Abstract
Flavonoids are low molecular weight secondary plant metabolites with a myriad of functions. As flavonoids affect auxin transport (an important growth-controlling hormone) and are biologically active in eukaryotes, flavonoid mutants were expected to have undescribed architectural phenotypes. The Arabidopsis thaliana transparent testa (tt) mutants are compromised in the enzymatic steps or transcriptional regulators affecting flavonoid synthesis. tt mutant seedlings were grown on hard-slanted agar (a stress condition), under varying light conditions, and in soil to examine the resulting growth patterns. These tt mutants revealed a wide variety of architectural phenotypes in root and aerial tissues. Mutants with increased inflorescences, siliques, and lateral root density or reduced stature are traits that could affect plant yield or performance under certain environmental conditions. The regulatory genes affected in architectural traits may provide useful molecular targets for examination in other plants.
Keywords: Aglycone, Arabidopsis, auxin, ethylene, flavonoid, plant architecture
Introduction
Flavonoids are secondary metabolites found throughout the plant kingdom. They have diverse roles in plants and animals. In plants, flavonoids are allelopathic (Bais et al., 2004), modulate reactive oxygen levels (reviewed in Taylor and Grotewold, 2005), inhibit auxin transport (reviewed in Peer and Murphy, 2007), act in defence (Treutter, 2005), provide flower colouring (Mol et al., 1998), are required for pollen viability in some species (Coe et al., 1981; Mo et al., 1992; Taylor and Jorgenson, 1992), signal to symbiotic organisms (Redmond et al., 1986; Djordjevic et al., 1987; Wasson et al., 2006), and afford UV protection (Li et al., 1993). Flavonoids may exert their effects indirectly, for example, through the modulation of plant hormones or reactive oxygen species, although direct effects are possible. The evidence for direct affects comes from studies with pollen fertility. Adding the aglycone kaempferol to infertile petunia pollen devoid of flavonoids rapidly restores fertility (Mo et al., 1992) and affects gene transcription (Guyon et al., 2000). Flavonoids also have other roles in plants. They modulate the extent of somatic embryogenesis (Imin et al., 2007) and accumulate in the progenitor cells of root organs in legumes (Morris and Djordjevic, 2006; Mathesius et al., 1998, 2000). It has recently been shown that Arabidopsis seedlings are capable of selectively taking up exogenously applied aglycones, and that the synthesized flavonoid compounds are capable of long distance cell-to-cell movement (Buer et al., 2007, 2008).
Flavonoids are also active in animal systems where they have antioxidant properties (Kandaswami and Middleton, 1994), modulate angiogenesis (Fotsis et al., 1993), interact with kinases (O'Prey et al., 2003), cause apoptosis (Kuntz et al., 1999), and are oestrogenic (Miksicek, 1993). These findings show that flavonoids are bioactive, and the molecular targets include multi-drug resistance transporters (now ABCB transporters; Verrier et al., 2008) in plants and animals (Taylor and Grotewold, 2005).
In Arabidopsis, the flavonoid pathway is well characterized at the genetic, enzymatic, and product levels, and the genes involved are sequenced (Fig. 1). Previous studies explored the irregularities that flavonoid-pathway mutations cause, especially in relation to seed colour and viability (Debeaujon et al., 2000). All the mutants used in this study have defective proanthocyanidin accumulation in the seed that generates the transparent testa (tt) phenotype (Koornneef, 1990). A few architectural phenotypes of flavonoid-pathway mutants are known. These include a delay in the gravity response in tt4, a mutant that cannot make flavonoids (Buer and Muday, 2004). The transparent testa glabra (ttg) mutants lack trichomes (Larkin et al., 1994) and have root hair differences (Galway et al., 1994). Recently, cell shape irregularities were found in rol1 mutants (Ringli et al., 2008), although this mutant has not been used in this study. Brown et al. (2001) concluded that the flavonoid-induced branching phenotype found in the tt4(2YY6) mutant of Arabidopsis was due to the lack of flavonoids, but subsequent experimentation showed that this was due to a second mutation, max4 (Bennett et al., 2006). To our knowledge, no systematic assessment of architectural phenotypes in the transparent testa mutants has been conducted.
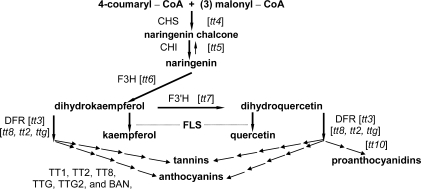
Fig. 1.
The flavonoid pathway in Arabidopsis. Shown are the locations of the transparent testa mutations included in these experiments (in brackets). Mutations tt1, tt2, tt8, and ttg are in regulatory genes involved in several points of the pathway. The gene products affected by these regulatory mutations are TT2 which is a R2R3 repeat MYB transcription factor; TT8 which is a bHLH transcription factor; and TTG which encodes a WD40 repeat gene. These proteins operate as a complex to induce BANYLUS expression to form anthocyanins as downstream products of the pathway. TT1 is a WIP family zinc finger transcription factor and TTG2 is a WRKY type transcription factor that acts downstream of TTG1. The mutated genes and the affected products are: TT3 (DFR: dihydroflavonol reductase); TT4 (CHS: chalcone synthase), TT5 (CHI: chalcone isomerase), TT6 (F3H: flavonol 3-hydroxylase), TT7 (F3′H: flavonol 3′-hydroxylase), and TT10 an enzyme for biflavonol conversion and oxidizing procyanidins to proanthocyanidins in the seed testa. The figure is adapted from Buer et al. (2007), (www.plantphysiol.org), Copyright American Society of Plant Biologists.
Flavonoid accumulation in plants can be visualized using diphenyl boric acid 2-amino ethyl ester (DPBA), a flavonoid-specific dye (Peer et al., 2001; Buer and Muday, 2004; Buer et al., 2007). In Arabidopsis, DPBA forms a highly fluorescent complex with quercetin and kaempferol generating golden and green fluorescence, respectively (Buer et al., 2007; Fig. 2). Other flavonoid intermediates and background phenolics produce greatly reduced fluorescence with DPBA compared to quercetin and kaempferol (Buer et al., 2007).
