Abstract
Cold shock domain proteins (CSPs) are highly conserved from bacteria to higher plants and animals. Bacterial cold shock proteins function as RNA chaperones by destabilizing RNA secondary structures and promoting translation as an adaptative mechanism to low temperature stress. In animals, cold shock domain proteins exhibit broad functions related to growth and development. In order to understand better the function of CSPs in planta, detailed analyses were performed for Arabidopsis thaliana CSPs (AtCSPs) on the transcript and protein levels using an extensive series of tissue harvested throughout developmental stages within the entire life cycle of Arabidopsis. On both the transcript and protein levels, AtCSPs were enriched in shoot apical meristems and siliques. Although all AtCSPs exhibited similar expression patterns, AtCSP2 was the most abundantly expressed gene. In situ hybridization analyses were also used to confirm that AtCSP2 and AtCSP4 transcripts accumulate in developing embryos and shoot apices. AtCSPs transcripts were also induced during a controlled floral induction study. In vivo ChIP analysis confirmed that an embryo expressed MADS box transcription factor, AGL15, interacts within two AtCSP promoter regions and alters the respective patterns of AtCSP transcription. Comparative analysis of AtCSP gene expression between Landsberg and Columbia ecotypes confirmed a 1000-fold reduction of AtCSP4 gene expression in the Landsberg background. Analysis of the AtCSP4 genomic locus identified multiple polymorphisms in putative regulatory cis-elements between the two ecotypes. Collectively, these data support the hypothesis that AtCSPs are involved in the transition to flowering and silique development in Arabidopsis.
Keywords: Cold shock domain proteins, floral development, RNA binding proteins, silique development
Introduction
Cold shock domain proteins (CSPs) are DNA/RNA binding proteins, which are conserved from bacteria to animals and higher plants (Makino et al., 1996; Yamanaka et al., 1998; Karlson and Imai, 2003). Bacterial cold shock proteins function as transcription anti-terminators and RNA chaperones, and affect the transcription and subsequent translation of proteins (Jiang et al., 1997;Bae et al., 2000). Plant CSPs also exhibit nucleic acid binding and chaperone activity (Karlson et al., 2002; Nakaminami et al., 2006; Kim et al., 2007a, b). A wheat CSP (WCSP1) localizes to the ER network, nuclear envelope, nucleus, and Cajal bodies and exhibits similar in vitro and in vivo functions as bacterial cold shock proteins (Nakaminami et al., 2006). Collectively, these data suggest that plant CSPs share a similar function with prokaryotic counterparts and may function at the transcriptional and/or translational level in planta.
In other eukaryotic systems, many CSPs have been identified and extensively studied. A Xenopus Y-box protein, FRGY2 accumulates during oogenesis and embryogenesis and functions during nucleolar disassembly (Tafuri and Wolffe, 1990; Gonda et al., 2003, 2006). The rabbit P50 protein was identified as a component of ribonucleoprotein particles (Evdokimova et al., 1995) and it functions to maintain mRNA stability via binding to the cap region of mRNA and, consequently, inhibits translation (Evdokimova et al., 1995; Nekrasov et al., 2003). The post-transcriptional regulator from C. elegans, LIN-28 has an essential role in the timing of developmental transitions (Moss et al., 1997; Seggerson et al., 2002). YB-1 (dbpB) is a well-studied multi-functional human CSD protein (Sakura et al., 1988; Horwitz et al., 1994; Wolffe, 1994) which is involved in the regulation of transcription and translation, DNA repair, stress response, and RNA masking (Kohno et al., 2003; Evdokimova et al., 2006). YB-1 also regulates gene expression of cell cycle-related genes and plays an important role for embryo development in mouse (Jurchott et al., 2003; Lu et al., 2005; Uchiumi et al., 2006). Collectively, these aforementioned studies enabled us to hypothesize that plant CSPs may also play an important role related to developmental transitions such as flowering and silique development in planta.
Throughout the various stages of plant development, a myriad of regulatory genes are expressed. For example, in the vegetative stage, leaf development is largely regulated by KNOX1, a homeotic transcription factor (Bharathan and Sinha, 2001; Hake et al., 2004). To generate the inflorescence, dramatic alterations occur between the vegetative and reproductive stages within apical meristems. Floral development is regulated by many genes including homeotic genes. As described in the ABC model of floral development, APETALA2 (AP2), APETALA1 (AP1), APETALA3 (AP3), PISTILLATA (PI), and AGAMOUS (AG) are integral components for regulating the development of various floral organs (Bowman et al., 1991; Coen and Meyerowitz, 1991). Seed development and embryogenesis have also been extensively studied on a molecular level. Among the numerous genes associated with embryogenesis, AGAMOUS-like 15 (AGL15) is a well-studied MADS domain transcription factor that is implicated to have an important role during embryogenesis (Heck et al., 1995; Rounsley et al., 1995). Developmental related transcription factors regulate specific downstream gene targets and may trigger a response in plants which promotes transitions in development such as the initiation of flowering and embryogenesis. Consequently, the identification of downstream targets for developmental-related transcription factors is of great interest.
Arabidopsis contains four CSP homologues (Karlson and Imai, 2003). In this study the gene family is referred to as: AtCSP1 (CSDP1; At4g36020), AtCSP2 (AtGRP2/CSDP2; At4g38680), AtCSP3 (At2g17870), and AtCSP4 (AtGRP2b; At2g21060). AtCSPs contain an N-terminal cold shock domain and either seven or two C-terminal glycine-rich regions for AtCSP1/AtCSP3 and AtCSP2/AtCSP4, respectively. Similar to bacterial CSPs and winter wheat WCSP1 (Nakaminami et al., 2006), two independent studies recently demonstrated that Arabidopsis thaliana CSPs (AtCSPs) exhibit DNA/RNA binding and RNA chaperone activity (Kim et al., 2007a; Sasaki et al., 2007). Fusaro et al. (2007) reported that AtCSPs are localized to the shoot apical meristem and ovules. These data suggested that AtCSPs are related to flowering and silique development. However, the precise functional relationship of AtCSPs to these aforementioned developmental stages is poorly understood. In the present study it was confirmed that AtCSPs gene expression patterns are strongly correlated to various stages of plant development by using qRT-PCR, in situ hybridization, and Western blot analysis. In good accordance to public microarray data, AtCSPs are highly expressed in shoot apices and in the early stages of floral tissue, especially in siliques, which contain developing embryos. Consistent with the study of AtGRP2 (Fusaro et al., 2007), these correlative data strongly suggest that AtCSPs may have an important functional role related to flowering and silique development.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotypes Columbia and Landsberg erecta were used in this study. Depending upon the experiment, plants were grown at 20 °C under continuous light or long-day conditions (16/8 h light/dark) or short-day conditions (8/16 h light/dark) in a growth chamber (Percival incubator, model; CU-36L3X and AR36LC9, Percival Scientific). For gene expression and protein blot analyses, seedlings were grown on MS medium and harvested at different developmental time points. For the analysis of various floral or silique developmental stages, plants were grown on MS plates for 2 weeks and transferred to soil. Plants were then subsequently grown in the same growth chamber.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from each tissue using the Plant RNA Purification Reagent (Invitrogen, Carlsbad, CA) according to the supplier's instructions. Five-hundred nanograms of isolated total RNA was subsequently converted to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA) with a half-scale reaction. qRT-PCR reactions were performed with an Applied Biosysytems 7500 real-time PCR system using either Taq-Man Universal PCR Master Mix or SYBR Green reagents (Applied Biosystems, Foster City, CA). Sequences, with the following GenBank accession numbers, served for the design of the pre-developed TaqMan assay reagents or primers and probe, purchased from Applied Biosystems: NM_180280.1 (ACT2), NM_104305.3 (eIF4A2), NM_119769.2 (AtCSP1), NM_120029.2/S47408.1 (AtCSP2), NM_127341.3 (AtCSP3), and NM_127676.2 (AtCSP4).
