experiment (0 h). S. Typhimurium caused a large drop in TEER, whereas S. Typhi, under any of four growth conditions tested (as indicated), caused a minor drop in TEER (t test for static low-salt growth conditions, S. Typhi versus S. Typhimurium; *P<0.005). Data shown are from three independent experiments in which each condition was tested in triplicate. Means±sd are shown. (c) Z-stack projections showing the distribution of S. Typhimurium and S. Typhi on polarized T84 monolayers following gentamicin treatment, visualized by immunofluorescence microscopy. Salmonella are stained red, dying cells green, and host cell nuclei blue. Mid-exponential high-salt bacterial growth conditions were used for these assays. Images are representative of data from two independent experiments. Bars, 50 μm.
We compared the ability of S. Typhi and S. Typhimurium to invade T84 cells from the basolateral surface. Increased invasion was observed for S. Typhi applied basolaterally, compared with apical invasion, whereas S. Typhimurium was invasive to similar extents via the two routes of infection (Fig. 3a). The increased bacterial load was accompanied by a lower TEER for basolateral S. Typhi invasion of T84 cells compared with apical infection (Fig. 3b). Similar results for basolateral compared with apical infection of T84 cells were seen with S. Typhi strain Ty2, the parent strain of BRD948 (data not shown). In each case, invasion and the accompanying drop in TEER were largely invA-dependent (Fig. 3a,b). We note that apical and basolateral invasion of polarized cells was very similar for experiments that utilized the aroA S. Typhimurium mutant (Fig. 3a, b, mid-exponential cultures) and the double aroA htrA (Fig. 2a, b, static cultures) mutant, supporting previous observations that htrA mutation does not compromise invasion by S. Typhimurium. Deletion of the tviB gene of S. Typhi did not result in enhanced invasion of polarized T84 cells at the apical or basolateral surfaces (Fig. 3c), nor was there a further decrease in TEER compared with that observed for the isogenic strain wild-type for tviB BRD948 (data not shown).
Transcriptional expression of S. Typhi surface structure determinants
Having observed differences in invasion for S. Typhi following culture under a variety of conditions, we next determined whether transcription of genes encoding S. Typhi-specific surface structures was modulated by these growth conditions. Transcription of invA, tviB, pilS, staA and tcfA was determined using semi-quantitative RT-PCR (Fig. 4). 16S rRNA is abundant regardless of culture conditions (Fey et al., 2004), and was therefore used as a control. The mRNA products for all of these genes were detectable, although staA mRNA was in low abundance and consequently the appearance of a non-specific amplification product made the interpretation of the data problematic. However, in each case, and particularly in the case of the invA gene, expression was lowest in stationary-phase cultures compared with that under the other culture conditions, which may in part explain the decreased invasion after growth to stationary phase (Fig. 1d). Expression of invA was increased, and that of tviB decreased, by high-salt culture conditions (particularly in exponential phase), consistent with earlier studies (Arricau et al., 1998; Pickard et al., 1994; Virlogeux et al., 1996). Expression of pilS was greatest in static cultures and showed an increase in the presence of elevated NaCl concentration, which is contrary to a study using reporter constructs that showed the highest pil operon expression in 100 mM NaCl stationary-phase cultures and inhibition at higher salt concentrations (Lee et al., 2006). Transcription of staA was greater at elevated NaCl concentrations, particularly for static cultures. Static high-salt conditions also enhanced tcfA transcription. Additional incubation, for 2 h in DMEM+10% FCS at 37°C, of high-salt-medium mid-exponential phase-cultured S. Typhi strain BRD948 had no effect on expression of invA, tviB, pilS, staA or tcfA (data not shown).
Fig. 4.
