Abstract
While most dicot plants produce little ethylene in their vegetative stage, many monocots such as rice liberate a relatively large amount of ethylene with cyanide as a co-product in their seedling stage when etiolated. One of the known functions of β-cyanoalanine synthase (CAS) is to detoxify the co-product cyanide during ethylene biosynthesis in higher plants. Based on a tryptic peptide sequence obtained from a partially purified CAS activity protein preparation in etiolated rice seedlings, the full-length putative rice CAS-encoding cDNA sequence (OsCAS), which is homologous to those O-acetylserine sulphydrylase (OASS) genes, was cloned. Unlike most of the CAS genes reported from dicots, the transcription of OsCAS is promoted by auxins but suppressed by ethylene. To address the function and the subcellular localization of this gene product in planta, a binary vector construct consisting of this gene appended with a yellow fluorescent protein-encoding sequence was employed to transform Arabidopsis. Specific activities on CAS and OASS of the purified recombinant protein from transgenic Arabidopsis were 181.04 μmol H2S mg−1 protein min−1 and 0.92 μmol Cys mg−1 protein min−1, respectively, indicating that OsCAS favours CAS activity. The subcellular localization of OsCAS was found mostly in the mitochondria by immunogold electron-microscopy. Chemical cross-linking and in-gel assay on a heterodimer composed of functional and non-functional mutants in a yeast expression system on OsCAS suggested that OsCAS functions as a homodimer, similar to that of OASS. Despite the structural similarity of OsCAS with OASS, it has also been confirmed that OsCAS could not interact with serine-acetyltransferase, indicating that OsCAS mainly functions in cyanide detoxification.
Keywords: Cyanide, β-cyanoalanine synthase, ethylene, rice, serine acetyltransferase
Introduction
Cyanide is a toxic molecule generated in plants either by hydrolysis of cyanogenic glycosides in ‘cyanogenic’ species or from oxidation of 1-aminocyclopropane-1-carboxylic acid (ACC) during phyto-hormone ethylene biosynthesis. Most of the plants do not contain cyanogenic glycosides, but most of the higher plant species generate cyanide along with ethylene from ACC by the action of ACC oxidase (ACO) (Yip and Yang, 1988). Cyanide exerts its toxicity by inhibiting vital functions in cells including respiration (Grossmann, 1996; Solomonson, 1982).
Cyanide detoxification in plants can be handled by two enzymes including β-cyanoalanine synthase (CAS, EC 4.4.1.9) and rhodanese (EC 2.8.1.1). CAS has been demonstrated widely in higher plants (Blumenthal et al., 1968; Miller and Conn, 1980), insects (Ogunlabi and Agboola, 2007), and bacteria (Dunnill and Fowden, 1965; Castric and Strobel, 1969); while rhodanese has been found from animals sources (Jones, 1998), bacteria (Westley, 1973), and also in a few higher plant species (Tomati et al., 1972; Chew, 1973). When feeding higher plants with either 14C-labelled cyanide or 14C-labelled ACC at C-1, radioactive carbon would first be incorporated into β-cyanoalanine and then converted to asparagine by a hydratase (Peiser et al., 1984). Later, Yip and Yang (1988) demonstrated the importance of CAS in cyanide metabolism during active ethylene biosynthetic conditions such as in fruit ripening, organ senescence, auxin-induction, and in various stress conditions, etc. Thus, it is widely accepted that cyanide detoxification in higher plants is mainly by CAS activity.
Purification and kinetic studies on CAS from plants were carried out by Hendrickson and Conn (1969). They demonstrated that CAS is a pyridoxal-5′-phosphate (PLP)-dependent enzyme which also possesses cysteine synthase activity but favours CAS activity. By contrast, a closely related group of PLP-dependent O-acetylserine sulphydrylases (OASS: EC 4.2.99.8) are homologous with CAS (Hatzfeld et al., 2000; Maruyama et al., 2001), but favours cysteine synthase activity (equivalent to OASS activity) and also CAS activity. That a specific lysine residue facilitates PLP binding on spinach OASS was determined by complementation in cysteine-deficient bacteria with SpOASS mutants on the lysine residue (Saito et al., 1993). Hence, it is highly possible that the same lysine residue on CAS protein may form a Schiff base, facilitating the β-replacement in the CAS reaction.
Subcellular localization of various OASS has been found in organelles including cytosol, chloroplast, and mitochondrion in plants by immuno-detection on protein isolated from subcellular fractionation in spinach (Takahashi and Saito, 1996) and Arabidopsis (Hesse et al., 2004), in vitro import assay to mitochondria in Arabidopsis (Hesse et al., 2004), or OASS activity assay on extracts from different subcellular fractionations in Datura innoxia Mill. (Kuske et al., 1996). CAS has been determined in mitochondria exclusively by mitochondrial fractionation in barley leaves (Wurtele et al., 1985) and blue lupin seedlings (Akopyan et al., 1975). Due to the main difference in subcellular localizations among these two proteins, it is hypothesized that CAS should localize better to mitochondria for effective removal of cyanide so as to protect the oxidative phosphorylation process; and that the optimal pH for CAS activity is around 8.5 which is also the pH in the matrix of mitochondria. Some authors have suggested that CAS could be regarded as OASS-like protein located in mitochondria, but in Arabidopsis two different genes coding for CAS and OASS co-exist in the mitochondria (Jost et al., 2000). Therefore, it is inappropriate to assign CAS and OASS to a subcellular location based on activity assays and immunoassays on the subcelluar fractions, as both types of proteins possess overlapping structural and functional properties. A better approach to determine their subcellular location in plant cells would be to visualize OASS/CAS fused with a fluorescent protein.
Beside the overlapping structural and functional properties of CAS and OASS, both proteins are supposed to work in sequence in vivo. CAS removes cyanide by combining it with cysteine to form β-cyanoalanine, the displaced sulphide being recycled back to cysteine by the action of OASS. It is widely accepted that cysteine synthesis is a highly regulated process that is catalysed by a cysteine synthase complex comprising of serine acetyltransferase (SAT) and OASS in bacteria and plants (Kredich, 1971; Nakamura et al., 1987; Droux et al., 1992, 1998; Liszewska et al., 2005). It is not known whether CAS could also interact with SAT, and whether certain OASS/CAS gene family member(s) could possess a dual function in vivo. Based on the OASS/CAS mutant study in Arabidopsis, K Saito's group speculated that some OASS proteins in cytoplasm can function in cyanide detoxification (Watanabe et al., 2008).
Although the importance of CAS in cyanide detoxification during ethylene biosynthesis has been postulated for almost two decades (Yip and Yang, 1988), there have been very few studies on CAS in plant species with high ethylene biosynthetic rate except one recently on apple fruit (Han et al., 2007). A high rate of ethylene biosynthesis (2 nmol g−1 h−1) is found in rice seedlings under etiolated growth; this is comparable to ripening apple (2–5 nmol g−1 h−1). In these tissues, ample capacity of CAS activity was also found. By following the CAS activity in the etiolated rice (Oryza sativa) seedlings with a chromatograph scheme and based on a tryptic sequence of the partially purified CAS preparation, it has been possible to isolate OsCAS used in this study. The aim is to establish the relevance of this gene-encoding protein in cyanide detoxification in rice. By visualizing the recombinant OsCAS protein expressed in Arabidopsis, the evidence that OsCAS encodes an authentic CAS is provided here by satisfying three consensus criteria: (i) a high CAS to OASS activity ratio with mM to sub-mM range Km for cyanide (Hatzfeld et al., 2000; Maruyama et al., 2001; Han et al., 2007); (ii) a severe inhibition on CAS activity at [HCN] above 10 mM (Jost et al., 2000; Warrilow and Hawkesford, 2000); and (iii) CAS should localize to mitochondria (Wurtele et al., 1985). To delineate CAS from OASS in the OASS/CAS gene families further, chemical cross-linking and mutagenesis experiments have been conducted on the products of these genes. The native assembly and amino acid residues important for OsCAS activity were examined.
Materials and methods
Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana; Columbia ecotype 0) plants were allowed to germinate on MS (Murashige, 1962) agar plates with an antibiotic selection. Resistant seedlings were transferred to soil and kept in a growth chamber at 22 °C under a 16/8 h photoperiod at 264 μmol m−2 s−1. After 3 weeks, leaves were harvested at a stage before flowering and material was used for identification of OsCAS transgenic lines and confirmation of authentic OsCAS protein function in these transgenic lines.
RNA extraction and the northern blotting on transgenic lines
Total RNA was extracted using the hot phenol extraction method (Katharina, 1995). Twenty micrograms of total RNA was separated in a 1.2% formaldehyde gel in MOPS buffer pH 7 and 5% formaldehyde and then blotted onto a piece of Hybond-N+ membrane (GE Healthcare). After cross-linking under an ultraviolet lamp, the blot was prehybridized with denatured salmon sperm DNA at 42 °C for 6 h with rotation. During prehybridization, radioactive probe was synthesized in vitro by using the Random Primed DNA Labelling Kit (Roche) with radioactive [32P]dCTP following the manufacturers’ instruction. The purified denatured probe was added on the blot with rotation overnight. After washing steps, the blot with an X-ray film (Fujifilm) was locked in a cassette and this cassette was kept in –80 °C for several days. Film was developed by an X-ray processor (Kodak X-OMAT).
