Abstract
Bud mutations often arise in citrus. The selection of mutants is one of the most important breeding channels in citrus. However, the molecular basis of bud mutation has rarely been studied. To identify differentially expressed genes in a spontaneous sweet orange [C. sinensis (L.) Osbeck] bud mutation which causes lycopene accumulation, low citric acid, and high sucrose in fruit, suppression subtractive hybridization and microarray analysis were performed to decipher this bud mutation during fruit development. After sequencing of the differentially expressed clones, a total of 267 non-redundant transcripts were obtained and 182 (68.2%) of them shared homology (E-value ≤1×10−10) with known gene products. Few genes were constitutively up- or down-regulated (fold change ≥2) in the bud mutation during fruit development. Self-organizing tree algorithm analysis results showed that 95.1% of the differentially expressed genes were extensively coordinated with the initiation of lycopene accumulation. Metabolic process, cellular process, establishment of localization, response to stimulus, and biological regulation-related transcripts were among the most regulated genes. These genes were involved in many biological processes such as organic acid metabolism, lipid metabolism, transport, and pyruvate metabolism, etc. Moreover, 13 genes which were differentially regulated at 170 d after flowering shared homology with previously described signal transduction or transcription factors. The information generated in this study provides new clues to aid in the understanding of bud mutation in citrus.
Keywords: Bud mutation, candidate genes, cDNA microarray, Citrus, real-time RT-PCR, suppression subtractive hybridization (SSH)
Introduction
Mutations have proved to be a key resource for functional genomics studies in model plant species (Chatelet et al., 2007). Besides the mutants artificially generated in model plants, naturally occurring bud mutants are extensively found for most species (Koornneef et al., 2004). These can be of particular scientific value for citrus.
Bud mutations (bud sports), a consequence of genetic variation of somatic cells leading to the occurrence of phenotypic alteration in plants, arise often in citrus (Raghuvanshi, 1962). Mutations occurred spontaneously in buds and limbs, representing the main natural source of new cultivars (Spiegel-Roy and Goldschmidt, 1996). When these bud sports are vegetatively propagated by clonal techniques, the new phenotype is generally maintained, leading to a new variety (Marcotrigiano, 1997). Mutants are generally detected by the growers themselves in branches of trees showing altered horticultural traits, such as maturity and flowering time or fruit characteristics (Bernet and Asins, 2003). To date, many bud mutants with elite characteristics such as early ripening and red-flesh in citrus fruit have been discovered (Zhang and Deng, 2006).
Genetic improvement in some woody perennial plants such as citrus, apples, and grapes by hybridization has been inefficient, long lasting and time consuming due to their heterozygosity and long juvenility (Asins et al., 1999; Aradhya et al., 2003; Kenis and Keulemans, 2005). Improvement in citrus has been largely the result of selection of naturally occurring bud mutants. Thus, exploring mutants is one of the most important breeding methods to obtain new cultivars with superior traits in citrus. Most cultivars of clementine mandarin (Citrus clementina Hort. ex Tan.), satsuma mandarin (C. unshiu Marc), and sweet orange [C. sinensis (L.) Osbeck] have resulted from bud mutations (Cameron and Frost, 1968; Asins et al., 1999).
Although bud mutations have been important to the citrus industry, the molecular basis behind the generation of sports in citrus is not well understood. Most molecular genetic approaches to study bud mutations in citrus are limited to the detection of genetic variations of bud mutants from their original cultivars by molecular markers, such as RFLPs, RAPDs, AFLPs, ISSR, and SCAR (Moore, 2001; Fanizza et al., 2003; Tao et al., 2006; Mase et al., 2007). However, minor genetic variation existing between the bud mutant and its original cultivars could not be efficiently distinguished by these kinds of markers (Deng et al., 1995; Fang and Roose, 1997; Breto et al., 2001). Several mechanisms that might be the molecular basis of bud mutations have been hypothesized, such as transposon activity, gene mutation, and DNA methylation (Breto et al., 2001). Transposable elements, first recognized by Barbara McClintock in maize (McClintock, 1951), have been identified in many species including citrus (Rico-Cabanas and Martinez-Izquierdo, 2007), grape (Kobayashi et al., 2004), and apple (Yao et al., 2001), and, in grape and apple, the mobility of the transposable elements can be responsible for changes in fruit colour. The deletion of two regulatory genes of the berry locus was responsible for the colour change of grape berries of two bud mutants (Walker et al., 2006). A spontaneous epigenetic mutation in a gene encoding an SBP-box transcription factor resulted in the Colorless non-ripening (Cnr) mutant in tomato (Manning et al., 2006).
Despite such understanding of the mechanism of bud mutations, little information is available on the whole genome level regarding the candidate genes linked to the altered phenotype in mutant fruits of citrus. Until recently, transcriptomic and proteomic profiling and metabolite analysis of a stay-green mutation in the Navel Negra citrus mutant was conducted, and elevated Chl levels and photooxidative stress were associated with the mutant (Alos et al., 2008). A spontaneous bud mutation in sweet orange [C. sinensis (L.) Osbeck] ‘Hong Anliu’, which results in fruits with lycopene accumulation, low citric acid, and high sucrose was reported in a previous study (Liu et al., 2007). To identify differentially expressed genes linked to this bud mutation, techniques combining suppression subtractive hybridization (SSH) and microarray were used. First, by means of SSH (Diatchenko et al., 1996), two libraries of differentially expressed clones were obtained. Then, these clones were printed on a microarray and subsequently used for a global comparison between the mutant fruits and the wild type, and the results were validated with real-time reverse transcriptase polymerase chain reaction (RT-PCR).
Materials and methods
Accession numbers
All the EST sequences generated in this study were deposited in GenBank with accession numbers from FE659063 to FE659327, plus FE660221 and FE660222.
Microarray data and experimental information from this work were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE10729.
Plant material and sample preparation
‘Anliu’ sweet orange [Citrus sinensis (L.) Osbeck] and its red-flesh mutant, ‘Hong Anliu’, cultivated at the Institute of Citrus Research located in Guilin, Guangxi Province, China, were used in the present investigation. Both of them were of the same age, grown in the same orchard and subjected to standard cultivation practices. Fruits of each genotype were collected from three different trees, 10 representative fruits from each tree, for a total of 30 fruits per genotype. These samples were collected at five time points from August to December: 120, 150, 170, 190, and 220 d after flowering (DAF) [fig. 1 in Liu et al., 2007). Sampled fruits were frozen in liquid nitrogen immediately, and kept at –80 °C until analysed.
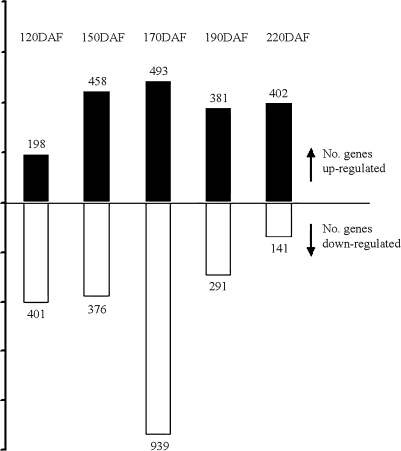
Fig. 1.
Profile of gene expression during fruit development. Number of SSH cDNA clones significantly up- or down-regulated in the sweet orange mutant during fruit development. DAF, Days after flowering.