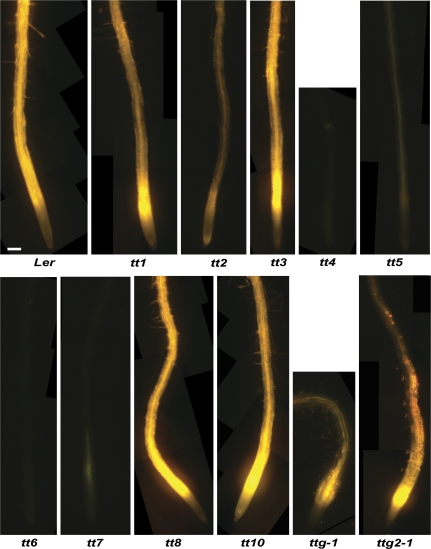
Fig. 2.
DPBA-flavonoid fluorescence in roots of transparent testa mutants varies according to the aglycone that accumulates in the organ. Most seedlings accumulate quercetin that fluoresces a bright golden yellow complexed with DPBA. The mutants tt4 and tt6 have very dim fluorescence from background sinapate esters and naringenin, respectively. Naringenin-DPBA fluoresces over 400 times less strongly than quercetin- or kaempferol-DPBA complexes (Buer et al., 2007). The tt7 mutant shows a dim fluorescence from dihydroquercetin-DPBA and tt5 shows fluorescence from the spontaneous reaction forming naringenin and subsequent downstream products. Several mutants have obvious DPBA fluorescence in the root hairs. Seedlings were analysed for fluorescence 5 d following germination. Seedlings were grown without sucrose in the medium. The bar=100 μm.
Due to the biological activity of flavonoids in plants and animals, we reasoned that there should be more morphological phenotypes other than seed colour and the few previously described architectural phenotypes in the tt mutants. Advantage was taken of the many mutants available in the flavonoid pathway in Arabidopsis in the Landsberg erecta ecotype. These included mutants with genetic lesions affecting enzymatic gene products and mutants in several regulatory genes (Table 1; Fig. 1). Most genes in the flavonoid pathway are single copy (Winkel-Shirlely, 2001), simplifying experimentation. A wide range of new shoot and root morphological phenotypes for the transparent testa mutants is reported here, and it is shown that environmental conditions influence the observed phenotypes.
Table 1.
The transparent testa mutants from Arabidopsis used in these experiments
| Line | Enzyme lesion | Locusa | Mutagen | Reference |
| Ler | Wild type | – | – | – |
| tt1-1 | WIP zinc finger protein | At1G34790 | X-rays | Sagasser et al., 2002 |
| tt2-1 | MYB domain protein 123 | At5G35550 | X-rays | Nesi et al., 2001 |
| tt3-1 | Dihydroflavonol 4-reductase | At5G42800 | X-rays | Shirley et al., 1995 |
| tt4-1 | Chalcone synthase | At5G13930 | EMS | Koornneef, 1990 |
| tt5-1 | Chalcone isomerase | At3G55120 | fast neutrons | Shirley et al., 1995 |
| tt6-1 | Flavonone 3-hydroxylase | At3G51240 | EMS | Pelletier and Shirley, 1996 |
| tt7-1 | Flavonoid 3′-hydroxylase | At5G07990 | EMS | Schoenbohm et al., 2000 |
| tt8-1 | bHLH domain protein | At4G09820 | unk | Nesi et al., 2000 |
| tt10-1 | Laccase-like protein | At5G48100 | EMS | Pourcel et al., 2005 |
| ttg-1 | WD40 repeat protein | At5G24520 | EMS | Walker et al., 1999 |
| ttg2-1 | WRKY TF | At2G37260 | unk | Ishida et al., 2007 |
All seeds were obtained from the Ohio Biotechnology Resource Center, University of Ohio. Abbreviations: TF, transcription factor; EMS, ethylmethane sulphonate; unk, unknown.
Locus locations are from TAIR.
Materials and methods
Chemicals
All chemicals are from Sigma unless otherwise noted. Dihydrokaempferol was obtained from TransMIT, Marburg, Germany.
Plant growth conditions
All seeds are from the Arabidopsis Biotechnology Resource Centre (Ohio State University). Seeds were stored in airtight glass vials after drying at 28 °C and exhibited no loss in germination for months. Seedlings of Landsberg erecta (Ler) and the transparent testa mutants were grown in a single line ∼1 cm apart in unwrapped 150 mm Petri dishes. For each experiment, there were two plates of each mutant with 18 seedlings per plate. The plates were placed at a 45º angle in continuous light in an incubator at 22–23 °C. The MS medium with Gamborg's B5 vitamins (Sigma M0404) was hardened with Type ‘M’ agar (Sigma; at 1.5%) without sucrose unless otherwise stated. The different day lengths other than continuous light were long days (16 h day) and short days (8 h day). All seedlings were analysed at 5 d following germination unless otherwise stated.
To test if high CO2 atmospheres or ethylene caused root ‘nodules’, seedlings were grown in 5000 ppm CO2 obtained by supplementation in a growth cabinet and high ethylene atmospheres by wrapping plates with Nescofilm.
The soil medium for pot growth was the Research School of Biological Sciences, Controlled Environment Facilities, Arabidopsis mix. Pots were 8×8 cm. Feeding naringenin to tt4 seedlings occurred in pots and the seedlings were watered with 10 μM naringenin three times a week, beginning at the basal leaf stage and continuing until pollen formation.
The long-term root growths were cleared by placing root tissue in 100% ethanol overnight, and then an overnight incubation in lactic acid (88%).
DPBA fluorescence
DPBA fluorescence analysis was as previously described by Buer et al. (2007). All experiments were performed on 5-d-old seedlings grown without sucrose unless otherwise noted.
Measurements
Root hair density was determined by counting the root hairs in the focal plane on one side of the root and dividing this number by the length of the root across the counted distance. All measurements were made above the root hair initiation site. Lateral root density was determined by counting the number of lateral roots on both sides of the root and dividing by the root length across the measurement. All measurements were made using ImageJ software freely available from NIH on 600 dpi TIFF images generated on a Canon flatbed scanner. Root skew was measured by the angle scribed from the root/shoot junction to the root tip as measured by ImageJ. At least 25 roots were averaged from three separate experiments. Vertically down has been defined as –90 degrees.