Genomic DNA extraction from transgenic lines and Southern blot analysis
Genomic DNA was extracted from 3 g of leaves from transgenic lines with cetyltrimethylammonium bromide (Allen et al., 2006) and then digested with the two restriction enzymes SacI and EcoRV at 37 °C overnight. After agarose gel electrophoresis at a constant 40 V, digested genomic DNA in gel was denatured in alkali prior to blotting. Probe synthesis with radioactive labelling, hybridization, and imaging were as described in the preceding section.
Protein extraction from Arabidopsis transgenic lines
Leaves were ground in liquid nitrogen and resuspended in extraction buffer containing 50 mM TRIS-HCl (pH 8.5), 5 mM EDTA, 10 μM pyridoxal-5-phosphate, and 30% glycerol, the homogenate was kept on ice for 2 min with vortex and centrifuged at 17 900 g for 10 min sequentially. The supernatant was filtered through two layers of nylon net filters NY80 (Millipore) and the filtrate was saved for enzyme activity determination and OsCAS–yellow fluorescent protein (YFP) purification. All steps were performed at 4 °C unless otherwise specified.
Measurement of ethylene production from rice seedlings by gas chromatography
Ten rice seedlings (∼3 cm from apex of seedling) were harvested and collected in a sealed 6 ml glass tube filled with 200 μl water. The tube was incubated either in light or dark for 1 h and then 1 ml of gas from the tube was injected into the gas chromatography (Hewlett-Packard 5890 Series II; Hewlett-Packard, Wilmington, DE, USA) according to Yip and Yang (1988). The concentration of ethylene was presented in parts per million (ppm) and the rate of ethylene production from seedlings was determined as a unit of nmol g−1 h−1.
N-terminal protein sequencing and protein identification by tryptic mass fingerprinting
N-terminal sequencing of mature OsCAS–YFP protein from transgenic lines was conducted on a protein sequencer (Hewlett-Packard G1000A). Purified OsCAS–YFP protein separated in 10% SDS–PAGE was blotted on Immobilon™-P (Millipore) according to the user guide from Millipore and then the membrane was stained in Coomassie blue. The protein band of OsCAS–YFP was excised and saved for N-terminal protein sequencing.
Immuno-precipitated protein bands were identified by tryptic mass fingerprinting according to Luk et al. (2005). Peptide masses were determined by a MALDI–TOF MS (Voyager-DE™ STR Biospectrometry™ Workstation; Applied Biosystems, Foster City, CA, USA). Peptide mass lists were searched against the online database of the National Center for Biotechnology with limitation on species by program MS–FIT (http://prospector.ucsf.edu). Program parameters were selected as follows: monoistopic peptide masses adopted, mass tolerance set as 100 ppm, and allowance of one missed cleavage.
In-gel CAS activity assay, supershift in gel CAS activity assay, CAS activity assay, OASS activity assay, and SAT activity assay
Twenty micrograms of protein extract from Arabidopsis transgenic lines were incubated with a series dilution of polyclonal anti-green fluorescent protein (GFP) antibodies (Invitrogen) on ice for 30 min and then centrifuged at 17 900 g for 1 min for supershift in-gel CAS activity assay or no antibody was mixed with protein extract from these lines for original in-gel CAS activity assay. Supernatants were separated in a 10% native PAGE. The in-gel CAS activity assay in slab gel was conducted, based on the methods described by Akopyan et al. (1975), after electrophoresis, except CAS activity staining was done at 35 °C for 30 min. CAS activity and OASS activity assays were performed as described by Yip and Yang (1988) and Lunn et al. (1990), while the SAT activity assay was conducted following a spectrophotometric assay of Bonner et al. (2005). Determination of enzymatic activities was repeated at least in triplicate.
Protein purification and quantification of OsCAS–YFP from Arabidopsis transgenic lines
Native purification of OsCAS–YFP was conducted using uMAC™ epitope tag protein isolation kit with polyclonal anti-GFP microbeads (Miltenyl Biotec) following the manufacturer's instructions. To perform each immuno-purification, protein extract from 1 g of leaves was required. Protein extract was incubated with anti-GFP microbeads in ice for 30 min and the protein–antibody bead complex was loaded in a column under a strong magnetic force. After column washing steps, purified OS CAS–YFP was eluted in native elution buffer containing 50 mM TRIS-HCl (pH 8.5), 5 mM EDTA, 10 μM PLP, and 30% glycerol without magnetic interaction. The brown eluate was collected for CAS activity assay or denative electrophoresis.
Protein quantification of pure OsCAS–YFP in eluate was estimated by a PharoxFX™ molecular imager system (BIO-RAD), based on Coomassie blue staining on SDS–PAGE with bovine serum albumin as a standard.
Immunoblotting
Protein samples from either total protein from Arabidopsis or purified OsCAS–YFP eluate were separated in a 10% (w/v) acrylamide gel by SDS–PAGE (Laemmli, 1970). Proteins were subsequently transferred to Hybond ECL nitrocelluolose membrane (Amersham Biosciences) using Hoefer Semiphor (Pharmacia Biotech) at a maximum current of 100 mA for 1 h. Rabbit polyclonal anti-GFP antibodies (Invitrogen) and donkey ECL horseradish peroxidase-conjugated anti-rabbit IgG antibodies (Amersham Biosciences) were both applied at 1:5000 dilution. Signals were captured on a sheet of autoradiography film after incubation with detection reagents (Amersham Biosciences).
Generation of OsCAS–YFP overexpression Arabidopsis transgenic lines
A full-length CAS encoding sequence was amplified with primers 5′-GGATCCATGGAGAGGCTCTGATGAGCCTCATGA-3′ and 5′-CTCGAGTTAGTCCACTGGCACTGGCTGATGGCCT-3′ from pCR OsCAS and then cloned into pCR-Blunt II-TOPO vector. This full-length sequence was recovered by restriction enzyme digestion with SalI and KpnI and this fragment was subcloned into XhoI- and KpnI-digested pBA002-EYFP vector. Expression of OsCAS tagged with EYFP at the C-terminal was driven by the CaMV 35S promoter in vector pBA002-EYFP. The construct was mobilized into Agrobacterium tumefaciens ABI strain by freeze–thaw transformation (Chen et al., 1994) and introduced to Arabidopsis ecotype (Col-0) in planta by a floral dip (Clough and Bent, 1998).
Chemical cross-linking and immunoblotting of Os CAS–YFP complex in crude leaf extract
Various concentrations of bis(sulphosuccinimidyl) suberate (BS3) (Pierce) were added to 2 mg ml−1 of Arabidopsis crude leaf protein extracted in conjugated buffer containing 100 mM sodium carbonate (pH 8.5), 5 mM EDTA, 10 μM PLP, and 30% glycerol. The cross-linking reaction was carried out at 0 °C for 2 h and was quenched subsequently by adding 50 mM TRIS (pH 8.0) for 30 min at room temperature. Cross-linked protein was precipitated from the reaction mixture by adding 1 volume of 20% of trichloroacetic acid for 45 min at –20 °C. After high-speed centrifugation at 35000 g for 15 min, protein precipitate was washed by 1 volume of acetone containing 1 mM phenylmethylsulphonyl fluoride and 2 mM EDTA twice and lyophilized for 1 h. Protein precipitate was dissolved in 1× electrophoresis sample buffer at room temperature overnight. A protein sample (40 μg lane−1) was analysed in a 10% (w/v) acrylamide gel by SDS–PAGE (Laemmli, 1970). Proteins in PAGE were transferred to Hybond ECL nitrocelluolose membrane (Amersham Biosciences) and detected by a rabbit polyclonal anti-GFP antibody.
Cryo-EM immunolocalization
Immunogold labelling of Arabidopsis cells was carried out as described previously (Tse et al., 2004). Briefly, transgenic Arabidopsis seeds (OsCAS–YFP line 4 and YFP line 1) were subjected to germination on an MS agar plate with BASTA for 3–4 d before root tips (about 0.2 cm in length) were collected and fixed in 2 ml of fixative solution containing 0.25% (v/v) glutaraldehyde and 1.5% (v/w) paraformaldehyde in 50 mM phosphate buffer, pH 7.4, for 15 min at room temperature, and then transferred to 4 °C for an additional 16 h. After washing with phosphate buffer at room temperature, cells were dehydrated in an ethanol series and then embedded in Lowicryl HM20 resin. Ultra-thin sections were then prepared from the Lowicryl HM20 blocks, followed by blocked in blocking buffer PBS (80 mM disodium hydrogen orthophosphate, 20 mM sodium dihydrogen orthophosphate, 100 mM sodium chloride) with 3% BSA. Treated samples were then incubated with GFP antibodies (at 10 μg ml−1 diluted with PBS containing 1% BSA) for 1 h, followed by washing with buffer (PBS containing 1% BSA) before incubation with gold-conjugated secondary antibodies (at 1:50 diluted with PBS containing 1% BSA). Labelled samples were post-stained with uranylacetate before the stained specimens were examined in a JOEL JEM-1200EX II transmission electron microscope (JOEL, Tokyo, Japan) operating at 80 kV as described (Tse et al., 2004). The GFP antibodies used in this study were prepared by injecting recombinant GFP into rabbits at the animal house of The Chinese University of Hong Kong and affinity-purified.