Two mRNA pools were built for the construction of SSH libraries: R (the mutant ‘Hong Anliu’) and CK (the wild type ‘Anliu’). Pool R and pool CK were enriched for equal amount of mRNA at each time point from the mutant and wild-type fruits, respectively.
Total RNA and mRNA isolation
Total RNA was extracted from fruits following Liu et al. (2006). Isolated RNA was treated with DNase I at 37 °C for 1 h to remove genomic DNA contamination. For SSH, equal amounts of total RNA for each sample from ‘Hong Anliu’ and wild type were mixed and the mRNA was purified from the mixed total RNA using PolyATract® mRNA Isolation System I (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Construction of subtracted cDNA library
The cDNA reversely transcribed from 2 μg of the mixed mRNA mentioned above was used for SSH with the PCR-selected cDNA subtraction kit (BD Biosciences Clontech, San Jose, CA, USA). Both forward (mutant as tester and wild type as driver) and reverse (wild type as tester and mutant as driver) SSH libraries were constructed following the manufacturer's instructions.
Amplification of cDNA inserts
The three thousand cDNA clones, which were randomly picked from each subtracted SSH library, were cultured in 384-well plates overnight at 37 °C and used as templates. PCR amplification was conducted following Ouyang et al. (2007). PCR products were precipitated with anhydrous ethanol–sodium acetate (25:1), resuspended in 40 μl sterile water, and run on 1.2% agarose gel and examined by Bio-Rad UV spectroscopy (Bio-Rad Laboratories, Washington, DC, USA) to ensure both the quality and quantity.
cDNA microarray slides preparation
The PCR products were precipitated again by adding 100 μl of anhydrous ethanol and resuspended in 15 μl of 50% dimethylsulphoxide at a final concentration of 0.1–0.5 μg μl−1 and then spotted onto amino-silaned glass slides (CapitalBio Corp., Beijing, China) with a SmartArrayer™ microarrayer (CapitalBio Corp.). Each clone was printed in triplicate. After printing, the slides were baked for 1 h at 80 °C and stored dry at room temperature till use. Prior to hybridization, the slides were rehydrated over 65 °C water for 10 s, snap dried on a 100 °C heating block for 5 s, and UV cross-linked at 250 mJ cm−2. The unimmobilized PCR products were washed off with 0.5% SDS for 15 min at room temperature, and SDS was removed by dipping the slides in anhydrous ethanol for 30 s. The slides were spun dry at 1000 rpm for 2 min. Eight sequences derived from intergenic regions in yeast genome, showing no significant homology to all existing citrus sequences, were spotted multiple times onto the microarray as exogenous controls. Total citrus RNA was spiked with a mixture of these exogenous control RNAs to validate the semi-quantitative microarray result.
Preparation of fluorescent dye-labelled cDNA and hybridization
The relative gene expression profiles of ‘Hong Anliu’ fruits at 120, 150, 170, 190, and 220 DAF compared with those of wild type at the corresponding stages were investigated by microarray analysis. An aliquot of 5 μg total RNA was used to produce Cy5/Cy3-labelled cDNA employing an RNA amplification combined with Klenow enzyme labelling strategy according to a previously published protocol (Guo et al., 2005).
Cy5/Cy3-labelled cDNA was hybridized with the microarray at 42 °C overnight. Each hybridization was performed in duplicate by dye swap. After that, the arrays were washed with 0.2% SDS, 2× SSC at 42 °C for 5 min, and then with 0.2% SSC for 5 min at room temperature.
Microarray data analysis
Arrays were scanned with a confocal laser scanner, LuxScan™ 10K (CapitalBio Corp.), and the resulting images were analysed with SpotData Pro 2.0 software (CapitalBio Corp.). Spots with fewer than 50% of the signal pixels exceeding the local background value for both channels (Cy3 and Cy5) plus two standard deviations of the local background were removed. cDNA spots with less than four out of a total of six data points in each replicated hybridization were removed. A spatial and intensity-dependent (LOWESS) normalization method was employed (Yang et al., 2002). Normalized ratio data were then log transformed. Differentially expressed genes were identified using a t-test, and multiple test corrections were performed using false discovery rate (FDR) (Benjamini and Hochberg, 1995). Genes with FDR <0.01 and a fold change ≥2 were identified as differentially expressed genes.
EST sequence analysis
All the clones differentially expressed in at least one of five stages were single-pass sequenced (AuGCT Biotechonology Co. Ltd, Beijing, China). The software SeqClean was used for performing vector removal, poly(A) removal, trimming of low quality segments at the 5′ and 3′ ends, and cleaning of low complexity regions. RepeatMasker was used to mask repeats (Smit, 2007). Reading assembly was performed with the CAP3 program (Huang and Madan, 1999), using the read quality and defaults parameters. Cluster analysis was performed by the self-organizing tree algorithm (SOTA) (Herrero et al., 2001), using linear correlation coefficient as the distance between genes. The tree was allowed to grow up using a variability threshold of 40% as the training condition.
The Blast2Go (Conesa et al., 2005) program was used for the gene ontology (GO) data mining.
Quantitative real-time PCR verification
Total RNA was extracted from ‘Anliu’ and ‘Hong Anliu’ fruits collected at five different development stages according to Liu et al. (2006). Primer pairs were designed with the Primer Express software (Applied Biosystems, Foster City, CA, USA). Primer sequences are provided in Table S3 available in Supplementary data at JXB online. Real-time PCR verification was performed according to Liu et al. (2007).
Results
Construction of SSH libraries and overall features of the mutant-responsive expression profile
To isolate genes differentially expressed in the mutant ‘Hong Anliu’ sweet orange compared with its wild type ‘Anliu’ during fruit development, forward (mutant as tester and wild type as driver) and reverse (wild type as tester and mutant as driver) subtractions were conducted between fruits of the red-flesh mutant and its wild type. Two thousand nine hundred and eighty-nine clones were randomly picked from each SSH library. The average insert size of the SSH clones was around 0.4 kb. The clones from the two SSH libraries were amplified and used for microarray analysis. RNA samples of mutant and wild-type fruits collected at 120, 150, 170, 190, and 220 DAF were used for microarray hybridization.
In total, 2394 differentially expressed cDNA clones (fold change ≥2 and FDR <0.01) were identified. However, only one clone (GenBank accession no. FE659117) was constitutively down-regulated, and no clone was constitutively up-regulated in the mutant versus its wild type during fruit development. The number of up- or down- regulated clones was largest at 170 DAF when lycopene began to accumulate in the juice sacs of the mutant ‘Hong Anliu’ (Fig. 1). At this time point, there are more clones down-regulated than up-regulated in mutant compared with the wild type, and the number of down-regulated clones was twice that up-regulated. The gene expression profiles of up- and down-regulated seemed to be symmetrically opposite each other except for the time point of 170 DAF. The number of up-regulated genes increased from 120 DAF to 170 DAF, and then it remained almost the same from 190 DAF to 220 DAF. On the contrary, the number of down-regulated genes remained almost the same from 120 DAF to 150 DAF, and then it decreased from 170 DAF to 220 DAF.
EST analysis
All the clones which showed differential expression for at least one time point out of five between the mutant ‘Hong Anliu’ and wild type were selected. Single-run sequencing of the selected clones yielded 698 readable sequences longer than 100 bp. Of these, 526 were grouped into 96 contiguous sequences (contigs) and 171 were single sequences (singletons) with the CAP3 program (Huang and Madan, 1999). Thus, in total, 267 independent sequences were obtained. Sequence redundancy was 61.6%. BLASTX analysis showed that 68.3% of the independent sequences (116 singletons and 66 contigs) exhibited high sequence homology with known proteins in the NCBI non-redundant protein sequences database (E-value ≤1×10−10) and that 85 (55 singletons and 30 contigs) were classified as no hits (E-value >e−10). Of the 267 genes, 255 (95.1%) showed differential expression at 170 DAF.
