Abstract
NAD+-dependent sorbitol dehydrogenase (NAD-SDH, EC 1.1.1.14), a key enzyme in sorbitol metabolism, plays an important role in regulating sink strength and determining the quality of apple fruit. Understanding the tissue and subcellular localization of NAD-SDH is helpful for understanding sorbitol metabolism in the apple. In this study, two NAD-SDH cDNA sequences were isolated from apple fruits (Malus domestica Borkh cv. Starkrimson) and named MdSDH5 and MdSDH6. Immunohistochemical analysis revealed that NAD-SDH is distributed in both the flesh and the vascular tissue of the fruit, and the vascular tissue and mesophyll tissue in the young and old leaves, indicating that it is a ubiquitous protein expressed in both sink and source organs. Immunogold electron microscopy analysis demonstrated that NAD-SDH is localized mainly in the cytoplasm and chloroplast of the fruit and leaves. The chloroplast localization of NAD-SDH was confirmed by the transient expression of MdSDH5-GFP and MdSDH6-GFP in the mesophyll protoplast of Arabidopsis. NAD-SDH was also found in electron opaque deposits of vacuoles in young and mature leaves. These data show that NAD-SDH has different subcellular localizations in fruit and leaves, indicating that it might play a different role in sorbitol metabolism in different tissues of apple.
Keywords: Apple, chloroplast, fruit, leaf, localization, sorbitol dehydrogenase
Introduction
Sorbitol metabolism is a major determinant of fruit yield and quality in woody members of the Rosaceae family including Malus, Pyrus, and Prunus. In these plants, sorbitol is the primary product of photosynthesis in the leaf and also the major translocated form of carbohydrate from source to sink tissues (Bieleski and Redgwell, 1985; Yamaki and Ishikawa, 1986; Loescher and Everard, 1996; Noiraud et al., 2001). However, there is little sorbitol in the fruit, which indicates that sorbitol is metabolized rapidly after unloading there (Yamaki and Ishikawa, 1986). The sorbitol in sink tissue is oxidized to fructose via NAD+-dependent sorbitol dehydrogenase (NAD-SDH) (Loescher et al., 1982; Yamaki and Ishikawa, 1986; Yamaguchi et al., 1996; Beruter et al., 1997). NAD-SDH-involved sorbitol metabolism plays an important role in establishing fruit sink strength during the fruit set phase (Nosarzewski and Archbold, 2007) and determining apple fruit quality (Teo et al., 2006). NAD-SDH has been purified and characterized from apple fruit (Yamaguchi et al., 1994; Yamada et al., 1998; Park et al., 2002). It belongs to the alcohol dehydrogenase family which contains a zinc-containing alcohol dehydrogenase signature, a zinc-binding site (structural zinc binding site and catalytic zinc binding site), and a NAD-binding pocket (Yamada et al., 2001; Ito et al., 2005). NAD-SDH catalyses a reversible reaction (D-sorbitol+NAD+↔D-fructose+NADH+H+) and, in vivo, favours the conversion of sorbitol to fructose rather than the reverse reaction (Yamaguchi et al., 1994).
Previous data have shown that sorbitol is not available for remetabolization in the mature apple leaf and it is only loaded into the transport pathway (Bieleski, 1982; Zhou et al., 2001; McQueen and Minchin, 2005). In addition, NAD-SDH has been found primarily in non-green callus, root, seedling, immature leaf, and fruit (Negm and Loescher, 1979, 1981; Yamaguchi et al., 1994). However, some NAD-SDH isoforms, such as MdSDH1 and NAD-SDH1, are expressed at high levels in source leaves (Park et al., 2002; Nosarszewski et al., 2004). Therefore, the question of whether there is NAD-SDH in the apple source leaves is a subject needing further investigation.
It is thought that sorbitol is synthesized in the cytosol of source tissues and metabolized in the cytosol of sink tissues in the apple (Teo et al., 2006). In addition, sorbitol is also transported into the vacuoles of the apple fruit (Yamaki, 1987). Recently, Nadwodnik and Lohaus (2008) found that there was sorbitol in the chloroplast in addition to the vacuole and cytosol of the mesophyll cell in peaches (Prunus persica). However, it is unknown whether the sorbitol is involved in metabolism in the chloroplast and vacuole or if these compartments are used only as a storage space for sorbitol. Therefore, determining the subcellular localization of NAD-SDH in these cells is very important for understanding the sites of sorbitol metabolism in apples.
In this study, two MdSDH genes from apple (Malus domestica Borkh cv. Starkrimson) fruits were cloned that are both expressed in the leaf and fruit of the apple. Biochemical analysis revealed that they all represent the NAD-SDH activity detected in vitro. NAD-SDH was found in both the sink organs (young leaves and fruit) and the source organs (mature leaves). In leaves, NAD-SDH was localized in the cytoplasm, chloroplasts, and vacuoles. By contrast, NAD-SDH was localized only in cytoplasm and chloroplast of the fruit, but not in the vacuoles. The possible function of NAD-SDH in the chloroplasts and vacuoles is discussed.
Materials and methods
Plant material
Apple (Malus domestica Borkh. cv. Starkrimson) fruits and leaves were sampled from a commercial orchard in the western suburb of Beijing. Fruits were collected at 30, 60, and 90 d after full bloom (DAFB). The young leaves were sampled when they had just germinated and were still curly. The leaves on bearing shoots were sampled from the same tree at 60 DAFB as mature leaves. Samples were picked for immediate use or frozen in liquid nitrogen and kept at –80 °C until use. For immunohistochemistry and subcellular immunogold labelling experiments, samples were fixed immediately.
Clone of MdSDH5 and MdSDH6 genes
RT-PCR and nested PCR were used to isolate cDNA of sorbitol dehydrogenase as described by Yu et al. (2006). Single-stranded cDNA was synthesized from total RNA of apple fruit using PowerScript reverse transcriptase and SMART III Oligonucleotide/CDSIII3′ as the primer (CLONTECH). Degenerate primers (forward primer: 5′-GA(G/A)AACATGGCTG(C/T)(T/C)TGGCT-3′; reverse primer: 5′-GG(T/C)GC(T/C)CC(G/A)AAAGCACGAGC-3′) were designed based on the conserved region of NAD-SDH in Rosaceae plants, Malus domestica (GenBank nos AB016256, AF323505, AF323506, AY053504); Eriobotrya japonica (AB042810); Prunus persica (AB025969); and Prunus cerasus (AY037946). Two sequences (Nos 1 and 2) were obtained having a higher identity with above-mentioned NAD-SDH.