Context sequences and amplicon lengths can be viewed in Supplementary Table S1 at JXB online. Nucleotide sequences of gene-specific primers for SYBR Green analysis can be viewed in Supplementary Table S2 at JXB online. ACT2 and eIF4A2 transcript levels were analysed as internal controls and used for normalization of AtCSP transcript abundance for stages of vegetative–floral and silique development, respectively. Expression levels for both internal control genes were not altered within their respective developmental time-coursed studies. The relative amount of expression levels of transcript was estimated by using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Western blot analysis
Total proteins were extracted from all tissues as previously described by Karlson et al. (2002). Each protein isolate (20 μg per lane) was separated by SDS-PAGE and used for subsequent Western blot analysis according to standard procedures. Anti-WCSP1 primary antibodies (1:3000) and an alkaline phosphatase conjugated anti-rabbit secondary antibody (Rockland, Gilbertsville, PA) were used (1:3000) for the detection of AtCSPs with the 1-Step NBT/BCIP (Pierce, Rockford, IL) or Phototope-Star Detection Kit (New England BioLabs, Ipswich, MA). The signal intensity of detected bands from the Western blot analysis was analysed using ImageJ software (National Institutes of Health, http://rsb.info.nih.gov/ij/download.html).
Photomicrographs of stages of embryo development
Developmental stages of Columbia and Landsberg seeds were confirmed by Nomarski microscopy. Seeds were cleared prior to visualization using a mixture of chloral hydrate/glycerol/water (8:2:1 by vol.). Samples were incubated for at least 2 h at room temperature. Microscopy was performed using a Nikon Eclipse E600 microscope system. The following stages were visualized: early developing embryo (Si1), early globular to mid-globular (Si2), mid-globular to early heart (Si3), early heart to late heart (Si4), mid-torpedo to late torpedo (Si6), walking-stick to early curled cotyledon stage (Si8), and green cotyledons (Si10).
Chromatin immunoprecipitation (ChIP)
Plant tissue used for chromatin immunoprecipitation (ChIP) was either ∼3.5 g of Columbia wild-type or agl15-3 loss-of-function somatic embryo culture prepared as previously described by Ikeda-Iwai et al. (2002) or 5–10 g of embryonic culture tissue as previously described by Harding et al. (2003). The tissue was fixed in MC buffer (10 mM potassium phosphate, pH 7.0, 50 mM NaCl, and 0.1 M sucrose) containing 1% formaldehyde on ice under vacuum for 1 h. Cross-linking was stopped by the addition of cold glycine to 0.125 M and incubation for 10–30 min before being washed in MC buffer and flash frozen. Nuclei were isolated from fixed tissue (Bowler et al., 2004) and chromatin immunoprecipitation was performed (Wang et al., 2002) using a polyclonal antibody raised against AGL15 (Heck et al., 1995) or preimmune serum as a control.
Association of AGL15 with suspected target DNA fragments in vivo was tested using an enrichment test. Specifically, oligonucleotide primers that amplify select DNA fragments that are believed to be bound by AGL15 and oligonucleotide primers that amplify a control DNA fragment not expected to be bound by AGL15 (e.g. TUB2 and EF1α) are used in multiplex PCR. In the input (total) DNA, target PCR product and control PCR product should be present at approximately equal levels (assuming an equal efficiency of PCR). After selection of DNA by ChIP, target fragments should be enriched over controls and the PCR products should reflect this. PCR reactions were performed using KlenTaq1 (Ab Peptides, St Louis, MO) using oligonucleotides specific for EF1α (At1g07920), TUB2 (At5g62690), AtCSP2 (At4g38680), and AtCSP4 (At2g21060) (see Supplementary Table 2 at JXB online). PCR products were analysed by agarose gel electrophoresis and gel images were captured using a ChemiImager (Alpha Innotech Corporation, San Leandro, CA). In some cases, the colour of the image was inverted for clarity.
In situ hybridization
Plant materials were fixed in 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4, overnight at 4 °C, dehydrated through a graded ethanol series. Subsequently, fixed tissue were passed through a graded t-butanol series and were finally embedded in Paraplast Plus (Sherwood Medical).
Microtome sections (8 μm thick) were mounted on glass slides treated with silane. Hybridization and immunological detection of the hybridized probes were performed according to the previously described method of (Kouchi and Hata, 1993). Digoxigenin-labelled RNA derived from the 3′ non-coding region of AtCSP2 or AtCSP4 were used to exclude cross-hybridization among these genes. DNA fragments specific for AtCSP2 or AtCSP4, which consisted of the 3′ non-coding region was amplified by PCR using the primers (AtCSP2, 5′-AAGCTGCTACAGCTGTGGCGAGTCGGG-3′ and 5′-TAGGGATCCATTTAAATGAAACAGAGCAACAAGTAGAG-3′, and AtCSP4, 5′-AAGCTGCTACAGCTGTGGAGAGTCTGG-3′ and 5′-TAGGGATCCATTTAAATCCAATCCAGTTTCTTTCTC-3′.
Genomic DNA isolation and sequence analysis
Genomic DNA was extracted from young leaves of Landsberg erecta wild-type plants using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). The AtCSP4 promoter and ORF regions were amplified using the following primer sets: 5′-GAGCTCTCTTCTTCCCACTC-3′ and 5′-CCTCACTTCAATCACACTCA-3′. These fragments were subcloned into the pGEM-T easy vector (Promega, Madison, WI) and subsequently confirmed with bi-directional sequence analysis. Putative cis-element regulatory regions were identified within the promoter region by using PLACE (http://www.dna.affrc.go.jp/PLACE/) (Higo et al., 1999).
Results
Tissue specific expression pattern for AtCSPs
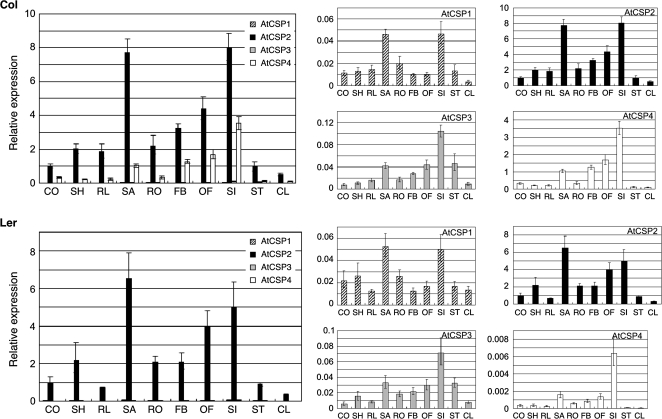
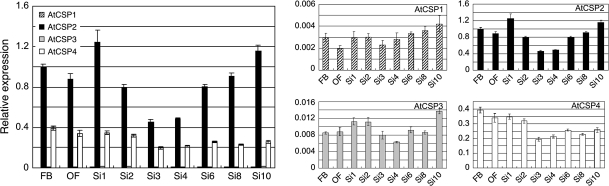
Preliminary comparative gene expression patterns were performed for all AtCSPs using the Genevestigator tool (https://www.genevestigator.ethz.ch/) (data not shown). Due to promising data trends, we were interested in characterizing the relationship between AtCSPs and developmental transitions in Arabidopsis. Tissue-specific expression analysis of AtCSPs was performed first using quantitative real-time PCR (qRT-PCR) in Columbia wild-type plants. Total RNAs were extracted from 1-week-old cotyledons, 2-week-old shoots, rosette leaves, shoot apices, and roots; floral buds and opened flowers from 4-week-old plants; siliques, stems, and cauline leaves collected 3 d after anthesis. Analysis with qRT-PCR revealed similar expression patterns among all AtCSP homologues, however, their relative levels of transcript abundance differed (Fig. 1). In this study and all subsequent qRT-PCR analyses for Columbia wild-type, AtCSP2 showed the highest level of expression followed by AtCSP4. Relative to AtCSP2 and AtCSP4, both AtCSP1 and AtCSP3 had very low levels of expression. AtCSP transcript accumulation was similar in shoots and roots. Relative to expression levels in rosette leaves, transcripts for all AtCSPs were enriched in shoot apices during vegetative growth stage. This difference was especially pronounced for AtCSP2. In the reproductive stage of plant development, AtCSPs expression was higher in floral tissues and siliques relative to other tissues. A similar comparative analysis was performed in Landsberg wild-type plants (Fig. 1). Interestingly, the relative expression level of AtCSP4 was reduced 1000-fold in the Landsberg ecotype. In order to begin to understand this dramatic difference in AtCSP4 gene expression, its genomic locus was isolated and comparative sequence analysis was performed with the Columbia and Landsberg AtCSP4 gene locus.