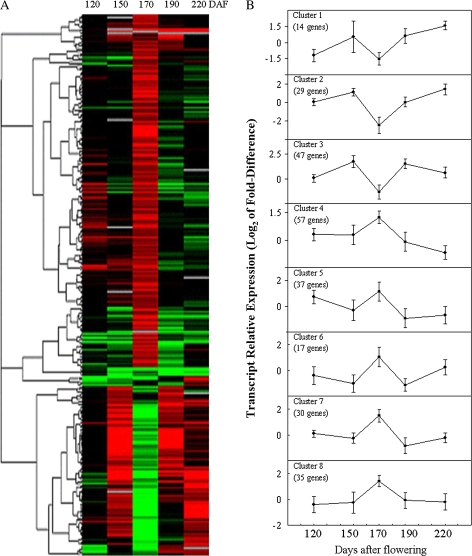
The relative expression profiles of the 267 genes were subjected to cluster analysis using the SOTA algorithm (Herrero et al., 2001). To group relative gene expression profiles on the basis of similar trends and not of similar expression levels, the Pearson correlation coefficient was used as the distance function. Figure 3 shows hierarchical clustering of transcript accumulation and eight relative expression patterns observed in the mutant versus its wild type at five time points, and demonstrates that gene expression changes are highly coordinated during fruit development. As expected, 95.1% of the prominent expression patterns observed in this study correlated well with the initiation of lycopene accumulation.
Fig. 3.
Cluster analysis of expression profiles of differentially expressed gene in the mutant versus its wild type during fruit development. (A) Hierarchical clustering of transcript accumulation between five time points (120, 150, 170, 190, and 220 DAF) during fruit maturation. For each stage, the log2 value of the ratio between ‘Hong Anliu’ and its wild type was represented. (B) SOTA algorithm was used for cluster analysis. There are eight clusters according to SOTA analysis. Data are average relative expression values ±standard deviation. The number of differentially expressed genes in each cluster is also shown. (This figure is available in colour at JXB online.)
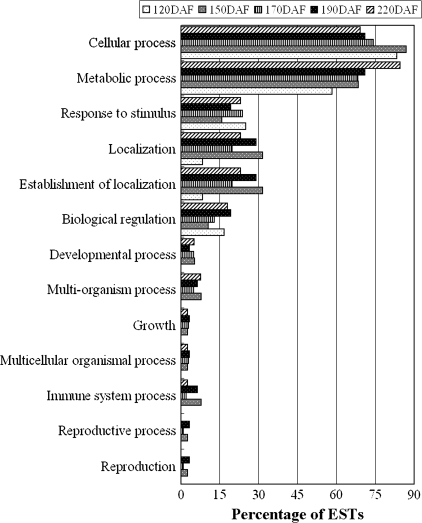
GO categories were assigned to 267 non-redundant genes with BLASTX hit using Blast2GO (Conesa et al., 2005). Table 1 showed a selected list of genes with putative functions that could be important for this mutant. The complete list is given in Table S1 available in Supplementary data at JXB online. Interestingly, 31% of the ESTs were found to be potentially new genes having no similarity in the public databases, while hypothetical proteins having no defined biological process annotation constituted 22% of the EST set. Figure 2 shows the percentage distributions of GO terms (2nd level GO terms) according to the biological process part of GO consortium during fruit development. Metabolic process and cellular process were the major 2nd level terms annotated to the biological process GO category. The percentage distributions of metabolic process increase, while the percentage distributions of cellular process decrease during fruit development.
Table 1.
Selected list of relevant candidate genes for the formation of the phenotype of the red-flesh bud mutant grouped in functional categories
| Biological process | GenBank Accession no. | Description | BLAST E-value | n |
| Cellular metabolic process | ||||
| Organic acid metabolic process | FE659316 | 12-Oxophytodienoate reductase | 1E-84 | 1 |
| Organic acid metabolic process | FE659229 | Glutamate decarboxylase | 1E-68 | 1 |
| Organic acid metabolic process | FE659194 | Glyoxysomal malate dehydrogenase | 1E-14 | 1 |
| Organic acid metabolic process | FE659103 | Phosphoenolpyruvate carboxykinase | 1E-36 | 2 |
| Organic acid metabolic process | FE659289 | Stearoyl-acyl carrier protein desaturase | 1E-16 | 1 |
| Organic acid metabolic process | FE659140 | Malonyl-acyl carrier protein transacylase | 1E-30 | 3 |
| Aromatic compound metabolic process | FE659159 | Flavonol synthase | 1E-58 | 1 |
| Primary metabolic process | ||||
| Lipid metabolic process | FE659078 | Lipoxygenase | 1E-38 | 2 |
| Lipid metabolic process | FE659085 | Myo-inositol-1-phosphate synthase | 1E-58 | 2 |
| Lipid metabolic process | FE659245 | Beta-carotene hydroxylase | 1E-27 | 1 |
| Lipid metabolic process | FE659242 | Aspartic proteinase | 1E-79 | 1 |
| Localization | ||||
| Transport | FE659068 | Lipid transfer protein | 1E-14 | 2 |
| Transport | FE659304 | Glycosyl hydrolase family 17 protein | 1E-12 | 1 |
| Transport | FE659183 | Sugar transporter | 1E-43 | 1 |
| Transport | FE659246 | Glucose-6-phosphate translocator | 1E-20 | 1 |
| Transport | FE659238 | Iron inhibited ABC transporter 2 | 1E-29 | 1 |
| Transport | FE659222 | ABC transporter | 1E-15 | 1 |
| Transport | FE659240 | ABC transporter | 1E-20 | 1 |
| Transport | FE659184 | Cytochrome c | 1E-25 | 1 |
| Macromolecule metabolic process | ||||
| Biopolymer metabolic process | FE659260 | Ubiquitin-conjugating enzyme e2 | 1E-66 | 1 |
| Biopolymer metabolic process | FE659110 | UBC36 ubiquitin-protein ligase | 1E-19 | 2 |
| Biopolymer metabolic process | FE659206 | Aldose 1-epimerase family protein | 1E-48 | 1 |
| Biopolymer metabolic process | FE659105 | 2-Oxoglutarate dehydrogenase e2 subunit | 1E-32 | 2 |
| Biopolymer metabolic process | FE659179 | Pyruvate kinase | 1E-20 | 1 |
| Biopolymer metabolic process | FE659122 | UDP-glucose pyrophosphorylase | 1E-58 | 2 |
| Biopolymer metabolic process | FE659182 | Soluble acid invertase | 1E-121 | 1 |
| Biopolymer metabolic process | FE659239 | Nucleotide sugar epimerase | 1E-55 | 1 |
| Biopolymer metabolic process | FE659287 | Glyoxalase i | 1E-90 | 1 |
| Transcription | FE659120 | Abscisic stress ripening protein | 5E-171 | 9 |
| Transcription | FE659326 | WRKY-type transcription factor | 1E-29 | 1 |
| Transcription | FE659307 | NAC domain protein | 1E-53 | 1 |
| Transcription | FE659294 | Zinc finger protein | 1E-28 | 1 |
| Transcription | FE659300 | Zinc finger transcription factor-like protein | 1E-30 | 1 |
| Transcription | FE659156 | S-adenosyl-l-homocysteine hydrolase | 1E-49 | 9 |
| Transcription | FE659308 | C-repeat binding factor | 1E-10 | 1 |
| Transcription | FE659195 | Agamous-like protein | 1E-38 | 1 |
| Transcription | FE659124 | Homeobox protein expressed | 1E-92 | 7 |
| Translation | FE659323 | Elongation factor 1-expressed | 1E-25 | 1 |
| Translation | FE659121 | Ribosomal protein l19 | 1E-35 | 7 |
| Nitrogen compound metabolic process | ||||