5′-RACE PCR was done with SMART™ III Oligonucleotide primer as the forward primer (5′ -AAGCAGTGGTATCAACGCAGAGTGGCCATTATGGCCGGG-3′) and the specific sequence primers (reverse primer1: 5′-TGTTCTTGAAGTGGTGAACATCACT-3′; reverse primer2: 5′-CCAACAGCCTTCAGCCGAACTCTAA-3′ based on the No. 1 sequence; reverse primer1: 5′-CACCGATGATCAGGACAGTTGTCTC-3′; reverse primer2: 5′-AGTTGTCTCGGGACCAACATTGGCT-3′ based on the No. 2 sequence) as reverse primers. 3′-RACE PCR was performed with the specific sequence primers (forward primer1: 5′-AAGAAACAAATGCCTTGGTCGTGGG-3′; forward primer2: 5′-ATAGGACTTGTTACACTGCTAGCCG-3′ based on the No. 1 sequence and forward primer1: 5′-TTGGTCCCGAGACAACTGTCCTGAT-3′ forward primer2: 5′-GGGCCTATTGGTCTCGTTTCAGTTT-3′ based on the No. 2 sequence) as forward primer and CDS III3′ as reverse primer (CDS III/3′ : 5′-ATTCTAGAGGCCGAGGCGGCCGACATG-3′). The products of 5′ and 3′ RACE were cloned into the pMD-18T vector and sequenced. Then three fragments were spliced, and a pair of new primers in the 5′and 3′ ends, respectively, were designed (forward primer: 5′-TAATTACGGCCGGGGGACAACAAGGGAGCT-3′; reverse primer: 5′-GCGGCATTAAGAGAAGCGAAGGGTTTGAAC-3′ based on the No. 1 sequence and forward primer: 5′-GCATTACGGCCGGGGATCAACAAATCAAAC-3′; reverse primer: 5′-CACCGAGGCGGCCGACATGTTTTTTTTTTT-3′ based on the No. 2 sequence). Two full-length cDNAs were obtained by PCR amplification encoding putative NAD-SDH from apple fruit and registered in GenBank as AY849315 and AY849316, and the products named as MdSDH5 and MdSDH6).
Expression of MdSDH5 and MdSDH6 in E. coli, purification and enzyme activity assay
The entire ORF sequences of MdSDH5 and MdSDH6 were amplified by PCR using pfu DNA polymerase (TAKARA, Dalian Division) with specific oligonucleotide primers (primer for MdSDH5: forward primer 5′-CCTGAATTCGGAAAGGGAGGCATGTCTGA-3′; reverse primer 5′-GGCCTCGAGTTACAGGTTAAACATGACCT-3′ and primer for MdSDH6: forward: 5′-TATGGATCCGGCAAGGGAGGCCAATCCTG-3′; reverse primer: 5′-GCGCCCGGGTTACAATTTGAACATCACCT-3′) and constructed in pGEX-4T-1 plasmid (Amersham Pharmacia Biotech). The PCR products containing EcoRI/XhoI (for MdSDH5) and BamHI/SmaI (for MdSDH6) sites were cloned into the pGEX-4T-1 vector for the expression of GST fusion proteins under the inducement of isopropyl-β-D-thiogalactopyranoside (IPTG). BL21(DE3) cells harbouring the recombinant vector pGEX-4T-1/MdSDH5 and pGEX-4T-1/MdSDH6 were cultured to A600=0.6 at 37 °C, and were induced with 0.2 mM IPTG for 3 h at 37 °C. Cells were pelleted and resuspended in sample buffer, and the cell extract was analysed by SDS–PAGE.
Purification and the enzyme activity assay were performed as described by Ramos et al. (2003). Pellets from a 0.5 l culture were collected by centrifugation, resuspended in 20 mM TRIS-HCl (pH 7.9) buffer with 5 mM imidazole and 500 mM NaCl, and then disrupted by sonication. The lysate was treated with DNaseI for 30 min, and centrifuged at 13 000 g at 4 °C. The pellet was suspended in the same buffer with 6 M guanidine hydrochloride and incubated at 4 °C for 1 h. The solution was diluted with reducing buffer (10 mM DTT in 50 mM TRIS-HCl, pH 8.5, 6 M guanidine hydrochloride), incubated at room temperature for 0.5 h, diluted again with oxidation buffer (50 mM TRIS-HCl, pH 8.5, 5 mM cysteine, 1 mM cystine, 100 mM ZnSO4, and 6 M guanidine hydrochloride). After incubation at room temperature for 0.5 h, the solution was then dialysed in the same buffer with several changes to slow removal of the guanidine hydrochloride at 4 °C for 24 h. The dialysed products were purified by Glutathione Sepharose 4B Column (Amersham Pharmacia Biotech) and analysed by SDS-PAGE. The protein solution was concentrated and the protein concentration was determined by the method of Bradford (1976).
The enzyme activity was determined as described by Yamaguchi et al. (1994) on a spectrophotometer (model UV) by following the reduction of NAD in the presence of sorbitol and by following the oxidation of NADH in presence of fructose at 340 nm (by following the increase in absorbance of NADH at 340 nm). The reaction mixture contained 68 mM TRIS-HCl (pH 9.0), 1.0 mM NAD, and 400 mM sorbitol for the reduction of NAD and 100 mM TRIS-acetate (pH 6.0), 0.05 mM NADH, and 400 mM fructose for the oxidation of NADH. More than three repetitions of the experiments were conducted for each sample.
Expression, purification and antiserum preparation of MdSDH6
The DNA sequence corresponding to the C-terminal fragment of 153 amino acids of MdSDH6 was amplified by using the primers (forward primer 5′-AATGGATCCCTCGTTTCAGTTTTAGCCGC-3′ reverse primer 5′-ATACCCGGGCGCTTCTTCCACCTCCTTC-3′). After digestion with BamHI/SmaI, the PCR product was then cloned into vector pGEX-4T-1 downstream of the GST. The ligated products were used to transform E. coli competent cells. After being analysed as described by Yu et al. (2006), the purified fusion protein (10 mg) was used for standard immunization protocols in rabbits and the polyclonal antiserum was extracted and affinity-purified firstly by HiTrapTM Protein-GHP (Amersham Pharmacia Biotech) and then by an immunosorbent column coupled with GST-MdSDH6 fusion protein. The affinity-purified antiserum was evaluated by Western blotting.
RT-PCR analysis for MdSDH5 and MdSDH6 expression
RT-PCR was used for expression analysis of MdSDH5 and MdSDH6 at the transcription level. Single-stranded cDNA was synthesized from total RNA of apple fruit, leaves using PowerScript reverse transcriptase and SMART III Oligonucleotide/CDSIII3′ as the primer (Clontech). PCR amplification was performed using gene-specific primers (forward primer 5′-GGACAACAAGGGAGCTCATCTA-3′, reverse primer 5′-GAATACCAACACTTAAGGGC-3′ for MdSDH5 and forward primer 5′-CGTGTATTCTGTGTCTTCTGTG-3′, reverse primer 5′-CGGAGATCATGGCTTCTTTAAT-3′ for MdSDH6) and 25 cycles. The apple elongation factor gene (EF-1a, GenBank AJ223969) was used as the control (forward primer: 5′-ATTGTGGTCATTGGYCAYGT-3′; reverse primer: 5′-CCTATCTTGTAVACATCCTG-3′). The RT-PCR products were detected on a 1.0% (w/v) agarose gel electrophoresis.