Statistical analysis
All statistical comparisons are with the wild type unless indicated otherwise. Statistically significant differences were determined with Student's t–test assuming equal or unequal variances determined by F–test.
Results
DPBA-flavonoid fluorescence differs between transparent testa mutants
DPBA was used to compare flavonoid accumulation in all the transparent testa mutants (Fig. 2). For the mutants affected in the enzymatic steps of flavonoid synthesis, the results follow that of the known biochemical pathway (Fig. 1). The wild-type roots are golden coloured due to DPBA-quercetin fluorescence. By contrast, tt4 and tt6 show very dim fluorescence due to background sinapate esters and naringenin accumulation, respectively. Mutations downstream of quercetin production showed the expected fluorescence of quercetin-DPBA conjugation. The chalcone isomerase mutant, tt5, showed dim golden fluorescence. This is most likely due to the previously described low-level spontaneous conversion of naringenin chalcone to naringenin (Mol et al., 1985; Cisak and Mielczarek, 1992) and the subsequent formation of quercetin. The examination of several mutants affected in regulatory genes (tt8, ttg, and ttg2) showed the strong golden fluorescence of quercetin. However, tt2 (a mutant with a perturbation in a regulatory MYB transcription factor; Table 1) had reduced golden fluorescence compared to the wild type, indicating that this mutation affects flavonoid accumulation in tissues in addition to the testa.
Root hair morphology differences among the transparent testa mutants
As ttg has root hair differences compared to the wild type (Galway et al., 1994), other transparent testa mutants were examined for alterations to root hairs at 5 d after germination. The measurements included: the distance from the root tip to the root-hair initiation zone, root hair density, root hair length, and root hair DPBA fluorescence (Table 2). Several mutants differed from the wild type. The ttg mutant was similar to the previously reported literature (Galway et al., 1994), except that the distance from the root tip to the root hair initiation zone was greater under our conditions. Considerable variation occurred among the other mutants in relation to the distance from the root tip to the root hair initiation zone, as well as root hair density, and the presence or absence of flavonoid compounds in the root hairs themselves. The tt2 and tt7 mutants are nearly devoid of root hairs and tt6 and ttg had very long root hairs compared to the wild type. By contrast, tt2 and ttg2 had significantly shorter root hairs. The tt7, ttg, and tt2 mutants were affected in most or all root hair parameters examined, whereas, the tt1 mutant was unaffected. The ttg and ttg2 mutants lack flavonoid fluorescence in the root hairs. This is due to the absence of flavonoid fluorescence in the epidermal cell layer of these mutants.
Table 2.
Root hair phenotypes exist between the transparent testa mutants
| Line | Distance to root hair (μm) | Root hair length (μm) | Root hair density (no. μm−1) | Root hair DPBA fluorescence |
| Ler | 853.3±52.9 | 175.5±7.1 | 0.013±0.001 | + |
| tt1-1 | 1014.0±51.0 | 186.1±9.3 | 0.013±0.0003 | + |
| tt2-1 | 1097.9±78.2a | 87.6±10.2b | 0.0093±0.0008b | + |
| tt3-1 | 1072.8±44.9a | 143.6±20.6 | 0.011±0.0007 | + |
| tt4-1 | 1135.9±79.5a | 159.0±12.6 | 0.013±0.001 | — |
| tt5-1 | 1299.2±74.6a | 156.2±18.8 | 0.011±0.0007 | — |
| tt6-1 | 1153.2±92.4a | 415.4±53.9b | 0.012±0.0005 | — |
| tt7-1 | 1382.1±108.8a | 297.1±16.7a | 0.0085±0.0005b | — |
| tt8-1 | 1273.0±126.9a | 111.9±9.9b | 0.012±0.001 | + |
| tt10-1 | 919.7±39.5 | 117.6±11.4 | 0.015±0.001a | + |
| ttg-1 | 1599.1±69.5b | 331.2±21.2b | 0.027±0.004a | — |
| ttg2-1 | 804.0±158.5 | 104.1±16.0a | 0.015±0.001 | — |
All seedlings grew on MS media without sucrose, 1.5% agar, and plates at 45º with continuous light and analysed after 5 d following germination. All statistical comparisons are with the wild type. Statistically significant differences were determined with Student's t–test assuming equal or unequal variances determined by F–test.
P <0.05.
P <0.001.
Root morphology differences in the transparent testa mutants
As sucrose can have hormone-like properties (Smeekens, 2000), the transparent testa mutants were analysed for any root architecture phenotypes using slanted hard-agar plates without sucrose and continuous light. Several root phenotypes emerged (Fig. 3; Table 3). The mutants tt1, tt3, tt4, tt8, and tt10 were altered in the skewing angle of root elongation across the agar surface. The tt4 and tt10 mutants had looping roots, but the looping occurred later in growth in the tt10 seedlings. Several mutants had more (tt6) or fewer (tt4 and tt8) lateral roots compared to the wild type, which was measured as lateral root density (the number of emerged lateral roots mm−1 root length). The tt4 mutant produced significantly fewer lateral roots than the wild type under the conditions used here. This result differed from those previously reported using MS salts, 1.5% sucrose, and a vertical orientation (Brown et al., 2001). However, when lateral root density of tt4 was analysed using the previously reported conditions (with sucrose), the results were congruent (Table 4; Brown et al., 2001).
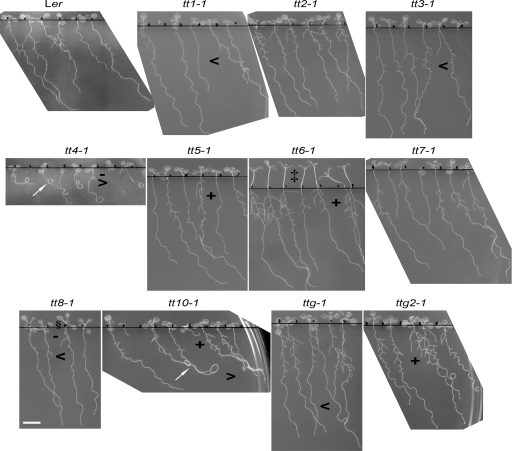
Fig. 3.