Protein expression in budding yeast
Recombinant protein was overexpressed in yeast transformant (strain INVSc1) with a pYES2 vector (Invitrogen) inserting a coding DNA sequence (for OsCAS expression, the N-terminal 35 amino acid residues encoding sequence was deleted). Protein expression was induced in minimal medium with galactose at 30 °C overnight with continuous shaking. Yeast cells were disrupted in breaking buffer containing 50 mM TRIS-HCl (pH 8.5), 5 mM EDTA, 10 μM PLP, and 30% glycerol by vortexing with glassbeads at 4 °C. These steps followed the procedures given in the manufacturer's manual (Invitrogen).
Miscellaneous molecular techniques
A list of miscellaneous molecular techniques including polymerase chain reaction, restriction enzyme digestion, ligation, and plasmid transformation into Escherichia coli, etc. were conducted according to Sambrook and Russell (2001). Silver staining on SDS–PAGE was carried out following the Vorum silver-staining method (Mortz et al., 2001), while recombinant His-tagged protein in yeast was purified by a nickel column according to the instructions in the manufacturer's manual (Invitogen).
Results
Alignment of the amino acid sequence of OsCAS with those of CAS in other plant species
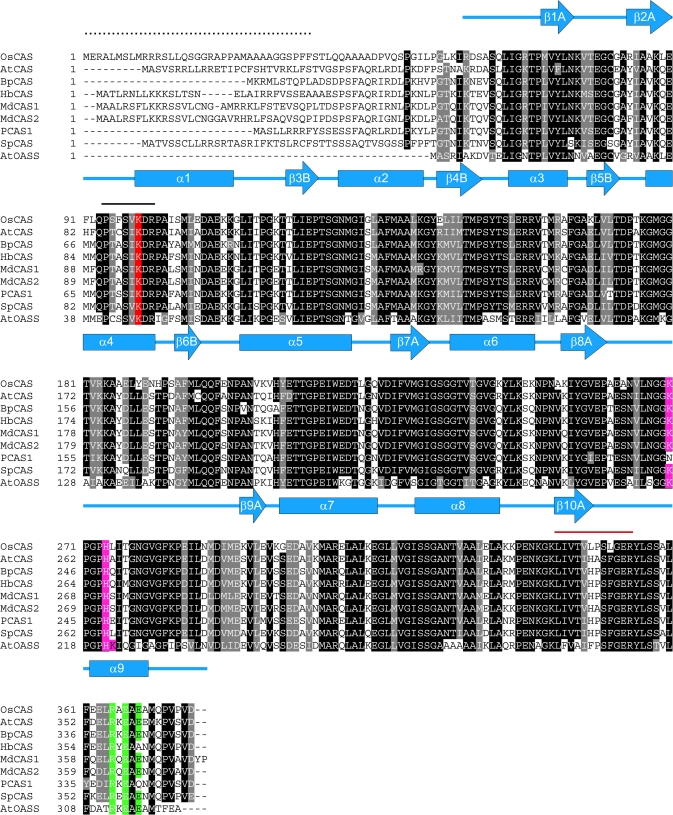
By following the increase in CAS activity through chromatography (starting from 31.02 μmol min−1 to 4.56 μmol min−1 after Mono Q column with a purification fold of 98%), CAS proteins were successfully purified in near homogeneity from etiolated rice seedlings. A tryptic peptide (LIVTVLPSLGER), a digestion product from this protein on a denative 2-D gel, could be resolved by MS/MS equipped with a nanospray source. Searching the database from the translated rice genome, this dodecapeptide was found to be a complete match with a protein encoded by a putative rice CAS gene (OsCAS) but not with other members in the rice OASS/CAS gene family. This putative OsCAS also contained a highly conserved PLP-binding domain (PXXSV/IKDR) as do other members in the rice OASS/CAS gene family in which the lysine residue may form a Schiff base with the PLP co-factor. By using primers according to the nucleotide sequence in the database, a full-length 1134 bp cDNA encoding OsCAS was amplified by PCR. The gene-encoding sequence and protein sequence of OsCAS have been deposited on Genbank® with accession numbers AY720933 and AAV48542, respectively. This translated protein consists of 378 amino acids and its sequence shows 69.3–71.2% homology with those CAS sequences from other plant species in the polypeptide alignment (Fig. 1). The N-terminal amino acid sequence of the mature OsCAS protein harvested from those OsCAS heterologous expression Arabidopsis lines has been sequenced by Edman degradation. The result revealed that the first 35 amino acid residues from the N-terminal of OsCAS were cleaved (data not shown), hence the molecular mass of the mature OsCAS monomer is 36.5 kDa. This cleaved peptide possibly serves as the mitochondrial targeting peptide based on the result of EM immunolocalization described in the following results.
Fig. 1.
Alignment of sequences of OsCAS and other CAS from plant species with AtOASS from Arabidopsis. Comparison of the derived amino acid sequence of OsCAS was made with those of the Arabidopsis AtCAS (Hatzfeld et al., 2000), spinach SpCAS (Hatzfeld et al., 2000), potato PCAS1 (Maruyama et al., 2001), birch BpCAS (GeneBank accession number AY154650), latex (Hevea brasilienis) HbCAS (GeneBank accession number AAP41852), Fuji apple MdCAS1, MdCAS2 (Han et al., 2007), and Arabidopsis AtOASS (EMBL number CAA56593). Amino acid residues that are conserved in at least eight of the nine sequences are shown in black. Secondary structures of AtOASS are represented in α-helices (blue rectangles) and β-stands (blue arrows). The solid line in black shows the conserved pyridoxal 5′ phosphate binding site while the red line covers a peptide from the partially purified rice CAS fraction sequenced by MS/MS. The mitochondrial target sequence from OsCAS which was determined by N-terminal Edman degradation on a mature OsCAS protein purified from a heterologous expression of Arabidopsis plant is indicated by dots. Amino acid residues which were chosen in this mutagenesis study are highlighted: Lysine in the PLP-binding domain is shaded in red while conserved amino acid residues for SAT interaction and dimerization are shaded in purple and green, respectively.
Expression of OsCAS transcript in etiolated seedlings under different treatments
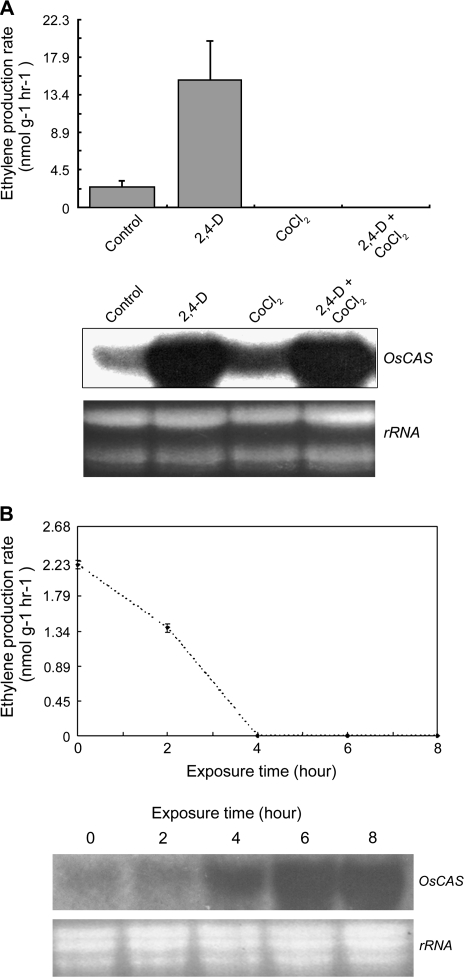
Yip and Yang (1988) demonstrated the importance of CAS in cyanide metabolism during active ethylene biosynthesis based on elevation of the cyanide level in plant tissues incubated with both the ethylene precursor (ACC) and CAS inhibitor (aminooxyacetic acid). CAS acts downstream of ACC synthase and ACO to remove the co-product cyanide. Auxins are known to induce ethylene biosynthesis by promoting ACC synthase (Kim et al., 1992) and ACO gene transcription (Petruzzelli et al., 2000); while ethylene has been shown to have both promotion and inhibition effects on gene transcription in some members of these two gene families. Hence, auxins and ethylene effects on OsCAS transcription were examined. It was found that ethylene biosynthesis in etiolated rice seedlings could be further induced by infiltration with 10 mM 2,4-D (a synthetic auxin). The ethylene biosynthetic rate was induced to about 7-fold over the control in seedlings treated with 2,4-D while the rate was suppressed to a non-detectable level in seedlings treated with cobalt chloride (an inhibitor of ACO), and with both 2,4-D and cobalt chloride (Fig. 2A, upper panel). The OsCAS transcripts from seedlings with different treatments were analysed by northern blotting with 32P radioactive XhoI–OsCAS–XhoI probe. The OsCAS transcript from untreated seedlings was low but detectable, it was greatly induced in seedlings treated with 2,4-D and 2,4-D plus cobalt chloride, although the ethylene biosynthetic rates of the seedlings in these treatments were completely different (Fig. 2A, lower panel). The OsCAS transcripts from seedlings treated with cobalt chloride were induced slightly despite a drop of ethylene biosynthetic rate to a non-detectable level.