Transient expression of MdSDH5 and MdSDH6 in Arabidopsis protoplasts
The full-length ORF of MdSDH5 cDNA was PCR amplified by using primers 5′-ATGGGAAAGGGAGGCATGTC-3′ (forward primer) and 5′-CAGGTTAAACATGACCTTAA-3′ (reverse primer). The PCR products were then fused upstream of the enhanced green fluorescence protein (GFP) at the EcoRI/SalI sites in the cauliflower mosaic virus 35S-EGFP vector (p-EZS-NL vector, Dr Ehrhardt, http://deepgreen.stanford.edu). Through the specific primers (5′-ATGGCTGCTTGGCTTGTTGA-3′ as forward primer and 5′-TCAATTTGAACATCACCTTG-3′ as the reverse primer), the full-length ORF of MdSDH6 cDNA was PCR amplified and ligased to XbaI and BamHI sticky ends of pBI221. Protoplasts were isolated from the leaves of 3–4-week-old plants of Arabidopsis (ecotype Columbia) and transiently transformed by using PEG according to Yoo et al. (2007). Fluorescence of GFP was observed by a confocal laser scanning microscope (LSM 510 meta) after incubation at 23 °C for 20 h.
Preparation of subcellular fractions, purity assay and immunoblotting
Subcellular fractions of apple fruit and leaf were prepared essentially according to the method described by Duan et al. (2003). All steps were performed at 4 °C. Plasma membranes and endomembranes were isolated from the microsomes by an aqueous polymer two phase system consisting of Dextran T500 and polyethylene glycol (PEG) 3350. A 60% and a 32% Percoll system was used to isolate the chloroplasts of apple fruit as described by Quick et al. (1995). Protein concentrations of fractions were determined using bovine serum albumin as the standard (Bradford, 1976). The purity of the fractions was evaluated by measuring the activity of the marker enzymes (Shen et al., 2004).
Western blot was essentially as described by Harper et al. (1994). The protein samples were separated on a 12% SDS-polyacrylamide gel. After electrophoresis, proteins in the gels were electrophoretically transferred to a nitrocellulose membrane (0.45 μm, Amersham Pharmacia). The membranes were blocked for 2 h at room temperature with 3% (w/v) BSA and 0.05% (v/v) Tween 20 in a TRIS-buffered saline (TBS) (10 mM TRIS-HCl, pH 7.5 and 150 mM NaCl), and then incubated with gentle shaking for 2 h at room temperature in the rabbit polyclonal antibodies (all diluted 1:1000 in the blocking buffer). After being washed three times for 10 min each in the TBS1 (0.05% (v/v) Tween 20 in TBS buffer), the membranes were incubated with the alkaline phosphatase-conjugated antibody raised in goat against rabbit IgG (diluted 1:1000 in the blocking buffer) at room temperature for 1 h, and then washed three times for 10 min with TBST2 (50 mM TRIS-HCl (pH 7.5) buffer containing 150 mM NaCl and 0.1% (v/v) Tween 20). The alkaline phosphatase reaction was detected by 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium.
Immunohistochemistry
The treatment of the tissues for immunohistochemistry was done essentially as described by Peng et al. (2003). The fruits were separated and cut into small cubes (about 2–10 mm3) and were immediately fixed with a solution of 4% (w/v) paraformaldehyde in 100 mM precooled PBS (pH 7.0). The fixed samples were dehydrated in a graded ethanol series, replaced with xylene, and embedded in paraffin. Microtome sections, 7 μm thick, were placed on glass slides coated with 1% poly-L-lysine solution. After removing the paraffin with xylene, the sections were rehydrated in an ethanol series. The leaves were prepared as frozen sections and immediately fixed with 4% (w/v) paraformaldehyde. After the sections were incubated in blocking buffer [PBS solution with 0.1% Tween-20 (v/v), 1.5% glycine, 5% BSA (w/w)] overnight at 4 °C, they were then labelled with the purified anti-MdSDH rabbit antiserum diluted 200-fold in a PBS solution containing 0.1% Tween-20 (v/v), 1.5% glycine, 0.8% BSA for 2 h at 37 °C. After extensive rinsing with PBS, the samples were incubated in goat anti-rabbit IgG-FITC antibody diluted 100-fold in a PBS solution (containing 0.1% Tween-20 (v/v), 1.5% glycine, 0.8% BSA) for 1 h at 37 °C. The specimens were rinsed in PBS and mounted with 80% glycerol in PBS and observed under a confocal microscope. The fluorescence of FITC was pseudocoloured in green, and the autofluorescence of chloroplast in red.
Subcellular immunogold labelling
The method of subcellular immunogold labelling was essentially as described by Wu et al. (2004). The fruits were cut into small cubes (about 2 mm3) and were immediately fixed with a solution of 4% (w/v) glutaraldehyde in 100 mM precooled phosphate buffer (pH 7.2), and incubated in the same solution for 6 h at 4 °C. After an extensive rinsing with the precooled phosphate buffer, the tissue cubes were post-fixed in 0.1% (w/v) OsO4 overnight at 4 °C. The samples were rinsed with the precooled phosphate buffer and dehydrated through a graded ethanol series. After infiltration for 48 h with Spurr's epoxy resin at 4 °C, the samples were polymerized at 58 °C for 12 h.
All the following procedures were performed at room temperature unless otherwise stated. The ultrathin sections on nickel grids were etched with 560 mM sodium metaperiodate (NaIO4) for 50 min, and 0.1 M HCl for 30 min, respectively. After a rinse with TRIS-buffered saline with Tween 20 (TBST) consisting of 10 mM TRIS (pH 7.4), 500 mM NaCl, and 0.3% (v/v) Tween 20 for 5 min, the sections were incubated in TBST buffer containing 2% (w/v) bovine serum albumin (BSA) for 1 h. Washed with TBST buffer containing 2% (w/v) BSA, they were incubated in rabbit antibody against sorbitol dehydrogenase of apple (diluted 1:50 in TBST buffer containing 0.1% BSA) at 37 °C for 3 h. Following extensive washes with TBST buffer containing 2% (w/v) BSA, the sections were incubated in goat anti-rabbit IgG antibody conjugated with 10 nm gold (diluted 1:100 in TBST buffer containing 0.1% BSA) for 1 h at 37 °C. The sections were rinsed consecutively with TBST buffer containing 2% BSA, TBST buffer and double-distilled water, and were stained with uranyl acetate. After extensively washing with double-distilled water, they were examined with a JEM-100S electron microscope.
