Abstract
Rice (Oryza sativa) allelic sugary1 (sug1) mutants defective in isoamylase 1 (ISA1) accumulate varying levels of starch and phytoglycogen in their endosperm, and the activity of a pullulanase-type of a debranching enzyme (PUL) was found to correlate closely with the severity of the sug1 phenotype. Thus, three PUL-deficient mutants were generated to investigate the function of PUL in starch biosynthesis. The reduction of PUL activity had no pleiotropic effects on the other enzymes involved in starch biosynthesis. The short chains (DP ≤13) of amylopectin in PUL mutants were increased compared with that of the wild type, but the extent of the changes was much smaller than that of sug1 mutants. The α-glucan composition [amylose, amylopectin, water-soluble polysaccharide (WSP)] and the structure of the starch components (amylose and amylopectin) of the PUL mutants were essentially the same, although the average chain length of the B2-3 chains of amylopectin in the PUL mutant was ∼3 residues longer than that of the wild type. The double mutants between the PUL-null and mild sug1 mutants still retained starch in the outer layer of endosperm tissue, while the amounts of WSP and short chains (DP ≤7) of amylopectin were higher than those of the sug1 mutant; this indicates that the PUL function partially overlaps with that of ISA1 and its deficiency has a much smaller effect on the synthesis of amylopectin than ISA1 deficiency and the variation of the sug1 phenotype is not significantly dependent on the PUL activities.
Keywords: Amylopectin, debranching enzyme, isoamylase, mutant, pullulanase, rice endosperm, starch biosynthesis
Introduction
It is generally accepted that ADP-glucose pyrophosphorylase, starch synthase, and starch branching enzymes are involved in starch biosynthesis in higher plants. The fourth kind of enzymes, namely, starch debranching enzymes, are also thought to play an essential role in starch biosynthesis; a recent analysis of sugary1 mutants, which accumulate highly branched phytoglycogen instead of starch in maize (Pan and Nelson, 1984; James et al., 1995), rice (Nakamura et al., 1989, 1996), barley (Burton et al., 2002), and Chlamydomonas (Mouille et al., 1996), showed that these mutants were caused by an isoamylase deficiency.
In plants, there are two types of starch debranching enzymes, isoamylase (ISA, EC 3.2.1.68) and pullulanase (PUL or limit dextrinase, EC 3.2.1.41). At least three ISA genes are present in rice, maize, potato, and Arabidopsis, whereas only one PUL gene is present in these plants (Dinges et al., 2003). The two types of debranching enzymes greatly differ from each other in substrate specificity: ISA can debranch amylopectin and glycogen, while PUL can debranch pullulan and amylopectin (Nakamura, 1996). James et al. (1995) identified the Sugary1 (Su1) gene in maize by transposon-tagging su1 mutants and showed that the gene encodes an ISA-type starch debranching enzyme. In rice, both the sugary1 (sug1) locus and the ISA1 gene are located on chromosome 8 (Yano et al., 1984; Fujita et al., 1999), while the PUL gene is located on chromosome 4 (Nakamura et al., 1996). The reduction of ISA1 activity to about only 6% by using antisense technology resulted in modified amylopectin with more abundant short side chains and increased accumulation of soluble α-glucans (Fujita et al., 2003). In transgenic rice generated by the introduction of the wheat ISA1 gene into sug1 rice, phytoglycogen synthesis was substantially replaced by starch synthesis in the endosperm (Kubo et al., 2005). These reports strongly suggest that sugary1 mutations in maize and rice are caused by ISA1 deficiency and ISA1 plays a crucial role in amylopectin biosynthesis.
On the other hand, the physiological function of PUL, another starch debranching enzyme, remains to be understood. It is generally accepted that the main function of PUL is starch degradation in germinated grain, although substantial PUL activity has been detected in the developing rice (Nakamura et al., 1996) and maize (Beatty et al., 1999) endosperm. A study of a knockout mutant of maize showed that PUL is necessary for normal rates of starch degradation in leaves and the endosperm (Dinges et al., 2003). The possibility remains open that another type of debranching enzyme also participates in starch degradation in the cereal endosperm (Smith et al., 2005).
The analysis of rice allelic sug1 mutant lines, which accumulate different levels of phytoglycogen instead of starch in the endosperm, showed that PUL activity is closely related to the amount of starch accumulation in the endosperm, while ISA activity by the native-PAGE/DBE staining method is lacking or extremely low in the developing rice endosperm of all sug1 mutant lines (Nakamura et al., 1997; Kubo et al., 1999). The reduction of PUL activity in sugary1 mutants was observed in maize (Beatty et al., 1999) and rice but not in Arabidopsis (Zeeman et al., 1998) and Chlamydomonas (Dauvillée et al., 2000). These reports suggest that PUL, as well as ISA1, has a distinct role in amylopectin biosynthesis, at least in maize and rice, and that the high PUL activity is responsible for the formation of amylopectin-like α-glucans instead of phytoglycogen in the starch region of sug1 mutation lines (Kubo et al., 1999). On the other hand, it is known that some starch mutants, such as the rice floury-2 mutant, as well as sug1 mutants in rice, have pleiotropic effects on PUL activity (Kawasaki et al., 1996).
To understand the physiological function of PUL, a maize PUL mutant line, zpu1-204 was isolated using the gene-tagging method (Dinges et al., 2003). The structure and composition of starch in the endosperm in maize zpu1-204 are very similar to those of the wild type, although amylopectin from the mutant leaves contains significantly fewer chains with 8 ≤DP ≤15 than that from leaves of the wild type because of the pleiotropic reduction of starch branching enzyme IIa (BEIIa) activity. The developing zpu1-204 endosperm accumulates branched malto-oligosaccharides not found in the wild type. Furthermore, in a background deficient in the ISA (su1) mutant, zpu1-204 showed a significant accumulation of phytoglycogen in the kernel that was not seen in the wild type. These results indicate that the maize PUL partially compensates for the defect in ISA, suggesting a function of PUL during starch synthesis as well as during degradation (Dinges et al., 2003).
In the present study, to understand the physiological functions of PUL on starch biosynthesis, rice PUL-deficient mutant lines were isolated, and the structure and composition of the endosperm starch in the PUL mutants were comprehensively compared with those of the wild type. Moreover, double mutant lines containing both PUL and ISA1 mutations were generated to understand the compensation of PUL for the defect of ISA1 and to discuss the role of PUL in the rice plant.
Materials and methods
Plant materials
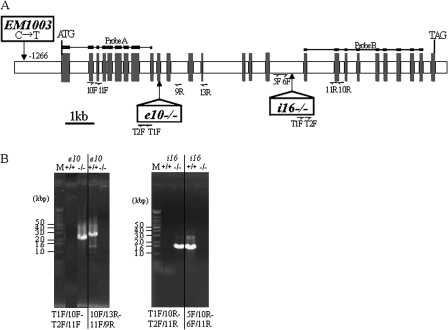
Three PUL mutant lines were used in this study: the mutant lines (e10–/– and i16–/–) containing the Tos17 insertion at exon 10 and intron 16 of the OsPUL gene (Francisco et al., 1998; AB012915), respectively; and EM1003, a product of N-methyl-N-nitrosourea (MNU) mutagenesis of the rice cultivar, Taichung65 (T65, Satoh and Omura, 1979), which mutated from C to T at 1266 bp before the start codon (Fig. 1A). The PUL activity band in native-PAGE/PUL and DBE activity staining assays was lacking in these mutants (Fig. 2B, D). e10+/+ and i16+/+ (which have no Tos17 insertions in the OsPUL gene but had a genetic background common to the mutant lines, e10–/– and i16–/–, respectively) and the parental cultivars Nipponbare (Nip) and T65 were used as control plants.
Fig. 1.
Sites of Tos17 insertion and mutation site by MNU in the OsPUL gene and determination of the Tos17 rice mutant line genotype by PCR. (A) Structure of the OsPUL gene. The exons and introns are depicted as grey and white boxes, respectively. ATG and TGA indicate the translation initiation and stop codons, respectively. The sites of Tos17 insertion and mutation by MNU in mutant lines are indicated with a vertical arrow. Horizontal half arrows show the sites of primers (T1F, T2F, 5F, 6F, 10F, 11F, 9R, 10R, 11R, and 13R) for PCR for genotype determination (B) and mutant line screening. The primers T1F and T2F were designed from the Tos17 sequence, while 5F, 6F, 10F, 11F, 9R, 10R, 11R, and 13R were designed from the OsPUL gene sequence. The region used as probes for Southern blotting to screen mutant lines is indicated. (B) Determination of genotype [homozygous for Tos17 insertion (–/–, left panel) or wild homozygous (+/+, right panel)] in Tos17 mutant lines by nested PCR. Primer pairs are indicated below the photographs. ‘T1F/10F-T2F/11F’ means that the primer pair T1F/10F was used for the 1st PCR and T2F/11F for the 2nd PCR. M, molecular markers.