Fig. 2.
Ethylene production rates and expressions of OsCAS transcripts in etiolated rice seedlings with 2,4-D, cobalt chloride, or both treatments. Ethylene biosynthetic rates on rice seedlings treated with different chemicals at 6 h after treatments (A), or transferred from dark to light (B) (upper panel). Expressions of OsCAS transcripts from seedlings were analysed by northern blotting with the 32P radioactive-labelled OsCAS probe (lower panel). Twenty micrograms of the total RNAs from seedlings were loaded per lane to show the equal loading. Measurement of ethylene production and the northern blotting of the hormone-treated sample were conducted in triplicate.
The increase in OsCAS expressions in rice seedlings could be related to a decline in endogenous ethylene biosynthesis. Rice seedlings, etiolated for 7 d, were transferred from dark to light in a constant temperature (25 °C). The seedlings were harvested to determine the ethylene biosynthetic rates and analyse OsCAS transcripts at 2 h intervals. The ethylene biosynthetic rates of these seedlings declined from about 2 nmol g−1 h−1 at 0 h to a non-detectable level at 4 h and the rate stayed at this level until 8 h (Fig. 2B, upper panel). The accumulation of OsCAS transcripts in rice seedlings was elevated accompanying the decline of ethylene biosynthetic rates. The transcripts began to accumulate from 4 h when the ethylene biosynthetic rate in the seedlings reached a non-detectable level (Fig. 2B, lower panel).
Generation of OsCAS–YFP overexpression in Arabidopsis and studies of CAS activity in transgenic lines
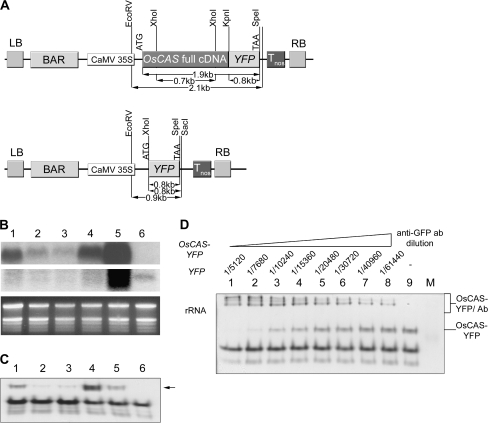
To facilitate studies on enzyme kinetics, structural properties, and subcellular localization of OsCAS encoding protein, Arabidopsis OsCAS–YFP overexpression lines (OsCAS–YFP lines) were generated. Expression of a 1.1 kb full coding sequence of OsCAS without a stop codon appended with a 0.8 kb YFP coding sequence was driven by a CaMV promoter. For the control transgenic line (YFP line), a 0.8 kb YFP coding sequence was driven by the CaMV promoter (Fig. 3A). Seeds from these lines were allowed to germinate on MS basal agar plates (Murashige and Skoog, 1962) under BLASTA selection and then resistant seedlings were transplanted into soil. Leaves from different lines were harvested 1 week after transplantation. Successful transformation was confirmed by genomic Southern blotting with either 32P-labelled YFP or the XhoI–OsCAS–XhoI probe (Fig. S1 in Supplementary data available at JXB online).
Fig. 3.
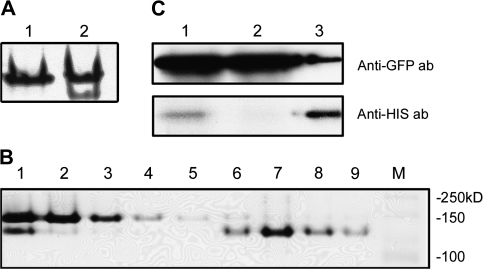
Expression of OsCAS in its heterologous expression Arabidopsis lines. (A) Schematic diagram of the binary vectors pBA OsCAS–YFP and pBA YFP. LB and RB refer to the left and right borders of the T-DNA, respectively. CaMV35S, Cauliflower mosaic virus 35S promoter; Tnos, terminator of nopaline synthetase gene; BAR, gene conferring BLASTA resistance. (B) Transcripts of OsCAS and YFP were analysed from 20 μg total RNA from leaves of these transgenic lines (lanes 1–5) and YFP line (lane 6) with OsCAS (first panel) or YFP probe (second panel), revealed by northern blotting. (C) An in-gel CAS activity assay was performed on 20 μg of leaf extracts from OsCAS–YFP transgenic lines (lanes 1–5) and YFP line (lane 6). A strong additional CAS activity band (arrowed) was found in the OsCAS–YFP transgenic line 4. To elucidate this additional CAS band due to OsCAS–YFP further, 20 μg of leaf proteins from line 4 were applied in a supershift in-gel assay (D). Ab, Antibody.
The expression levels of OsCAS–YFP and YFP transcripts were analysed in these lines. Twenty micrograms of total RNA from Arabidopsis leaves were separated in a 1.5% formaldehyde gel. Two different regions from constructs including 0.7 kb XhoI–OsCAS–XhoI from pBA OsCAS–YFP and 0.8 kb XhoI–YFP–SpeI from pBA YFP were selected as probes for quantification of the expression levels of these transcripts. Northern blotting on these OsCAS lines (Fig. 3B, lanes 1–5) showed that a 1.76 kb transcript of OsCAS–YFP was detected with various expression levels by hybridization with the 32P-labelled XhoI–OsCAS–XhoI probe, except the YFP line (Fig. 3B, lane 6). The northern blot analysis by hybridization to 32P-labelled XhoI–YFP–SpeI probe showed that the presence of YFP transcripts, 0.72 kb in size, in the YFP line. The northern blot analysis confirms that the transcripts of OsCAS–YFP and YFP are expressed in OsCAS–YFP lines and YFP line, respectively.
To verify the CAS activity band due to heterologous expression of OsCAS in these transgenic lines, in-gel CAS activity assay was conducted. An extra CAS activity band was found in OsCAS–YFP lines compared with the YFP line (Fig. 3C). A strong enzyme-stained band in all lines denoted as endogenous Arabidopsis CAS activity while the uppermost CAS activity-stained band is possibly due to the protein encoded by OsCAS–YFP (Fig. 3C, lanes 1–5). This additional band was detected in all OsCAS–YFP lines, and OsCAS–YFP line 4 was selected for downstream experiment for its strong CAS activity.
To confirm that this new CAS activity band was due to OsCAS–YFP in these transgenic lines, ‘super-shift in-gel CAS activity assay’ was conducted on crude leaf extract from OsCAS–YFP line 4. This enzyme assay method is similar to the in-gel CAS activity assay except that incubation was with different dilutions of polyclonal anti-GFP antibody prior to sample separation in the non-denative polyacrylamide gel. In-gel CAS activity assay was analysed on crude leaf extract from OsCAS–YFP line 4 incubated with different concentrations of polyclonal anti-GFP antibody (Fig. 3D). Intensities of the free form of CAS (Fig. 3D; indicated across the top, labelled OsCAS–YFP) decreased gradually when incubated without antibody (Fig. 3D, lane 9) to with antibody in a dilution of 1/10240. This free form was completely shifted to an upper position at antibody dilutions of 1/5120 and 1/7680. The decrease in intensity of free form CAS was accompanied by an increase in intensity of the complex form (Fig. 3D; indicated across the top, labelled OsCAS–YFP/anti-GFP ab dilution). Three complexes of different molecular mass were observed in crude extracts incubated with antibody in 1/5120 to 1/10240 dilutions. Hence, by selecting OsCAS–YFP line 4 as a representative, this assay confirmed that OsCAS protein from these lines possesses CAS activity in vitro.
Immuno-pull down of OsCAS–YFP from the transgenic line and kinetic studies on the immuno-purified protein
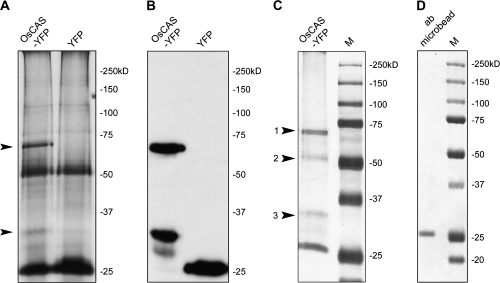
To characterize the enzyme activity of OsCAS–YFP protein in vitro, it was purified from a crude extract from leaves of transgenic line 4 by employing immuno-pull down with anti-GFP antibody immobilized on metal beads with washing by native extraction buffer. The pull-down proteins from the transgenic OsCAS–YFP line 4 and YFP line were separated in a 10% PAGE and visualized by silver staining. Two protein bands with molecular masses of 64 kDa and 28 kDa, respectively, were observed in the protein extract from OsCAS–YFP line 4 compared with the YFP line (Fig. 4A); these bands cross-reacted with anti-GFP antibody (Fig. 4B). An almost identical protein pattern was observed in pull-down protein from the transgenic OsCAS–YFP line 4 with strong stringency washing buffer. Four proteins with different molecular masses from this pull-down fraction were separated in a 10% PAGE (Fig. 4C) and three of these were identified by searching the molecular masses of tryptic-digested peptides from a protein against the database of translated rice genome encoding sequences. These three proteins with molecular masses about 64, 52, and 28 kDa were beta-cyanoalanine synthase (accession no. AY720933) with a molecular mass of 40 kDa, a large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (accession no. BAA84393) with a molecular mass of 53 kDa, and a green fluorescent protein RSGFP variant (accession no. AAA79304.1) with a molecular mass of 29 kDa, respectively (Table S1 in Supplementary data available at JXB online). The identity of the protein with a molecular mass 64 kDa should be OsCAS–YFP as the result of database searching and cross-reactivity with anti-GFP antibody, while the protein with a molecular mass 25 kDa was possibly the light chain of anti-GFP antibody (Fig. 4D). In this pull-down experiment, it was revealed that among these proteins OsCAS–YFP was in the majority while the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase and YFP were in the minority. By contrast, YFP is a unique band in an immuno-precipitated protein from the YFP transgenic line 1.