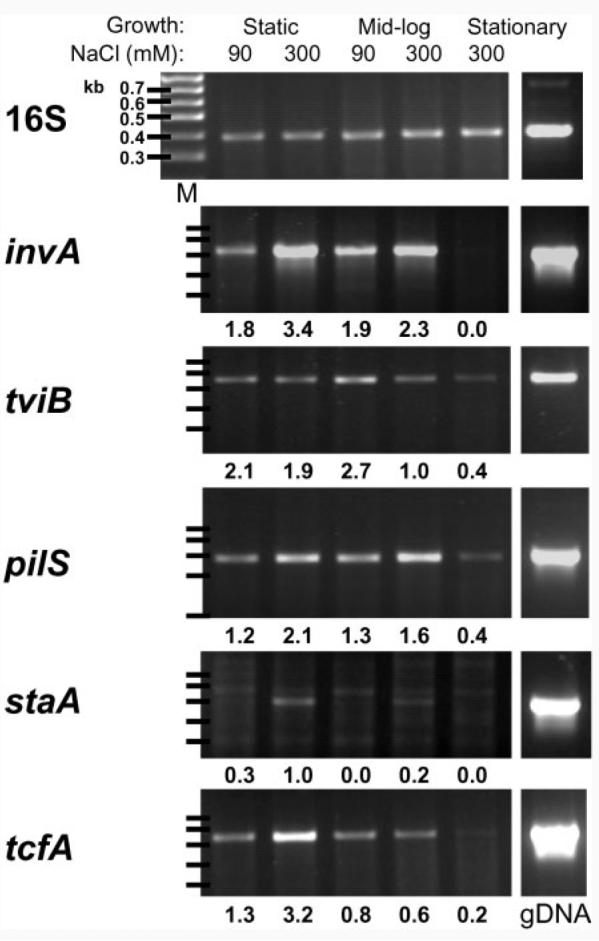
Expression of S. Typhi fimbriae determined with RT-PCR. Growth conditions and the salt concentration for S. Typhi BRD948 from which RNA was extracted are indicated. gDNA, genomic DNA; M, markers. The gene names are indicated to the left of each dataset. The number of thermal cycles and sequences of the primers used to generate each PCR product are noted in Methods. Density measurements, after background level subtraction and expressed relative to that of 16S, are shown below each band. Results for RNA samples from one experiment are shown and are representative of three independent experiments.
S. Typhi fimbriae have no detectable effect on adhesion to or invasion of epithelial cells
We next determined whether the S. Typhi pil, sta and tcf genes contribute to epithelial cell invasion following in vitro culture. To this end we constructed S. Typhi BRD948 variant strains in which pilS, staA or tcfA (putative major subunits) was replaced by the aph or cat gene. The ability of these mutant strains to adhere to, and invade, INT-407 tissue culture cells was compared with that of strain BRD948. We compared adhesion and invasion following culture under two conditions: (1) mid-exponential-phase culture in elevated NaCl, which gave maximal invasion and relatively high pilS and invA expression; (2) static cultures in high-salt medium, conditions under which there was low tviB and high TTSS (invA) and fimbrial (pilS, staA and tcfA) gene transcription. Invasion of the pilS, staA and tcfA mutants was similar to that of BRD948 (wild-type) after growth in high-salt LB broth to mid-exponential phase or static overnight (Fig. 5a, c).
S. Typhi and mutant derivatives lacking pilS, staA or tcfA were also tested for adhesion. No deficiency in adhesion for strains containing mutations in pilS, staA or tcfA, compared with BRD948, was detected using either the 4 °C adhesion assay (Fig. 5b, d) or CD-treated tissue culture cells (Fig. 5e). The distributions of pilS, tcfA and staA mutant S. Typhi in the adhesion assays and following gentamicin treatment were similar to that of BRD948 (data not shown). The preferential distribution of cell-associated extracellular bacteria towards the periphery of INT-407 cells in the 4 °C adhesion assay system, rather than on the apical surface as was seen after CD treatment, was also observed for the S. Typhi mutants tested (Fig. 1c; data not shown). Furthermore, no effect of S. Typhi pilS on apical or basolateral invasion of T84 cells was detectable (Fig. 3c).