The specificity and reliability of the immunogold labelling were tested by two negative controls. In the first one, rabbit preimmune serum was used instead of the rabbit antiserum to test the specificity of the antiserum. In the second one, the antiserum was omitted to test possible unspecific labelling of the goat anti–rabbit IgG antibody–gold conjugate. More than three repetitions of the experiments were conducted for each sample.
Results
Gene cloning, protein expression and enzymatic activity assay of MdSDH5 and MdSDH6
Several MdSDH (Malus domestica sorbitol dehydrogenase) genes, with unique sequences, have been cloned from the apple fruit. To obtain the genuine MdSDH gene sequences from the apple (Malus domestica Borkh cv. Starkrimson), two cDNA fragments were cloned from the apple fruit by RT-PCR using a pair of degenerate primers based on the major motifs of known NAD-SDH sequences. Sequence analysis revealed that these two cDNA fragments had high identity with the known NAD-SDH genes from Rosaceae. The full-length cDNA sequences were obtained by 5′- and 3′-RACE. The two new cDNA sequences were named MdSDH5 (GenBank no. AY849315) and MdSDH6 (GenBank no. AY849316). The full-length MdSDH5 cDNA is 1549 bp and it encodes a 371 amino acid polypeptide, while the MdSDH6 cDNA is 1429 bp and it encodes a polypeptide of 368 amino acids. The predicted molecular mass of both MdSDH5 and MdSDH6 is approximately 40 kDa. Sequence alignment and phylogenetic analysis revealed that MdSDH5 and MdSDH6 share high sequence identity with other apple NAD-SDH (Fig. 1).
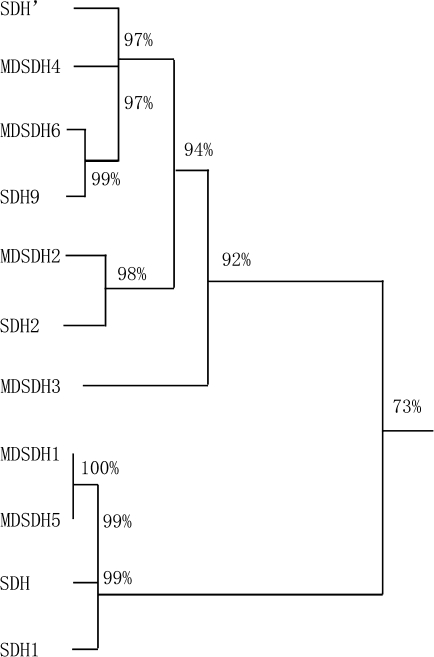
Fig. 1.
Phylogenetic tree of deduced amino acid sequences of MdSDH5*, MdSDH6* and other apple NAD-SDH, MdSDH1 (AAL37293), MdSDH2* (AAL37294), MdSDH3* (AAL37295), MdSDH4 (AAL37296), SDH* (BAA36481), SDH′ (AAL23440), SDH1 (AAP69749), SDH2 (AAP69750), and SDH9 (AAP69753). Asterisks show those isoforms proved to have the activity of sorbitol dehydrogenase in vitro.
To determine the NAD-SDH activity of MdSDH5 and MdSDH6, the open reading frames of MdSDH5 and MdSDH6 were each expressed as GST fusion proteins in E. coli. The induced proteins were analysed by SDS-PAGE, which revealed that these two proteins are both 66 kDa, consistent with the expected molecular mass for MdSDH5 or MdSDH6 (40 kDa) plus GST (26 kDa). The NAD-SDH activity of these two fusion proteins was then analysed in vitro. Their sorbitol oxidation activity was 4–6-fold higher than their fructose reduction activity (Table 1). This result indicates MdSDH5 and MdSDH6 preferentially catalyse the conversion of sorbitol to fructose rather than the reverse, similar to the other isoforms of NAD-SDH from the apple and other Rosaceae plants (Yamaguchi et al., 1994). Based on these results, it is concluded that the cDNAs of MdSDH5 and MdSDH6 cloned from the apple (Malus domestica Borkh cv. Starkrimson) fruit both encode functional sorbitol dehydrogenase.
Table 1.
The enzyme activity of the purified MdSDH5/GST, MdSDH6/GST proteins
| Reaction | Enzyme activity (μmol min−1 mg−1 protein)a |
||
| No enzyme | MdSDH5/GST | MdSDH6/GST | |
| NADH production | 0.00 | 0.78 | 0.61 |
| NADH oxidation | 0.00 | 0.18 | 0.10 |
All the data are the means of three independent repetitions.
The preparation of antibodies of MdSDH6
Since several NAD-SDH genes were cloned from the apple, this suggests that there are multiple NAD-SDH isoforms. To study the distribution and subcellular localization of the NAD-SDH proteins in the apple, antibodies that would potentially recognize as many NAD-SDH proteins as possible were prepared. Because the conserved amino acid sequences of the alcohol dehydrogenase family proteins were primarily found at the N-terminus, the C-terminal 153-amino acids of MdSDH6 were selected for protein expression and antibody preparation (Fig. 2A). This segment contains three predicted antigenic determinants (http://immunax.dfci.harvard.edu/Tools/antigenic.pl) shared by other known NAD-SDH isoforms (Fig. 2B). Western blot analysis revealed that the anti-MdSDH6 serum recognized both the MdSDH6 and MdSDH5 expressed in E. coli (Fig. 3A). This suggests that the anti-MdSDH6 serum can recognize other NAD-SDH family members. Moreover, only one signal was detected by immunoblotting crude protein preparations from the fruit and leaves with the MdSDH6 antiserum (Fig. 3B), which indicates that the anti-MdSDH6 serum only recognizes NAD-SDH protein. The reliability of the immunolabelling experiment with the MdSDH6 antiserum was further demonstrated by control experiments (omission of antiserum or using the preimmune serum instead of antiserum, data not shown).