Root phenotypes of the various transparent testa mutants. Growing seedlings on 1.5% slanted agar induced several different phenotypes in the seedlings. All seedlings grew without sucrose in the medium under continuous light. The scans were performed at 9 d following germination. Interesting phenotypes were looped roots in tt4 and tt10 (arrow), increased (+) or decreased (–) lateral roots compared to wild type, greater root (>) or lesser (<) skewing, elongated hypocotyls in tt6 (‡), and elongated petioles in tt8 (§). The scale bar=8 mm.
Table 3.
Summary of phenotype differences in transparent testa mutants
| Mutation | Hypocotyl length (mm) | Petiole length (mm) | Skew (º)a | Looping roots (%) | Lateral roots (no.) | Lateral root density (no. mm−1 root length) |
| L. erecta | 1.6±0.07 | 1.2±0.04 | –65.0±2.5 | 0 | 9.5±0.7 | 0.17±0.01 |
| tt1-1 | n.m. | n.m. | –71.5±1.5b | – | 8.6±0.8 | 0.18±0.02 |
| tt2-1 | n.m. | n.m. | –68.4±1.1 | – | 8.9±0.5 | 0.24±0.01c |
| tt3-1 | n.m. | n.m. | –79.7±0.9c | – | 8.3±0.5 | 0.17±0.009 |
| tt4-1 | n.m. | n.m. | –53.1±2.5b | 75.0±8.3 | 1.4±0.3cd | 0.05±0.01cd |
| tt5-1 | n.m. | n.m. | –68.9±1.6 | – | 11.5±0.7 | 0.22±0.01b |
| tt6-1 | 7.8±0.2c | n.m. | –69.0±1.6 | – | 11.9±0.8b | 0.25±0.01c |
| tt7-1 | n.m. | n.m. | –62.4±1.5 | – | 10.3±0.8 | 0.20±0.02 |
| tt8-1 | n.m. | 1.5±0.05c | –77.4±1.2c | – | 5.5±0.6c | 0.13±0.01b |
| tt10-1 | n.m. | n.m. | –42.7±1.3c | 38.9±5.6 | 9.1±0.7 | 0.24±0.02b |
| ttg-1 | n.m. | n.m. | –76.8±1.4c | – | 10.2±1.0 | 0.20±0.02 |
| ttg2-1 | n.m. | n.m. | –62.2±1.0 | – | 11.0±0.7 | 0.23±0.01c |
Growth conditions were MS salts, no sucrose, continuous light, on 1.5% agar, and plates tilted at 45º. Seedlings were scanned 9 d following germination. n.m., not measured
The angle the roots grow across the plate. The view is from the bottom of the plate, through the agar. Vertically down is –90º.
P <0.05 Results of Student's t–test for equal or unequal variance determined by F–test. All comparisons are with Landsberg erecta, the wild type.
P <0.001.
These data are contrary to previous literature, but the growth conditions are entirely different. This is generally tested with at least 1.5% sucrose in the media. See Table 4.
Table 4.
Growth conditions affect lateral rooting in tt4
| Ecotype/mutation | Lateral root density (No. mm−1) |
| L. erecta | 0.43±0.022 |
| tt4-1 | 0.57±0.015a |
| Col | 0.43±0.010 |
| tt4-2 | 0.53±0.013a |
Growth conditions: MS salts, 1.5% sucrose, and vertical orientation. Lateral root density was scored 9 d following germination.
Statistical comparisons are between wild type and mutant within the same ecotype. Comparisons were analysed with F–test for variance and two-tailed Student's t–test depending on results of F–test. P <0.001; n=20.
transparent testa mutants show phenotypes in the aerial tissues
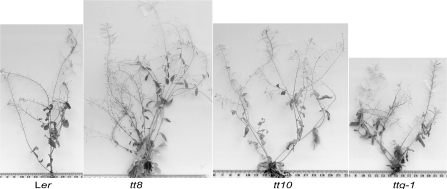
The mutants were examined for aerial phenotypes including inflorescence height, the number of inflorescences, and silique density. After germinating seeds on 9 cm plates, seedlings were transplanted to pots, and grown under long-day conditions. Table 5 summarizes the resulting phenotypes and representative images of the inflorescences are in Fig. 5. Interestingly, tt8, tt10, and ttg had significantly increased inflorescence production compared to the wild type. The tt8 and ttg2 mutants showed increased inflorescence height and ttg showed decreased inflorescence height and increased silique density. The tt4 mutant did not show increased aerial branching compared to the wild type, which is consistent with the published results (Bennett et al., 2006).
Table 5.
Summary of the aerial phenotype differences in selected transparent testa mutants
| Mutation | Inflorescence height (mm) | 1º Inflorescences (no.) | Silique density (no. mm−1) |
| L. erecta | 281.4±10.7 | 1.5±0.1 | 0.17±0.01 |
| tt3-1 | 268.8±11.9 | 1.3±0.1 | 0.24±0.009b |
| tt4-1 | 246.7±24.0 | 1.3±0.2 | 0.26±0.02 |
| tt5-1 | 266.7±10.9 | 1.3±0.3 | 0.27±0.01 |
| tt8-1 | 348.0±15.3b | 4.5±0.3b | 0.25±0.01 |
| tt10-1 | 263.1±13.2 | 3.5±0.4b | 0.30±0.02 |
| ttg-1 | 226.4±6.6b | 3.5±0.3b | 0.37±0.03a |
| ttg2-1 | 335.0±23.6a | 2.3±0.5 | 0.31±0.03 |
Growth conditions: pots under long days and sampling was after 9 weeks, following senescence.
P <0.05, results of Student's t–test for equal or unequal variance determined by F–test. All comparisons are with Landsberg erecta, the wild type. n ≥20.
P <0.001, results of Student's t–test for equal or unequal variance determined by F–test. All comparisons are with Landsberg erecta, the wild type. n ≥20.
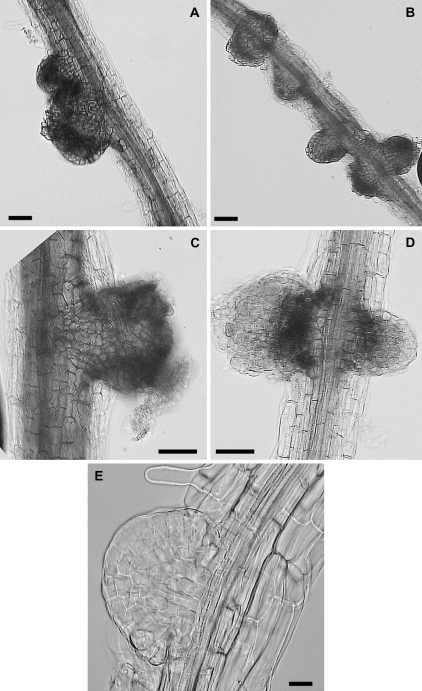
Fig. 5.