Discussion
Comparison of the genome nucleotide sequences of S. Typhi and S. Typhimurium reveals distinct coding capacities for a number of virulence determinants, including Vi capsule biosynthesis and fimbrial biogenesis. These differences may contribute to the observed disease syndrome and host range of these serovars (Townsend et al., 2001). We compared the interactions of S. Typhi and S. Typhimurium with an INT-407 epithelial cell line to define differences between these closely related pathogens. Culture conditions affected S. Typhi invasion of epithelial cells to a greater extent than that of S. Typhimurium. This may be explained, at least in part, by the effects of factors present in serotype Typhi that are absent in serotype Typhimurium. For example, the Vi polysaccharide capsule is encoded on a Typhi-specific pathogenicity island (SPI-7) and is downregulated, concomitant with upregulation of SPI-1 TTSS expression. This is particularly the case in the presence of elevated salt concentration (Tartera & Metcalf, 1993). We observed more invasion of INT-407 cells by S. Typhimurium compared with S. Typhi, in agreement with some studies (Mills & Finlay, 1994), but contradicting data from others (Weinstein et al., 1998). Also, an earlier study reported increased invasion following culture of S. Typhimurium SL1344 in elevated-salt conditions (Galan & Curtiss, 1990), which we did not observe in our study. It is not clear why these data differ, but bacterial culture conditions, choice of tissue culture cell lines or the bacterial strains used may play a role. Differences in invasion of tissue culture cells by S. Typhi and S. Typhimurium may reflect the relatively small intestinal involvement of S. Typhi, particularly in the early stages of the pathogenesis of typhoid fever, compared with the profound colonization and invasion of the epithelial brush border associated with the pathogenesis of S. Typhimurium gastroenteritis.
Earlier studies of adhesion by S. Typhi to epithelial cells have employed assays in which a standard invasion assay was performed but gentamicin treatment was omitted. In this format, total bacterial counts (intracellular and extracellular) were enumerated (Zhang et al., 2000; McCormick et al., 1995). We have used three different types of assay in an attempt to uncouple the phenomenon of adhesion from that of invasion. We observed greater adhesion for S. Typhimurium compared with S. Typhi, and this was not affected by bacterial growth conditions. Most dramatically, and consistent with earlier observations using MDCK cells (Lee & Falkow, 1990), we found that stationary-phase S. Typhi cultures had a low capacity for invasion, yet retained their ability to adhere to INT-407 cells (Fig. 1). The effects of environmental stimuli on S. Typhi invasion of epithelial cells reflect differences in invasive capacity rather than changes in adhesion. No such decrease in invasion by stationary-phase cultures of S. Typhimurium was observed.
We observed a modest increase in adhesion at 4 °C by a tviB mutant following culture in high-salt medium with static growth (Fig. 5d), suggesting that the Vi capsule inhibits adhesion as well as invasion. Increased invasion of S. Typhi tviB mutants (Fig. 5a, c) has been reported previously (Miyake et al., 1998; Zhao et al., 2001) and increased IL-8 secretion by epithelial cells has also been attributed to Vi capsule mutants (Sharma & Qadri, 2004; Raffatellu et al., 2005a), but this is believed to be the first evidence for negative effects of Vi capsule on adhesion to epithelial cells (Fig. 5d). One study reported that the S. Typhi Vi capsule may actually promote adhesion to Caco-2 epithelial cells in suspension, by interaction with prohibitin (Sharma & Qadri, 2004). Furthermore, Vi capsule has been reported to block agglutination of Saccharomyces cerevisiae, probably due to mechanical masking of type I fimbriae (Miyake et al., 1998). Additional surface structures may also be masked by Vi capsule polysaccharide.