| Amine metabolic process | FE659309 | S-adenosylmethionine decarboxylase | 1E-28 | 1 |
| Cell communication | ||||
| Signal transduction | FE659190 | Aux1-like permease | 1E-15 | 1 |
| Signal transduction | FE659263 | Calmodulin | 1E-68 | 1 |
| Signal transduction | FE659279 | WD-40 repeat family protein | 1E-15 | 1 |
| Response to stimulus | ||||
| Response to hormone stimulus | FE659093 | Stem-specific protein expressed | 1E-30 | 3 |
| Response to jasmonic acid stimulus | FE659089 | Dehydroascorbate reductase | 1E-39 | 3 |
| Response to oxidative stress | FE659293 | Monodehydroascorbate reductase | 1E-35 | 1 |
| Response to protein stimulus | FE659161 | KDA class i heat shock protein | 1E-38 | 1 |
| Response to water | FE659261 | Dehydrin | 1E-28 | 1 |
| Response to stress | FE659086 | Late embryogenesis-abundant protein | 1E-30 | 12 |
| Response to cold | FE659301 | BAP2 (bon association protein 2) | 1E-12 | 1 |
| Response to DNA damage stimulus | FE659119 | Thiazole biosynthetic enzyme | 1E-170 | 15 |
The complete list of genes is given in Table S1 in Supplementary data available at JXB online. For each gene, the EST GenBank accession numbers and the putative molecular function are given. The putative molecular functions were assigned according to the biological process categories of GO annotation. n is the number of sequenced clones in the libraries.
Fig. 2.
Distributions of differentially expressed ESTs according to the biological process part of GO (2nd level GO terms) consortium during fruit development. The total numbers of unique ESTs annotated for the biological process are 124. Since a gene product could be assigned to more than one GO term, the percentages in each main category will add up to >100%.
Metabolic pathways involved in the formation of the phenotype of the mutant fruit
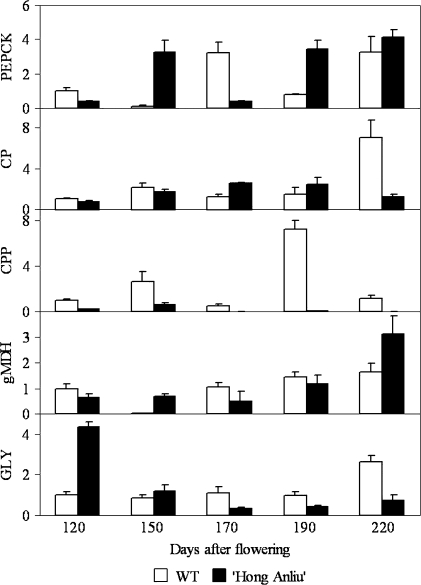
Thirty-nine different metabolic pathways were altered by the bud mutation, most of the genes in these pathways were differentially expressed at 170 DAF, and the identities of these genes are listed in Table S2 available in Supplementary data at JXB online. Pyruvate metabolism, glycolysis/gluconeogenesis, pentose and glucuronate interconversions, and carbon fixation were among the most altered metabolic pathways. Figure 4 showed relative expressions of selected genes which took part in pyruvate metabolism by real-time PCR. Cysteine protein precursor, the only gene which was consecutively down-regulated in the bud mutation during fruit development, showed a low expression in ‘Hong Anliu’ at the green stage (120–150 DAF). After the green stage, the expression of cysteine protein precursor in ‘Hong Anliu’ was barely detectable (170–220 DAF).
Fig. 4.
The relative expression of pyruvate metabolism in the mutant ‘Hong Anliu’ versus its wild type (WT) by real-time PCR. Five differentially expressed genes in the SSH libraries were thought to be involved in pyruvate metabolism. All EST clones shown have been sequence verified. PEPCK, Phosphoenolpyruvate carboxykinase; CP, cysteine protease; CPP, cysteine protease precursor; gMDH, glyoxysomal malate dehydrogenase; GLY1, glyoxalase1.
Several genes in pathways closely related to the altered phenotypes were also found to be differentially expressed (Table 2). Beta-carotene hydroxylase (EC 1.14.13.–), a gene encoding a key enzyme in the carotenoid biosynthesis pathway, was affected by the mutation. The expression of this gene was up-regulated at 170 DAF in mutant ‘Hong Anliu’ compared with its wild type, and was not significantly affected at the other four time points. Three genes involved in the citrate cycle were regulated by the mutation (Table 2). Cysteine protease (EC 3.4.22.–) was up-regulated at 170 DAF, and then down-regulated at 190 and 220 DAF, although it did not reach 2-fold. Phosphoenolpyruvate carboxylase (EC 4.1.1.49) and lipoic transsuccinylase (EC 2.3.1.61) were both up-regulated at 150 DAF and 190 DAF, and were not significantly altered at the other three time points. Several genes encoding key enzymes in the starch and sucrose metabolism and glycolysis were significantly altered; they included UDP-glucose pyrophosphorylase (EC 2.7.7.9), soluble acid invertase (EC 3.2.1.26), pyruvate kinase (EC 2.7.1.40), aldose 1-epimerase (EC 5.1.3.3), and fructose-bisphosphate aldolase (EC 4.1.2.13). Among these, UDP-glucose pyrophosphorylase was significantly down-regulated throughout all the five time points; and the other four were up-regulated at 170 DAF.
Table 2.
List of differentially expressed genes involved in carotenoid, organic acid, and sugar metabolic pathway according to the KEGG pathway database (KEGG = Kyoto Encyclopedia of Genes and Genomes; http://www.genome.jp/kegg/pathway.html)
| KEGG pathways | EC* | Putative function | ‘Hong Anliu’/wild type |
||||
| 120 DAF† | 150 DAF | 170 DAF | 190 DAF | 220 DAF | |||
| Citrate cycle (TCA cycle) | |||||||
| 3.4.22.– | Cysteine protease | –1.04‡ | 1.30 | 2.29 | –1.69 | –1.46 | |
| 4.1.1.49 | Phosphoenolpyruvate carboxylase | –1.01 | 2.91 | –1.97 | 2.41 | –1.25 | |
| 2.3.1.61 | Lipoic transsuccinylase | 1.41 | 2.38 | 1.57 | 3.37 | –1.03 | |
| Starch and sucrose metabolism | |||||||
| 2.7.7.9 | UDP-glucose pyrophosphorylase | –1.53 | –5.48 | –3.04 | –13.83 | –9.64 | |
| 3.2.1.26 | Soluble acid invertase | 1.40 | –1.46 | 2.57 | –1.61 | 1.08 | |
| Glycolysis | |||||||
| 2.7.1.40 | Pyruvate kinase | 1.17 | –2.67 | 3.30 | –1.74 | –1.21 | |
| 5.1.3.3 | Aldose 1-epimerase | 1.15 | –1.14 | 4.10 | –2.10 | –1.32 | |
| 4.1.2.13 | Fructose-bisphosphate aldolase | 1.47 | –1.05 | 2.08 | 1.21 | –1.44 | |
| Carotenoid biosynthesis | |||||||
| 1.14.13.– | Beta-carotene hydroxylase | –1.32 | 1.02 | 2.03 | 1.13 | –1.33 | |
Significant differences (FDR <0.01 and fold change >=2) in relative level are shown in bold.