Aberrant root outgrowths on tt4 roots at 40 d. (A–D) Aberrant growths that resemble root nodules are shown. Typically, each plant produced 100s of root outgrowths averaging 1 per mm root length, and often the structures were clustered in groups (B). A cleared root outgrowth is shown in (E). Cleared roots indicated that initiation is from the pericycle. Scale bars: (A)–(D) 50 μm; (E) 20 μm.
The effect of exogenous addition of flavonoids on the tt6 long hypocotyl and tt4 looping phenotypes
Growing seedlings on slanted hard-agar plates revealed two additional aerial differences. These included significantly longer hypocotyls on the tt6 mutant seedlings and longer petioles on the tt8 mutant seedlings (Fig. 4). The longer hypocotyls of tt6 plants were also apparent when plants were grown on vertical plates and in the presence of sucrose.
Fig. 4.
The aerial phenotypes in some transparent testa mutants have multiple inflorescences. The multiple inflorescences of tt8, tt10, and ttg-1 are compared to the wild type and clearly have more inflorescences. The photographs were taken 9 weeks following transplanting to pots. The scale is provided by a ruler shown at the bottom of each panel with major gradations in cm.
An attempt was made to reverse the long hypocotyl phenotype in tt6 by adding aglycones downstream of the lesion to the seedlings via the medium. The tt6 hypocotyls grew longer with dihydrokaempferol addition, and the lateral root density increased with the addition of dihydrokaempferol or dihydroquercetin (Table 6). Adding kaempferol or quercetin did not affect hypocotyl length.
Table 6.
The long hypocotyl phenotype and lateral root density in tt6 increases with the addition of DHK
| Ecotype/mutant | MS | DMSO | N | DHK | DHQ | K | Q |
| Hypocotyl length (mm) | |||||||
| Ler | 1.9±0.1 | 2.0±0.1 | 2.0±0.1 | 1.9±0.1 | 1.7±0.1 | 1.7±0.1 | 1.9±0.1 |
| tt6 | 8.5±0.2a | 8.4±0.1a | 8.6±0.2a | 9.2±0.2ab | 8.6±0.2a | 8.8± .2a | 8.7±0.2a |
| Lateral root density (no. mm−1) | |||||||
| Ler | 0.17±0.01 | n.t.c | n.t. | 0.23±0.02 | 0.24±0.01 | n.t. | n.t. |
| tt6 | 0.25±0.01 | n.t. | n.t. | 0.29±0.02c | 0.35±0.01b | n.t. | n.t. |
Seedlings were grown on hard-slanted agar without sucrose and continuous light. Statistical comparisons are between wild type and mutant unless stated otherwise. Comparisons were analysed with F–test for variance and two-tailed Student's t–test depending on the results of the F–test. Data were pooled from two independent experiments, n ≥20.
P <0.001
Comparison between tt6 on MS and DHK, P <0.01. The other data points were not different from tt6 on MS.
Not tested.
The tt4 mutant showed a root looping phenotype in the no sucrose, slanted-agar assay. Adding naringenin to the medium of the tt4 mutant reversed the looping phenotype with increasing concentration (Table 7); however, the phenotype did not completely rescue the phenotype. Growing tt4 seedlings in long day conditions (16 h light) reduced the looping phenotype, and short days (8 h light) abolished it completely. The looping phenotype was also sensitive to normal experimental manipulations. Moving the plates to make daily measurements, for example, abolished root looping. Thus, for all the slanted-plate experiments reported here, the plates remained stationary until scanned.
Table 7.
The looping phenotype in tt4 is reversed by adding naringenin to the medium under continuous light
| Treatment | Ler (% looping roots) | tt4-1 (% looping roots) |
| MS control | 0.43±0.03 | 97.7±2.1a |
| DMSO control | 0.38±0.03 | 89.8±8.3a |
| N (5×10−7 M) | 0.81±0.03 | 74.3±7.7a |
| N (5×10−6 M) | 0.22±0.01 | 55.6±2.9a |
| N (5×10−5 M) | 0.31±0.01 | 9.6±2.9a |
Combined results from five experiments of two replicates of nine seedlings within each experiment. The seedlings grew on hard-slanted agar without sucrose and were stationary until scanned.
Comparisons are between wild type and mutant within each category. Statistical analysis is from two-tailed Student's t–test assuming unequal variance according to F–test of variance, P <0.0001.
Aberrant outgrowths form on tt4 and wild-type roots after prolonged periods in agar supplemented with sucrose
The growth of tt4 seedlings in agar in the presence and absence of naringenin over long periods resulted in aberrant root growths that visually resembled root nodules (Fig. 5). These root outgrowths required at least 40 d to appear. Adding naringenin to the medium retarded their formation. However, after 90 d, the growths appeared on the wild-type roots as well. These root outgrowths formed in addition to normal appearing lateral roots and probably represent aborted lateral roots or, perhaps, determinate lateral roots. Observation of the cleared root outgrowths viewed under higher magnification indicated that they initiate from the pericycle (Fig. 5E). The possibility was checked that these root growths were the product of elevated ethylene or CO2 due to inadequate aeration of the medium. These root outgrowths did not form under high carbon dioxide or ethylene atmospheres (data not shown).