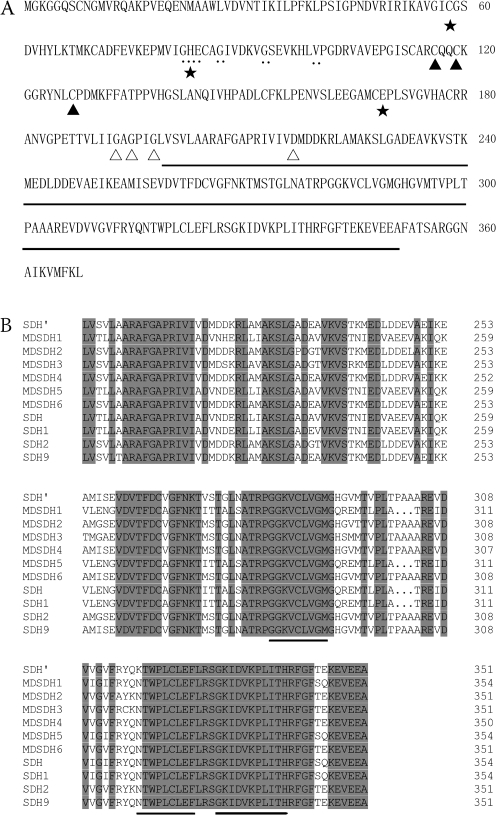
Fig. 2.
Sequence analysis of MdSDH6 and the predicted antigen determinants at the C-terminal shared by all apple NAD-SDH. (A) Sequence analysis of MdSDH6 shows the selected C-terminal of MdSDH6 (underlined) for antibodies preparation and the conserved domains of alcohol dehydrogenase family, zinc-containing alcohol dehydrogenase signature (..), structural zinc binging site (filled triangle), NAD-binding pocket (open triangle) and catalytic zinc binding site (star). (B) C-terminal sequence alignment of all known apple NAD-SDH isoforms (MdSDH1, MdSDH2, MdSDH3, MdSDH4, MdSDH5, MdSDH6, SDH, SDH′, SDH1, SDH2, and SDH9) showing the predicated antigenic determinants (underlined) and the same amino acids (black highlighting).
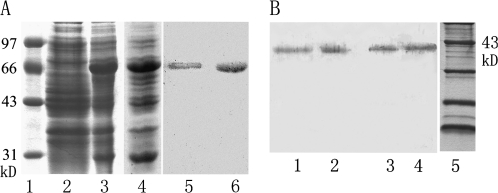
Fig. 3.
Protein expression and western-blot of different fractions probed with the anti-MdSDH6 serum. (A) Coomassie-stained gel of the molecular weight standard (lane 1), the total proteins of E. coli uninduced control (lane 2), expressing MdSDH5/GST (lane 3) and MdSDH6/GST (lane 4). MdSDH5/GST (lane 5) (20 μg) and MdSDH6/GST (lane 6) (20 μg) expressed in E. coli were recognized by anti-MdSDH6 serum. These data show that anti-MdSDH6 serum can recognize not only MdSDH6 but also MdSDH5, which suggest that this antibody can recognize other NAD-SDH members in apple. (B) Twenty μg crude protein of young apple fruit (lane 1, 30 DAFB), mature apple fruit (lane 2, 90 DAFB), young leaf (lane 3), and mature leaf (lane 4) were subjected to Western blot analysis with the anti-MdSDH6 serum. Lane 5 is the molecular weight standard.
Distribution of NAD-SDH in source and sink tissues
The Western blot analysis with the anti-MdSDH6 antibody revealed NAD-SDH protein in the young and mature apple fruit, and young and mature leaves (Fig 3B). To determine the exact localization of the NAD-SDH protein in apple fruit and leaves, an immunohistochemical analysis of fixed samples was performed with the anti-MdSDH6 serum and a FITC-coupled goat anti-rabbit IgG secondary antibody. At 30 d after full bloom (DAFB), the early developmental stage, NAD-SDH was localized in both the flesh cells and the vascular bundle cells of the fruit, except for the vessel elements (Fig. 4A). This distribution pattern was similar at all developmental stages of the fruit (Fig. 4B). In young leaf, the immunofluorescence was detected predominantly in the vascular tissues, but also and at low levels in mesophyll tissue (Fig. 4C). By contrast, NAD-SDH was localized in both vascular tissue and mesophyll tissue of the mature leaf (Fig. 4D). Furthermore, the analysis of gene expression patterns of MdSDH5 and MdSDH6, determined by RT-PCR analysis, showed that both MdSDH5 and MdSDH6 were expressed not only in the fruit and young leaf but also in the mature leaf (Fig. 4F).
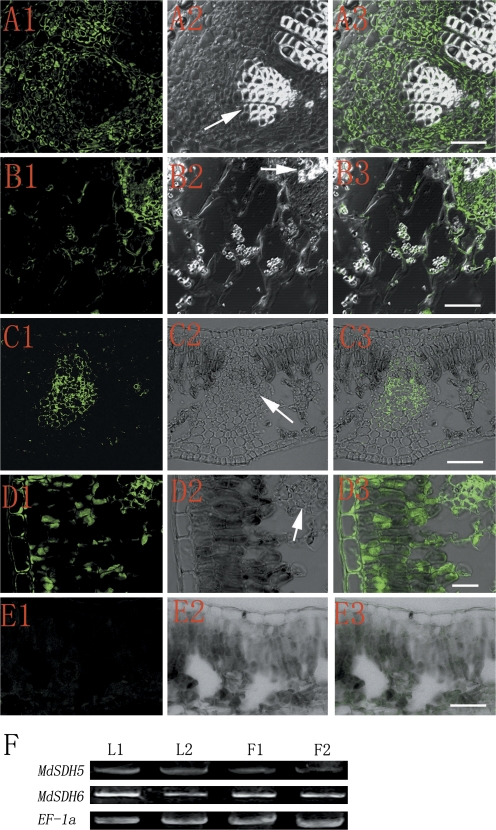
Fig. 4.
Tissue distribution of apple NAD-SDH in fruits and leaves. (A–E) Immunohistochemical assay of NAD-SDH by anti-MdSDH6 serum with FITC-goat anti-rabbit IgG in fruit (embedded in paraffin) 30 DAFB (A) and 60 DAFB (B), young leaf (C), and mature leaf (D). (E) The control using preimmune serum in the mature leaf. Arrows indicate the xylem in (A) and (B), and vascular bundle in (C) and (D). (C), (D), and (E) are the frozen sections. (F) RT-PCR assay for the expression of MdSDH5 and MdSDH6 in young (F1) and mature fruit (F2), young (L1) and mature leave (L2). The specific primers are described in the Materials and methods. The annealing temperature for PCR reactions is 52 °C for MdSDH5 and MdSDH6, and 56 °C for EF-1a.