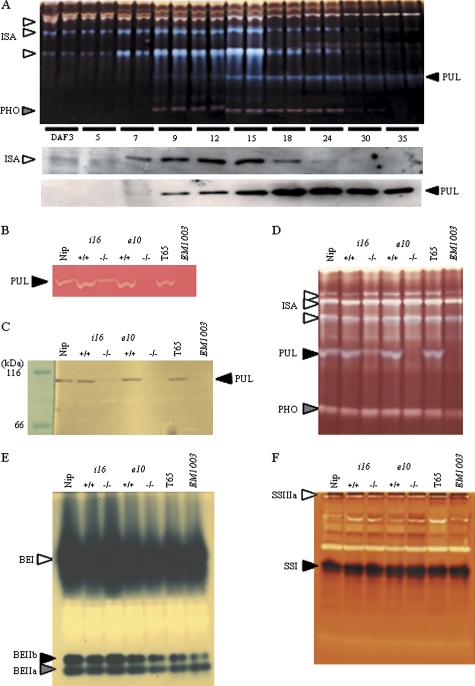
Fig. 2.
Native-PAGE/activity staining and immunoblotting of developing endosperm in rice PUL (pullulanase) mutant lines and the wild type. (A) Native-PAGE/debranching enzyme (DBE) activity staining (upper panel) and immunoblotting (lower panels) of rice developing endosperm of the wild type (T65) from day after flowering (DAF)-3 to -35. Two lanes of native-PAGE in each DAF were from the soluble fraction of the two independent developing rice endosperms. The ISA (isoamylase), PUL, and PHO (phosphorylase) bands are indicated by arrowheads. (B) Native-PAGE/PUL activity staining. PUL activity bands are indicated by arrowheads. (C) Immunoblotting using antiserum raised against purified rice PUL (Nakamura et al., 1996). (D) Native-PAGE/DBE activity staining. The ISA, PUL, and PHO activity bands are indicated by arrowheads. (E) Native-PAGE/branching enzyme (BE) activity staining. The BEI, BEIIa, and BEIIb activity bands are indicated by arrowheads. (F) Native-PAGE/starch synthase (SS) activity staining. The SSIIIa and SSI activity bands are indicated by arrowheads. Nip, Nipponbare; e10–/– and i16–/–,PUL mutant lines of Nip by Tos17 insertion; T65, Taichung 65; EM1003: PUL mutant of T65 induced by chemical mutagenesis. e10+/+ and i16+/+ are the control lines of e10–/– and i16–/–, respectively.
The two ISA1 mutant lines used in this study were EM653 and EM914, which are mild and severe types in the sug1 phenotype (Nakamura et al., 1997; Wong et al., 2003), respectively, a product of NMU mutagenesis of a rice cultivar, T65.
For establishing double mutants of PUL and ISA1, e10–/– and EM1003 were crossed to EM653 and EM914, respectively, and the resulting double heterozygotes (F1) were self-pollinated. sug1 phenotype or PUL-less F2 seeds were self-pollinated again, and double mutant F3 seeds shown to be deficient in PUL and ISA1 activity or proteins with native-PAGE/DBE staining or Western blotting, respectively, were used.
Rice plants were grown during the summer months in an experimental paddy field at Akita Prefectural University and Kyushu University under natural environmental conditions.
Tos17 and MNU mutagenesis and screening for PUL-deficient mutant lines (e10–/–, i16–/–, and EM1003)
Mutagenesis with Tos17 and pool sampling were performed as described by Hirochika (2001) and Kumar and Hirochika (2001). To screen for PUL-deficient mutant lines, DNA fragments carrying the Tos17 transposon from DNA pools constructed using the three-dimensional sampling method from approximately 40 000 Tos17-containing plants were subjected to nested PCR using combinations of the transposon-specific primers T1F (for 1st PCR) and T2F (for 2nd PCR, Fig. 1A) and the OsPUL-specific primers 10F (for 1st PCR), 11F (for 2nd PCR), 10R (for 1st PCR), and 11R (for 2nd PCR, Fig. 1A). The PCR products were hybridized with 1.4 kb (probe A) and 0.9 kb (probe B) of the BamHI and EcoRI fragments of OsPUL cDNA (Fig. 1A), and positive products were gel-purified and sequenced to identify those containing the real OsPUL sequence and the location of Tos17 in the OsPUL gene.
The screening of PUL mutants by MNU mutagenesis was carried out by the detection of deletion in the corresponding protein band in the SDS-PAGE/CBB staining analysis of mutant lines induced from the MNU treatment of fertilized egg cells in a rice cultivar, T65. Of 1270 mutant lines, two mutants derived from the independent MNU treatment were lacking in the 100 kDa PUL bands on SDS-PAGE/CBB staining. One mutant line (EM1003) was used in this study.
Determination of mutation sites in the PUL gene of EM1003
To establish the mutation sites in the PUL gene of EM1003, the nucleotide sequences of the genomic DNA from EM1003 were performed. Genomic DNA was prepared from seedling leaves (200 mg each) of cv. T65 and the mutant line EM1003 by the CTAB method of Murray and Thompson (1980). PCR amplifications of genomic DNA preparation using the primers listed in Supplementary Table S1 at JXB online were performed. Templates for the sequence were prepared by using LA Taq polymerase (Takara) following the established protocol. The genomic DNA sequences were determined using the ABI Dye Terminator Ready Reaction kits in an automated ABI 3130 DNA Sequencer. DNA sequence analysis was performed using EditView 1.0.1 and Auto Assembler 2.1. Sequences were compared between a wild type and mutant with CLUSTALW (htpp://www.ddbj.nig.ac.jp/search/clustalw-e.html) of the DNA Data Bank of Japan (DDBJ).
Native-PAGE/activity staining, enzyme assay, and immunoblotting
Native-PAGE/activity staining of DBE and BE was performed using the methods of Fujita et al. (1999) and Yamanouchi and Nakamura (1992), respectively. Native-PAGE/ activity staining of PUL was performed on 7.5% acrylamide slab gels including 1.3% (w/v) red pullulan (Magazyme). SS activity staining was performed on 7.5% (w/v) acrylamide slab gels containing 0.8% (w/v) oyster glycogen (G8751, Sigma) according to Nishi et al. (2001) with the modification that 0.5 M citrate was included in the reaction mixture to enhance the SS activity (Fujita et al., 2006). The assay for AGPase was performed following the methods of Nakamura et al. (1989). Immunoblotting was performed according to the methods of Fujita et al. (1999) using the antiserum raised against the purified PUL (Nakamura et al., 1996).
Analysis of starch granules of endosperm
The estimation of α-glucan of rice seeds, the pasting properties of endosperm starch measured by a rapid visco-analyser (RVA), X-ray diffraction measurement, measurement of the thermal properties of endosperm starch by differential scanning calorimeter (DSC), and the observation of starch granules by scanning electron microscopy (SEM, JEOL-5600) were performed as described previously (Fujita et al., 2003, 2006).
The molecular weight of amylopectin was determined by HPSEC-MALLS-RI according to the method of Fujita et al. (2003).
Preparation of starch granules or amylopectin for gel filtration
Starch granules were prepared from polished rice following the cold-alkali method (Yamamoto et al., 1973, 1981). Rice amylopectin was isolated from rice starch granules using n-butanol according to the method of Schoch (1954) as modified by Takeda et al. (1986).
Molecular size separation of starch and amylopectin by Sephacryl S-1000SF chromatography was performed by the method of Kubo et al. (1999). After chromatography, an aliquot of each fraction was used for the measurement of carbohydrate content by the phenolic sulphuric method and for the measurement of the λmax value of the glucan–iodine complex (Fujita et al., 2007).
Fluorescent labelling of debranched starch and amylopectin with 2-aminopyridine followed by HPSEC was performed as previously described (Hanashiro et al., 2002). The molar- and weight-based chain-length distribution was determined from an elution profile obtained by a fluorescence detector and a refractive index (RI) detector, respectively. The ratio of the detector responses of RI/fluorescence (RI/F) was used to calculate the DP using the RI/F value of standard amylose with known DPn (AS-10, DPn=57.7, purchased from International Polysaccharide Engineering Inc., Osaka, Japan). The RI/F ratio of a specimen was taken either as an area of a given fraction or as the height of a given elution position. The apparent amylose content was determined from the RI profile of debranched starch. The actual amylose content was calculated after subtracting the peak area corresponding to an extra-long chain (ELC) of amylopectin from that of apparent amylose of starch.