Fig. 1.
Tissue-specific AtCSP gene expression. Plants were grown under continuous light conditions in a growth chamber. Total RNA was extracted and converted into cDNA to serve as a template for qRT-PCR analysis with gene specific Taq-man probe sets. After normalization to the ACT2 control gene, AtCSP transcript levels are presented as relative values to the expression of AtCSP2 in cotyledons (which was set to a value of ‘1’). Data are shown as the means of three replicate reactions with standard deviations illustrated as vertical bars. CO, cotyledon; SH, shoot; RL, rosette leaves; SA, shoot apices; RO, root; FB, floral bud; OF, opened flower; SI, silique; ST, stem; CL, cauline leaf.
Comparative sequence analysis for the AtCSP4 genomic locus in Columbia and Landsberg ecotypes
Microarray analysis previously conducted between Columbia and Landsberg assessed DNA polymorphisms between these ecotypes. Hybridization of labelled genomic DNA onto AtGenome 1 arrays found that less than 1% of all loci contained six or more single-feature polymorphisms (Schmid et al., 2003). AtCSP4 (At2g21060) was predicted to be a highly polymorphic loci in Landsberg (Schmid et al., 2003). In order to investigate the differential expression pattern of AtCSP4 between Landsberg and Columbia in detail, the promoter region and the 5′ and 3′ UTRs were compared in both wild-type strains.
Several sequences differed in promoter, mRNA and 3′ DNA regions between Landsberg and Columbia. Comparisons revealed 96.5%, 98.7%, and 91.6% identities in 1.3 kb promoter region, 0.86 kb mRNA region including UTRs and a 0.15 kb 3′ DNA region between AtCSP4 and the subsequent gene (At2g21050), respectively (Fig. 2). On a DNA level, a few conserved DNA polymorphisms were found in the ORF region, however, none of these resulted in the alteration of translatable amino acids. Interestingly, the adjacent At2g21050 gene locus was 100% identical between Landsberg and Columbia ecotypes. The 1.3 kb promoter region of both AtCSP4 genes was analysed using the PLACE (http://www.dna.affrc.go.jp/PLACE/ web site (Higo et al., 1999). Several discrepancies in DNA sequences altered multiple putative cis-element sequences. The DNA sequence alignment of Landsberg and Columbia, and the unique cis-elements, were summarized (Fig. 2; Table 1). For example, GAREAT, which is a GA-responsive element, was unique to the Landsberg promoter region, whereas QELEMENTZMZM13, which is a pollen-specific enhancing factor, was only present in Columbia (Table 1). Although gibberellin responsive elements were found in both promoter regions, the location and sequences of their elements differed. On the other hand, the RY/G-box, which is related to abscisic acid response via the ABI3 transcription factor, was only found in Landsberg. Another interesting putative cis-element unique to Landsberg was SEBFCONSSTPR10A, which is a binding site of the silencing element binding factor (SEBF) (Boyle and Brisson, 2001). In Columbia, binding sites for a TCP-domain transcription factor (SITEIIATCYTC) are present. Collectively, these discrepancies may affect the differential transcription of the AtCSP4 gene locus between Columbia and Landsberg ecotypes.
Fig. 2.
Comparative promoter analysis for AtCPS4 between Landsberg (upper line) and Columbia (lower line) ecotypes. The genomic sequence for AtCSP4 was cloned from Landsberg and sequenced for the detection of DNA polymorphisms. Multiple disparities were detected within the promoters and UTR regions. Red, Landsberg specific cis-element; blue, Columbia specific cis-element; green, ATG and stop codon; light red, 5’ and 3’ UTR; yellow, ORF of AtCSP4.
Table 1.
Comparative promoter analysis for AtCSP4 between Landsberg and Columbia ecotypes
| Factor or Site Name | Location (Str.) | Sequence | Description |
| Landsberg | |||
| GAREAT | −1095 −1089 (+) | TAACAAR | GA-responsive element |
| MYBGAHV | −1095 −1089 (+) | TAACAAA | Central element of GA response complex (GARC) |
| SEBFCONSSTPR10A | −1030 −1042 (+) | YTGTCWC | Binding site of the silencing element binding factor (SEBF) |
| RYREPEATGMGY2 | −805 −799 (−) | CATGCAT | RY repeat motif |
| RYREPEATLEGUMINBOX | −805 −799 (−) | CATGCAY | RY repeat or legumin box |
| RYREPEATBNNAPA | −804 −799 (−) | CATGCA | RY/G box, which contain the two RY repeats and the G-box |
| BOXLCOREDCPAL | −387 −381 (+) | ACCWWCC | Box-L-like, which interact with MYB transcription factor |
| HEXAMERATH4 | −344 −338 (+) | CCGTCG | Hexamer motif |
| RYREPEATBNNAPA | −280 −275 (+) | CATGCA | RY/G box, which contain the two RY repeats and the G-box |
| Columbia | |||
| QELEMENTZMZM13 | −1203 −1198 (−) | AGGTCA | Q(quantitative)-element, pollen-specific enhancing factor |
| SORLREP3AT | −1095 −1087 (−) | TGTATATAT | Sequences Over-Represented in Light-Repressed Promoters |
| TATCCAOSAMY | −823 −818 (−) | TATCCA | Sugar and hormone regulation factor |
| NRRBNEXTA | −662 −655 (−) | TAGTGGAT | NRR (negative regulatory region) |
| GARE1OSREP1 | −467 −461 (−) | TAACAGA | GA-responsive element |
| SITEIIATCYTC | −347 −342 (−) | TGGGCY | Site II element, which interact with a TCP-domain transcription factor |
| UP1ATMSD | −285 −277 (+) | GGCCCAWWW | Up1, which up-regulate genes after main stem decapitation |
| SITEIIATCYTC | −285 −280 (−) | TGGGCY | Site II element, which interact with a TCP-domain transcription factor |
| TATABOX2 | −156 −150 (−) | TATAAAT | TATA box |
Landsberg and Columbia specific cis-elements in promoter region of AtCSP4 were summarized and briefly described.