Low levels of invasion of S. Typhi into polarized T84 cells relative to non-polarized cells, such as INT-407, were observed, and neither high-salt growth conditions nor deletion of tviB increased invasion. In contrast, application of bacteria to the basolateral surface of the cells enhanced invasion by S. Typhi, but not by S. Typhimurium, suggesting that a factor that facilitates S. Typhi invasion is present on the basolateral and not on the apical surface of T84 cells. Differences in Salmonella invasion of polarized compared with non-polarized cells have been observed previously. For instance, S. Typhimurium causes activation of Rac1, but not Cdc42, during apical invasion of polarized MDCK cells, whereas both of these Rho family GTPases are activated during invasion of non-polarized cells (Criss et al., 2001). Also, different SPI-1 effectors are essential depending on whether S. Typhimuirum is invading polarized or non-polarized cells (Raffatellu et al., 2005b). Interestingly, basolateral invasion of polarized MDCK cells by S. Typhimurium is dependent on actin, the Arp2/3 complex and a Rho family GTPase, yet Cdc42, Rac, RhoA or RhoG, which are activated during apical invasion of MDCK cells, are not required. This suggested a subtle mechanistic difference to basolateral invasion (Criss & Casanova, 2003). We observed SPI-1 dependence for basolateral invasion by both S. Typhi and S. Typhimurium (Fig. 3), but have not investigated the S. Typhi basolateral invasion mechanism further. CD treatment leads to depolarization, so could not be used to assay for adhesion to polarized cells; attempts to assay for apical bacterial adhesion to T84 cells at 4 °C resulted in low levels of adhesion for both S. Typhi and S. Typhimurium (data not shown). The low level of apical adhesion to polarized cells for both salmonellae at 4 °C suggested that in order for invasion to proceed, the epithelial cells and/or the bacteria require adaptations that cannot occur at 4 °C. Such adaptations could include changes in S. Typhi LPS and elaboration of the human cystic fibrosis transmembrane conductance regulator reported elsewhere in polarized MDCK cells (Lyczak et al., 2001; Lyczak & Pier, 2002). It is also possible that receptor recruitment is compromised due to reduced membrane dynamics at 4 °C. However, counter to this argument, we noted that adhesion to INT-407 cells after 1 h at 4 °C was only around half a log lower (cell-associated c.f.u.) than after 2 h incubation in the presence of CD at 37 °C (compare Fig. 1a with Fig. 1b and Fig. 5b with Fig. 5e). Also, 4 °C incubation (for 15–20 min) is often used to study adhesion of opsonized particles prior to phagocytosis without apparently compromising receptor recruitment (Caron & Hall, 1998). Reduced membrane dynamics could contribute to the different S. Typhi INT-407 cell adhesion patterns observed at 4 °C (around the cell periphery) compared with CD-treated cells (more evenly distributed) (see Fig. 1c). For polarized cells an increase in TEER was observed at 4 °C (data not shown), which could be due to changes in channel activity and/or membrane dynamics.
Transcription of staA and tcfA fimbrial gene mRNA did not correlate with the maximal invasion conditions. Transcription was greatest following static culture in media containing elevated salt concentrations (Fig. 4), while invasion was greatest following culture to mid-exponential phase in elevated salt concentrations (Fig. 1d). pilS transcription was increased by elevated salt concentration in both static and mid-exponential cultures (Fig. 4). A 2006 study using reporter constructs has shown the highest expression of pilS in stationary phase with 100 mM NaCl and control of expression by an overlapping set of regulators that control the tviB locus, including OmpR/EnvZ, RcsB/C and TviA (Lee et al., 2006). We found that pilS, tviB and tcfA mRNA was detectable, but not highly expressed (compared with 16S rRNA) in stationary phase (Fig. 4), although our analysis was semi-quantitative. Increased levels of anti-TcfB antibodies in typhoid convalescent patients suggest that Tcf is elaborated in the host. Transcription of invA was also greatest following static culture in media containing elevated salt concentrations. However, the reduction in tviB transcription at elevated salt concentrations was greater for exponential-phase cultures than for static cultures (Fig. 2), suggesting that reduced Vi capsule promotes maximal invasion.
Motility facilitates invasion of epithelial cells in vitro for both S. Typhimurium and S. Typhi (Jones et al., 1992; Liu et al., 1988). For S. Typhimurium, invasion of aflagellate mutants is restored by centrifugation of bacteria onto the epithelial cells, suggesting that flagella are required primarily to enable contact between bacteria and cells through motility (Jones et al., 1992). In contrast, centrifugation does not restore invasion of S. Typhi aflagellate mutants (Liu et al., 1988). These data suggest that S. Typhi flagella, in addition to facilitating movement towards cells, may support adhesion and/or invasion in some more direct manner. However, later studies have found that mutations in flagella genes, in particular fliZ or fliA (an alternative sigma factor), can reduce SPI-1 gene expression in S. Typhi, and to a lesser extent in S. Typhimurium, making motility and invasion difficult to study in isolation, and providing an explanation for the invasion defects of S. Typhi flagella mutants (Lucas et al., 2000; Iyoda et al., 2001; Eichelberg & Galan, 2000).