Chain-length distribution of endosperm amylopectin by capillary electrophoresis
The chain-length distributions of α-glucans from endosperm were analysed using the capillary electrophoresis methods of O'Shea and Morell (1996) and Fujita et al. (2001) in a P/ACE MDQ Carbohydrate System (Beckman Coulters, CA, USA).
Preparation of the starch pastes and measurement of dynamic viscoelasticity
Each starch suspension was weighed in a beaker to give a 4% concentration (w/w on the basis of the anhydrate), and demineralized water was added to bring the total to 20 g. The starch suspensions were allowed to swell for 15 min, after which they were heated for 20 min in an autoclave (ES-215, Tomy Co., Ltd.) at 105 °C. Each starch paste was measured with a Rheo-Stress I rheometer (Haake Co.), in which the diameter of the upper disc was 60 mm and the gap between the two plates was 0.052 mm. The measurements were carried out under conditions of constant stress in the frequency range of 0.01–10 s−1 and a temperature of 50 °C (Kawabata et al., 1996).
Cloning of the OsPUL cDNA and construction of the expression vector in Escherichia coli
The mRNA was isolated from the developing endosperm of rice Nipponbare cultivar according to the manufacturer's protocol of the RNeasy plant kit (Qiagen). 2.6 kbp of the mature protein coding sequence of OsPUL was prepared with a one-step RT-PCR system (Invitrogen) using forward primer 5′-GCCGCCGTGGCTTCCCAG-3’ and reverse primer 5′-TCAACATCTAGGTTGAACGAAC-3′. The PCR product was subcloned into a pGEM T-easy vector (Promega).
For the construction of the expression vector in Escherichia coli, the DNA fragment encoding the OsPUL mature protein was generated with a high-fidelity PCR system of KOD-Plus- (Toyobo), forward primer 5′-CACCGCCGCCGTGGCTTCCCAG-3′, and reverse primer 5′-TCAACATCTAGGTTGAACGAAC-3’ using cDNA of OsPUL as a template. The DNA fragment was subcloned into the pENTR™/TEV/D-TOPO vector (Invitrogen) by means of TOPO cloning according to the manufacturer's protocol (Invitrogen). The expression vector containing the coding sequence of OsPUL was constructed by means of GATEWAY technology (Invitrogen). The expression vector pET15b-OsPUL was created in the LR reaction with OsPUL inserted into pENTR™/TEV/D-TOPO and pET15b (Novagen) connected to the cassette for DNA recombination in vitro (Invitrogen).
Induction of OsPUL protein in E. coli
The constructed plasmid was transformed into E. coli BL21 (DE3) star strain (Invitrogen). The transformed cell was incubated in 10 ml of a TB (Terrific Broth) medium containing 100 μg ml−1 of ampicillin overnight at 37 °C. The 10 ml preculture was inoculated into 1.0 l of the TB medium containing 100 μg ml−1 of ampicillin at 37 °C. When OD600 of cell culture reached approximately 0.4–0.5, the cell culture was preincubated at 15 °C for 30 min, and the OsPUL protein was induced by the addition of isopropylthio-β-galactoside to a final concentration of 0.1 mM followed by incubation at 15 °C for 20 h. The cells were collected by centrifugation and stored at –80 °C until use.
Purification of recombinant OsPUL (rOsPUL) overproduced in E. coli
Purification of rOsPUL was achieved with 5 ml of HisTrap HP connected to the FPLC system (GE Healthcare) following TSKgel DEAE-5PW (7.5×75 mm) connected to the HPLC system (Tosoh). Cells were suspended in a binding buffer (20 mM imidazol-HCl (pH 7.4), 20 mM Na-phosphate (pH 7.4), 5% glycerol, 20 mM 2-mercaptoethanol, 0.5 M NaCl), lysed by sonication, and centrifuged at 10 000 g for 30 min at 0 °C. The supernatant was applied to the HisTrap HP column equilibrated with a binding buffer. After washing the column with the binding buffer, rOsPUL was eluted with a linear gradient of 0.05–0.5 M imidazol at a flow rate of 2 ml min−1. The fractions containing rOsPUL protein were collected, and the fractions were desalted and concentrated by ultrafiltration (Amicon Ultra-4, Millipore). The concentrated preparation was applied to the TSKgel DEAE-5PW column equilibrated with medium A [50 mM imidazol-HCl (pH 7.4), 8 mM MgCl2, 50 mM 2-mercaptoethanol)]. The rOsPUL was eluted with a linear gradient of 0–0.5 M NaCl at a flow rate of 1 ml min−1. The rOsPUL protein was collected and concentrated by ultrafiltration; then the buffer in the rOsPUL preparation was substituted for 50 mM imidazol-HCl (pH 7.4), 8 mM MgCl2, 1 mM DTT, 0.5 M NaCl, and 12.5% glycerol. The rOsPUL preparation was stored at –80 °C until use.
PUL assay
The PUL assay was performed by the method described by Utsumi and Nakamura (2006) with a few modifications. The purified enzyme preparation containing 2 μg proteins was incubated with 50 mM Na-acetate (pH 5.0) and 6 mg of substrates in a total volume of 1 ml at 30 °C for 10 min. At selected time intervals, aliquots (200 μl) were taken and added to an equal volume of 0.1 M sodium carbonate to terminate the reaction. The amount of reducing equivalent was determined according to the method of Fox and Robyt (1991) using maltose as a standard. Assays were performed in ranges where the enzymatic activity linearly proceeded with increases in protein amount and duration to 15 min.
Preparation of β-limit dextrin and phosphorylase-limit dextrin
β-limit dextrin (β-LD) of the amylopectin from EM21 (waxy rice) was prepared according to Inouchi et al. (1987). Phosphorylase-limit dextrin (φ-LD) was prepared by degradation with phosphorylase a from rabbit muscle (Sigma). Three grams of amylopectin from EM21 was added to 80 mM Na-phosphate (pH 6.8), phosphorylase a (Sigma, 92 U), 0.25 mM AMP, 8 mM MgCl2, and 0.02% NaN3 in 200 ml of total volume and incubated at 30 °C for 24 h with dialysing against 2.0 l of 80 mM Na-phosphate (pH 6.8), 0.25 mM AMP, 8 mM MgCl2, and 0.02% NaN3. This procedure was repeated three times, and the final preparation was washed with ethanol, acetone, and diethylether and dried in a draught.
Results
Production of the PUL mutant lines
To date, no PUL-deficient mutants have been isolated in rice, which seems attributable to the fact that PUL mutants have no morphological traits in their seeds, in contrast to sug1 mutants, which show a shrunken seed morphology (Nakamura et al., 1997). For the first trial to isolate PUL mutants, the gene tagging method was used. Two lines containing a Tos17 insertion in the rice PUL gene (OsPUL) were isolated by PCR screening of a Tos17 knockout rice population of approximately 40 000 lines (see Materials and methods). The PUL gene is composed of 26 exons and 25 introns (Fig. 1A; Francisco et al., 1998). Tos17 was inserted into exon 10 (e10–/–) or intron 16 (i16–/–) in the line used in this study (Fig. 1A). The genotype of the lines was determined to be either homozygous for the Tos17 insertion (–/–) or wild homozygous (+/+) using nested-PCR (see Materials and methods; Fig. 1B). For instance, in line e10+/+, the PCR reaction using the T1F/10F and T2F/11F primer pairs had no product (Fig. 1B, left panel, lane e10+/+), while the 10F/13R and 11F/9R primer pairs generated a c. 2.8 kbp band (Fig. 1B, left panel, lane e10+/+). In line e10–/–, the PCR product of the T1F/10F and T2F/11F primer pairs was detected as a 2.5 kbp band (Fig. 1B, left panel, lane e10–/–), while the 10F/13R and 11F/9R primer pairs gave no product (Fig. 1B, left panel, lane e10–/–). The same PCR results were obtained in the i16 lines (Fig. 1B, right panel). The e10–/– and i16–/– lines were used as the PUL mutant lines, and the e10+/+ and i16+/+ lines or Nipponbare, the wild-type parent (Nip), as its control.
Another PUL mutant line (EM1003) lacking the 100 kDa PUL band on SDS-PAGE was isolated from a population of the MNU-treated rice cv. Taichung 65 (T65) (see Materials and methods). Most of the mutations induced by MNU have been reported to be the GC to AT transition not only in mammals and microbes but also in plants (Suzuki et al., 2008). Analysis of the genomic DNA sequence of the PUL gene showed that there was no nucleotide substitution between the wild-type and the mutant line except for the C to T transition at 1266 bp upstream from the initiation codon in EM1003 (Fig. 1). It is likely that PUL deficiency in EM1003 is caused by this replacement in the promoter region of the PUL gene.