Gene and protein expression during vegetative stages and floral transition
According to public microarray data and our tissue-specific gene expression analysis, all AtCSPs are enriched in shoot apices. Therefore, we were interested in determining expression patterns in relation to the transition to reproductive development. To clarify this, a detailed analysis of gene and protein expression was performed during vegetative stages of growth and after the transition to flowering. Plants were grown on MS plates under continuous light conditions. Cotyledons, rosette leaves, and shoot apices of different developmental stages (see Supplementary Fig. S1 at JXB online) were harvested for analysis.
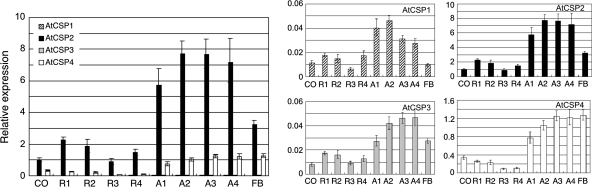
AtCSPs gene expression was enriched in the shoot apex relative to cotyledons or rosette leaves (Fig. 3). A similar gene expression pattern was exhibited among all AtCSP homologues except AtCSP1. AtCSPs expression levels were maintained at a high level or increased during development in shoot apices and then dropped in floral buds. In the Columbia background, AtCSP1 expression progressively decreased in shoot apices, however, its level of expression was relatively low. The gene expression pattern for the most abundantly expressed gene was similar in the Landsberg background (see Supplementary Fig. S2A at JXB online). As previously mentioned, AtCSP4 gene expression was suppressed in vegetative stage samples in the Landsberg background.
Fig. 3.
AtCSP gene expression in vegetative growth and floral transition. Columbia wild-type plants were grown under continuous light conditions in a growth chamber. Total RNA was extracted from 1-week-old cotyledons (CO), 1-week-old rosette leaves (R1) and shoot apices (A1), 2-week-old rosette leaves (R2) and shoot apices (A2), 3-week-old rosette leaves (R3) and shoot apices (A3) before bolting, and 3-week-old rosette leaves (R4) and shoot apices (A4) after bolting and floral buds (FB). Total RNA was isolated and converted into cDNA template for qRT-PCR analysis with gene specific Taq-man probe sets. Transcript levels are presented as relative values to the expression level of the AtCSP2 gene in cotyledons (set to the value of 1), after being normalized to the ACT2 gene. Data are shown as the mean of three replicates with error bars indicating the standard deviation.
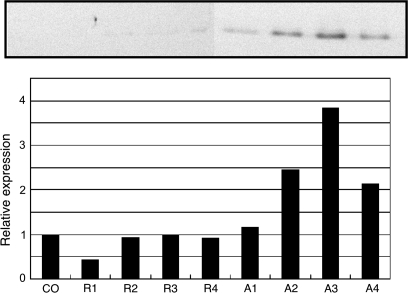
In a subsequent experiment, a comparative analysis with total protein extracts from shoot apices and rosette leaf tissues was performed throughout a developmental time-line. A protein band corresponding to the putative molecular mass for AtCSP2 (19 kDa) and AtCSP4 (19 kDa) was detected with a polyclonal antibody specific for the cold shock domain of winter wheat WCSP1 (Fig. 4). Unfortunately due to their identical predicted size, it was not possible to distinguish AtCSP2 from AtCSP4. It is likely that AtCSP1 (30 kDa) and AtCSP3 (29.5 kDa) bands were not detected due to their very low expression levels. Western blot analysis of purified recombinant AtCSP proteins confirmed that the antibody recognizes all four AtCSPs (data not shown). In relative comparison between shoot apices and rosette leaves, AtCSP2 and 4 accumulate in shoot apices and progressively increase during the developmental time-line, peaking at 3 weeks, just prior to bolting in the Columbia background. Subsequent to bolting, the protein levels decreased. In the Landsberg background, a similar pattern was detected and this protein most likely corresponded to AtCSP2 since AtCSP4 gene expression was suppressed (see Supplementary Fig. S2B at JXB online). These data are in good agreement with our qRT-PCR results and provide correlative evidence suggesting that shoot apical accumulation of AtCSP2 and 4 may be involved in leaf development and/or floral transition.
Fig. 4.
Analysis of AtCSP protein accumulation during floral transition. Total proteins were extracted from 1-week-old cotyledon (CO), 1-week-old rosette leaves (R1) and shoot apices (A1), 2-week-old rosette leaves (R2) and shoot apices (A2), 3-week-old rosette leaves (R3) and shoot apices (A3) before bolting, and 3-week-old rosette leaves (R4) and shoot apices (A4) after bolting. Protein extracts were separated by SDS-PAGE. Signals were detected by Western blot analysis using a polyclonal antibody specific for the cold shock domain (anti-WCSP1). Due to their similar size, AtCSP2 and AtCSP4 cannot be distinguished from one another with Western blot analysis. A band similar to the predicted size range for AtCSP2 and AtCSP4 accumulated during the transition to flowering. Comparative analyses in rosette leaves confirmed that this trend was specific to shoot apices. It should be noted that no corresponding protein bands were detected in the putative size range for AtCSP1 and 3.
AtCSPs expression during floral induction
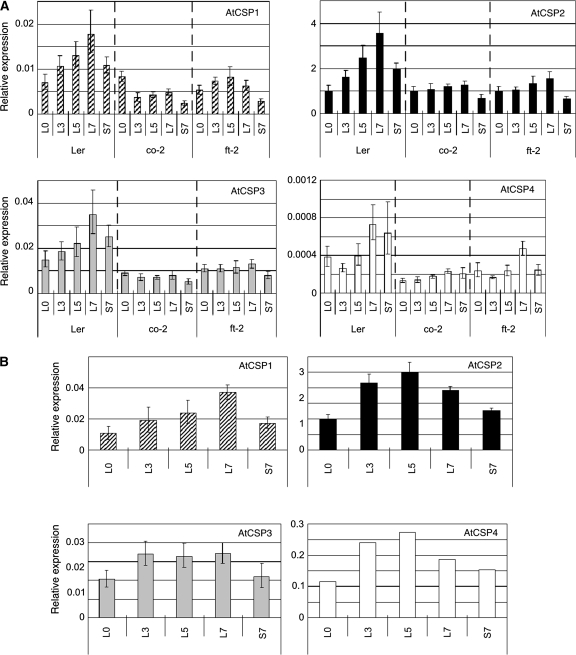
In order to confirm if AtCSPs are induced during flowering via the photoperiodic pathway, an experiment was performed that was designed to stimulate flowering under controlled conditions. Plants were grown under short day (SD) conditions for 30 d in order to maintain them in a vegetative state. They were subsequently shifted to an inductive long day (LD) condition and monitored for gene expression patterns for up to 7 d after the shift. If AtCSP transcripts are triggered to accumulate during the induction of flowering, an increase in gene expression under LD conditions should be observed. As a negative control, replicate plants were maintained for an additional 7 d under non-inductive SD conditions. Two well-characterized late-flowering mutants (co-2 and ft-2) were also used to provide additional evidence linking our gene expression patterns to floral induction and the photoperiodic signalling pathway for flowering. Total RNA was extracted from shoot apices for gene expression analysis. All of the samples were harvested at the same time of day. ACT2 was used as an internal control gene since no changes in transcription were noted for this gene under the tested experimental conditions. A flowering related control gene (SOC1) was used as a positive control to confirm that our shift to LD conditions truly induced the flowering response (see Supplementary Fig. S3 at JXB online). As shown in Fig. 5A, the expression level of AtCSPs increased after transfer to LD conditions in wild-type relative to the SD control. It is important to note that a similar induction was not observed in either mutant background.