EC, Enzyme code.
DAF, Days after flowering.
‘–’ means the value of wild type/‘Hong Anliu’.
Candidate regulatory genes for the formation of the phenotype of mutant fruit
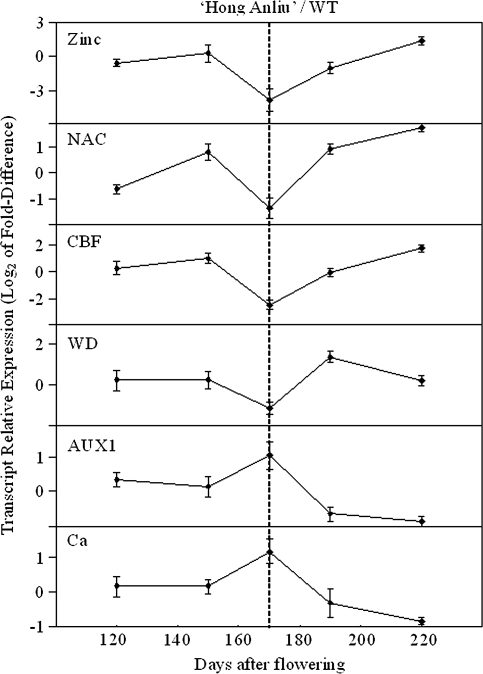
Among the 267 genes described above, 13 (4.9%) were assigned to the categories of transcription factor (10 loci, E-value ≤1×10−10) and signal transduction (3 loci, E-value ≤1×10−10). Figure 5 depicts expression for six candidate regulatory genes, including three putative transcription factors: zinc-finger protein, NAC domain transcription factor, and C-repeat binding factor. All of the genes shown in Fig. 5 were in accordance with the initiation of lycopene accumulation. Candidate regulatory genes likely to encode signal transduction factors have also been identified. Three examples of genes belonging to this functional category are shown in Fig. 5, including aux1-like permease, wd-40 repeat family protein, and calmodulin.
Fig. 5.
Candidate regulatory genes for the bud mutation. Since the expression profile of each gene in the same gene family was similar, one representative gene of each family was chosen. Six candidate regulatory genes identified in this study were shown. Columns show the relative expression of ‘Hong Anliu’ versus its wild type by real-time PCR. Only ESTs with substantial sequence homology were considered (E-value <1×10−10), and all EST clones shown have been sequence verified. The vertical dashed line denotes the stage when dramatic lycopene accumulation was initiated in the fruit. Zinc, Zinc-finger protein; NAC, NAC-domain protein; CBF, C-repeat binding factor; WD, WD-40 repeat family protein; AUX1, aux1-like permease; Ca, Calmodulin.
Verification of microarray data
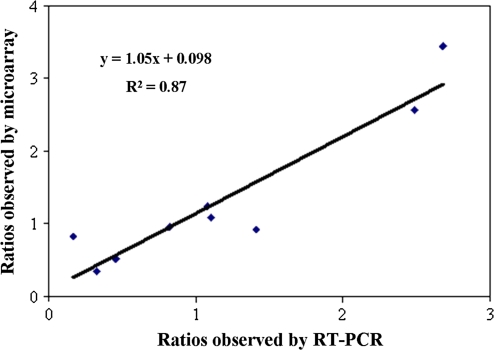
To validate expression profiles obtained using microarray analysis, quantitative RT-PCR was performed on 10 genes using gene-specific primer pairs (Table S3 available in Supplementary data at JXB online). Transcript abundance patterns were compared at five time points between mutant ‘Hong Anliu’ and its wild type ‘Anliu’ (Fig. 6). Linear regression [(microarray value) = a(RT-PCR value)+b] analysis showed coefficients of variation of 0.87. This confirmed the differential expression of all six selected genes.
Fig. 6.
Comparison of gene expression ratios observed by the microarray and by quantitative real-time RT-PCR. Data were from 10 probe sets at five time points between mutant ‘Hong Anliu’ and its wild type ‘Anliu’. The microarray log2 (expression ratio) values (y-axis) are plotted against the log2 (expression ratio) obtained by quantitative real-time RT-PCR (x-axis).
Discussion
Reported here is a collection of differentially expressed genes and metabolic pathways caused by the bud mutation of sweet orange based on SSH, microarray comparison, and sequence homology, since this set of approaches has proved to be an efficient way to enrich and identify differentially expressed genes (Diatchenko et al., 1996; Yang et al., 1999; Derory et al., 2006; Ouyang et al., 2007; Terol et al., 2007; Xu et al., 2007). The results identified a list of genes probably associated with the novel traits of this mutant fruit.
In the mutant ‘Hong Anliu’, the expression levels of hundreds of genes have been altered (fold change ≥2) compared with its progenitor ‘Anliu’ during fruit development (Fig. 1). Similar results have been found in several researches on mutant–progenitor pairs. Through microarray analysis, 564 and 268 genes were found to be differentially expressed in Navel Negra [Citrus sinensis (L.) Osbeck], an abnormal brown-coloured falvedo mutant, versus Washington navel orange (wild type) at mature green stage and ripe stage, respectively (Alos et al., 2008). Two hundred and sixty genes were differentially expressed in blood orange ‘Moro’ compared with common orange ‘Cadenera’ through SSH and reverse northern analysis (Licciardello et al., 2008). In tomato (Solanum lycopersicum=Lycopersicon esculentum), mutation of an ethylene receptor [Never-ripe (Nr)], which reduces ethylene sensitivity and inhibits ripening, altered the expression of 322 genes that were differentially expressed in developing tomato pericarp (Alba et al., 2005); 477 unique genes were differentially expressed between a Colorless non-ripening (Cnr) and wild-type fruits through microarray and differential screen analysis (Eriksson et al., 2004). According to current research, it is common that expression of hundreds of genes is altered by natural mutations.
As for all the differentially expressed genes identified in our SSH libraries during fruit development, most (95.1%) of them showed differential expression at the time point of 170 DAF when a large amount of lycopene started to accumulate, indicating that 170 DAF might be a key developmental stage for the formation of novel traits of the mutant ‘Hong Anliu’ fruits. However, few genes were constitutively up- or down-regulated (fold change ≥2) in the bud mutation during fruit development. In our SSH libraries, only one gene (cysteine protease precursor, EST accession no. FE659117) was constitutively down-regulated by the bud mutation. This result was similar to that of a stay-green mutation in the Navel Negra citrus mutant. Although 11 distinct genes differentially expressed between the Navel Negra citrus mutant and its wild type during all three developmental stages, only one gene (SGR gene homology) showed constitutive down-regulation (Alos et al., 2008).
All these differentially expressed genes were involved in many biological processes such as organic acid metabolism, lipid metabolism, transport, and pyruvate metabolism, etc. Moreover, 13 genes shared homology with previously described signal transduction or transcription factors which might be of particular interest.