All F1 seeds by crossing e10–/– to EM1003 lacked the 100 kDa PUL band by immunoblotting, indicating that these are PUL allelic mutant lines (data not shown).
Debranching enzyme activities at different stages during seed development in wild-type rice
The activities of both debranching enzymes, ISA and PUL, have been detected in the developing rice endosperm (Kubo et al., 1999). To determine the changes in the activities of these debranching enzymes in developing stages, Native-PAGE/DBE activity staining of the soluble fraction of the wild-type (T65) developing endosperm from DAF-3 to DAF-35 was performed (Fig. 2A, upper panel). Both debranching enzymes were detected as blue bands on the Native-PAGE gel containing potato amylopectin stained with an iodine solution. ISA was detected as three major blue bands with low mobility. The blue bands with the lowest and the second-lowest mobility are attributed to the OsISA1 and OsISA2 hetero-oligomer, and the third band, to the OsISA1 homo-oligomer (Utsumi and Nakamura, 2006). Three ISA activity bands were detected at the early stages, from DAF-7 to DAF-15, but gradually diminished at the later stages (Fig. 2A, upper panel). By contrast, PUL was detected as the fastest blue band. The PUL activity band was detected during a wide range of seed development from DAF-9 to DAF-35, although the highest activity was detected from DAF-15 to DAF-24 (Fig. 2A, upper panel). The changes of the protein levels of ISA1 and PUL in the soluble fraction of developing endosperm detected by immunoblotting were closely related to those of both enzyme activities (Fig. 2A, lower panels).
PUL activities and pleiotropic effects on starch biosynthesis enzymes in the PUL mutant lines
To evaluate the effect of the insertion of Tos17 into the OsPUL gene of e10–/– and i16–/– and mutation of the OsPUL gene in EM1003, the activities of PUL in the soluble fraction from developing endosperms at DAF-15 were examined by native-PAGE/PUL activity staining using a gel containing red pullulan (Fig. 2B). A white band on the red pullulan gel was dependent on the degradation of pullulan by PUL activities in the wild types. No band was detected in the soluble fraction, from the e10–/– and EM1003 developing endosperm, while the wild type, Nipponbare, and T65, and the control lines, e10+/+ and i16+/+, all had PUL activity bands. One of the PUL mutants, i16–/–, showed a faint band, and its activity was estimated to be 6% of the wild type on the basis of the methods of Nelson (1944) and Somogyi (1952) (data not shown). The PUL activity band was also detected as a blue band on the native-PAGE gel containing potato amylopectin (Fig. 2D). The immunoblotting analysis using antiserum raised against the purified rice PUL (Nakamura et al., 1996) showed a single band of approximately 100 kDa in the soluble fraction of the wild-type (Nipponbare and T65) and control lines (e10+/+ and i16+/+), but no band was found in the e10–/– and EM1003 lines (Fig. 2C), indicating that both the e10–/– and EM1003 lines are null mutants for PUL. By contrast, i16–/– showed a faint 100 kDa band, and the relative amounts of PUL protein in the mutant were estimated to be roughly 6% of those of the wild type (data not shown), indicating that i16–/– is a leaky mutant.
To test whether the deficiency in PUL activity has pleiotropic effects, the activities of other enzymes involved in starch biosynthesis were measured in the PUL mutant lines. In the other debranching enzyme, the ISA activity was found not to decrease by the native-PAGE/DBE activity staining method in the PUL mutant lines, although a complete loss or significant reduction of PUL activity occurred (Fig. 2D). The activity levels of BEI, BEIIa, BEIIb, Pho, and SSI, and SSIIIa detected by native-PAGE/BE, DBE, and SS activity staining, respectively, in PUL mutants were not different from those of the wild type (Fig. 2D, E, F). The AGPase activities in e10–/–, i16–/–, and EM1003 (78, 83, and 92% of those of the control, respectively) were slightly lower than those of the wild type (data not shown).
Seed morphology and morphology and crystallinity of starch granules of PUL mutant lines
The seed morphology of the PUL mutant lines, e10–/– , i16–/–, and EM1003, was almost the same as that of their wild type (data not shown). Their dehulled grain weight and starch content were not significantly different from those of their wild type (Table 1).
Table 1.
Dehulled grain weight and carbohydrate content in dry seeds of rice PUL mutant and their wild type
| Lines | Genotype | Dehulled grain weighta | Starch (mg)a, h | WSP (mg)a, h | WSP (%)i | ||
| PUL | ISA | ||||||
| Nipponbare | +/+ | +/+ | 20.37±0.03b | (100)g | 12.06±0.17 | 0.73±0.14 | 5.7 |
| e10–/– | −/− | +/+ | 20.12±0.02b | (98.8) | 12.12±0.60 | 0.77±0.16 | 6.0 |
| i16–/– | −/− | +/+ | 19.70±0.03b | (96.7) | 13.01±1.33 | 0.79±0.10 | 5.7 |
| T65 | +/+ | +/+ | 22.85±0.16b | (100) | 13.92±0.90 | 0.50±0.02 | 3.5 |
| EM1003 | −/− | +/+ | 22.42±0.29c | (98.1) | 14.16±0.72 | 0.50±0.12 | 3.4 |
| EM653 | +/+ | −/− | 14.95±0.51d | (65.4) | 7.56±0.59 | 1.37±0.14 | 15.3 |
| #4001 | −/− | −/− | 11.94±0.08e | (52.3) | 3.38±0.37 | 1.30±0.25 | 27.7 |
| EM914 | +/+ | −/− | 11.80±0.11f | (51.6) | 0.41±0.03 | 2.96±0.03 | 87.9 |
| AMF60 | −/− | −/− | 10.41±0.07f | (45.6) | 0.16±0.04 | 3.06±0.09 | 95.0 |
Mean endosperm−1 ±SE.
n=50.
n=174.
n=30.
n=14.
n=56.
Per cent of wild type.
n=3.
Per cent of total α-glucans.
Scanning electron micrograph (SEM) observations of the isolated starch granules from the PUL mutant lines showed that the mutant endosperms contained granules of similar shape and size to those of their wild type (data not shown).
The X-ray diffraction patterns of the starch granules from the PUL mutant lines, e10–/–, i16–/–, and EM1003, were the same as those of their wild types (Nipponbare and T65), and all of them exhibited a typical A-type diffraction pattern, indicating that their starches share quite a similar crystalline structure (data not shown).
Analysis of the carbohydrate content, starch component, and amylopectin fine structure in the endosperm of the PUL mutant lines
To investigate whether or not PUL mutant lines contain WSP in the endosperm, α-glucan samples were separated into soluble and insoluble fractions by suspending powdered endosperm in water at ambient temperature followed by centrifugation at 600 g for 20 min at 20 °C. The endosperms of e10–/–, i16–/–, and EM1003 contained WSP by 6.0%, 5.7%, and 3.4% of total α-glucan, respectively, and the WSP contents were not significantly different from those of their wild type (those of Nipponbare and T65 were 6.0% and 3.5%, respectively; Table 1). This result regarding the WSP content was contrary to the observation for the sug1 mutant lines, which showed a significant increase (those of EM653 and EM914 were 15.3% and 87.9%, respectively; Table 1) from their wild type. The Suc and Glc contents in the endosperm of the PUL mutant lines were also not significantly different from those of their wild type (data not shown).
To test whether the reduction of PUL activity influences the structure of endosperm starch, the endosperm starch of the PUL mutants and the wild type were subjected to size-exclusion chromatography using Sephacryl S-1000SF (Fig. 3A). Judging from the λmax values of the α-glucan–iodine complex, fractions containing most, if not all, of the amylopectin and amylose were eluted at 70–120 ml and 130–200 ml, respectively (Fig. 3A). The patterns of gel filtration of the starch from the PUL mutant lines (e10–/–, Fig. 3A; EM1003, data not shown) were similar to those of the wild type, indicating that the amylose contents of the endosperm starch in the PUL mutant lines were the same as those of their wild type.
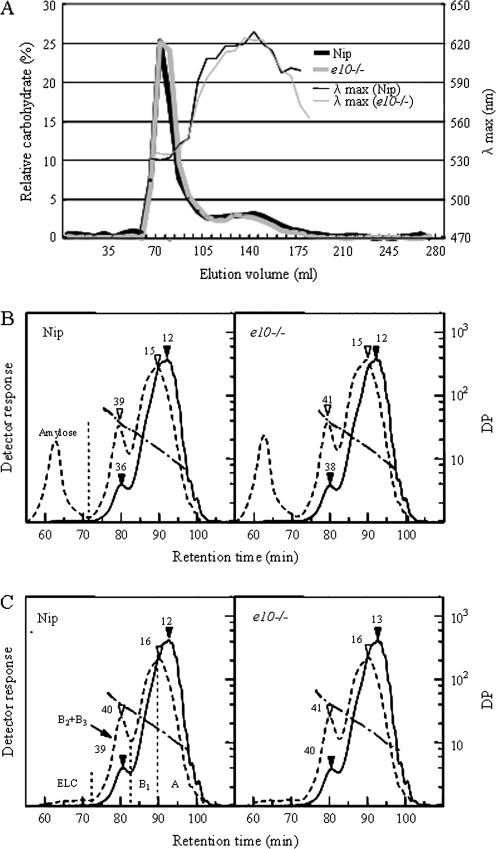
Fig. 3.