Fig. 5.
Analysis of AtCSPs gene expression during floral induction. Plants were grown under short-day conditions for 30 d and then transferred to long days for the initiation of flowering. An additional subset of plants was maintained for an additional 7 d under short-day conditions (S7) as a negative control. Total RNA was extracted from shoot apices and converted into cDNA template for quantification with qRT-PCR analysis with gene-specific Taq-Man probe sets. Transcript levels are presented as relative values to L0 of AtCSP2 gene expression level (the value of 1), after being normalized to ACT2 transcript levels. Data are shown as the means with standard deviations from three replicate reactions. Analysis was carried out using late flowering mutants (A) and Columbia wild-type (B).
In Columbia wild-type, developmentally regulated expression patterns were observed for AtCSP4 in the vegetative stages of development (Fig. 3). Since the flowering mutant backgrounds used in the floral induction experiment were in the Landsberg background, it was not possible to ascertain whether or not AtCSP4 was also induced during floral induction (since AtCSP4 expression is 1000-fold reduced in the Landsberg ecotype). Therefore, a floral induction experiment was also performed in the Columbia wild-type background. All AtCSPs showed a positive response during the induction to flowering in the Columbia ecotype, with AtCSP2 exhibiting the highest relative expression levels (Fig. 5B). Thus, these data provide additional evidence to suggest that AtCSPs are also induced during floral induction in the Columbia wild-type and may function in relation to flowering in planta.
Gene and protein expression during stages of silique development
Analysis of public microarray data showed changes in AtCSP transcript accumulation during the progression of silique development (data not shown). In mammals, a knock-out of a cold shock domain protein (YB-1) causes severe damage to embryos or in some cases embryo lethality (Lu et al., 2005, 2006; Uchiumi et al., 2006). Thus, we were interested in characterizing AtCSPs in relation to various stages of silique development by using qRT-PCR and Western blot analyses. Each silique developmental stage was harvested upon the time-course as reported from the Weisshaar group at AtGenExpress (http://www.genomforschung.uni-bielefeld.de/GF-research/AtGenExpress-SeedsSiliques.html). In addition, for each tissue harvest, their exact stage of development was visualized and documented with microscopic analysis (see Supplementary Fig. S4 at JXB online) as described in the Materials and methods. In this experiment eIF4A2 (At1g54270) was used instead of ACT2 as a normalizing control for qRT-PCR analysis.
With the exception of AtCSP4, qRT-PCR analysis revealed similar expression patterns for all AtCSPs in Columbia wild-type (Fig. 6). The expression of AtCSP2, which is the major transcribed gene among AtCSPs, was shown at a high level in the earliest stage of silique development (Fig. 6; Si1). Expression of AtCSP2 decreased during the middle stages of silique development and subsequently increased again during the later stages containing walking-stick and green cotyledon-stage embryos (Fig. 6; Si8 and Si10). Expression of AtCSP1, AtCSP2, and AtCSP3 transcripts increased during later stages of silique development. On the other hand, AtCSP4 transcript accumulated in floral buds, opened flowers, and an early stage of silique development (Fig. 6; FB-Si2). Its transcript levels subsequently decreased in the late stages of silique development. With the exception of AtCSP4, similar patterns of gene expression for AtCSP1, AtCSP2, and AtCSP3 were observed in the Landsberg background (see Supplementary Fig. S5A at JXB online). In Landsberg, AtCSP4 mRNA accumulated in the middle and late stages of silique development; however its relative expression level was extremely low relative to Columbia. Collectively, these correlative data suggest that AtCSPs may be involved with silique development with AtCSP2 and AtCSP4 potentially functioning at the early stages and AtCSP1, 2, and 3 possibly functioning in the later stages of development in Columbia.
Fig. 6.
AtCSP gene expression during stages of silique development. Columbia wild-type seedlings were grown in soil for 5–9 weeks under continuous light conditions in a growth chamber. Total RNA was extracted from floral buds (FB), opened flowers (OF), and silique stages 1–10 (S1–S10). Total RNA was converted into cDNA to serve as the template for qRT-PCR with gene-specific Taq-Man probe sets. After normalization to eIF4A2 RNA levels, AtCSP transcript levels are presented as relative values to the AtCSP2 gene expression level in floral bud tissue (the value of 1). Data are shown as the means from three replicates with standard deviations displayed as error bars.
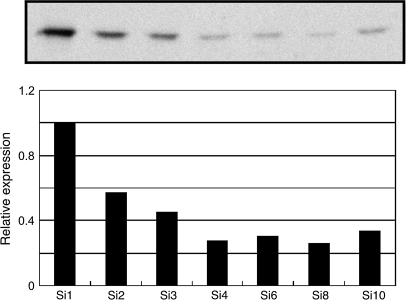
Similar to the vegetative stage analysis, a strong protein band corresponding to the putative size of AtCSP2 and 4 (19 kDa) was detected. However, there were no additional bands detected corresponding to AtCSP1 (30 kDa) and AtCSP3 (29.5 kDa) presumably due to their very low level of gene expression. Accumulation of AtCSP2/AtCSP4 protein was highest in the earliest stage of silique development (Fig. 7; Si1). Protein levels for AtCSP2/AtCSP4 progressively decreased with silique development and slightly increased the during late stages of silique development. This protein accumulation pattern was in good agreement with qRT-PCR analysis. A similar pattern of 19 kDa protein accumulation was observed in the Landsberg background (see Supplementary Fig. S5B at JXB online). In Landsberg, it is assumed that the 19 kDa band is predominantly AtCSP2, since AtCSP4 expression is 1000-fold reduced relative to Columbia wild-type. These data support the hypothesis that AtCSPs may function during silique development in Arabidopsis.
Fig. 7.
Analysis of AtCSPs protein expression during stages of silique development. Total proteins were extracted from silique stage 1–10 (S1–S10) and separated by SDS-PAGE. Signals were detected by Western blot analysis using a polyclonal anti-WCSP1 antibody. AtCSP2 and 4 cannot be distinguished from one another with Western blot analysis. A band similar to the predicted size range for AtCSP2 and 4 was detected and found to accumulate early in silique development. It should be noted that no corresponding protein bands were detected in the putative size range for AtCSP1 and 3.
Chromatin immunoprecipitation analysis with AGL15 antisera
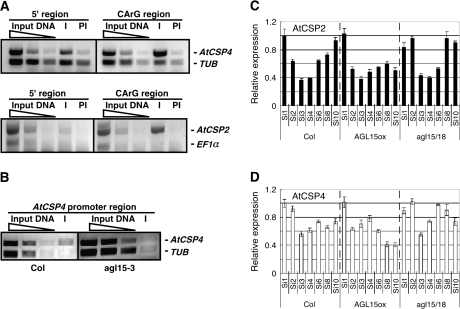
AGL15 is a well-studied MADS domain protein that accumulates to its highest amounts during embryogenesis. Along with the closely related family member AGL18, it has been implicated in the control of flowering time (Adamczyk et al., 2007) and somatic embryogenesis (Thakare et al., 2008). AGL15 preferentially binds to CArG motifs of a form C(A/T)8G in vitro (Tang and Perry, 2003) and activates or represses gene expression (Wang et al., 2002; Zhu and Perry, 2005; Hill et al., 2008). Based on promoter analysis of AtCSPs, putative binding sites for AGL15 were identified in the promoter region of both AtCSP2 and AtCSP4. To investigate whether AtCSP2 and AtCSP4 are potentially directly regulated by AGL15 during silique development, it was first confirmed whether AGL15 directly binds to the promoter region of AtCSP2 and AtCSP4 using an enrichment assay. If AGL15 can bind AtCSP2 and AtCSP4 promoter regions, co-purified AtCSP2 and AtCSP4 promoter fragments should be detected as enriched over non-bound control DNA fragments by multiplex PCR on DNA selected by chromatin immunoprecipitation (ChIP) using an AGL15 specific antibody. The oligonucleotide primer sets were designed to amplify regions of the AtCSP2 and AtCSP4 promoter containing potential binding sites for AGL15 or 5’ regions in the promoter which lacked binding sites. β-tubulin (At5g62690) (Snustad et al., 1992) or EF1α (At1g07920), that are not expected to be bound by AGL15, were used as control DNA fragments. AtCSP2 and AtCSP4 promoter and control regions were both amplified in input DNA (Fig. 8). Relative to the 5′ region and control fragments amplification, AtCSP2 and AtCSP4 promoter fragments containing potential binding sites were enriched in DNA selected by ChIP (Fig. 8A).