We did not observe altered adhesion or invasion of strains of S. Typhi lacking the staA or tcfA genes. It is important to note that, to date, elaboration of fimbriae encoded by the sta and tcf operons has not been described. While we cannot discount a role for these hypothetical fimbriae in natural infections, we can confirm that these genes are dispensable for adhesion and invasion under the growth conditions described here. We also did not observe any significant effects of mutations in pilS, which encodes the major subunit of a type IV fimbria in S. Typhi (Fig. 5). These data are in contrast to those of earlier studies that report a role for pilS in epithelial cell invasion and in combined adhesion and invasion assays (Zhang et al., 2000; Tsui et al., 2003). The contradictory observations may in part be explained by differences in the strains used. In this study we used a Vi capsule-expressing S. Typhi strain, while a Vi capsule-negative derivative of Ty2 was used in earlier studies. In addition, only static cultures were used by Zhang et al. (2000), the pilS mutant was analysed in competition with the wild-type rather than in single-strain infections, and bacteria were centrifuged onto the cells, whereas we allowed the bacteria to attach to and invade the cells without centrifugation. These differences in strain background and infection assay protocol may account for the differences in results. Our data suggest that pilS may be less important for adhesion in the presence of Vi capsule than was previously thought, even where Vi capsule expression has been suppressed by elevated NaCl concentration.
ACKNOWLEDGEMENTS
This work was supported by The Wellcome Trust.
Abbreviations
- CD
cytochalasin D
- SPI
Salmonella pathogenicity island
- TEER
trans-epithelial electrical resistance
- TTSS
type III secretion system
- Vi capsule
virulence capsule
REFERENCES
- Arricau N, Hermant D, Waxin H, Ecobichon C, Duffey PS, Popoff MY. The RcsB–RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol. 1998;29:835–850. doi: 10.1046/j.1365-2958.1998.00976.x. [DOI] [PubMed] [Google Scholar]
- Avogadri F, Martinoli C, Petrovska L, Chiodoni C, Transidico P, Bronte V, Longhi R, Colombo MP, Dougan G, Rescigno M. Cancer immunotherapy based on killing of Salmonella-infected tumor cells. Cancer Res. 2005;65:3920–3927. doi: 10.1158/0008-5472.CAN-04-3002. [DOI] [PubMed] [Google Scholar]
- Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- Chatfield SN, Strahan K, Pickard D, Charles IG, Hormaeche CE, Dougan G. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb Pathog. 1992;12:145–151. doi: 10.1016/0882-4010(92)90117-7. [DOI] [PubMed] [Google Scholar]
- Criss AK, Casanova JE. Coordinate regulation of Salmonella enterica serovar Typhimurium invasion of epithelial cells by the Arp2/3 complex and Rho GTPases. Infect Immun. 2003;71:2885–2891. doi: 10.1128/IAI.71.5.2885-2891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss AK, Ahlgren DM, Jou TS, McCormick BA, Casanova JE. The GTPase Rac1 selectively regulates Salmonella invasion at the apical plasma membrane of polarized epithelial cells. J Cell Sci. 2001;114:1331–1341. doi: 10.1242/jcs.114.7.1331. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Liou SR, Plunkett G, III, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall G, Gaines S, Landy M, Tigertt WD, Sprinz H, Trapani RJ, Mandel AD, Benenson AS. Studies on infection and immunity in experimental typhoid fever. I. Typhoid fever in chimpanzees orally infected with Salmonella typhosa. J Exp Med. 1960;112:143–166. doi: 10.1084/jem.112.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg K, Galan JE. The flagellar sigma factor FliA σ28 regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun. 2000;68:2735–2743. doi: 10.1128/iai.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey A, Eichler S, Flavier S, Christen R, Hofle MG, Guzman CA. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl Environ Microbiol. 2004;70:3618–3623. doi: 10.1128/AEM.70.6.3618-3623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., III Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., III Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesemann J, Laufs R. Double immunofluorescence microscopic technique for accurate differentiation of extracellularly and intracellularly located bacteria in cell culture. J Clin Microbiol. 1985;22:168–175. doi: 10.1128/jcm.22.2.168-175.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog. 2001;30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- Jones BD, Lee CA, Falkow S. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FK, Morris C, Hackett J. The Salmonella enterica serovar Typhi Vi capsule and self-association pili share controls on expression. FEMS Microbiol Lett. 2006;261:41–46. doi: 10.1111/j.1574-6968.2006.00338.x. [DOI] [PubMed] [Google Scholar]