(A) Elution profiles by gel filtration chromatography through Sephacryl S-1000SF of Nipponbare (Nip, black line) and e10–/– (grey line). (B, C) Chain-length distributions of starch (B) and amylopectin (C) from Nip (left) and e10–/– (right). Solid line, fluorescence; short dashed line, refractive index; dashed and dotted line, DP; number with arrowhead, peak DP.
For further analysis of the structure of e10–/– endosperm starch, the chain-length distributions of endosperm amylopectins purified from Nipponbare and e10–/– were determined by fluorescent labelling/HPSEC (Fig. 3B). The chain-length distributions of debranched starches (Fig. 3B) were very similar in the two starches. The peak that was eluted first (retention time of c. 55–70 min) was taken as the apparent amylose, and its content was calculated as a percentage of the total peak area found in the first peak. The apparent amylose content was virtually identical (Nipponbare, 16.8%; e10–/–, 17.3%), which was in agreement with the results by Sephacryl S-1000SF chromatography described above. The actual amylose content, which was obtained by subtracting the area corresponding to extra-long chains (ELC) of amylopectin (described below; Fig. 3C) from the apparent amylose fraction in starch (Fig. 3B), was, again, almost the same (Nipponbare, 15.5%; e10–/–, 15.1%). In the analysis of debranched amylopectins (Fig. 3C), the two amylopectins showed very similar chain-length distributions on both the molar and weight bases. The peak DPs were also almost the same in the two amylopectins. As shown in Fig. 3C (left panel), the chain-length distributions were fractionated at the inflection points of the fluorescence profile into four fractions, A, B1, B2+B3, and ELC, and the percentage and number-average chain length (CLn) of each fraction were determined (Table 2). Although most of the values listed in Table 2 were essentially the same for Nipponbare and e10–/– amylopectins, a statistically significant difference was found in the weight% of the ELC fraction and the mole% and CLn of the B2+B3 fraction. In the e10–/– amylopectin, the weight% of the ELC fraction was increased, and the mole% of the B2+B3 fraction was decreased. Because the weight% of the B2+B3 fraction was unchanged, the decreased mole% resulted in a longer CLn, by c. three residues on average. A straightforward explanation for the difference in CLn is presence of short stubs with CL of c. 3, resulted from PUL-deficiency. This possibility was tested by the following experiments. Amylopectin was debranched successively with Pseudomonas isoamylase and recombinant rice PUL (rOsPUL), and the chain-length distribution was then examined by fluorescent labelling/HPSEC. The additional debranching with rOsPUL was expected to release such short chains, which isoamylase hardly attacks. After additional debranching with rOsPUL, small peaks of maltose and maltotriose were detected from the two amylopectins, and the peak area of these peaks was apparently the same in e10–/– and Nipponbare (data not shown). In another experiment, ISA-debranched amylopectin was treated with β-amylase, and the resulting hydrolysate was subjected to fluorescent labelling/HPSEC. Short branches that are resistant to ISA action should cause incomplete hydrolysis in the subsequent β-amylase treatment. Chromatograms of the β-amylolysates were basically identical in e10–/– and Nipponbare amylopectins (data not shown). These results exclude the possibility that the presence of short stubs of DP 2–3 causes the slightly greater length of the B2-3 chains of e10–/– amylopectin.
Table 2.
Chain-length distributions and average chain lengths of rice amylopectins
| Lines | Amount by mole (%)a |
Molar ratio of (A+B1)/(B2+B3) | Amount by weight (%)a |
Average chain-length |
||||||||
| A | B1 | B2+B3b | A | B1 | B2+B3 | ELCb | A | B1 | B2+B3b | Whole | ||
| Nipponbare | 68.7 | 22.6 | 8.7 | 10.6 | 47.5 | 29.5 | 21.0 | 2.0 | 12.1 | 22.9 | 43.1 | 17.5 |
| e10–/– | 69.0 | 22.6 | 8.4 | 11.1 | 46.9 | 29.7 | 20.6 | 2.8 | 12.2 | 23.6 | 45.8 | 18.1 |
The values are the averages of three or four independent measurements. Relative SDs were less than 2.4% except for weight% of ELC in Nipponbare (17.9%).
The values are based on the results by HPSEC (Fig. 3C).
Significantly different (P<0.05) by t-test between Nipponbare and e10–/–.
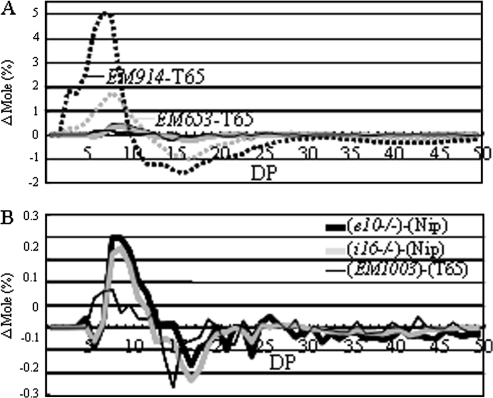
To evaluate the effects of the deficiency of the PUL activity on the short-chain composition of the endosperm amylopectin, the chain-length distributions of endosperm amylopectin in the two PUL mutant lines were further examined in detail using capillary electrophoresis. The mole% of each chain length from DP 3–60 can be estimated by this method. The short chains of amylopectin with DP ≤12 was slightly increased in the e10–/–, i16–/–, and EM1003 lines compared with those of the wild type (Fig. 4B), but the extent of the changes was much smaller than those of sug1 mutant lines (EM914, EM653; Fig. 4A; Nakamura et al., 1997; Wong et al., 2003). In the case of SSI mutant lines having different levels of SSI activities, the extent of the differences in chain-length distribution was closely related to the remaining SSI activities (Fujita et al., 2006). However, such a relationship between the remaining PUL activity and the extent of changes in chain-length distribution of amylopectin was not observed.
Fig. 4.
(A) Differences in the chain-length distribution patterns of endosperm amylopectin in the mature endosperm of PUL mutant lines (e10–/–, i16–/–, and EM1003), ISA mutant lines (EM914 and EM653), and the wild-type parent cultivars ‘Nip’ and ‘T65’. (B) Magnification of the pattern of PUL mutant lines in (A). Values for mole% of PUL mutant lines in (A) and (B) for each DP are averages of three (e10–/– and i16–/–) or two (EM1003) seeds arbitrarily chosen from a single homozygous plant. Relative SDs of the mole% of each chain length from DP6-30 was less than 8.3%.
The weight-average molecular weight of amylopectin in e10–/– endosperm starch (1.19×108 g mol−1) determined by the HPSEC-MALLS-RI method was not significantly different from that of Nipponbare (1.21×108 g mol−1).
Thermal and viscoelastic properties of endosperm starch in PUL mutant lines
To evaluate the physico-chemical properties of endosperm starch in PUL mutant lines, the gelatinization temperature of endosperm starch was analysed by differential scanning calorimetry (DSC). The onset temperature (To) of the gelatinization of endosperm starch in e10–/– and i16–/–, the peak temperature (Tp), and the conclusion temperatures (Tc) in e10–/–, i16–/–, and EM1003 were slightly (1–3 °C) lower than those of the control lines. There were no significant differences in the To of EM1003 (Table 3). The gelatinization enthalpies of endosperm starch in PUL mutants were also slightly lower than those of control lines (Table 3). These results are, for the most part, derived from a slight enrichment of short chains in rice PUL mutants, although the other reasons can not be excluded.
Table 3.
Thermal properties of endosperm starch as determined by DSC
| Lines | T0 (°C)a | Tp (°C)b | Tc (°C)c | ΔH (J/g)d |
| Nipponbare | 56.8±0.4 | 63.6±0.4 | 69.0±0.3 | 5.5±0.2 |
| i16–/– | 53.6±1.4 | 61.0±0.5 | 66.8±0.3 | 4.9±0.8 |
| (i16–/–)-Nipponbare | –3.2 | –2.6 | –2.2 | –0.6 |
| e10–/– | 54.9±1.4 | 61.6±0.3 | 67.9±1.2 | 4.1±0.6 |
| (e10–/–)–Nipponbare | –1.9 | –2.0 | –1.1 | –1.4 |
| T65 | 57.6±0.4 | 67.4±0.7 | 76.8±0.5 | 9.2±1.1 |
| EM1003 | 57.9±0.5 | 66.2±0.6 | 74.0±0.6 | 7.1±0.3 |
| EM1003–T65 | +0.3 | –1.2 | –2.8 | –2.1 |
The values are the averages of three replications (mean ±SE). n=3.