Fig. 8.
ChIP analysis of the AtCSP2 and AtCSP4 promoter region in 35S::AGL15 overexpression embryo culture background (A), Columbia and agl15-3 null mutant somatic embryos system (B). AGL-15 antibodies were used to immunoprecipitate chromatin for subsequent PCR amplification of two regions within the promoter of AtCSP2 and AtCSP4. Upper bands display amplicons specific for AtCSP2 and AtCSP4 promoter regions, whereas lower bands represent the amplified control gene. In comparison to preimmune immunoprecipitated fractions (PI), note the enriched AtCSP2 and AtCSP4 amplicon band in the immune (I) fraction corresponding to the CArG region. (C, D) AtCSP2 and AtCSP4 gene expression during the stages of silique development in Columbia wild-type, 35S::AGL15 overexpression and agl15/18 mutant background. Plants were grown in soil for 5–9 weeks under continuous light conditions. Total RNA was extracted from silique stages 1–10 (S1–S10). Total RNA was converted into cDNA to serve as the template for qRT-PCR with gene specific Taq-Man probe sets. Transcript levels are presented as relative values to silique stage 1 of each gene expression level (the value of 1), after being normalized to the eIF4A2 RNA levels. Data are shown as the means from three replicate reactions with standard deviations presented as error bars.
To clarify whether AGL15 does indeed bind to the AtCSP2 and AtCSP4 promoter regions, the same assay was performed using the AGL15 knock-out mutant (agl15-3). AGL15 binds to its own promoter region and can therefore be used as a positive control in this assay. An AGL15 fragment was amplified in input DNA and recovered by the ChIP reaction in the Columbia background, however, this fragment was not recovered in the mutant background (data not shown). Similarly, the AtCSP4 fragment was not enriched in the mutant background (Fig. 8B). Collectively, these data suggest that AGL15 can bind promoter regions of AtCSP2 and AtCSP4 and may affect the gene expression of AtCSP2 and AtCSP4 during silique development.
As a means to determine whether gene expression of AtCSP2 and AtCSP4 is affected by AGL15, qRT-PCR analysis was performed to quantify expression levels of AtCSP2 and AtCSP4 in AGL15 overexpression or agl15/18 knock-out lines during various stages of silique development. The preparation of silique samples was performed as previously described and all stages were confirmed with microscopy (data not shown). AGL15 expression levels were quantified first from silique samples harvested from overexpression or knock-out lines. AGL15 expression was clearly elevated and suppressed in the overexpression and knock-out lines, respectively (data not shown). Relative to Columbia wild-type, AtCSP2 and AtCSP4 expression patterns were altered in both the AGL15 overexpression and knock-out mutant background (Fig. 8C, D). AtCSP2 expression decreased in the late stages of silique development in the AGL15 overexpressed backgrounds (Fig. 8C; Si8 and Si10). Conversely, AtCSP2 expression levels were slightly elevated in the AGL15 double knock-out mutant, agl15/agl18 (Fig. 8C; Si8) as well as at the early stage of silique development (Fig. 8C; Si2). In addition, AtCSP4 expression patterns were altered in both AGL15 overexpressed and knock-out mutant backgrounds. Collectively, these data suggest that promoters for both AtCSP2 and AtCSP4 are bound by AGL15 and that AGL15 affects their transcription patterns during various stages of silique development.
In situ hybridization of AtCSP2 and AtCSP4 in developing floral and silique tissue
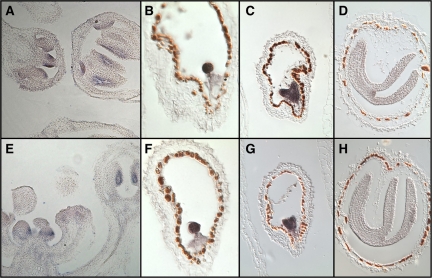
According to the analysis of AtCSP gene and protein expression, AtCSPs are correlated to floral and silique development. In addition, both a ChIP enrichment assay and expression analysis linked AtCSP2 and AtCSP4 to the AGL15 MADS domain transcription factor during silique development. Thus we were interested in performing in situ hybridization analysis for AtCSP2 and AtCSP4 to confirm qRT-PCR expression patterns and to visualize transcripts within developing floral and silique tissues.
In developing floral tissue, both AtCSP2 and AtCSP4 were localized in a ring-like pattern within young flower buds and tapetum tissue (Fig. 9A, E). Expression was also detected in unfertilized ovules within siliques (data not shown). The localization of AtCSP2 and 4 expression was investigated next in developing siliques (Fig. 9B–D, F–H). Both genes were highly expressed during the early stages of embryo development (Fig. 9B, F) and the expression gradually reduced as embryos proceeded to the walking stick embryo stage. Interestingly mRNA expression of AtCSP2 at the walking stick stage was enriched in the apical meristem region (Fig. 9D).
Fig. 9.
In situ hybridization of AtCSP2 and AtCSP4 gene expression during stages of floral and embryo development. Expression patterns for AtCSP2 (A–D) and AtCSP4 (E–H) were monitored within inflorescence tissues (A, E), embryo of globular (B, F), heart (C, G), and bent (D, H) stages, respectively.
Discussion
Two recent studies on Arabidopsis AtCSPs described a functional relationship to plant development (Fusaro et al., 2007; Kim et al., 2007a). In order to better understand how AtCSPs may function in relation to plant development, gene expression, protein accumulation, and localization analyses were performed using a variety of developmental stages. In this study, a detailed analysis describing the relationship of AtCSPs to flowering and silique development is provided. As shown by expression analysis, AtCSP2 is the most abundantly expressed gene followed by AtCSP4. On the other hand, AtCSP1 and AtCSP3 are expressed at a considerably lower level. According to these analyses, AtCSPs expression was enriched in the tissues which normally possess very high activity of cell division, like shoot apices, floral buds, and the early stages of embryos (Figs 1, 3, 4, 6, 7, 9). In addition, a floral induction study was performed and it was shown that AtCSPs are up-regulated during the transition to flowering (Fig. 5). Collectively, these observations support the hypothesis that AtCSPs may function during various stages of plant development and from the transition of vegetative to the reproductive stage.