Root application of naringenin restores pollen fluorescence in tt4
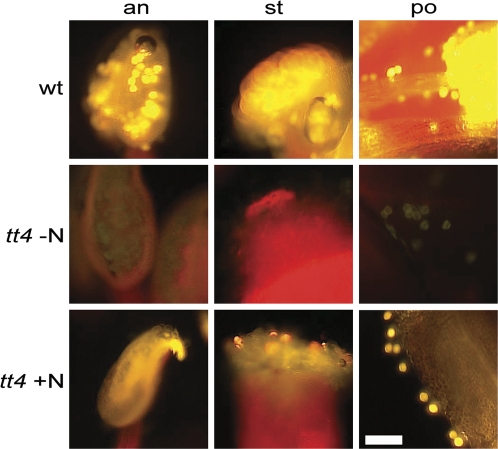
It was demonstrated previously that certain flavonoids are capable of root-to-shoot movement in Arabidopsis (Buer et al., 2007, 2008). It was determined if naringenin addition to the agar medium could restore the wild-type seed colour to tt4 mutants. Naringenin addition did not complement the seed colour. Work by Hsieh and Huang (2007) indicated that flavonoids accumulate in Brassicaceae pollen. It was ascertained if feeding naringenin to tt4 would restore pollen fluorescence. Figure 6 shows that flavonoid fluorescence is restored to tt4 pollen by feeding 10 μM naringenin to seedlings growing in soil under long-day conditions. These results are consistent with reciprocal grafts where wild-type cross-feeding occurs in wild-type/tt4 combinations (Buer et al., 2008).
Fig. 6.
Feeding naringenin to tt4 restores flavonoid fluorescence to pollen. tt4 seedlings were watered three times weekly with 10 μM naringenin. Following pollen formation, flower tissue was analysed for flavonoid fluorescence using DPBA. The wild-type fluorescence is similar to that reported by Peer et al. (2001). Abbreviations: an, anther; st, stigma; po, pollen. Bar=100 μm.
Discussion
New architectural phenotypes in transparent testa mutants
Although extensively studied, the transparent testa mutants have little information available on specific architectural phenotypes resulting from flavonoid level perturbations. This study examined the regulatory and enzymatic mutants affected in the flavonoid pathway from the Landsberg erecta ecotype and showed new phenotypes in these mutants. Mutations in the various genes of the pathway led to differential fluorescence in the mutants caused by alterations in flavonoid accumulation. The type and intensity of the DPBA-flavonoid fluorescence observed was due to the presence or absence of flavonoids or the intermediates that accumulate because of pathway alterations (Peer et al., 2001). The mutants tt3, tt8, ttg, and tt10 are blocked from producing anthocyanins and accumulated more quercetin than the wild type except for tt2. Since TT2 occurs in the same part of the flavonoid pathway, one would expect the accumulation of quercetin to be equal in these mutants. The TT2 gene expresses strongly and transiently in immature seed and works synergistically with TT8, TTG1, and BANYLUS to synthesize the downstream products of the flavonoid pathway (Nesi et al., 2001). Although, Nesi et al. (2001) did not detect TT2 transcript in seedlings, leaves, or stems at 4 d, or in roots on 10 d, these results clearly show that, relative to the control, flavonoid levels are affected in the roots of the tt2, tt8, and ttg1 mutants at 5 d. It is possible that Nesi et al. (2001) missed TT2 expression in roots under their time constraints. It is also possible that TT2 expresses at very low levels or in a few select cells.
Several new architectural defects were discovered in the tt mutants compared to the wild type. These differences included the root looping phenotypes in tt4 and tt10, increased lateral root density in tt5, tt6, and tt10, decreased lateral root density in tt4 if grown on hard slanted agar, and the long hypocotyl phenotype in tt6. The regulatory mutant, tt8, had a long petiole phenotype. Differential effects in root hair length or density or the distance from the root tip to the root hair initiation zone were found in all mutants except tt1, with tt2, tt7, and ttg1 affected in all three measures. The tt8, tt10, and ttg mutants also had increased inflorescence production compared with the wild type and there were differences in silique density, inflorescence height, and number in ttg. The absence of flavonoid fluorescence in ttg and ttg2 root hairs is because flavonoid fluorescence is absent from the root epidermal cell layer (Fig. 2). Specific localization of flavonoid fluorescence was noted previously in tt4 (Buer et al., 2007), so such an occurrence in other transparent testa mutants is possible.
The distance from the root tip to when root hairs began elongating in ttg was considerably greater in our experiments than reported by Galway et al. (1994). This may be an ethylene-induced difference; indeed the plates were wrapped with Parafilm (Schiefelbein and Somerville, 1990), and ethylene accumulation occurs under these circumstances (Buer et al., 2003), while our experiments were in unwrapped plates. The root hair density in the ttg mutant in our experiments was about double the density compared to the wild type, similar to previous work (Galway et al., 1994).
Flavonoid interactions with auxin, ethylene, and touch perception
One possible explanation for these phenotypes is the effect that certain flavonoids (especially quercetin and kaempferol) exert on auxin levels in the plant through modulation of auxin transport (Jacobs and Rubery, 1988). Auxin is thought to regulate lateral root formation (reviewed in Fukaki et al., 2007) and indeed, Dubrovsky et al. (2008) show that localized auxin levels are the critical determinant in setting up stem cell niches for lateral root formation. A connection with root looping and slanting across the agar surface and auxin fluxes is also apparent (Lucas et al., 2008), and the formation of lateral roots at the apex of root waves depends on auxin (De Smet et al., 2007). Root hair formation is also related to auxin transport and ethylene levels (reviewed in Bibikova and Gilroy, 2003), and previous reports describe root hair phenotypes in flavonoid mutants (Galway et al., 1994; Bharti and Khurana, 2003; Mo et al., 1992).
As discussed above, growing seedlings on hard-slanted agar caused several phenotypes, and there are several postulates as to why these differences occur. It is suggested that the regular waving, looping, and skewing is a combination of thigmotropism and a positive response to gravitropism (Okada and Shimura, 1990) or root–gel interactions (Thompson and Holbrook, 2004). Ethylene is an influencing factor (Buer et al., 2003), as well as nutrient conditions in the medium including sucrose (Buer et al., 2000). A new mutant in Arabidopsis with resistance to auxin also has perturbed root phenotypes (Fortunati et al., 2008). Complicating matters are experiments that show an interaction between ethylene and flavonoids (Buer et al., 2006). Many mutants have aberrant root phenotypes, but it is still unclear how these growth patterns are manifested (Oliva and Dunand, 2007), but many of them occur in auxin transport mutants. The lack of auxin transport inhibition during the oscillations from positive to negative gravitropic responses possibly caused the root looping phenotype in tt4. Adding naringenin to the medium nearly reversed the looping phenotype from tt4 suggesting a direct role for flavonoids in this phenotype. Ringli et al. (2008) also noted auxin-dependent and independent effects in plant growth and cell shape in flavonoid glycoside mutants. A recent report implicated Multidrug Resistance-Like4 (MDR4) (now ABCB4; Verrier et al., 2008) regulation by flavonols during gravitropic responses showing that ABCB4 and TT4 are epistatic (Lewis et al., 2007).