Fig. 4.
Detection and identification of immuno-pull-down proteins from OsCAS–YFP and YFP transgenic lines. Immuno-pull-down proteins from these two transgenic lines washed with low stringency buffer were applied for silver staining (A) and immuno-detection of YFP-tagged protein, respectively (B). Arrows point out the differences in patterns of immuno-pull-down proteins between those lines in (A). To further reduce the background of immuno-pull-down proteins, immuno-pull-down proteins from OsCAS–YFP lines washed with high stringency buffer supplied in the kit were visualized by Coomassie blue staining. Four immuno-pull-down proteins were detected and identified (C, labelled with numbers). In a negative control, immobilized anti-GFP antibody was applied for Coomassie blue staining (D).
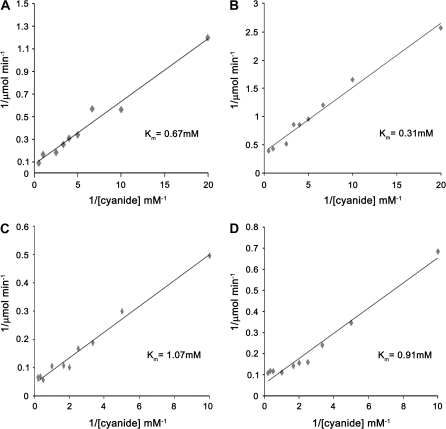
CAS/OASS activity and kinetic parameters with different substrates on immuno-purified OsCAS–YFP were determined on purified OsCAS–YFP protein from OsCAS–YFP overexpression transgenic line 4 according to the enzyme assay methods for CAS from Yip and Yang (1988) and for OASS from Lunn et al. (1990) (Table 1). The specific enzyme activity of CAS on OsCAS–YFP was calculated as 181 μmol H2S min−1 mg−1 protein, while that of OASS on OsCAS–YFP was 0.92 μmol Cys min−1mg−1 protein. OsCAS–YFP showed much stronger activity of CAS than that of OASS with a ratio of 197. Regarding kinetic parameters with different substrates of OsCAS–YFP, the Km for potassium cyanide was 0.67 mM and the Km for cysteine was 1.07 mM, as revealed by double-reciprocal plots (Fig. 5).
Table 1.
OASS/CAS activities and kinetic parameters of OsCAS in rice, OsCAS–YFP in transgenic Arabidopsis, and OsCAS–YFP yeast transformant
| Organisms | OASS-specific activity (μmol Cys min−1 mg−1 protein) | CAS-specific activity (μmol H2S min−1 mg−1 protein) | Activity ratio of CAS/OASS | Km for CAS reaction (mM) | Inhibition on CAS activity | |
| Cys | KCN | Cyanide IC50 (mM)a | ||||
| Rice | N.D.b | 13.3c | / | 0.84 | 0.27 | / |
| Arabidopsis | 0.92±0.06 | 181±13.89 | 197 | 1.07±0.20 | 0.67±0.15 | 20 |
| Yeast | 1.84±0.77 | 111.3±5.11 | 60.5 | 0.91±0.11 | 0.31±0.01 | 40 |
Km values were calculated by linear regression of double-reciprocal plots. Measurements of enzyme activities and determinations of Km values were repeated at least three times.
The concentration of cyanide required for 50% inhibition of CAS activity. IC50 was determined as 20 mM for spinach CAS previously by Warrilow and Hawkesford (2000).
N.D., Not detected.
CAS specific activity was determined on partially purified fraction (750-fold) from etiolated rice seedling extract.
Fig. 5.
Double reciprocal plots for determining Km values of cyanide (A, B) and cytsteine (C, D) on heterologous OsCAS. Kinetic assays were conducted on OsCAS from transgenic Arabidopsis (A, C) and yeast (B, D).
Localization of OsCAS–YFP protein from OsCAS–YFP overexpression in Arabidopsis
To determine the transit peptide of OsCAS, a comparison was made between the N-terminal from OsCAS full-length encoding sequence (accession number AAV48542) and mature OsCAS from the transgenic line of which the N-terminal sequencing had been identified by a protein sequencer (Hewlett-Packard protein sequencer G1000A). The peptide sequence from 1 to 35 of OsCAS may serve as a transit peptide as mature OsCAS starts from peptide sequence 36 of the full-length encoding sequence. This transit peptide cleavage pattern of OsCAS (F↓STLQQ…) is classified as mitochondrial targeting sequence class II according to Zhang et al. (2001). This class is predominant, occupying 44.4% of all experimentally determined mitochondrial targeting sequences available (Zhang et al., 2001). In addition, a transit peptide has been identified targeting mitochondria from the N-terminus of a full-length amino acid from OsCAS in silico by the two topology prediction programs iPSORT (Bannai et al., 2002) (http://hc.ims.u-tokyo.ac.jp/iPSORT/) and MitoProt (Manuel and Claros, 1996) (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html); these results also suggest that OsCAS may be targeted to mitochondria in silico.
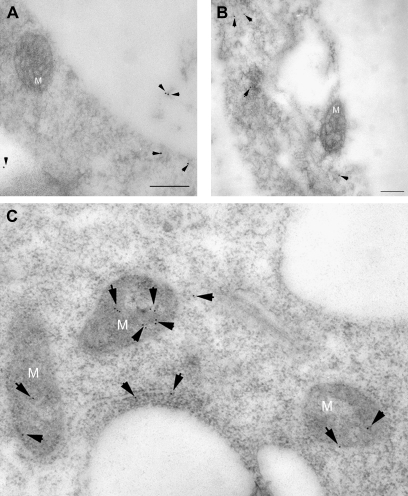
To determine the subcellular localization of OsCAS in transgenic Arabidopsis, root tip cells were prepared from germinating transgenic and YFP lines, followed by chemical fixation and preparation of ultra-thin sections for immunogold labelling with GFP antibodies. GFP antibodies labelled mitochondria predominantly (Fig. 6C), and these gold particles were evenly distributed in the mitochondrion matrix, although gold particles was also found in the cytoplasm (3 dots in the cytoplasm and 12 dots inside mitochondria in Fig. 6C). It is noted that these particles in the cytoplasm may indicate YFP instead of OsCAS–YFP as a start codon is located just upstream of the encoding sequence of the YFP gene in the binary construct pBA002 overexpressing OsCAS. From the control YFP line, anti-GFP antibody labelled cytoplasm in cells without any import to mitochondria (Fig. 6A, B). This demonstrated that OsCAS mainly targeted mitochondria.
Fig. 6.
Immunogold EM localizations of OsCAS fusion protein in root tip cells of the transgenic OsCAS–YFP line and its control YFP line. Immunogold particles were observed in cytoplasm in the control YFP line without signal in mitochondria (A, B) while those particles were confined in mitochondria predominantly in the transgenic OsCAS–YFP line (C). The black arrows indicate gold particles. M, Mitochondrion. Scale bar=200 nm.
Elucidation of the native form of OsCAS–YFP and some amino acid residues which are important for CAS activity
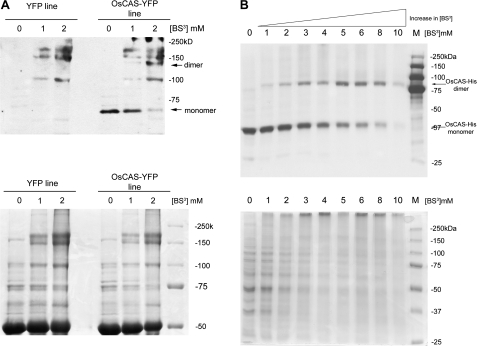
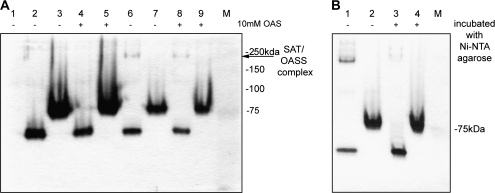
To elucidate the native form of OsCAS in planta, the native form of OsCAS–YFP was ‘fixed’ by incubating various concentrations of cross-linking agent BS3 with extracts from OsCAS–YFP line 4 or the YFP transgenic line. After incubation with crude extracts from both transgenic lines and BS3, proteins were separated in SDS–PAGE prior to visualization by Coomassie blue staining. YFP cross-reacting bands were revealed by western blotting with the anti-GFP antibody. Molecular masses of protein bands were estimated by a software Quantity One version 4.6.1 from Bio-Rad Laboratories. In the western blotting, an anti-GFP cross-reacting band with a molecular mass about 64 kDa appeared in the OsCAS line 4 when no cross-linker was applied (Fig. 7A, upper panel). Following an increase in concentrations of the cross-linking reagent, this anti-GFP cross-reacting band shifted from 64 kDa to about double its original mass of roughly 130 kDa; and the intensity of this 130 kDa band increased following the increase of cross-linker BS3 from 1 mM to 2 mM. This new band was found in OsCAS–YFP extracts with BS3 but not in YFP extracts with BS3. As several known signals were detected in the OsCAS transgenic line and the control Arabidopsis lines, a chemical cross-linking experiment with different concentrations of BS3 from the OsCAS–HIS (consecutive histidine residues) overexpressing yeast was conducted (Fig. 7B, upper panel). A shift in anti-HIS cross-reacting band with a molecular mass of ∼40–80 kDa followed the gradual increase of the concentration in BS3. It is noted that several unknown signals were cross-reacted with anti-GFP antibody, but not anti-HIS antibody, hence these bands may be raised by non-specificity of anti-GFP antibody. As a ratio between the OsCAS complex form with either YFP or HIS6 peptide and its monomer was close to 2, this indicates that the native OsCAS is possibly in a dimeric form.