Animal cold shock domain proteins are known to regulate the gene expression of cell cycle-related genes and play important roles for embryo development (Jurchott et al., 2003; Lu et al., 2005; Uchiumi et al., 2006). Lu et al. reported transcriptomic and proteomic analyses using YB-1 mutant lines (Lu et al., 2005). According to this study, the mutation in the cold shock domain protein did not have a major effect on gene and protein expression under normal conditions, however, the expression of several genes was altered under stress conditions. Specifically, they were cell cycle related genes, p16Ink4a and p21Cip1, which are G1-specific cell cycle inhibitors, and YB-1 exerted negative regulation on the transcriptional level. Therefore, YB-1 has been implicated in cell proliferation under stress conditions. In animals, these data suggested that cold shock domain proteins are involved in the regulation of the cell cycle and development. Our observation of enrichment of AtCSPs in cell tissues which commonly exhibit high cell division are agree with these examples. Unfortunately, no significant alterations of embryo development were observed relative to wild-type (data not shown) from analysis of a homozygous insertion line for AtCSP4 (GABI-KAT; GK-623B08.01). Since the AtCSP4 knock-out line grew normally, and its expression is drastically reduced in expression in Landsberg, it is assumed that its functional role can be complemented by AtCSP2. In the future, it will be of great interest to determine whether AtCSP mutants may have altered cell cycles and altered floral and/or silique development.
Animal YB-1 has been well studied and is important for both early and late stages of embryo development. YB-1 knock-out mice exhibited embryonic and perinatal lethality due to abnormal patterns of cell proliferation. MSY4, which is a homologue of YB-1, is also expressed during embryogenesis and is important for embryo development (Jurchott et al., 2003; Lu et al., 2005, 2006; Uchiumi et al., 2006). In this study, it is confirmed that AtCSPs are localized in embryos and especially high levels of expression were detected at the globular and late stages of embryo development (Figs 6, 7, 9; see Supplementary Fig. S5 at JXB online). Interestingly, RNAi analysis of AtCSP2 was shown to affect embryo development and altered flowering time (Fusaro et al., 2007). The authors suggested that RNAi plants exhibited an early flowering phenotype due to an alteration in the translation of flowering-related proteins. In this study, AtCSPs were highly expressed during floral transition and were induced under conditions conducive for stimulating flowering. Taken together, these data suggest that AtCSPs may exhibit preferential target specificity related to floral transition. AtCSPs may bind and maintain the stability of target genes and function to regulate translation due to RNA chaperone activity.
AGL15, which is a well-studied MADS-domain transcription factor, regulates gene expression (Fernandez et al., 2000; Perry et al., 1999; Wang et al., 2004) and is functionally redundant with AGL18 in relation to somatic embryogenesis and the control of flowering time (Thakare et al., 2008, Adamczyk et al., 2007). In this study, chromatin immunoprecipitation was used to confirm that AGL15 could bind promoter regions of both AtCSP2 and AtCSP4. In addition, when a time-course of silique development was analysed in mutant backgrounds of AGL15, alterations of gene expression patterns for both AtCSP2 and AtCSP4 were observed. Taken together with our gene and protein expression studies, these data provide additional evidence supporting a putative role of AtCSPs in silique development and flowering in Arabidopsis.
Cold shock domain proteins are highly conserved from bacteria to higher plants and animals. In previous studies of a winter wheat cold shock domain protein (WCSP1) (Nakaminami et al., 2006), WCSP1 exhibited nucleic acid binding activity and functioned as an RNA chaperone similar bacterial cold shock proteins (Bae et al., 2000). The functional significance of RNA chaperone activity is the ability of cold shock domain proteins to melt stabilized secondary structures of nucleic acids and subsequently to maintain them in a single-stranded state. Under stress conditions, this is important since RNA typically adopts secondary structures. In bacteria, cold shock domain proteins relax mRNA secondary structures and play a crucial role for subsequent translation of proteins under low temperature stress (Jiang et al., 1997; Horn et al., 2007). Previous nucleic acid binding assays of plant CSD proteins utilized lambda phage DNA as the source of nucleic acids. Thus, plant CSPs appear to bind nucleic acids with apparent non-specificity. However, it is important to note that specific targets of cold shock domain proteins do exist. For example, YB-1 binds to a sequence called the Y-box sequence in the promoter of major histocompatibility class II (MHCII) promoters (Kohno et al., 2003). In Chlamydomonas, a cold shock domain protein (NAB1) binds LHCBM genes, which are light-harvesting antenna of photosystem II (Mussgnug et al., 2005). In this study it was not investigated whether AtCSPs have preferential nucleic acid targets. However, the expression patterns and localization of AtCSPs makes it tempting to speculate that AtCSPs may target a subset of genes that are related to plant development. In the future, it will be necessary to utilize knock-out or RNAi mutants to determine if mutations in AtCSPs have an effect on the post-transcriptional level for any genes related to plant development.
Although other plant CSPs have been correlated to abiotic stress conditions (Karlson et al., 2002; Kim et al., 2007a, b), high levels of AtCSPs expression were clearly detected in meristem tissues under non-stress conditions. Therefore, plant cold shock domain proteins probably possess multi-functions containing an RNA chaperone activity that may be important under both stress and non-stress conditions. In the future, more research would help to determine if AtCSPs exhibit preferential binding to specific gene targets, thereby affecting their translation through an RNA masking activity or perhaps affecting mRNA stability (Evdokimova et al., 2001, 2006).
Our confirmation for the dramatic suppression of AtCSP4 gene expression in the Landsberg background was consistent with the previous observation which designated the At2g21060 locus as one of a few highly polymorphic loci between Landsberg and Columbia ecotypes (Schmid et al., 2003). In order to understand further the striking difference between expression of the AtCSP4 gene between Landsberg and Columbia, the AtCSP4 genomic locus from Landsberg was cloned and comparative sequence analysis between Landsberg and Columbia were performed. According to this analysis, two different cis-elements of interest (SEBFCONSSTPR10A and SITEIIATCYTC) existed in Landsberg and Columbia, respectively. The SEBFCONSSTPR10A element represents the binding site of the silencing element binding factor (SEBF). In the Landsberg ecotype, this element sequence is ‘TTGTCAC’ whereas in Columbia is ‘TTGTCAT’. Interestingly, the last nucleotide of this element, ‘C’ is a critical nucleotide for the binding of SEBF (Boyle and Brisson, 2001). Thus, this difference in nucleotide sequences may represent one of the potential mechanisms for altering AtCSP4 gene expression between Landsberg and Columbia ecotypes. Another putative cis-element of interest is the SITEIIATCYTC binding site for TCP-domain transcription factors (Welchen and Gonzalez, 2005, 2006). This element is frequently present in more then one copy and it is important to note that two SITEIIATCYTC elements are found in all AtCSPs promoter regions in the Columbia background. However, in the case of the AtCSP4 Landsberg locus, no SITEIIATCYTC elements were identified in our genomic sequence analysis. The TCP domain protein TCP2 or 3 has been reported to localize in shoot apices, vegetative leaf primordia, and floral organ primordia (Cubas et al., 1999) and Cytc-1 genes regulated by a TCP domain transcription factor are also expressed in meristematic regions (Welchen and Gonzalez, 2005). In good accordance to these aforementioned examples, temporal and spatial expression patterns of AtCSPs are very similar to those of the TCP domain genes. Thus, it is possible that the difference in this putative cis-element may also affect the transcription of AtCSP4 in planta. Transgenic studies in the Landsberg ecotype with over-expression of AtCSP4 or transfer of the entire Columbia AtCSP4 gene locus, will enable us to decipher any functional effect of the reduced AtCSP4 expression in Landsberg wild-type. In addition, a future comparative ChIP analysis between Landsberg and Columbia ecotypes using antibodies specific for transcription factors of interest will allow us to understand potential mechanisms of transcriptional regulation in greater detail.