Subcellular localization of NAD-SDH in fruit flesh parenchyma cells
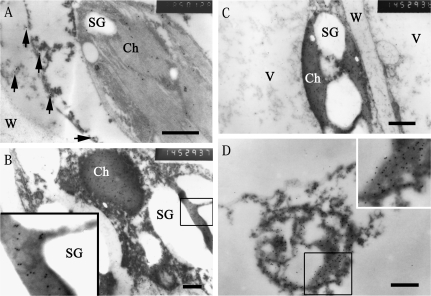
Immunogold labelling with MdSDH6 antiserum showed that NAD-SDH was localized in the cytosol and chloroplast of flesh parenchyma cells at 30 and 60 DAFB (Fig. 5A–C). Compared with 30 DAFB, the volume and amount of the starch store increased at 60 DAFB in the chloroplast, as shown in Fig. 5B and C. At 90 DAFB, the late developmental stage of the fruit, when the fruit flesh parenchyma cell was being degraded, the gold particles were still being found in the degrading cytoplasm even if cellular structure was difficult to discern (Fig. 5D). Furthermore, the amount of immunogold obviously increased with the development of the fruit (Fig. 5A–D), which shows that the expression of sorbitol dehydrogenase is regulated along with fruit development. By contrast, there was no signal in the vacuole or cell wall of flesh parenchymal cells at any developmental stage of the fruit (Fig. 5A–D). In addition, immunogold particles were observed in the companion cells of the vascular bundle with the same localization pattern as in the fruit flesh parenchymal cells (data not shown).
Fig. 5.
Subcellular localization of NAD-SDH in fruit flesh parenchyma cells. (A, B, C) Transmission electron microscopy of fruit flesh parenchyma cells with anti-MdSDH6 serum and goat anti-rabbit IgG antibody conjugated with 10 nm gold. The gold particles are distributed mainly in chloroplast and cytosol of fruit at 30 DAFB (A) and 60 DAFB (B, C). No immunogold particles in the cell wall and vacuole (C). (D) Organelles are indistinguishable at 90 DAFB, and a lot of gold particles are still present in the cytoplasm. Short arrows in (A) indicate the gold particles in the cytosol. Inlets in (B) and (D) are the amplified portions of the boxed-in area of (B) and (D), respectively. Ch, chloroplast; SG, starch grain; V, vacuole; W, cell wall. Bars = 50 μm.
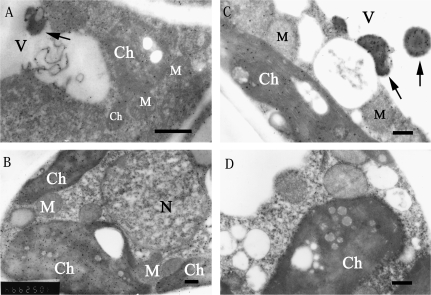
In young leaves, which are also a sink organ, labelled with the anti-MdSDH6 serum, the gold particles are localized predominantly in the cytosol, less so in the chloroplasts, and not in the cell wall or mitochondria (Fig. 6A). By contrast to the fruit, lots of gold particles were observed in electron opaque deposits of vacuoles in young leaves (Fig. 6A). Since the mRNA and protein of MdSDH6 were found in the mesophyll cells of apple mature leaves, the subcellular localization of NAD-SDH in this tissue was determined. In the mature leaves, immunogold labelling was observed primarily in the chloroplasts, cytosol and in electron opaque deposits in the vacuoles (Fig. 6B, C). However, the amount of immunogold particles in chloroplasts increased more obviously in mature leaves than in young leaves (Fig. 6A, B). The immunogold particles were also detected in vascular parenchymal cells and revealed the same subcellular distribution (data not shown). No gold particles were found in the controls labelled with preimmune serum (Fig. 6D), indicating the specificity and reliability of the immunolabelling.
Fig. 6.
Immunogold labelling of NAD-SDH in mesophyll cells of apple leaves. (A–C) Immunogold labelling of NAD-SDH by anti-MdSDH6 serum and gold-conjugated goat anti-rabbit IgG antibody.in young leaves (A), mature leaves (B, C). The gold particles were distributed in the cytosol, chloroplast, and electron opaque deposits of vacuole (arrows). (D) The preimmune serum control in mature leaf, no substantial signal was detected. Ch, chloroplast; M, mitichondria; N, nucleus; V, vacuole. Bars = 50 μm.
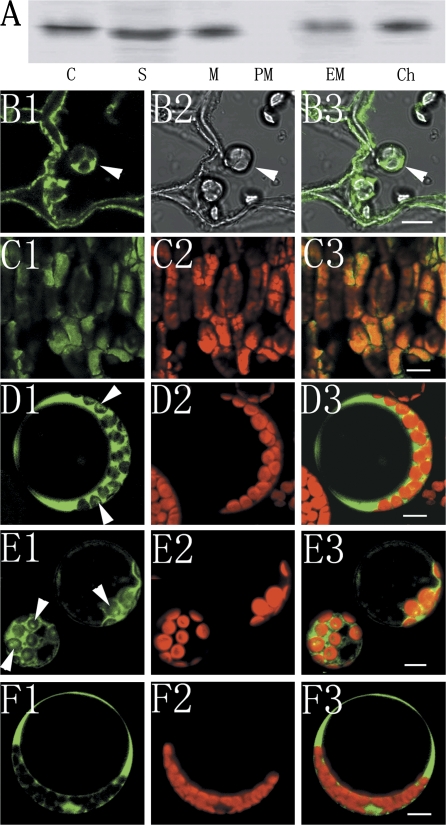
To confirm the chloroplast-localization of NAD-SDH, different subcellular fractions extracted from 60 DAFB apple fruit were analysed by immunoblotting. The signal was detected in crude protein, the soluble fraction, microsomal fraction (including all membrane structures of cells), the endomembrane (the microsomal fraction except for the plasma membrane) and the chloroplast fraction, but not in the plasma membrane fraction (Fig. 7A). It is consistent with the results of the anti-MdSDH6 serum immunogold labelling experiments. The immunohistochemical analysis also revealed a similar localization of NAD-SDH, with green fluorecense (SDH) being found in the chloroplasts of fruit flesh parenchymal cells at 60 DAFB and in the mature leaves (Fig. 7B, C). In addition, the transient expression of 35S::MdSDH5-GFP and 35S::MdSDH6-GFP was produced by polyethylene glycol-mediated transfection of the Arabidopsis mesophyll protoplast to test the chloroplast localization of NAD-SDH. Green fluorescence of MdSDH5/6-GFP fusion protein was found in chloroplasts in addition to the cytosol (Fig. 7D, E).
Fig. 7.