Fig. 7.
Immunodetection of YFP-tagged protein from transgenic lines and yeast transformants incubated with chemical cross-linker BS3. YFP tag or HIS6 tag fused with OsCAS from either 40 μg of crude extracts from the transgenic line (A) or 30 μg of crude protein from the yeast transformant (B), respectively, incubated with various concentrations of BS3, were immune-detected with anti-GFP (A) antibody or anti-HIS antibody (B). The arrows indicate monomeric and dimeric forms of OsCAS–YFP (upper panel). Equal loading of crude extracts were visualized by Coomassie blue staining (lower panel).
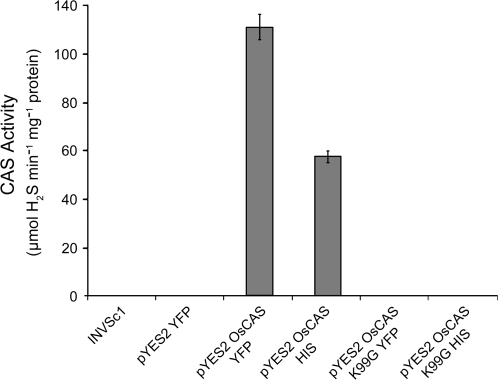
As the native form of OsCAS is in dimeric form, monomers in an OsCAS dimer may work independently for CAS activity. To investigate the feasibility that all catalytic domains for CAS activity come from an individual monomer, both functional OsCAS and non-functional OsCAS were overexpressed in yeast. For a non-functional OsCAS, a mutant OsCAS K99G–HIS was made in which critical lysine for PLP binding was mutated. CAS activity of pure OsCAS–YFP from yeast was 111.3±5.11 μmol H2S min−1 mg protein−1; no CAS activity was detected from OsCAS K99G–HIS (Fig. 8). While there was only one CAS activity band in yeast transformed with pYES2 OsCAS–YFP (Fig. 9A, lane 1), there were two CAS activity bands in yeast co-transformed with pYES2 OsCAS–YFP and pYES3 OsCAS K99G–HIS (Fig. 9A, lane 2). If the upper activity band in this lane was from homodimeric OsCAS–YFP in yeast transformed with pYES2 OsCAS–YFP, the lower activity band should come from the heterodimer made from OsCAS–YFP and OsCAS K99G–HIS in the co-transformed yeast. To address whether this activity band came from the heterodimer, this complex was purified by a nickel column and then each fraction in the purification was subjected to CAS in-gel assay. This complex was purified successfully (Fig. 9B, lane 7) and was recognized by both anti-HIS antibody and anti-GFP antibody (Fig. 9C). The western blotting strongly suggested that this complex is a heterodimer, consisting of a wild-type OsCAS–YFP and a mutant OsCAS K99G–HIS. As all domains for CAS activity are confirmed from an individual monomer, it can be concluded that dimer formation as well as PLP binding are essential for CAS activity.
Fig. 8.
CAS activities of various purified OsCAS mutants from heterologous expression in yeast strain INVSc1 were determined.
Fig. 9.
In-gel CAS activity assay and immunodetection on wild-type OsCAS–YFP homodimer and OsCAS–YFP/OsCAS K99G–HIS heterodimer. In-gel CAS activities on 20 μg of crude proteins from yeast transformants with wild-type OsCAS–YFP (A, lane 1) and co-expressions of OsCAS–YFP/OsCAS K99G–HIS (A, lane 2) were determined. In-gel CAS activity assay on fractions during the purification of the heterodimer through the nickel column from the yeast transformant with co-expressions of OsCAS–YFP/OsCAS K99G–HIS (B) were conducted. Twenty micrograms of fractions from crude extract (lane 1), flow through (lane 2), three washing steps (lanes 3–5), and four elution steps (lanes 6–9) were applied in the assay. Immunodetection of YFP- or HIS-tagged protein on fractions from crude (C, lane 1), flow through (C, lane 2), and purified heterodimer (C, lane 3) were detected by anti-GFP antibody and anti-HIS antibody, respectively.
Discussion
Auxin and ethylene regulate the expression of OsCAS transcripts in an antagonistic manner. Auxin serves to up-regulate OsCAS while ethylene serves to down-regulate OsCAS in etiolated rice seedlings. The promotion effect of auxins is much stronger than the suppression effect of auxin-induced ethylene (Kang et al., 1971; Yu et al., 1979) on OsCAS transcription. Etiolated monocot leaves possess a greater capability of ACC accumulation and thus ethylene production than dicot plants in their seedling stages (Jiao et al., 1987). This antagonistic regulation may have evolved in monocot plants and be crucial for fine-tuning the OsCAS expressions to metabolize the cyanide formed concurrently with high ethylene biosynthetic rates. Several previous reports from dicots showed that exogenous ethylene would promote CAS gene transcription and thus CAS activity. Auxinic herbicides, such as 2,4-D and quinclorac (Grossmann and Kwiatkowski, 1995), are known to be effective in killing dicot weeds in grain fields. It is possible that the difference in transcription regulation of CAS genes between monocot and dicot may involve this phenomenon.
Enzymology studies on recombinant CAS expressed in bacterial mutant NK3 strains including potato (Maruyama et al., 2001), Arabidopsis, and spinach (Hatzfeld et al., 2000) have been reported. It is noted that there is a significant difference in the kinetic parameters of spinach CAS by comparing the native CAS with its recombinant protein (Ikegami et al., 1988; Hatzfeld et al., 2000). This is the first report on heterologous CAS expression in plants and in yeast. It has not been possible to immuno-purify OsCAS from rice seedlings or transgenic Arabidopsis by the anti-peptide anti-serum raised against a specific peptide in OsCAS; however, the purification of the recombinant OsCAS from transgenic Arabidopsis lines has been successful using an immobilized GFP antibody column. It is important to note that a shorter tag, like HIS6 or MYC polypeptide, may hamper affinity of substrate binding of OsCAS instead of YFP protein (Km for cyanide on OsCAS with either HIS6 or MYC polypeptide almost increased at least ∼8-fold compared with values from OsCAS from seedlings or OsCAS–YFP; KW Lai and KM Kwan, unpublished results) the same as OASS (Wirtz and Hell, 2006); thus, YFP protein fused at the C-terminal of OsCAS facilitates purification with an immunoaffinity-column. In addition, significant reduction in CAS activity of OsCAS in a relatively high concentration of cyanide is in agreement with Warrilow and Hawkesford (2000) on the cyanide inhibitory effect on CAS activity of CAS members.
Visualization of OsCAS in root tip mitochondria by electron microscopy on immunogold labelling with GFP antibodies is in agreement with previous reports which identified subcellular localization of CAS activity by organelle fractionation methods (Akopyan et al., 1975; Wurtele et al., 1985). However, in those previous studies CAS activity in the mitochondrial fraction might have originated from mitochondrial OASS or other OASSs due to impure preparation. Hence, the present result is the first to reveal the mitochondrial localization of OsCAS in plant cells and at the same time provide evidence for its role in cyanide detoxification.
By combining results on the homodimeric native form from chemical cross-linking on OsCAS protein from transgenic lines and 35 amino acid residues from the N-terminal of the full-length OsCAS cleaved from mature OsCAS protein in those lines by Edman degradation, the molecular mass of OsCAS is calculated as 73 kDa. This 35 residue cleavage polypeptide may serve as a mitochondrial signal peptide facilitating targeting of OsCAS.
A protein model of Arabidopsis cytosolic OASS, which shares a similar amino acid sequence, suggests that a monomer consists of all critical sites and a PLP for facilitating its function without sharing these sites; however, native OsCAS or CAS family in other plant species are reported to be homodimeric in form. The importance of dimerization to its functions has not been addressed. CAS activity was detected from a dimer composed of a wild-type OsCAS monomer and an OsCAS monomer mutated at the PLP-binding lysine residue. Hence, the present results showed that one intact monomer with PLP cofactor binding within a dimer would be sufficient for CAS activity. It is speculated that dimerization may provide the necessary conformational change(s) on OsCAS monomer for its catalytic activity.