Daylength and the manual movement of plates affected the presence or absence of the root-looping phenotype in tt4. Because of this, the plates remained stationary during these experiments. It was noted that tt4 roots of pot-grown plants do not display a root-looping phenotype. Therefore, researchers should consider the use of more realistic growth conditions if the architectural phenotypes shown in this study are translated to other plant systems.
Analysis of flavonoid pathway mutants: fertile ground to improve plant yield or tailor plant architecture?
Some phenotypes are worth further investigation for the possibility of increasing yields or adapting plants to local soil conditions. For example, multiple inflorescences occurred in tt8, tt10, and ttg. Other mutants showed possible desirable yield-enhancing phenotypes such as shorter stature (ttg) and significantly increased silique density (tt3 and ttg). Other mutants showed increased lateral root density or root hair density, traits beneficial for growth in poor soil conditions (Lynch, 2007). The flavonoid regulatory mutants (tt1, tt2, tt8, and ttg) are difficult to interpret as they operate in concert at several points in the pathway and effects of the mutations are probably pleiotropic.
There may be some possible problems applying these phenotypes to agronomy. Many of the early flavonoid pathway mutants are UV sensitive (Li et al., 1993), which could be problematic for field-grown plants. However, the susceptibility to UV is compensated for by higher sinapate ester production in tt mutants (Sheahan, 1996). The tt8 and ttg regulatory mutants accumulate more quercetin than the wild type, and although UV sensitivity was not tested, it is probably not an issue because of this accumulation. Seed storage problems have been reported for the tt mutants (Debeaujon et al., 2000), which would be an issue related to agronomic uses.
The mutations responsible for these plant architecture phenotypes with the potential to affect yield occur in transcription factors that are the potential targets of the second generation of molecular biology (Busov et al., 2008; Century et al., 2008). Major advances in agronomy are necessary to increase yields to feed the projected nine billion-world population that will require food by 2050 (Cohen, 2003).
Low-level quercetin production in tt5
The individual mutants in the enzymatic steps within the flavonoid pathway are relatively straightforward, as they accumulate intermediates consistent with the corresponding lesions in the pathway. Since the mutated genes occur as single copies, they are expected to be solely responsible for the results found, unless flavonoid perturbations have pleiotropic effects or the mutations are leaky. The only mutant that gave a less definitive result was tt5 (chalcone isomerase) where there was evidence of trace amounts of golden fluorescence (indicative of quercetin formation). Previous experimentation with this tt5 allele showed naringenin chalcone accumulation at the root–shoot junction plus an unidentified peak with a chalcone and flavanone skeleton with a retention time equal to quercetin (Peer et al., 2001), and they speculated on a spontaneous isomerization of naringenin chalcone to form naringenin. Shirley et al. (1995) and Pelletier et al. (1999) showed kaempferol in seeds and flowers by thin-layer chromatography and HPLC, respectively, also supporting the hypothesis that the tt5 mutation does not lead to a complete metabolic block at chalcone isomerase. There is experimental evidence of the spontaneous isomerization of naringenin chalcone to naringenin, which occurs at physiological pH (Mol et al., 1985; Cisak and Mielczarek, 1992). This spontaneous isomerization is consistent with the low levels of golden fluorescence seen in experiments with tt5.
The long hypocotyl phenotype of tt6
The inability to reverse the tt6 long hypocotyl phenotype by adding dihydrokaempferol may indicate that naringenin accumulation caused the extension of the tissue. Supplementation with dihydrokaempferol would restore downstream flavonoid synthesis past the block in the pathway; however, it would not alleviate the accumulation of naringenin due to the mutation. Indeed, adding dihydrokaempferol to tt6 seedlings increased the hypocotyl length. There are indications the mutant may be leaky (Peer et al., 2001; Wiseman et al., 1998), but we did not see any downstream flavonoid products in our DPBA staining experiments. This phenotype requires further investigation.
The long-hypocotyl phenotype in tt6 is similar to that of the hy5 mutant in Arabidopsis (Lee et al., 2007), which lacks flavonoids in the root and has increased auxin transport (Sibout et al., 2006). HY5 is a transcriptional regulator that binds to the chalcone synthase promoter (Lee et al., 2007).
Phenotypes of tt4 mutants
Interestingly, day length was important for the looping phenotype seen in tt4 roots. The root looping decreased with decreasing day length and disappeared entirely when seedlings were grown under 8 h light regimes. Although light regulates flavonoid-producing enzymes, it is difficult to decipher how day length interacts with flavonoid levels and root gravi- and thigmotropism.
Recent experiments by Hsieh and Huang (2007) probably explain the inability to revert tt4 seed colour back to the wild type by adding exogenous naringenin to plants. They showed that flavonoids in the Brassicaceae end up in the pollen surface rather than the seed testa. The testa tissue derives from ovular tissue, and thus is maternal in origin (Debeaujon et al., 2000). Adding exogenous naringenin to tt4 seedlings from the basal leaf stage until pollen formation resulted in the restoration of flavonoid fluorescence in the pollen coat at a slightly lower level than the wild type under our conditions. This supports the long-distance movement of flavonoids in tt4 plants shown previously (Buer et al., 2007, 2008).
The root outgrowths during long-term seed colour complementation experiments are remotely similar to aberrant lateral roots created by drought in Brassicaceae (Vartanian et al., 1994). We believe the outgrowths are aborted lateral roots, as they arise from the pericycle. The seedlings also have normal lateral roots intermingled with these outgrowths (data not shown) and why some lateral roots would abort and not others is not understood.
Conclusion
Perturbation in flavonoid levels resulting from mutations directly or indirectly regulating the flavonoid pathway caused a wide range of morphological features of Arabidopsis plants. Overall, flavonoid effects on auxin movement are a unifying theme that may explain many of the phenotypes observed in this study. However, it is unclear whether flavonoids act directly as regulatory agents or indirectly through auxin accumulation or movement, or both (Peer and Murphy, 2007). The recent work by Santelia et al. (2008) supports modulation through PIN localization. The direct effect of flavonoids on plant growth and architecture is the focus of our further experiments within the transparent testa mutants.