Supplementary data
Readers are encouraged to view the supplementary data files that can be found at JXB online.
Supplementary Fig. S1. AtCSP expression analysis during vegetative growth and floral transition.
Supplementary Fig. S2. AtCSP gene expression in vegetative growth and floral induction.
Supplementary Fig. S3. Analysis of SOC1 gene expression during floral induction.
Supplementary Fig. S4. Analysis of AtCSP gene expression during stages of silique development.
Supplementary Fig. S5. AtCSP gene expression during stages of silique development.
Supplementary Table S1. List of context sequences and amplicon lengths for Applied Biosystems Taq-Man probes.
Supplementary Table S2. List of primers used for SYBR-green qRT-PCR and ChlP analysis.
Supplementary Material
Acknowledgments
We would like to thank Dr Ryozo Imai and Mr Kentaro Sasaki (National Agricultural Research Center for Hokkaido Region) for kindly providing the anti-WCSP1 antibody. We would also like to thank Dr Joseph Morton (West Virginia University) for graciously supporting our microscopy work. We would like to thank the ABRC for generously providing seeds for the co-2 and ft-2 flowering time mutants. We also thank Dr Donna Fernandez for providing agl15/18 double knock-out and AGL15 overexpression seeds. We thank GABI-KAT for the GK-623B08.01 T-DNA insertion line. This project was supported by a National Science Foundation grant (IBN-0416945) to DK. West Virginia Agriculture and Forestry Experiment Station Scientific Article No. 3028.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. The Plant Journal. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proceedings of the National Academy of Sciences, USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan G, Sinha NR. The regulation of compound leaf development. Plant Physiology. 2001;127:1533–1538. [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J. Chromatin techniques for plant cells. The Plant Journal. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Boyle B, Brisson N. Repression of the defense gene PR-10a by the single-stranded DNA binding protein SEBF. The Plant Cell. 2001;13:2525–2537. doi: 10.1105/tpc.010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. The Plant Journal. 1999;18:215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Evdokimova VM, Wei CL, Sitikov AS, Simonenko PN, Lazarev OA, Vasilenko KS, Ustinov VA, Hershey JW, Ovchinnikov LP. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. Journal of Biological Chemistry. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO Journal. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Anglesio MS, Sorokin AV, Ovchinnikov LP, Buckley J, Triche TJ, Sonenberg N, Sorensen PH. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Molecular and Cellular Biology. 2006;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC. The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. The Plant Cell. 2000;12:183–198. doi: 10.1105/tpc.12.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Bocca SN, Ramos RL, et al. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta. 2007;225:1339–1351. doi: 10.1007/s00425-006-0444-4. [DOI] [PubMed] [Google Scholar]
- Gonda K, Fowler J, Katoku-Kikyo N, Haroldson J, Wudel J, Kikyo N. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nature Cell Biology. 2003;5:205–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- Gonda K, Wudel J, Nelson D, Katoku-Kikyo N, Reed P, Tamada H, Kikyo N. Requirement of the protein B23 for nucleolar disassembly induced by the FRGY2a family proteins. Journal of Biological Chemistry. 2006;281:8153–8160. doi: 10.1074/jbc.M512890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. The role of knox genes in plant development. Annual Review of Cell and Developmental Biology. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiology. 2003;133:653–663. doi: 10.1104/pp.103.023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck GR, Perry SE, Nichols KW, Fernandez DE. AGL15, a MADS domain protein expressed in developing embryos. The Plant Cell. 1995;7:1271–1282. doi: 10.1105/tpc.7.8.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. The Plant Journal. 2008;53:172–185. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- Horn G, Hofweber R, Kremer W, Kalbitzer HR. Structure and function of bacterial cold shock proteins. Cell and Molecular Life Sciences. 2007;64:1457–1470. doi: 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Maloney KA, Ley TJ. A human protein containing a ‘cold shock’ domain binds specifically to H-DNA upstream from the human gamma-globin genes. Journal of Biological Chemistry. 1994;269:14130–14139. [PubMed] [Google Scholar]
- Ikeda-Iwai M, Satoh S, Kamada H. Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. Journal of Experimental Botany. 2002;53:1575–1580. doi: 10.1093/jxb/erf006. [DOI] [PubMed] [Google Scholar]
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. Journal of Biological Chemistry. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. Journal of Biological Chemistry. 2003;278:27988–27996. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- Karlson D, Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiology. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D, Nakaminami K, Toyomasu T, Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. Journal of Biological Chemistry. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- Kim JY, Park SJ, Jang B, Jung CH, Ahn SJ, Goh CH, Cho K, Han O, Kang H. Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. The Plant Journal. 2007b;50:439–451. doi: 10.1111/j.1365-313X.2007.03057.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Research. 2007a;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Molecular and General Genetics. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Molecular and Cellular Biology. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. Cold shock domain family members YB-1 and MSY4 share essential functions during murine embryogenesis. Molecular and Cellular Biology. 2006;26:8410–8417. doi: 10.1128/MCB.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y, Ohga T, Toh S, Koike K, Okumura K, Wada M, Kuwano M, Kohno K. Structural and functional analysis of the human Y-box binding protein (YB-1) gene promoter. Nucleic Acids Research. 1996;24:1873–1878. doi: 10.1093/nar/24.10.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Mussgnug JH, Wobbe L, Elles I, et al. NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. The Plant Cell. 2005;17:3409–3421. doi: 10.1105/tpc.105.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Karlson DT, Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proceedings of the National Academy of Sciences, USA. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov MP, Ivshina MP, Chernov KG, Kovrigina EA, Evdokimova VM, Thomas AA, Hershey JW, Ovchinnikov LP. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. Journal of Biological Chemistry. 2003;278:13936–13943. doi: 10.1074/jbc.M209145200. [DOI] [PubMed] [Google Scholar]
- Perry SE, Lehti MD, Fernandez DE. The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiology. 1999;120:121–130. doi: 10.1104/pp.120.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. The Plant Cell. 1995;7:1259–1269. doi: 10.1105/tpc.7.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H, Maekawa T, Imamoto F, Yasuda K, Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988;73:499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Kim MH, Imai R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochemical and Biophysical Research Communications. 2007;364:633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Developmental Biology. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed beta-tubulin genes. The Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafuri SR, Wolffe AP. Xenopus Y-box transcription factors: molecular cloning, functional analysis and developmental regulation. Proceedings of the National Academy of Sciences, USA. 1990;87:9028–9032. doi: 10.1073/pnas.87.22.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Perry SE. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. Journal of Biological Chemistry. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiology. 2008;146:1663–1672. doi: 10.1104/pp.108.115832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Fotovati A, Sasaguri T, et al. YB-1 is important for an early stage embryonic development: neural tube formation and cell proliferation. Journal of Biological Chemistry. 2006;281:40440–40449. doi: 10.1074/jbc.M605948200. [DOI] [PubMed] [Google Scholar]
- Wang H, Tang W, Zhu C, Perry SE. A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. The Plant Journal. 2002;32:831–843. doi: 10.1046/j.1365-313x.2002.01455.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. The Plant Cell. 2004;16:1206–1219. doi: 10.1105/tpc.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiology. 2005;139:88–100. doi: 10.1104/pp.105.065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation machinery. Plant Physiology. 2006;141:540–545. doi: 10.1104/pp.105.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Molecular Microbiology. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- Zhu C, Perry SE. Control of expression and autoregulation of AGL15, a member of the MADS-box family. The Plant Journal. 2005;41:583–594. doi: 10.1111/j.1365-313X.2004.02320.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.