Protein–protein interaction of plant OASS to SAT has been reported in early purification of the cysteine synthase complex from plants (Droux et al., 1992; Nakamura et al., 1987), a complex association in co-expression of Arabidopsis OASS/SAT in a bacterial system (Droux et al., 1998), and a yeast two-hybrid system in screening interactions between proteins encoding the tobacco expression library and bacterial OASS/SAT (Liszewska et al., 2005). Association/dissociation of this complex has been widely accepted as a regulation mechanism for cysteine synthesis in plants (Droux et al., 1992; Wirtz et al., 2001) and this interaction is generally accepted as a criterion for an authentic OASS (Jost et al., 2000). Because of the highly conserved amino acid sequence of OsCAS to those OASS, one may suspect that OsCAS may play dual roles in cyanide metabolism and cysteine synthesis by recycling sulphide liberated from the CAS reaction inside mitochondria, as there is no other rice OASS/CAS member predicted in mitochondrial in silico. This prompted us to conduct a simple experiment regarding the interaction of OsCAS to watermelon (Wt) SAT in yeast as this interaction is highly plastic among species (Droux et al., 1998; Liszewska et al., 2005). The results (Fig. 10) showed that OsCAS could not interact with WtSAT in yeast despite the interaction of SpOASS to WtSAT being demonstrated. In addition, there was no protein co-precipitated with OsCAS from transgenic Arabidopsis lines; these two lines of evidence might indicate that OsCAS could not interact with SAT in planta or in yeast. These findings agree well with the work of Bonner et al. (2005) on SAT interaction with AtOASS in Arabidopsis. The three critical amino acid residues facilitating SAT interaction on AtOASS have been examined and those amino acid residues in OsCAS and other reported CAS compared and it was discovered that these three amino acid residues are not fully conserved in those of OsCAS and in most of the CAS reported from other species. The present results thereby strongly suggested that OsCAS play a role in cyanide detoxification but are not involved in sulphide recycling inside the mitochondria.
Fig. 10.
Detection of a cysteine synthase complex in yeasts with co-expressions of SpOASS or OsCAS with WtSAT. In-gel CAS activity assay (A) was conducted on 100 μg crude protein from yeast transformants with pYES2 HIS–WtSAT (lane 1), pYES3 SpOASS–MYC (lanes 2 and 4), pYES3 OsCAS–MYC (lanes 3 and 5), co-expression of pYES2 HIS–WtSAT and pYES3 SpOASS–MYC (lanes 6 and 8), and co-expression of pYES2 HIS–WtSAT and pYES3 OsCAS–MYC (lanes 7 and 9). A ‘+’ sign represents an addition of OAS prior to loading and an arrow indicates a putative cysteine synthase complex. To demonstrate further that SAT is a component in this putative cysteine synthase complex, an in-gel CAS activity assay (B) was conducted on 100 μg crude protein (lanes 1 and 2) and protein pre-incubated with nickel agarose (lanes 3 and 4) from yeast transformant with co-expression of pYES2 HIS–WtSAT and pYES3 SpOASS–MYC (lanes 1 and 3)or co-expression of pYES2 HIS–WtSAT and pYES3 OsCAS–MYC (lanes 2 and 4). Pre-incubation extracts from yeast co-expressing SpOASS and WtSAT with nickel agarose would remove this additional CAS-stained band (lanes 1 and 3) and this demonstrated the interaction between SpOASS to WtSAT. SAT-specific activities from yeast transformants with co-expression of pYES2 HIS–WtSAT and either pYES3 SpOASS–MYC or pYES3 OsCAS–MYC were 86.32 μmol 5-thio-2-nitrobenzoate min−1 mg−1 protein and 74.00 μmol 5-thio-2-nitrobenzoate min−1 mg−1 protein, respectively, following the spectrophotometric assay from Bonner et al. (2005).
Contrary to CAS activity which was mostly found in mitochondria, cysteine synthesis have been predicted in different subcellular compartments including cytoplasm, chloroplasts, and mitochondria, each compartment using their own allocation of OASS and SAT to make the cysteine synthase complex. OASS and SAT are thereby encoded by two gene families and members are targeted by different compartments (Wirtz and Hell, 2006). Based on the results of the present search, the rice genome contains nine members in the OASS/CAS gene family. OsCAS is the sole sequence encoding CAS and the sole sequence encoding protein predicted (or proven in this study) to be localized to mitochondria. Also it was not possible to find a putative gene coding for mitochondrial SAT in the rice genome database search. This raises the question as to how cysteine can be synthesized in rice mitochondria—recycled from hydrogen sulphide as a result of CAS action—and whether cysteine can be imported to mitochondria from the cytoplasm in rice. A recent publication of Watanabe et al. (2008) reported the evidence on intracellular cysteine transport between cytosol, mitochondrion, and plastid in Arabidopsis OASS T-DNA knockout lines. Their report suggested that the cytosolic OASS would be more critical in cysteine synthesis than other OASS in other compartments. In rice, it is logical to propose that maintaining the mitochondrial cysteine pool while the ethylene production rate is high may depend on cysteine import from the cytosol. The likely mechanism is that hydrogen sulphide liberated from the CAS reaction is either transported or diffused from mitochondria to cytsoplasm (Wirtz and Hell, 2007) in which cysteine synthesis may take place and then be transported back to the mitochondria to sustain cyanide detoxification. However, identification of the mitochondrial cysteine permease or the import mechanism of cysteine to mitochondria has not been fully elucidated.
Supplementary data
Results regarding integrations of OsCAS–YFP and YFP encoding sequences in the Arabidopsis genome by Southern blotting (Fig. S1) and results of the protein identities for immuno-purified proteins by database searching (Table S1) have been supplemented and this material is available at JXB online.
Supplementary Material
Acknowledgments
We give credit to Mr King-Ming Wai, Miss Yee-Wai Cheung, and Mr Manda Yu, former graduate students in WKY's laboratory for their work on CAS protein purification and OsCAS isolation. We are grateful to Prof. Kazuki Saito in Chiba University for providing the vectors, a full-length spinach OASS cDNA and a full-length watermelon SAT cDNA, used in this work. This project is supported by a research grant (HKU 7544/06M) awarded to WKY from the Research Grant Council of Hong Kong SAR Government.
Glossary
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylic acid
- ACO
1-aminocyclopropane-1-carboxylate oxidase
- At
Arabidopsis thaliana
- BS3
bis(sulphosuccinimidyl) suberate
- CAS
β-cyanoalanine synthase
- GFP
green fluorescent protein
- HIS
consecutive histidine residues
- OASS
O-acetylserine sulphydrylase
- Os
Oryza sativa
- PLP
pyridoxal-5′-phosphate
- SAT
serine acetyltransferase
- Sp
spinach
- Wt
watermelon
- YFP
yellow fluorescent protein
References
- Akopyan TN, Braunstein AE, Goryachenkova EV. Beta-cyanoalanine synthase: purification and characterization. Proceedings of the National Academy of Sciences, USA. 1975;72:1617–1621. doi: 10.1073/pnas.72.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature Protocols. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Blumenthal SG, Hendrickson HR, Abrol YP, Conn EE. Cyanide metabolism in higher plants. 3. The biosynthesis of beta-cyanolanine. Journal of Biological Chemistry. 1968;243:5302–5307. [PubMed] [Google Scholar]
- Bonner ER, Cahoon RE, Knapke SM, Jez JM. Molecular basis of cysteine biosynthesis in plants: structural and functional analysis of O-acetylserine sulfhydrylase from Arabidopsis thaliana. Journal of Biological Chemistry. 2005;280:38803–38813. doi: 10.1074/jbc.M505313200. [DOI] [PubMed] [Google Scholar]
- Castric PA, Strobel GA. Cyanide metabolism by Bacillus megaterium. Journal of Biological Chemistry. 1969;244:4089–4094. [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze–thaw transformation and drug selection. Biotechniques. 1994;16:664–668. 670. [PubMed] [Google Scholar]
- Chew M-Y. Rhodanese in higher plants. Phytochemistry. 1973;12:2365–2367. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Droux M, Martin J, Sajus P, Douce R. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Archives of Biochemistry and Biophysics. 1992;295:379–390. doi: 10.1016/0003-9861(92)90531-z. [DOI] [PubMed] [Google Scholar]
- Droux M, Ruffet ML, Douce R, Job D. Interactions between serine acetyltransferase and O-acetylserine (thiol) lyase in higher plants: structural and kinetic properties of the free and bound enzymes. European Journal of Biochemistry. 1998;255:235–245. doi: 10.1046/j.1432-1327.1998.2550235.x. [DOI] [PubMed] [Google Scholar]
- Dunnill PM, Fowden L. Enzymatic formation of beta-cyanoalanine from cyanide by Escherichia coli extracts. Nature. 1965;208:1206–1207. doi: 10.1038/2081206a0. [DOI] [PubMed] [Google Scholar]
- Grossmann K. A role for cyanide, derived from ethylene biosynthesis, in the development of stress symptoms. Physiologia Plantarum. 1996;97:772–775. [Google Scholar]
- Grossmann K, Kwiatkowski J. Evidence for a causative role of cyanide, derived from ethylene biosynthesis, in the herbicidal mode of action of quinclorac in barnyard grass. Pesticide Biochemistry and Physiology. 1995;51:150–160. [Google Scholar]