Onset temperature.
Peak temperature.
Conclusion temperature.
Gelatinization enthalphy of starch.
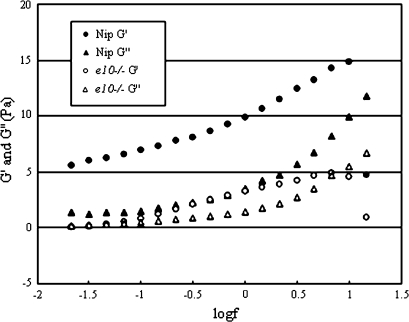
Figure 5 shows the frequency dependence of the storage modulus (G′) and loss modulus (G″) of endosperm starch in Nipponbare and e10–/–. These frequency dependence curves of storage moduli did not show a plateau, but the values of G′ were greater than those of G″ at all frequencies of the endosperm starch paste in Nipponbare. On the other hand, the values of G′ and G″ of the endosperm starch paste in the e10–/– showed no difference at lower frequencies, and these values were slightly increased with increasing frequency. The values of G′ and G″ for the endosperm starch paste in Nipponbare were higher than those of e10–/– at all frequencies. For a polymer system in which the molecular weight and fraction of functional groups are controlled, the changing value and the relationship of G′ and G″ of the frequency would resemble an infinite three-dimensional network (Winter, 1987). Singh et al. (2006, 2008) reported the structural and viscoelastic properties of normal, sugary, and waxy maize starches. These reports suggested that the values of G′ and G″ of these starch pastes reflected the starch structure, but their starch samples had quite different structures. Our results of the viscoelastic properties of endosperm starch pastes in Nipponbare and e10–/– suggest the possibility that these pastes did not form the same aqueous solution, although the differences were not marked. Because starch pastes are generally the function between water and thermal energy, the crystalline structure of these starch molecules collapses by the thermal motion of water molecules. Thus, a slight difference in starch structure would influence their rheological properties.
Fig. 5.
Frequency dependence of the storage modulus (G’) and loss modulus (G'’) for the 4wt% endosperm starch paste in Nipponbare and e10–/– at 50 °C.
Starch properties in double mutant (ISA×PUL mutant) lines
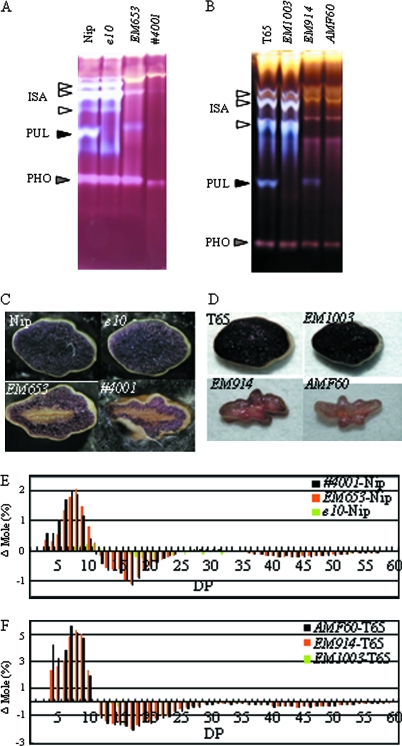
The PUL null mutant lines, e10–/– and EM1003, were crossed with a mild and severe type of the sug1 mutant, EM653 and EM914, respectively, to generate double mutant lines (#4001 and AMF60). The double mutants were analysed to determine if a defect in PUL has an effect on an ISA1-deficient (sug1) background. By native-PAGE/DBE activity staining, a low PUL activity band was observed in EM653 and EM914, but such a band was completely lost in the double mutants (Fig. 6A, B). Our previous studies (Nakamura et al., 1997; Kubo et al., 1999; Wong et al., 2003) indicated that a sugary-1 mutant line, EM914, accumulates almost only phytoglycogen instead of starch, although a small part of the endosperm is stained with an iodine solution, whereas another sugary-1 mutant line, EM653, accumulates sugary-amylopectin and amylose in an iodine-stained starch region within the outer region of the endosperm and only phytoglycogen in a non-stained-phytoglycogen region in the inner region of the endosperm, as shown in Fig. 6C, D. On the other hand, the cross-sections of seeds in the PUL mutant lines were completely stained with the iodine solution, as in the wild type. The cross-section of the #4001 seed was more shrivelled, and the iodine-stained starch region was smaller than that of EM653 (Fig. 6C). The cross-section of the AMF60 seed was also shrivelled in a manner similar to that of EM914, although the iodine-stained area was smaller than that of EM914 (Fig. 6D). The chain-length distribution of the total α-glucans of the double mutant endosperm showed that they had more short chains with DP ≤11 than those of the wild type, as shown by capillary electrophoresis. Moreover, the short chains with 3 ≤DP ≤7 of the total α-glucans of #4001 and AMF60 were increased compared to those of EM653 and EM914, respectively (Fig. 6E, F). The weights of the seeds of #4001 and AMF60 were smaller than those of their sug1 parents. The amounts of WSP of #4001 and AMF60 were 27.7% and 95.0%, respectively, which were also higher than those of the sug1 parents, EM653 (15.3%) and EM914 (87.9%), respectively (Table 1).
Fig. 6.
Characterization of the double mutant lines, #4001 (A, C, E) and AMF60 (B, D, F), in PUL mutant lines (e10–/– and EM1003) and ISA mutant lines (EM653 and EM914), respectively, and the wild type, Nipponbare (Nip), and Taichung65 (T65). (A, B) Native-PAGE/DBE activity staining. The ISA, PUL, and PHO activity bands are indicated by arrowheads. (C, D) Stereo micrographs of the cross-sections of mature endospern stained by iodine solution. (E, F) Differences in the chain-length distribution patterns of endosperm amylopectin in the nature endosperm between the double mutan lines #4001 (E) and AMF60 (F), the PUl mutant lines e10–/– (E) and EM914 (F), and the ISA mutant lines EM653 (E) and EM914 (F) and the wild-type parent cultivars.
Enzyme activity of rPUL using several kinds of substrates
ISA and PUL differ greatly in their substrate specificity (Nakamura, 1996). When pullulan was used as a substrate, the specific activity of rice PUL expressed in E. coli (rPUL) was 29.2 nmol maltose-equivalent μg−1 protein min−1. When amylopectin was used as a substrate, the specific activity of rPUL was only 5% of that toward pullulan. In contrast, the specific activities of rPUL toward the β-limit and φ-limit amylopectin treated by β-amylase or phosphorylase, respectively, were much higher than those toward non-treated amylopectin (Table 4). When β-limit phytoglycogen was used as a substrate, the specific activity was eight times higher than that toward non-treated phytoglycogen isolated from the rice sug1 mutant (EM914), although the specific activity of rPUL toward phytoglycogen was only 1% of that toward pullulan (Table 4).
Table 4.
The enzyme activity of rOsPUL toward several kinds of substrates
| Specific activity | ||
| (nmol maltose equivalent μg−1 protein min−1) | ||
| Pullulan | 29.2±4.0 | (100)a |
| Amylopectin | 1.4±0.7 | (5) |
| β-LD from amylopectin | 19.1±2.0 | (65) |
| φ-LD from amylopectin | 6.3±1.7 | (22) |
| Phytoglycogen | 0.3±0.4 | (1) |
| β-LD from phytoglycogen | 2.4±0.9 | (8) |
The values are the average of 12 independent measurements (means ±SE).
Per cent of activity when pullulan was used as the substrate.
Discussion
Isolation of rice PUL mutant lines and characterization of the structure and physicochemical properties of their endosperm starches
The PUL mutants of Arabidopsis (Atpu1, Wattebled et al., 2005) and maize (zpu1-204, Dinges et al., 2003) have been reported. The analysis of the Atpu1 single-mutant lines did not lead to a distinctive phenotype. However, Iso1 (isoamylase)/pullulanase-defective (Atisa2/Atpu1 double-mutant) lines display a 92% decrease in starch content. This suggests that the function of pullulanase in Arabidopsis partly overlaps with that of Iso1, although its implication remains negligible when Iso1 is present within the leaf cell (Wattebled et al., 2005). Developing zpu1-204 maize endosperm accumulates branched malto-oligosaccharides not found in the wild type. Furthermore, in a background deficient in ISA, zpu1-204 results in a significant accumulation of phytoglycogen in the kernel that is not seen in the wild type, indicating that PUL in maize also partly compensates for the defect in ISA and functions during starch biosynthesis as well as degradation (Dinges et al., 2003).