Chloroplast localization of NAD-SDH. (A) Immunoblotting analysis for NAD-SDH in different subcellular fractions. Twenty μg of crude protein (Lane C), soluble fraction (Lane S), microsomal fraction (Lane M), plasma membrane (Lane PM), endomembrane fraction (Lane EM), and chloroplast fraction (Lane Ch) separated from apple fruits were subjected to Western blot analysis with anti-MdSDH6 serum. (B, C) Immunohistochemistry analysis for NAD-SDH with anti-MdSDH6 serum and FITC-goat anti-rabbit IgG antibody. The FITC fluorescence representing SDH is present in chloroplasts of parenchyma tissue in fruit 60 DAFB embedded in paraffin (B) and mesophyll cells in frozen section of mature leaf (C). B1 and C1 represent the FITC fluorescence in green; B2 is the bright image; C2 is the chlorophyll fluorescence. (D, E, F) Transient expression of 35S::MdSDH5-GFP (D), 35S::MdSDH6-GFP (E), and 35S::GFP (F) in the protoplast of Arabidopsis thaliana. The fusion protein is localized in both the cytoplasm and chloroplasts, shown by GFP fluorescence image (D1, E1), Chlorophyll autofluorescence image (D2, E2) and merged image (D3, E3) of laser-scanning confocal microscopy. Arrows in (D) and (E) show the GFP fluorescence in chloroplasts. Bars=10 μm.
Discussion
NAD-SDH has been reported to be distributed primarily in sink organs (Negm and Loescher, 1979, 1981; Yamaguchi et al., 1994). However, Park et al. (2002) and Nosarszewski et al. (2004) found that the sorbitol dehydrogenase genes MdSDH1 and NAD-SDH1 are also expressed in mature leaves. Our present results reveal that the NAD-SDH protein is localized in both the flesh and the vascular tissue of the fruit, and in the vascular tissue and mesophyll tissue in the young and mature leaves, indicating that NAD-SDH is a ubiquitous protein in fruit and leaves of apple. In the mature leaves (source organs), a great deal of NAD-SDH protein is also localized. Loescher et al. (1982) found that NAD-SDH activity in mature leaves was lower than young leaves, but the NAD-SDH activity increased rapidly when leaves became senescent in autumn. Combined with our results, it could be suggested that there is a post-translation regulation mechanism for NAD-SDH in the leaves. When the leaf becomes senescent, NAD-SDH activity appears to be reactivated and, therefore, sorbitol can be hydrolysed to fructose to take part in metabolism. It was reported that NAD-SDH activity in fruit is regulated by NAD-SDH transcription (Yamada et al., 1999) or by sorbitol availability (Archbold, 1999; Teo et al., 2006; Zhou et al., 2006). The mechanism for the regulation of NAD-SDH activity in leaves needs further study.
The majority of soluble carbohydrate in apple leaves, spur and peduncle is sorbitol, but there is only a little in the fruit, which means that the sorbitol is metabolized rapidly after unloading. Despite the central importance of the NAD-SDH in sorbitol metabolism, fruit development and quality, up to now little has been known about the distribution of NAD-SDH in subcellular compartments. Study of the subcellular localization of NAD-SDH is essential for understanding the mechanisms of sorbitol metabolism. Our results show that NAD-SDH is distributed not only in the cytosol but also in the chloroplasts of leaf and fruit cells. Yamaki (1981) found that sorbitol-6-phosphate dehydrogenase, the key enzyme in sorbitol synthesis, is distributed predominantly in the chloroplasts of the apple cotyledon. Nadwodnik and Lohaus (2008) recently found that sorbitol was present in the chloroplasts in mature peach leaves, and the concentration of sorbitol in the chloroplast was 1.5-fold higher than in the cytosol. These results suggest that sorbitol can be synthesized and decomposed in the chloroplast. However, there has been little progress in understanding this process in chloroplasts. It was found that the amount of starch increases along with the elevation of NAD-SDH in the chloroplasts of apple leaves and fruit (Figs 5, 6). Therefore, one possible explanation for NAD-SDH function in the chloroplast is that NAD-SDH might be involved in regulating starch synthesis by converting sorbitol to fructose. In leaves of peach, the concentration of sorbitol in the stroma of the chloroplast is much higher than that of any other sugar, such as myo-inositol, glucose, fructose or sucrose (Nadwodnik and Lohaus, 2008). It has been suggested that sorbitol acts as an osmoregulatory substance to help maintain the osmotic balance of the chloroplast (Nadwodnik and Lohaus, 2008). If this is true, the NAD-SDH present in the chloroplast may take part in the regulation of the sorbitol concentration in the stroma to help maintain the osmotic balance of the chloroplast.
Generally, young leaves and fruits are considered to be sink organs, and mature leaves are the source organs. It was found that NAD-SDH was localized in the vacuoles of young and mature apple leaves, but not in the fruit. Immunoelectron microscopy revealed that NAD-SDH was localized in electron opaque deposits within the vacuoles of leaves. These electron opaque deposits in the vacuoles were often considered to be storage proteins (Olbrich et al., 2007). It is suggested here that the vacuolar NAD-SDH may be the store in young and mature apple leaves. Loescher et al. (1982) found that the NAD-SDH activity in apple mature leaves is very low and increases in autumn. Vacuolar NAD-SDH may be released from vacuoles in aged apple leaves, which contributes to the rapid elevation of its activity when leaves become senescence (Loescher et al., 1982). All of our data show that NAD-SDH has different subcellular localizations in fruit and leaves, and its functions in different sites are waiting for further study.
Acknowledgments
We thank Professor Da-Peng Zhang (Tsinghua University, China) for his kind guidance for this work. This work was supported by grants from the National Natural Science Foundation of China (No. 30770193).