- Han SE, Seo YS, Kim D, Sung SK, Kim WT. Expression of MdCAS1 and MdCAS2, encoding apple beta-cyanoalanine synthase homologs, is concomitantly induced during ripening and implicates MdCASs in the possible role of the cyanide detoxification in Fuji apple (Malus domestica Borkh.) fruits. Plant Cell Reports. 2007;26:1321–1331. doi: 10.1007/s00299-007-0316-9. [DOI] [PubMed] [Google Scholar]
- Hatzfeld Y, Maruyama A, Schmidt A, Noji M, Ishizawa K, Saito K. beta-Cyanoalanine synthase is a mitochondrial cysteine synthase-like protein in spinach and Arabidopsis. Plant Physiology. 2000;123:1163–1171. doi: 10.1104/pp.123.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson HR, Conn EE. Cyanide metabolism in higher plants. IV. Purification and properties of the beta-cyanolanine synthase of blue lupine. Journal of Biological Chemistry. 1969;244:2632–2640. [PubMed] [Google Scholar]
- Hesse H, Nikiforova V, Gakiere B, Hoefgen R. Molecular analysis and control of cysteine biosynthesis: integration of nitrogen and sulphur metabolism. Journal of Experimental Botany. 2004;55:1283–1292. doi: 10.1093/jxb/erh136. [DOI] [PubMed] [Google Scholar]
- Ikegami F, Takayama K, Tajima C, Murakoshi I. Purification and properties of [beta]-cyano-alanine synthase from Spinacia oleracea. Phytochemistry. 1988;27:2011–2016. [Google Scholar]
- Jiao XZ, Yip WK, Yang SF. The effect of light and phytochrome on 1-aminocyclopropane-1-carboxylic acid metabolism in etiolated wheat seedling leaves. Plant Physiology. 1987;85:643–647. doi: 10.1104/pp.85.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA. Why are so many food plants cyanogenic? Phytochemistry. 1998;47:155–162. doi: 10.1016/s0031-9422(97)00425-1. [DOI] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Wirtz M, Hopkins L, Hawkesford MJ, Hell R. Genomic and functional characterization of the oas gene family encoding O-acetylserine (thiol) lyases, enzymes catalyzing the final step in cysteine biosynthesis in Arabidopsis thaliana. Gene. 2000;253:237–247. doi: 10.1016/s0378-1119(00)00261-4. [DOI] [PubMed] [Google Scholar]
- Kang BG, Newcomb W, Burg SP. Mechanism of auxin-induced ethylene production. Plant Physiology. 1971;47:504–509. doi: 10.1104/pp.47.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katharina P, Reinhard K, Sacco de Vries K, Ton B. Isolation of total, poly(A) and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht: Kluwer Academic; 1995. D5/1–4. [Google Scholar]
- Kim WT, Silverstone A, Yip WK, Dong JG, Yang SF. Induction of 1-aminocyclopropane-1-carboxylate synthase mRNA by auxin in mung bean hypocotyls and cultured apple shoots. Plant Physiology. 1992;98:465–471. doi: 10.1104/pp.98.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredich NM. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. I. Effects of growth of varying sulfur sources and O-acetyl-L-serine on gene expression. Journal of Biological Chemistry. 1971;246:3474–3484. [PubMed] [Google Scholar]
- Kuske CR, Hill KK, Guzman E, Jackson PJ. Subcellular location of O-acetylserine sulfhydrylase isoenzymes in cell cultures and plant tissues of Datura innoxia Mill. Plant Physiology. 1996;112:659–667. doi: 10.1104/pp.112.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liszewska F, Gaganidze D, Sirko A. Isolation of Nicotiana plumbaginifolia cDNAs encoding isoforms of serine acetyltransferase and O-acetylserine (thiol) lyase in a yeast two-hybrid system with Escherichia coli cysE and cysK genes as baits. Acta Biochimica Polonica. 2005;52:117–128. [PubMed] [Google Scholar]
- Luk JM, Su YC, Lam SC, Lee CK, Hu MY, He QY, Lau GK, Wong FW, Fan ST. Proteomic identification of Ku70/Ku80 autoantigen recognized by monoclonal antibody against hepatocellular carcinoma. Proteomics. 2005;5:1980–1986. doi: 10.1002/pmic.200401084. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine(thiol)lyase in spinach leaves. Plant Physiology. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel G, Claros PV. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Maruyama A, Saito K, Ishizawa K. Beta-cyanoalanine synthase and cysteine synthase from potato: molecular cloning, biochemical characterization, and spatial and hormonal regulation. Plant Molecular Biology. 2001;46:749–760. doi: 10.1023/a:1011629703784. [DOI] [PubMed] [Google Scholar]
- Miller JM, Conn EE. Metabolism of hydrogen cyanide by higher plants. Plant Physiology. 1980;65:1199–1202. doi: 10.1104/pp.65.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortz E, Krogh TN, Vorum H, Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nakamura K, Hayama A, Masada M, Fukushima K, Tamura G. Measurement of serine acetyltransferase activity in crude plant extracts by a coupled assay system using cysteine synthase. Plant Cell Physiology. 1987;28:885–891. [Google Scholar]
- Ogunlabi OO, Agboola FK. A soluble beta-cyanoalanine synthase from the gut of the variegated grasshopper (Zonocerus variegatus L.) Insect Biochemistry and Molecular Biology. 2007;37:72–79. doi: 10.1016/j.ibmb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Peiser GD, Wang TT, Hoffman NE, Yang SF, Liu HW, Walsh CT. Formation of cyanide from carbon 1 of 1-aminocyclopropane-1-carboxylic acid during its conversion to ethylene. Proceedings of the National Academy of Sciences, USA. 1984;81:3059–3063. doi: 10.1073/pnas.81.10.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzelli L, Coraggio I, Leubner-Metzger G. Ethylene promotes ethylene biosynthesis during pea seed germination by positive feedback regulation of 1-aminocyclo-propane-1-carboxylic acid oxidase. Planta. 2000;211:144–149. doi: 10.1007/s004250000274. [DOI] [PubMed] [Google Scholar]
- Saito K, Kurosawa M, Murakoshi I. Determination of a functional lysine residue of a plant cysteine synthase by site-directed mutagenesis, and the molecular evolutionary implications. FEBS Letters. 1993;328:111–114. doi: 10.1016/0014-5793(93)80976-2. [DOI] [PubMed] [Google Scholar]
- Saito K, Yokoyama H, Noji M, Murakoshi I. Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. Journal of Biological Chemistry. 1995;270:16321–16326. doi: 10.1074/jbc.270.27.16321. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Solomonson L. Cyanide as a metabolic inhibitor. In: Vennesland B, Conn EE, Knowles CJ, Westley J, Wissing F, editors. Cyanide in biology. London: Academic Press; 1982. pp. 11–28. [Google Scholar]
- Takahashi H, Saito K. Subcellular localization of spinach cysteine synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiology. 1996;112:273–280. doi: 10.1104/pp.112.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomati U, Federici G, Cannella C. Rhodanese activity in chloroplasts. Physiological Chemistry and Physics. 1972;4:193–196. [PubMed] [Google Scholar]
- Tse YC, Mo B, Hillmer S, Zhao M, Lo SW, Robinson DG, Jiang L. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. The Plant Cell. 2004;16:672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow AG, Hawkesford MJ. Cysteine synthase [O-acetylserine (thiol) lyase] substrate specificities classify the mitochondrial isoform as a cyanoalanine synthase. Journal of Experimental Botany. 2000;51:985–993. doi: 10.1093/jexbot/51.347.985. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kusano M, Oikawa A, Fukushima A, Noji M, Saito K. Physiological roles of the beta-substituted alanine synthase gene family in Arabidopsis. Plant Physiology. 2008;146:310–20. doi: 10.1104/pp.107.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westley J. Rhodanese. Advances in Enzymology and Related Areas of Molecular Biology. 1973;39:327–368. doi: 10.1002/9780470122846.ch5. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Berkowitz O, Droux M, Hell R. The cysteine synthase complex from plants: mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein–protein interaction. European Journal of Biochemistry. 2001;268:686–293. doi: 10.1046/j.1432-1327.2001.01920.x. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. Journal of Plant Physiology. 2006;163:273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. The Plant Cell. 2007;19:625–639. doi: 10.1105/tpc.106.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele ES, Nikolau BJ, Conn EE. Subcellular and developmental distribution of beta-cyanoalanine synthase in barley leaves. Plant Physiology. 1985;78:285–290. doi: 10.1104/pp.78.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip WK, Yang SF. Cyanide metabolism in relation to ethylene production in plant tissues. Plant Physiology. 1988;88:473–476. doi: 10.1104/pp.88.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YB, Adams DO, Yang SF. Regulation of auxin-induced ethylene production in mung bean hypocotyls: role of 1-aminocyclopropane-1-carboxylic acid. Plant Physiology. 1979;63:589–590. doi: 10.1104/pp.63.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XP, Sjoling S, Tanudji M, Somogyi L, Andreu D, Eriksson LE, Graslund A, Whelan J, Glaser E. Mutagenesis and computer modelling approach to study determinants for recognition of signal peptides by the mitochondrial processing peptidase. The Plant Journal. 2001;27:427–438. doi: 10.1046/j.1365-313x.2001.01108.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.