Organic acid metabolic process
This bud mutation had a profound effect on the organic acid content of mutant (‘Hong Anliu’ sweet orange) fruits (Liu et al., 2007). Seven genes involved in organic acid metabolic process were differentially expressed in the mutant ‘Hong Anliu’ versus its wild type in our library. Glutamate decarboxylase (GenBank accession no. FE659229) is an enzyme catalysing the conversion of L-glutamate to γ-aminobutyric acid. In plants, glutamate decarboxylase is activated by acidic pH (Snedden et al., 1995, 1996; Shelp et al., 1999) and γ-aminobutyric acid accumulates in response to cytosolic acidification (Shelp et al., 1999). Thus, it is possible that glutamate decarboxylase could participate in regulating the cytosolic pH of the mutant fruit. Glyoxysomal malate dehydrogenase (FE659194), another organic acid-related gene, belongs to the glyoxylate cycle, which bypasses the two decarboxylative steps of the citric-acid cycle and redirects the carbon flow toward gluconeogenesis (Guex et al., 1995). A 12-oxophytodienoate reductase (FE659316) was involved in jasmonate biosynthesis (Schaller et al., 2000), which implied that jasmonic acid metabolism has been affected by the mutation. Phosphoenolpyruvate carboxykinase (EC 4.1.1.49, FE659103) in plants is a cytosolic enzyme that catalyses a reversible reaction which lies at an important crossroads involved in the metabolism of lipids, organic acids, and amino acids (Chen et al., 2004).
Lipid metabolic process
In citrus fruits, carotenoid accumulation was considered to be the result of coordination of the genes encoding key enzymes in the carotenoid biosynthesis pathway (Kato et al., 2004; Liu et al., 2007). A differentially expressed gene encoding beta-carotene hydroxylase (EC 1.14.13.–, FE659245) was present in our library. This gene showed 97 % similarity to ABB49053, which has been identified as a key member of carotenoid biosynthesis in higher plants (Sun et al., 1996). The lipoxygenase activity was inhibited by beta-carotene (Serpen and Gokmen, 2006). Plant lipoxygenases (EC 1.13.11.12, FE659078) are thought to be involved in the biosynthesis of lipid-derived signalling molecules and jasmonic acid (Bell et al., 1995), and play roles in conferring resistance against pathogens and early potato tuber development (Feussner and Wasternack, 2002). The organic acid and sugar content of the mutant ‘Hong Anliu’ were altered by the mutation. Aspartic proteinases (EC 3.4.23, FE659242) were most active at acidic pH (Milisavljevic et al., 2007). Myo-inositol 1-phosphate synthase (EC 5.5.1.4, FE659085) catalyses the formation of myo-inositol 1-phosphate from glucose-6-P. A synergistic effect of sugar and abscisic acid was found on myo-inositol-1-phosphate synthase expression (Yoshida et al., 2002).
Transport
Genes encoding transport-related enzymes are also represented in the library. ATP binding cassette (ABC) transporters are membrane proteins known for their function of translocating a broad range of substances across biological membranes, including lipids, sterols, and drugs (Terol et al., 2007). The library contains two up-regulated clones (FE659222 and FE659240) which show high similarity to ABC transporters (BAD93879, 85% similarity and 72% similarity, respectively). A differentially expressed lipid transfer protein (FE659068) which was found to catalyse transfer of carotenoids between lipohphorins of Bombyx mori (Tsuchida et al., 1998) was found in the library. This supports the hypothesis that carotenoid transfer leads to lycopene accumulation in the albedo and segment membrane of mutant fruits. A glucose-6-phosphate translocator (FE659246) was represented in the library, and the acute inhibition of this gene leads to increased de novo lipogenesis in rats (Bandsma et al., 2001). The sugar content of the mutant fruits was altered by the mutation, and a gene-encoding sugar transporter (FE659183) was identified in this study, the function of which was found to mediate carbon distribution within cells and between organs (Chiou and Bush, 1996).
Pyruvate metabolic pathway
It is worthy to note that five differentially expressed genes belonging to the pyruvate pathway, the metabolic pathway most altered by the bud mutation, were identified and their expressions were verified by real-time PCR (Fig. 4). The pyruvate pathway is closely related to all three major altered traits of the mutant ‘Hong Anliu’: carotenoid accumulation, low citric acid, and high sugar content. The key precursor of carotenoid biosynthetic pathway, isopentenyl diphosphate, is provided by the pyruvate pathway (Lichtenthaler et al., 1997). Pyruvate, an important intermediate compound of pyruvate pathway, is the output of the anaerobic metabolism of glucose known as glycolysis. Moreover, pyruvate could be converted into acetyl-coenzyme A, which is a substrate for citric acid metabolism (Ke et al., 2000). Since no gene sequence difference of all the related pathways was observed between the mutant ‘Hong Anliu’ and its wild type, it is likely that the pyruvate pathway might play an important role in the alteration of the traits of the mutant ‘Hong Anliu’ fruit.
Transcription regulation
Nine genes encoding transcription factors were identified by microarray analysis. Among the group of transcription factors, four genes belonging to the zinc-finger protein family of transcription factors (FE659294, FE659300, FE659303, and FE659177) were identified in our library. Some zinc-finger proteins are known to play a regulatory role by interacting with the cis elements of specific target genes. A type of Cys2/His2-type zinc finger (CX4–CX22–23HX1H) unique to plants was found within the conserved regions of the WRKY family of proteins (Takatsuji, 1998). Zinc-finger proteins, especially those members mediating stress responses, are uniquely expanded in plants (Ross et al., 2007). WRKY proteins (FE659326) were involved in the regulation of abscisic acid signalling in aleurone cells (Xie et al., 2005). Thus, it is possible that the abscisic acid metabolism of the mutant (‘Hong Anliu’ sweet orange) has been altered by the bud mutation.
Two genes belong to the NAC (petunia NAM, Arabidopsis ATAF1,2 and CUC2 genes) proteins (FE659187 and FE659307) which constitute one of the largest families of plant-specific transcription factors (Olsen et al., 2005). Genes from this family participate in various biological processes including development, defence, and biotic and abiotic stress (Hegedus et al., 2003; Olsen et al., 2005). An NAC gene, OsNAC2, was greatly induced in mutant plants of rice and overexpression of OsNAC2 contributes to tiller bud outgrowth (Mao et al., 2007). However, little is known about the functions of NAC genes in citrus. Recently, an NAC-line gene in ‘Navel’ orange fruit response to post-harvest stresses was cloned and characterized (Fan et al., 2007).
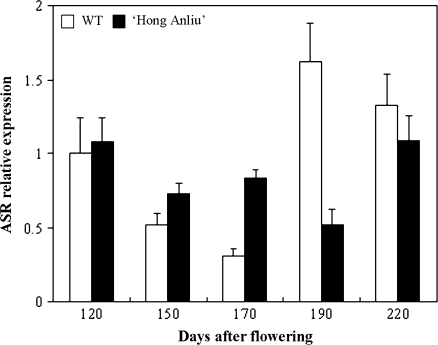
A putative transcription factor, ASR (FE659120), was found in our library. The expression of the ASR gene could be induced by abscisic acid, stress, and ripening (Iusem et al., 1993; Amitai-Zeigerson et al., 1994). A grape ASR gene was found to be involved in sugar and abscisic acid signalling (Cakir et al., 2003). Moreover, an ASR protein was identified in a low acid pummelo mutant fruit (Canel et al., 1995). It is notable that the ASR gene (similar to the Lycopersicon esculentum gene asr4; K Yu, unpublished data) in our library showed a different expression in the mutant ‘Hong Anliu’ compared with the wild type (Fig. 7). And the sugar and acid content were altered by the mutation. Thus it is hypothesized that the ASR might be involved in the alteration of the traits of the mutant ‘Hong Anliu’ fruit.