- Liu SL, Sanderson KE. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci U S A. 1995;92:1018–1022. doi: 10.1073/pnas.92.4.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Ezaki T, Miura H, Matsui K, Yabuuchi E. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun. 1988;56:1967–1973. doi: 10.1128/iai.56.8.1967-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DC, Savidge TC, Pickard D, Eckmann L, Kagnoff MF, Dougan G, Chatfield SN. Characterization of candidate live oral Salmonella typhi vaccine strains harboring defined mutations in aroA, aroC, and htrA. Infect Immun. 1999;67:700–707. doi: 10.1128/iai.67.2.700-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Pier GB. Salmonella enterica serovar Typhi modulates cell surface expression of its receptor, the cystic fibrosis transmembrane conductance regulator, on the intestinal epithelium. Infect Immun. 2002;70:6416–6423. doi: 10.1128/IAI.70.11.6416-6423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Zaidi TS, Grout M, Bittner M, Contreras I, Pier GB. Epithelial cell contact-induced alterations in Salmonella enterica serovar Typhi lipopolysaccharide are critical for bacterial internalization. Cell Microbiol. 2001;3:763–772. doi: 10.1046/j.1462-5822.2001.00154.x. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. other authors. [DOI] [PubMed] [Google Scholar]
- McCormick BA, Miller SI, Carnes D, Madara JL. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SD, Finlay BB. Comparison of Salmonella typhi and Salmonella typhimurium invasion, intracellular growth and localization in cultured human epithelial cells. Microb Pathog. 1994;17:409–423. doi: 10.1006/mpat.1994.1086. [DOI] [PubMed] [Google Scholar]
- Miyake M, Zhao L, Ezaki T, Hirose K, Khan AQ, Kawamura Y, Shima R, Kamijo M, Masuzawa T, Yanagihara Y. Vi-deficient and nonfimbriated mutants of Salmonella typhi agglutinate human blood type antigens and are hyperinvasive. FEMS Microbiol Lett. 1998;161:75–82. doi: 10.1111/j.1574-6968.1998.tb12931.x. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. other authors. [DOI] [PubMed] [Google Scholar]
- Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- Pickard D, Li J, Roberts M, Maskell D, Hone D, Levine M, Dougan G, Chatfield S. Characterization of defined ompR mutants of Salmonella typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect Immun. 1994;62:3984–3993. doi: 10.1128/iai.62.9.3984-3993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D, Wain J, Baker S, Line A, Chohan S, Fookes M, Barron A, Gaora PO, Chabalgoity JA. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J Bacteriol. 2003;185:5055–5065. doi: 10.1128/JB.185.17.5055-5065.2003. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Baumler AJ. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun. 2005a;73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Baumler AJ. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype Typhimurium invasion of epithelial cells. Infect Immun. 2005b;73:146–154. doi: 10.1128/IAI.73.1.146-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Qadri A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci U S A. 2004;101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartera C, Metcalf ES. Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells. Infect Immun. 1993;61:3084–3089. doi: 10.1128/iai.61.7.3084-3089.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K, Maloy S, Parkhill J. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. other authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui IS, Yip CM, Hackett J, Morris C. The type IVB pili of Salmonella enterica serovar Typhi bind to the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2003;71:6049–6050. doi: 10.1128/IAI.71.10.6049-6050.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlogeux I, Waxin H, Ecobichon C, Lee JO, Popoff MY. Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J Bacteriol. 1996;178:1691–1698. doi: 10.1128/jb.178.6.1691-1698.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein DL, O'Neill BL, Hone DM, Metcalf ES. Differential early interactions between Salmonella enterica serovar Typhi and two other pathogenic Salmonella serovars with intestinal epithelial cells. Infect Immun. 1998;66:2310–2318. doi: 10.1128/iai.66.5.2310-2318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Tsui IS, Yip CM, Fung AW, Wong DK, Dai X, Yang Y, Hackett J, Morris C. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect Immun. 2000;68:3067–3073. doi: 10.1128/iai.68.6.3067-3073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ezak T, Li ZY, Kawamura Y, Hirose K, Watanabe H. Vi-suppressed wild strain Salmonella typhi cultured in high osmolarity is hyperinvasive toward epithelial cells and destructive of Peyer's patches. Microbiol Immunol. 2001;45:149–158. doi: 10.1111/j.1348-0421.2001.tb01283.x. [DOI] [PubMed] [Google Scholar]