References
- Archbold DD. Carbohydrate availability modifies sorbitol dehydrogenase activity of apple fruit. Physiologia Plantarum. 1999;105:391–395. [Google Scholar]
- Beruter J, Feusi MES, Ruedi P. Sorbitol and sucrose partitioning in the growing apple fruit. Journal of Plant Physiology. 1997;151:269–276. [Google Scholar]
- Bieleski RL. Sugar alcohols. In: Loewus FA, Tanner W, editors. Encyclopaedia of plant physiology, N.S., Vol 13A, Plant carbohydrates. Berlin: Springer; 1982. pp. 158–192. [Google Scholar]
- Bieleski RL, Redgwell RJ. Sorbitol versus sucrose photosynthesis and translocation products in developing apricot leaves. Australian Journal of Plant Physiology. 1985;132:657–668. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Duan CQ, Shen YY, Liang XE, Zhang DP. Membrane-associated protein kinase activities in developing apple fruit. Physiologia Plantarum. 2003;118:105–113. doi: 10.1034/j.1399-3054.2003.00103.x. [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang JF, Lloyd SJ. Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry. 1994;33:7267–7277. doi: 10.1021/bi00189a031. [DOI] [PubMed] [Google Scholar]
- Ito A, Hayama H, Kashimura Y. Partial cloning and expression analysis of genes encoding NAD-dependent sorbitol dehydrogenase in pear bud during flower bud formation. Scientia Horticulturae. 2005;103:413–420. [Google Scholar]
- Loescher WH, Everard JD. Metabolism of carbohydrates in sinks and sources. In: Zamski E, Schafer AA, editors. Photoassimilate distribution in plants and crops: source–sink relationships. New York: Marcel Dekker Inc; 1996. pp. 185–207. [Google Scholar]
- Loescher WH, Marlow GC, Kennedy RA. Sorbitol metabolism and sink–source interconversions in developing apple leaves. Plant Physiology. 1982;70:335–339. doi: 10.1104/pp.70.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen JC, Minchin PEH. Brief look at sorbitol in 1-year-old shoots of apple (Malus domestica) New Zealand Journal of Crop and Horticultural Science. 2005;33:81–87. [Google Scholar]
- Nadwodnik J, Lohaus G. Subcellular concentrations of sugar alcohols and sugars in relation to phloem translocation in Plantago major, Plantago maritima, Prunus persica, and Apium graveolens. Planta. 2008;227:1079–1089. doi: 10.1007/s00425-007-0682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negm FB, Loescher WH. Detection and characterization of sorbitol dehydrogenase from apple callus tissue. Plant Physiology. 1979;64:69–73. doi: 10.1104/pp.64.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negm FB, Loescher WH. Characterization and partial purification of aldose-6-phosphate reductase (alditol-6-phosphate: NADP-1-oxidoreductase. from apple leaves. Plant Physiology. 1981;67:139–142. doi: 10.1104/pp.67.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R. Transport of polyols in higher plants. Plant Physiology and Biochemistry. 2001;39:717–728. [Google Scholar]
- Nosarzewski M, Archbold DD. Tissue-specific expression of SORBITOL DEHYDROGENASE in apple fruit during early development. Journal of Experimental Botany. 2007;58:1863–1872. doi: 10.1093/jxb/erm048. [DOI] [PubMed] [Google Scholar]
- Nosarszewski M, Clements AM, Downie AB, Archbold DD. Sorbitol dehydrogenase expression and activity during apple fruit set and early development. Physiologia Plantarum. 2004;121:391–398. [Google Scholar]
- Olbrich A, Hillmer S, Hinz G, Oliviusson P, Robinson DG. Newly formed vacuoles in root meristems of barley and pea seedlings have characteristics of both protein storage and lytic vacuoles. Plant Physiology. 2007;145:1383–1394. doi: 10.1104/pp.107.108985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Song KJ, Kim MY, Hwang JH, Shin YU, Kim WC, Chung W. Molecular clone and characterization of four cDNAs encoding the isoforms of NAD-dependent sorbitol dehydrogenase from the Fuji apple. Plant Science. 2002;162:513–519. [Google Scholar]
- Peng YB, Lu YF, Zhang DP. Abscisic acid activates ATPase in developing apple fruit especially in fruit phloem cells. Plant. Cell and Environment. 2003;26:1329–1342. [Google Scholar]
- Quick WP, Scheibe R, Neuhaus HE. Induction of hexose-phosphate translocator activity in spinach chloroplasts. Plant Physiology. 1995;109:113–121. doi: 10.1104/pp.109.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos OH, Carmona AK, Selistre-de-Araujo HS. Expression, refolding, and in vitro activation of a recombinant snake venom pro-metalloprotease. Protein Expression and Purification. 2003;28:34–41. doi: 10.1016/s1046-5928(02)00644-7. [DOI] [PubMed] [Google Scholar]
- Shen YY, Duan CQ, Liang XE, Zhang DP. Membrane-associated protein kinase activities in the developing mesocarp of grape berry. Journal of Plant Physiology. 2004;161:15–23. doi: 10.1078/0176-1617-01038. [DOI] [PubMed] [Google Scholar]
- Teo G, Suzuki Y, Uratsu SL, Lampinen B, Ormonde N, Hu WK, Dandekar AM. Silencing leaf sorbitol synthesis alters long-dispartitioning and apple fruit quality. Proceedings of the National Academy of Sciences, USA. 2006;103:18842–18847. doi: 10.1073/pnas.0605873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GL, Zhang XY, Shen YY, Zhang DP. Phloem unloading in developing walnut fruit is symplasmic in the seed pericarp and apoplasmic in the fleshy pericarp. Plant and Cell Physiology. 2004;45:1461–1470. doi: 10.1093/pcp/pch169. [DOI] [PubMed] [Google Scholar]
- Yamada K, Mori H, Yamaki S. Gene expression of NAD-dependent sorbitol. dehydrogenase during fruit development of apple (Malus pumila Mill. var. domestica Schneid. Journal of the Japanese Society for Horticultural Science. 1999;68:1099–1103. [Google Scholar]
- Yamada K, Niwa N, Shiratake K, Yamaki S. cDNA cloning of NAD-dependent sorbitol dehydrogenase from peach fruit and its expression during fruit development. Journal of Horticultural Science and Biotechnology. 2001;76:581–587. [Google Scholar]
- Yamada K, Oura Y, Mori H, Yamaki S. Cloning of NAD-development sorbitol dehydrogenase from apple fruit and gene expression. Plant and Cell Physiology. 1998;39:1375–1379. doi: 10.1093/oxfordjournals.pcp.a029345. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Kanayama Y, Yamaki S. Purification and properties of NAD-dependent sorbitol dehydrogenase from apple fruit. Plant and Cell Physioogy. 1994;35:887–892. [Google Scholar]
- Yamaguchi Y, Kanayama Y, Soejima J, Yamaki S. Changes in the amounts of the NAD+-dependent sorbitol dehydrogenase and its involvement in the development of apple fruit. Journal of the American Society for Horticultural Science. 1996;121:848–852. [Google Scholar]
- Yamaki S, Ishikawa K. Role of four sorbitol related enzymes and invertase in the seasonal alteration of sugar metabolism in apple tissue. Journal of the American Society for Horticultural Science. 1986;111:134–137. [Google Scholar]
- Yamaki S. Subcellular localization of NADP-dependent sorbitol-6-phosphate dehydrogenase in protoplast from apple cotyledons. Plant and Cell Physioogy. 1981;22:359–367. [Google Scholar]
- Yamaki S. ATP-promoted sorbitol transport into vacuoles isolated from apple fruit. Plant and Cell Physiology. 1987;28:557–564. [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yu XC, Li MJ, Gao GF, Feng HZ, Geng XQ, Peng CC, Zhu SY, Wang XJ, Shen YY, Zhang DP. Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiology. 2006;140:558–579. doi: 10.1104/pp.105.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Cheng L, Dandekar AM. Down-regulation of sorbitol dehydrogenase and up-regulation of sucrose synthase in shoot tips of the transgenic apple with decreased sorbitol synthesis. Journal of Experimental Botany. 2006;57:3647–3657. doi: 10.1093/jxb/erl112. [DOI] [PubMed] [Google Scholar]
- Zhou R, Sicher RC, Quebedeaux B. Diurnal changes in carbohydrate metabolism in mature apple leaves. Australian Journal of Plant Physiology. 2001;28:1143–1150. [Google Scholar]