In this report, three allelic PUL mutant lines were isolated using both reverse genetics and chemical mutagenesis in rice. The α-glucan composition and structure of the endosperm starch in the PUL mutants were comprehensively compared with those of the wild types to understand the function of PUL on starch biosynthesis. The amylose content (Fig. 3A), seed morphology, morphology, and crystallinity of starch granules (data not shown) of the PUL mutant lines were almost the same as those of the wild type. By contrast, slight changes in the chain-length distribution of the mutant lines were detected when measured by capillary electrophoresis (Fig. 4B), although the extent of the changes was much smaller than those in sug1 mutant lines (Fig. 4A). Such changes were not observed in zpu1-204 endosperm (Dinges et al., 2003). On the other hand, amylopectin from the mutant leaves contains significantly fewer chains with 8 ≤DP ≤15 than that from leaves of the wild type because of the pleiotropic reduction of BEIIa activity (Dinges et al., 2003). However, these were not observed in rice PUL mutant lines (data not shown; Fig. 2E), suggesting that pleiotropic effects of PUL mutants between maize and rice were dissimilar. Furthermore, the double mutant lines between the PUL and ISA1 mutant lines, #4001 and AMF60, also contained more WSP and short chains with DP ≤7 of amylopectin in the endosperm than those of the sug1 parents (Fig. 6; Table 1). These results indicate that PUL can partly compensate for the function of ISA1, although its contribution to amylopectin biosynthesis is much smaller than that of ISA1 in rice, as it is in Arabidopsis and maize. On the other hand, it was suggested that the B2 and B3 chains in amylopectin of e10–/– might have a stub of DP2–3 on the basis of two facts: first, that the average chain length in the B2 and B3 chains of e10–/– amylopectin was greater than that of the wild type (Table 2), and second, that rOsPUL showed much higher specific activity toward the β- or φ-limit dextrins of amylopectin (Table 4). These dextrins contain short branches with DP2-4, which are absent in non-treated amylopectin. However, attempts to identify such short stubs in amylopectin of e10–/– by sequential hydrolysis with isoamylase and rOsPUL or with isoamylase and β-amylase were unsuccessful. Therefore, the exact reason for the observed longer CLn of B2+B3 fraction of e10–/– amylopectin remains to be elucidated, and the longer chains might not be the direct consequence of PUL-deficiency but a change that occurred specifically in the mutant line e10–/–. The fact that the thermo-gelatinization (Table 3) and rheological properties (Fig. 5) of the PUL mutant were not the same as those of the wild type implies that there might be other differences in the structure of the endosperm starch than the slight abundance of short chains with DP ≤13 between the rice PUL mutant and the wild type.
Function of PUL on starch biosynthesis
The production of phytoglycogen and sugary-amylopectin instead of normal amylopectin in all of the sug1 mutant lines is caused by an extremely low ISA activity (less than 1% of that of the wild type) in the rice endosperm; however, the sugary1 phenotypes are not always related to the ISA activity level (Kubo et al., 1999). For example, partially purified ISA1 from EM41, which is a severe type of the sug1 mutant, has higher activity than that from EM5, which is mild type of mutant (Kubo et al., 1999). By contrast, the amount of the PUL protein and PUL activity is closely related to the magnitude of the starch region in allelic sug1 mutant lines (Nakamura et al., 1997; Kubo et al., 1999), although the mRNA expression level of the rice PUL gene in these lines is unaffected, as detected by Northern blotting (Kubo, 2001). These results suggested that the high PUL activity is responsible for the formation of amylopectin-like α-glucans instead of phytoglycogen in the starch region and that both types of starch debranching enzymes, ISA and PUL, are involved in the construction of the amylopectin fine structure in the rice endosperm (Kubo et al., 1999).
To determine whether or not the formation of starch is recovered by PUL activity, a mild type of sug1 rice mutant, EM653, which has relatively high PUL activity in the developing endosperm, was crossed with the PUL null mutant, e10–/– (Fig. 6A, C, E). If the above hypothesis was correct, the resulting double mutants would be expected to show the accumulation of phytoglycogen and the concomitant disappearance of starch regions in a cross-section of the endosperm, resulting in a severe sug1 phenotype. The double mutant #4001, however, accumulated an iodine-stained starch region within the outer region of the endosperm, although the accumulation of WSP in the #4001 endosperm was slightly higher than that in EM653 (Fig. 6C; Table 1). These results strongly suggest that the PUL activity was not related with the amount of starch regions in the rice endosperm. It is necessary to determine if the phenotype of the transgenic rice remains unaffected even when the PUL gene is introduced and over-expressed in a severe sug1 mutant line.
The issue of what affects the severity of the sug1 phenotype in allelic rice sug1 mutant lines remains to be resolved. One possibility is that the ISA activity level in vivo in the sug1 mutant lines is responsible for their sugary1 phenotype, although Kubo et al. (1999) reported that the ISA activity levels based on the method of native-PAGE/DBE activity staining were not related to the sugary1 phenotype. There are a few corroborations; in EM914, which contains a stop codon in exon 6 of the ISA1 gene, the mRNA expression was completely repressed, and its sugary1 phenotype was very severe, whereas EM653, which displayed a normal level of mRNA expression by RT-PCR, showed a mild sugary1 phenotype (Kubo, 2001). It seems that only less than 1% of the remaining ISA activity of the wild type might be sufficient to recover the formation of starch from phytoglycogen (Fujita et al., 2003) and the narrow gradation range from zero to 1% of the ISA activity level might be related to the sugary1 phenotype. It is necessary to identify the mutations of the sug1 allelic mutant lines and to evaluate the ISA activity, including the ratio of ISA1 to ISA2 by the mutations.
It is important to determine the reason that the amount of starch accumulation in the endosperm was closely related to the PUL activity levels in the sug1 rice mutant lines. It is more likely that the decrease in the amount of iodine-stained starch leads to a reduction in the PUL activity and not that the low PUL activity results in the reduction in the amount of iodine-stained starch. The lower PUL activity in rice sug1 mutants is recovered to the normal level, when the starch is synthesized by the introduction of wheat (Kubo et al., 2005) or rice ISA1 genes (Y Utsumi et al., unpublished data) into the mutants. Furthermore, the mRNA levels of the PUL gene in the rice sug1 mutant lines (Kubo, 2001) and maize su1 mutants (Beatty et al., 1999) were not different from those of the wild type, although the protein level of PUL was reduced as well as the PUL activity in the rice sug1 mutant lines (Nakamura et al., 1997). These results imply that PUL might be unstable and susceptible to degradation by proteases under the conditions in which phytoglycogen is predominantly accumulated.
The presumed function of ISA is the trimming of improper branches of amylopectin formed by branching enzymes to keep an integrated cluster structure of amylopectin during starch biosynthesis (Nakamura, 2002). PUL appears to compensate for the trimming function of ISA, since the short chains with DP ≤12 and DP ≤7 of amylopectin were increased in the PUL mutations on the background of the wild-type and sug1 mutants, respectively. Both debranching enzymes showed activity at different stages; high activities of ISA and PUL were observed at the early-to-middle stages and the middle-to-late stages, respectively, during the development of the rice endosperm (Fig. 1A). Moreover, both enzymes have different substrate specificity; PUL can debranch the short branched chains with DP ≤2–4 more easily than ISA. These results indicate that both debranching enzymes play a different role of debranching on starch biosynthesis.
The double mutant AMF60 contained trace amounts of starch-like α-glucans in the endosperm, which were stained with iodine (Table 1). This suggests that another debranching enzyme, ISA3, which is expressed mainly in the leaf (Ohdan et al., 2005), might also contribute to storage starch synthesis. More detailed analysis, including germination for understanding DBEs in plants, is needed.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primer list for sequencing analysis of OsPUL gene.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Jay-Lin Jane and Dr Jin-Hee Park (Iowa State University, USA) for help in measurement of the molecular mass of amylopectin, to Ms Aiko Nishi (Kyusyu University), Dr Akiko Kubo (Osaka Prefecture University) and Dr Hiroyuki Iwamoto (Fukuyama University), for helpful discussions, and to the Biotechnology Center in Akita Prefecture University for DNA sequencing.