Fig. 7.
Real-time PCR determination of ASR (EST accession no. FE659120) expression during development of wild-type (WT, open columns) and mutant (‘Hong Anliu’ filled columns) fruits. Columns and bars represent the means and standard error (n=3), respectively.
Other putative transcription factors related to this bud mutation, such as Agamous-like protein (FE659195), C-repeat binding factor (FE659308), and homeobox 2 protein (FE659124) were also identified in this study for the first time.
In conclusion, our SSH library contains a set of structural enzymes that are probably good candidates linked to the phenotype of the mutant fruit. Moreover, a number of interesting regulatory candidate genes was identified. However, much more work is needed to elucidate these genes’ functions, and further experiments aiming at understanding which gene(s) have played the key role for the mutant can be designed on the basis of this study.
Supplementary data
Table S1. List of unique ESTs from the subtractive library and their relative expression patterns in the mutant ‘Hong Anliu’ to wild type comparison at five time points.
Table S2. Metabolic pathways involved in the bud mutation ‘Hong Anliu’ sweet orange according to the KEGG pathway database.
Table S3. Primers used for amplifying candidate and control genes for real-time PCR.
Supplementary Material
Acknowledgments
The authors thank Dr Jude W. Grosser and Associate Prof. Yongzhong Liu for their critical reading of this manuscript. This work was supported by the National Natural Science Foundation of China (No. 30830078), the MATS program of the Ministry of Agriculture of China, and the Ministry of Education of China (IRT0548).
References
- Alba R, Payton P, Fei ZJ, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. The Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alos E, Roca M, Iglesias DJ, Minguez-Mosquera MI, Damasceno CMB, Thannhauser TW, Rose JKC, Talon M, Cercos M. An evaluation of the basis and consequences of a stay-green mutation in the navel negra citrus mutant using transcriptomic and proteomic profiling and metabolite analysis. Plant Physiology. 2008;147:1300–1315. doi: 10.1104/pp.108.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai-Zeigerson H, Scolnik PA, Bar-Zvi D. Genomic nucleotide sequence of tomato Asr2, a second member of the stress/ripening-induced Asr1 gene family. Plant Physiology. 1994;106:1699–1700. doi: 10.1104/pp.106.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aradhya MK, Dangl GS, Prins BH, Boursiquot JM, Walker MA, Meredith CP, Simon CJ. Genetic structure and differentiation in cultivated grape, Vitis vinifera L. Genetical Research. 2003;81:179–192. doi: 10.1017/s0016672303006177. [DOI] [PubMed] [Google Scholar]
- Asins MJ, Monforte AJ, Mestre PF, Carbonell EA. Citrus and Prunus copia-like retrotransposons. Theoretical and Applied Genetics. 1999;99:503–510. doi: 10.1007/s001220051263. [DOI] [PubMed] [Google Scholar]
- Bandsma RHJ, Wiegman CH, Herling AW, Burger H-J, ter Harmsel A, Meijer AJ, Romijn JA, Reijngoud D-J, Kuipers F. Acute inhibition of glucose-6-phosphate translocator activity leads to increased de novo lipogenesis and development of hepatic steatosis without affecting VLDL production in rats. Diabetes. 2001;50:2591–2597. doi: 10.2337/diabetes.50.11.2591. [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B–Methodological. 1995;57:289–300. [Google Scholar]
- Bernet GP, Asins MJ. Identification and genomic distribution of gypsy like retrotransposons in Citrus and Poncirus. Theoretical and Applied Genetics. 2003;108:121–130. doi: 10.1007/s00122-003-1382-1. [DOI] [PubMed] [Google Scholar]
- Breto MP, Ruiz C, Pina JA, Asins MJ. The diversification of Citrus clementina Hort. ex Tan., a vegetatively propagated crop species. Molecular Phylogenetics and Evolution. 2001;21:285–293. doi: 10.1006/mpev.2001.1008. [DOI] [PubMed] [Google Scholar]
- Cakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R. A grape ASR protein involved in sugar and abscisic acid signaling. The Plant Cell. 2003;15:2165–2180. doi: 10.1105/tpc.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JW, Frost HB. Genetics, breeding and nucellar embryony. Berkeley, CA: University of California Berkeley; 1968. [Google Scholar]
- Canel C, Bailey-Serres JN, Roose ML. Pummelo fruit transcript homologous to ripening-induced genes. Plant Physiology. 1995;108:1323–1324. doi: 10.1104/pp.108.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelet P, Laucou V, Fernandez L, Sreekantan L, Lacombe T, Martinez-Zapater JM, Thomas MR, Torregrosa L. Characterization of Vitis vinifera L. somatic variants exhibiting abnormal flower development patterns. Journal of Experimental Botany. 2007;58:4107–4118. doi: 10.1093/jxb/erm269. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Walker RP, Tecsi LI, Lea PJ, Leegood RC. Phosphoenolpyruvate carboxykinase in cucumber plants is increased both by ammonium and by acidification, and is present in the phloem. Planta. 2004;219:48–58. doi: 10.1007/s00425-004-1220-y. [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR. Molecular cloning, immunochemical localization to the vacuole, and expression in transgenic yeast and tobacco of a putative sugar transporter from sugar beet. Plant Physiology. 1996;110:511–520. doi: 10.1104/pp.110.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Deng ZN, Gentile A, Nicolosi E, Domina F, Vardi A, Tribulato E. Identification of in-vivo and in-vitro lemon mutants by RAPD markers. Journal of Horticultural Science. 1995;70:117–125. [Google Scholar]
- Derory J, Leger P, Garcia V, Schaeffer J, Hauser MT, Salin F, Luschnig C, Plomion C, Glossl J, Kremer A. Transcriptome analysis of bud burst in sessile oak (Quercus petraea) New Phytologist. 2006;170:723–738. doi: 10.1111/j.1469-8137.2006.01721.x. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YFC, Campbell AP, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson EM, Bovy A, Manning K, Harrison L, Andrews J, De Silva J, Tucker GA, Seymour GB. Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiology. 2004;136:4184–4197. doi: 10.1104/pp.104.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Gao X, Yang YW, Deng W, Li ZG. Molecular cloning and characterization of a NAC-like gene in ‘navel’ orange fruit response to postharvest stresses. Plant Molecular Biology Reporter. 2007;25:145–153. [Google Scholar]
- Fang DQ, Roose ML. Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theoretical and Applied Genetics. 1997;95:408–417. [Google Scholar]
- Fanizza G, Chaabane R, Ricciardi L, Resta P. Analysis of a spontaneous mutant and selected clones of cv. Italia. (Vitis vinifera) by AFLP markers. Vitis. 2003;42:27–30. [Google Scholar]
- Feussner I, Wasternack C. The lipoxygenase pathway. Annual Review of Plant Biology. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- Guex N, Henry H, Flach J, Richter H, Widmer F. Glyoxysomal malate-dehydrogenase and malate synthase from soybean cotyledons (Glycine max L.) – enzyme association, antibody-production and cDNA cloning. Planta. 1995;197:369–375. doi: 10.1007/BF00202659. [DOI] [PubMed] [Google Scholar]
- Guo Y, Guo HY, Zhang L, et al. Genomic analysis of anti-Hepatitis B virus (HBV) activity by small interfering RNA and lamivudine in stable HBV-producing cells. Journal of Virology. 2005;79:14392–14403. doi: 10.1128/JVI.79.22.14392-14403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Molecular Biology. 2003;53:383–397. doi: 10.1023/b:plan.0000006944.61384.11. [DOI] [PubMed] [Google Scholar]
- Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iusem ND, Bartholomew DM, Hitz WD, Scolnik PA. Tomato (Lycopersicon esculentum) transcript induced by water deficit and ripening. Plant Physiology. 1993;102:1353–1354. doi: 10.1104/pp.102.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Ikoma Y, Matsumoto H, Sugiura M, Hyodo H, Yano M. Accumulation of carotenoids and expression of carotenoid biosynthetic genes during maturation in citrus fruit. Plant Physiology. 2004;134:824–837. doi: 10.1104/pp.103.031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiology. 2000;123:497–508. doi: 10.1104/pp.123.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenis K, Keulemans J. Genetic linkage maps of two apple cultivars (Malus×domestica Borkh.) based on AFLP and microsatellite markers. Molecular Breeding. 2005;15:205–219. [Google Scholar]
- Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982–982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Licciardello C, Russo MP, Vale G, Recupero RG. Identification of differentially expressed genes in the flesh of blood and common oranges. Tree Genetics & Genomes. 2008;4:315–331. [Google Scholar]
- Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Letters. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu J, Liu YZ, Zhao XL, Deng XX, Guo LL, Gu JQ. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck) Journal of Experimental Botany. 2007;58:4161–4171. doi: 10.1093/jxb/erm273. [DOI] [PubMed] [Google Scholar]
- Liu YZ, Liu Q, Tao NG, Deng XX. Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck) Journal of Huazhong Agricultural University. 2006;25:300–304. [Google Scholar]
- Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature Genetics. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Mao CZ, Ding WN, Wu YR, Yu J, He XW, Shou HX, Wu P. Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytologist. 2007;176:288–298. doi: 10.1111/j.1469-8137.2007.02177.x. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano M. Chimeras and variegation: patterns of deceit. Hortscience. 1997;32:773–784. [Google Scholar]
- Mase N, Iketani H, Sato Y. Analysis of bud sport cultivars of peach (Prunus persica (L.) Batsch) by simple sequence repeats (SSR) and restriction landmark genomic scanning (RLGS) Journal of the Japanese Society for Horticultural Science. 2007;76:20–27. [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harbor Symposia on Quantitative Biology. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Milisavljevic MD, Timotijevic GS, Radovic SR, Konstantinovic MM, Maksimovic VR. Two types of aspartic proteinases from buckwheat seed – gene structure and expression analysis. Journal of Plant Physiology. 2007 doi: 10.1016/j.jplph.2007.03.016. doi:10.1016/j.jplph.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Moore GA. Oranges and lemons: clues to the taxonomy of Citrus from molecular markers. Trends in Genetics. 2001;17:536–540. doi: 10.1016/s0168-9525(01)02442-8. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Lo Leggio L, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Ouyang B, Yang T, Li HX, Zhang L, Zhang YY, Zhang JH, Fei ZJ, Ye ZB. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. Journal of Experimental Botany. 2007;58:507–520. doi: 10.1093/jxb/erl258. [DOI] [PubMed] [Google Scholar]
- Raghuvanshi SS. Cytogenetical studies in genus Citrus. 4. Evolution in genus Citrus. Cytologia. 1962;27:172–188. [Google Scholar]
- Rico-Cabanas L, Martinez-Izquierdo JA. CIRE1, a novel transcriptionally active Ty1-copia retrotransposon from Citrus sinensis. Molecular Genetics and Genomics. 2007;277:365–377. doi: 10.1007/s00438-006-0200-2. [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QXJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2007;49:827–842. [Google Scholar]
- Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW. 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta. 2000;210:979–984. doi: 10.1007/s004250050706. [DOI] [PubMed] [Google Scholar]
- Serpen A, Gokmen V. A proposed mechanism for the inhibition of soybean lipoxygenase by beta-carotene. Journal of the Science of Food and Agriculture. 2006;86:401–406. [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends in Plant Science. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Smit A. 2007. Repeat Masker. http://www.repeatmasker.org. [Google Scholar]
- Snedden WA, Arazi T, Fromm H, Shelp BJ. Calcium-calmodulin activation of soybean glutamate-decarboxylase. Plant Physiology. 1995;108:543–549. doi: 10.1104/pp.108.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden WA, Koutsia N, Baum G, Fromm H. Activation of a recombinant Petunia glutamate decarboxylase by calcium/calmodulin or by a monoclonal antibody which recognizes the calmodulin binding domain. Journal of Biological Chemistry. 1996;271:4148–4153. doi: 10.1074/jbc.271.8.4148. [DOI] [PubMed] [Google Scholar]
- Spiegel-Roy P, Goldschmidt EE. Biology of citrus. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- Sun ZR, Gantt E, Cunningham FX. Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. Journal of Biological Chemistry. 1996;271:24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger transcription factors in plants. Cellular and Molecular Life Sciences. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao NG, Wei J, Liu YZ, Cheng YJ, Deng XX. Copia-like retrotransposons in a precocious mutant of trifoliate orange [Poncirus trifoliata (L.) Raf] Journal of Horticultural Science & Biotechnology. 2006;81:1038–1042. [Google Scholar]
- Terol J, Conesa A, Colmenero JM, et al. Analysis of 13000 unique Citrus clusters associated with fruit quality, production and salinity tolerance. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida K, Arai M, Tanaka Y, Ishihara R, Ryan RO, Maekawa H. Lipid transfer particle catalyzes transfer of carotenoids between lipophorins of Bombyx mori. Insect Biochemistry and Molecular Biology. 1998;28:927–934. doi: 10.1016/s0965-1748(98)00036-8. [DOI] [PubMed] [Google Scholar]
- Walker AR, Lee E, Robinson SP. Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Molecular Biology. 2006;62:623–635. doi: 10.1007/s11103-006-9043-9. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou XL, Huang J, Ruas P, Thompson D, Shen QJ. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiology. 2005;137:176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BY, Su W, Liu JH, Wang JB, Jin ZQ. Differentially expressed cDNAs at the early stage of banana ripening identified by suppression subtractive hybridization and cDNA microarray. Planta. 2007;226:529–539. doi: 10.1007/s00425-007-0502-6. [DOI] [PubMed] [Google Scholar]
- Yang GP, Ross DT, Kuang WW, Brown PO, Weigel RJ. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Research. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JL, Dong YH, Morris BAM. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proceedings of the National Academy of Sciences, USA. 2001;98:1306–1311. doi: 10.1073/pnas.031502498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida KT, Fujiwara T, Naito S. The synergistic effects of sugar and abscisic acid on myo-inositol-1-phosphate synthase expression. Physiologia Plantarum. 2002;114:581–587. doi: 10.1034/j.1399-3054.2002.1140411.x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Deng XX. Advances in research of citrus cultivars selected by bud mutation and the mechanism of formation of mutated characteristics. Journal of Fruit Science. 2006;23:871–876. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.