References
- Beatty MK, Rahman A, Cao H, Woodman W, Lee EP, Myers AM, James MG. Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiology. 1999;119:255–266. doi: 10.1104/pp.119.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Jenner H, Carrangis L, et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. The Plant Journal. 2002;31:97–112. doi: 10.1046/j.1365-313x.2002.01339.x. [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Mestre V, Colleoni C, Slomianny MC, Mouille G, Delrue B, d'Hulst C, Bliard C, Nuzillard JM, Ball S. The debranching enzyme complex missing in glycogen accumulating mutants of Chlamydomonas reinhardtii displays an isoamylase-type specificity. Plant Science. 2000;157:145–156. doi: 10.1016/s0168-9452(00)00256-9. [DOI] [PubMed] [Google Scholar]
- Dinges JR, Colleoni C, James MG, Myers AM. Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. The Plant Cell. 2003;15:666–680. doi: 10.1105/tpc.007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JD, Robyt JF. Miniaturization of three carbohydrate analyses using a microsample plate reader. Analytical Biochemistry. 1991;195:93–96. doi: 10.1016/0003-2697(91)90300-i. [DOI] [PubMed] [Google Scholar]
- Francisco PB, Jr, Zhang Y, Park S-Y, Ogata N, Yamanouchi H, Nakamura Y. Genomic DNA sequence of a rice gene coding for a pullulanase-type of starch debranching enzyme. Biochemica et Biophysica Acta. 1998;1387:469–477. doi: 10.1016/s0167-4838(98)00154-x. [DOI] [PubMed] [Google Scholar]
- Fujita N, Hasegawa H, Taira T. The isolation and characterization of a waxy mutant of diploid wheat (Triticum monococcum L.) Plant Science. 2001;160:595–602. doi: 10.1016/s0168-9452(00)00408-8. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Francisco PB, Jr, Nakakita M, Harada K, Minaka N, Nakamura Y. Purification, characterization, and cDNA structure of isoamylase from developing endosperm of rice. Planta. 1999;208:283–293. doi: 10.1007/s004250050560. [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Suh S-D, Wong K-S, Jane J-L, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant and Cell Physiology. 2003;44:607–618. doi: 10.1093/pcp/pcg079. [DOI] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiology. 2006;140:1070–1084. doi: 10.1104/pp.105.071845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, et al. Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiology. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashiro I, Tagawa M, Shibahara S, Iwata K, Takeda Y. Examination of molar-based distribution of A, B, and C chains of amylopectin by fluorescent labeling with 2-aminopyridine. Carbohydrate Research. 2002;337:1211–1215. doi: 10.1016/s0008-6215(02)00110-6. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Contribution of the Tos17 retrotransposon to rice functional genomics. Current Opinion in Plant Biology. 2001;4:118–122. doi: 10.1016/s1369-5266(00)00146-1. [DOI] [PubMed] [Google Scholar]
- Inouchi N, Glover DV, Fuwa H. Chain length distribution of amylopectins of several single mutants and the normal counterpart, and sugary1 phytoglycogen in maize (Zea mays L.) Starch. 1987;39:259–266. [Google Scholar]
- James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. The Plant Cell. 1995;7:417–429. doi: 10.1105/tpc.7.4.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata A, Akuzawa S, Ishii Y, Yazaki T, Otsubo Y. Sol-gel transition and elasticity of starch. Bioscience, Biotechnology and Biochemstry. 1996;60:567–570. [Google Scholar]
- Kawasaki T, Mizuno K, Shimada H, Satoh H, Kishimoto N, Okumura S, Ichikawa N, Baba T. Coordinated regulation of the genes participating in starch biosynthesis by the rice floury-2 locus. Plant Physiology. 1996;110:89–96. doi: 10.1104/pp.110.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A. The analysis and regulation of amylopectin biosynthesis using rice sugary1 mutant lines. Japan: Chiba University; 2001. PhD thesis. [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiology. 1999;121:399–409. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Rahman S, Utsumi Y, et al. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiology. 2005;137:43–56. doi: 10.1104/pp.104.051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Hirochika H. Applications of retrotransposons as genetic tools in plant biology. Trends in Plant Science. 2001;6:127–134. doi: 10.1016/s1360-1385(00)01860-4. [DOI] [PubMed] [Google Scholar]
- Mouille G, Maddelein M-L, Libessart N, Talaga P, Decq A, Delrue B, Ball S. Pre-amylopectin processing: a mandatory step for starch biosynthesis in plants. The Plant Cell. 1996;8:1353–1366. doi: 10.1105/tpc.8.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Reserch. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y. Some properties of starch debranching enzymes and their possible role in amylopectin biosynthesis. Plant Science. 1996;121:1–18. [Google Scholar]
- Nakamura Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant and Cell Physiology. 2002;43:718–725. doi: 10.1093/pcp/pcf091. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Kubo A, Shimamune T, Matsuda T, Harada K, Satoh H. Correlation between activities of starch debranching enzyme and α-polyglucan structure in endosperms of sugary-1 mutants of rice. The Plant Journal. 1997;12:143–153. [Google Scholar]
- Nakamura Y, Umemoto T, Ogata N, Kuboki Y, Yano M, Sasaki T. Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: purification, cDNA and chromosomal localization of the gene. Planta. 1996;199:209–218. doi: 10.1007/BF00196561. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yuki K, Park SY, Ohya T. Carbohydrate metabolism in the developing endosperm of rice grains. Plant and Cell Physiology. 1989;30:833–839. [Google Scholar]
- Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. Journal of Biological Chemistry. 1944;153:375–380. [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. Journal of Experimental Botany. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3, 6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–688. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- Pan D, Nelson OE., Jr A debranching enzyme deficiency in endosperms of the sugary1 mutants of maize. Plant Physiology. 1984;74:324–328. doi: 10.1104/pp.74.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Omura T. Induction of mutation by treatment of fertilized egg cell with N-methyl-N-nitrosourea in rice. Journal of the Faculty of Agriculture, Kyushu University. 1979;24:165–174. [Google Scholar]
- Schoch LK. Purification of starch and the starch fractions. Methods in Enzymology. 1954;3:5–6. [Google Scholar]
- Singh N, Inouchi N, Nishinari K. Structural, thermal and viscoelastic characteristics of starches separated from normal, sugary and waxy maize. Food Hydrocolloids. 2006;20:923–935. [Google Scholar]
- Singh N, Isono N, Srichuwong S, Noda T, Nishinari K. Structural, thermal and viscoelastic properties of potato starches. Food Hydrocolloids. 2008;22:979–988. [Google Scholar]
- Smith AM, Zeeman SC, Smith SM. Starch degradation. Annual Review of Plant Biology. 2005;56:73–98. doi: 10.1146/annurev.arplant.56.032604.144257. [DOI] [PubMed] [Google Scholar]
- Somogyi M. Notes on sugar determination. Journal of Biological Chemistry. 1952;195:19–23. [PubMed] [Google Scholar]
- Suzuki T, Eiguchi M, Kumamaru T, Satoh H, Matsusaka H, Moriguchi K, Nagato Y, Kurata N. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Molecular Genetics and Genomics. 2008;279:213–223. doi: 10.1007/s00438-007-0293-2. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Hizukuri S, Juliano BO. Purification and structure of amylose from rice starch. Carbohydrate Research. 1986;148:299–308. [Google Scholar]
- Utsumi Y, Nakamura Y. Structural and enzymatic characterization of the isoamylase1 homooligomer and the isoamylase1-isoamylase2 heterooligomer from rice endosperm. Planta. 2006;225:75–87. doi: 10.1007/s00425-006-0331-z. [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, et al. Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiology. 2005;138:184–195. doi: 10.1104/pp.105.059295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K-S, Kubo A, Jane J-L, Harada K, Satoh H, Nakamura Y. Structures and properties of amylopectin and phytoglycogen in the endosperm of sugary-1 mutants of rice. Journal of Cereal Science. 2003;37:139–149. [Google Scholar]
- Winter HH. Can the gel point of a cross-linking polymer be detected by the G'–G'’ crossover? Polymer Engineering and Science. 1987;27:1698–1702. [Google Scholar]
- Yamamoto K, Sawada S, Onogaki I. Properties of rice starch prepared by alkali method with various condition. Denpun Kagaku. 1973;20:99–104. [Google Scholar]
- Yamamoto K, Sawada S, Onogaki I. Effects of quality and quantity of alkali solution on the properties of rice starch. Denpun Kagaku. 1981;28:241–244. [Google Scholar]
- Yamanouchi H, Nakamura Y. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant and Cell Physiology. 1992;33:985–991. [Google Scholar]
- Yano M, Isono Y, Satoh H, Omura T. Gene analysis of sugary and shrunken mutants of rice, Oryza sativa L. Japanese Journal of Breeding. 1984;34:43–49. [Google Scholar]
- Zeeman SC, Umemoto T, Lue W-L, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. The Plant Cell. 1998;10:1699–1711. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.