Acknowledgments
The Australian Research Council Centre of Excellence for Integrated Legume Research (project no. CEO348212) is acknowledged for the grant supporting this work. Discussions with colleagues in the Centre of Excellence are appreciated.
References
- Bais HP, Park S-W, Weir TL, Callaway RM, Vivanco JM. How plants communicate using the underground information superhighway. Trends in Plant Science. 2004;9:26–32. doi: 10.1016/j.tplants.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Bennett T, Sieberer T, Willett B, Booker J, Luschnig C, Leyser O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Current Biology. 2006;16:553–563. doi: 10.1016/j.cub.2006.01.058. [DOI] [PubMed] [Google Scholar]
- Bharti AK, Khurana JP. Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana (ecotype Estland) impaired in flavonoid biosynthetic pathway. Plant Science. 2003;165:1321–1332. [Google Scholar]
- Bibikova T, Gilroy S. Root hair development. Journal of Plant Growth Regulation. 2003;21:383–415. [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiology. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Masle J, Wasteneys GO. Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant and Cell Physiology. 2000;41:1164–1170. doi: 10.1093/pcp/pcd042. [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. The Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiology. 2007;145:478–490. doi: 10.1104/pp.107.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. Implications of long-distance flavonoid movement in Arabidopsis thaliana. Plant Signaling and Behavior. 2008;3:415–417. doi: 10.4161/psb.3.6.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Sukumar P, Muday GK. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiology. 2006;140:1384–1396. doi: 10.1104/pp.105.075671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Wasteneys GO, Masle J. Ethylene modulates root-wave responses in Arabidopsis. Plant Physiology. 2003;132:1085–1096. doi: 10.1104/pp.102.019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov VB, Brunner AM, Strauss SH. Genes for control of plant stature and form. New Phytologist. 2008;177:589–607. doi: 10.1111/j.1469-8137.2007.02324.x. [DOI] [PubMed] [Google Scholar]
- Century K, Reuber TL, Ratcliffe OJ. Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiology. 2008;147:20–29. doi: 10.1104/pp.108.117887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisak A, Mielczarek C. Practical and theoretical aspects of flavanone-chalcone isomerisations. Journal of the Chemical Society Perkin Transactions. 1992;2:1603–1607. [Google Scholar]
- Coe EH, McCormick SM, Modena SA. White pollen in maize. The Journal of Heredity. 1981;72:318–320. [Google Scholar]
- Cohen JE. Human population: the next half century. Science. 2003;302:1172–1175. doi: 10.1126/science.1088665. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Redmond JW, Batley M, Rolfe BG. Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO Journal. 1987;6:1173–1179. doi: 10.1002/j.1460-2075.1987.tb02351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proceedings of the National Academy of Sciences, USA. 2008;105:8790–8794. doi: 10.1073/pnas.0712307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunati A, Piconese S, Tassone P, Ferrari S, Miglaccio F. A new mutant of Arabidopsis disturbed in its roots, right-handed slanting, and gravitropism defines a gene that encodes a heat-shock factor. Journal of Experimental Botany. 2008;59:1363–1374. doi: 10.1093/jxb/ern047. [DOI] [PubMed] [Google Scholar]
- Fotsis T, Pepper M, Adlercreutz H, Fleischmann G, Hase T, Montesano R, Schweigerer L. Genistein, a dietary-derived inhibitor of in vitro angiogenesis. Proceedings of the National Academy of Sciences, USA. 1993;90:2690–2694. doi: 10.1073/pnas.90.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Okushima Y, Tasaka M. Auxin-mediated lateral root formation in higher plants. International Review of Cytology. 2007;256:111–137. doi: 10.1016/S0074-7696(07)56004-3. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Developmental Biology. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Guyon VN, Astwood JD, Garner EC, Dunker AK, Taylor LP. Isolation and characterization of cDNAs expressed in the early stages of flavonol-induced pollen germination in petunia. Plant Physiology. 2000;123:699–710. doi: 10.1104/pp.123.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Huang AHC. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. The Plant Cell. 2007;19:582–596. doi: 10.1105/tpc.106.049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Wu T, Rolfe BG. Factors involved in root formation in Medicago truncatula. Journal of Experimental Botany. 2007;58:439–451. doi: 10.1093/jxb/erl224. [DOI] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. The Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Kandaswami C, Middleton E., Jr Free radical scavenging and antioxidant activity of plant flavonoids. Advances in Experimental Medical Biology. 1994;366:351–376. doi: 10.1007/978-1-4615-1833-4_25. [DOI] [PubMed] [Google Scholar]
- Koornneef M. Mutations affecting the testa color in Arabidopsis. Arabidopsis Information Services. 1990;27:1–4. [Google Scholar]
- Kuntz S, Wenzel U, Daniel H. Comparative analysis of the effects of flavonoids on proliferation, cytotoxicity, and apoptosis in human colon cancer cell lines. European Journal of Nutrition. 1999;38:133–142. doi: 10.1007/s003940050054. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. The Plant Cell. 1994;6:1065–1076. doi: 10.1105/tpc.6.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis Multidrug Resistance-Like ABC transporter genes. The Plant Cell. 2007;19:1838–1850. doi: 10.1105/tpc.107.051599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ou-Lee T-M, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. The Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Godin C, Jay-Allemand C, Laplaze L. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. Journal of Experimental Botany. 2008;59:55–66. doi: 10.1093/jxb/erm171. [DOI] [PubMed] [Google Scholar]
- Lynch JP. Roots of the second green revolution. Australian Journal of Botany. 2007;55:493–512. [Google Scholar]
- Mathesius U, Bayliss C, Weinmann JJ, Schlaman HRM, Spaink HP, Rolfe BG, McCully ME, Djordjevic MA. Flavonoids synthesized in cortical cells during nodule initiation are early developmental markers in white clover. Molecular Plant–Microbe Interactions. 1998;11:1223–1232. [Google Scholar]
- Mathesius U, Weinmann JJ, Rolfe BG, Djordjevic MA. Rhizobia can induce nodules in white clover by ‘hijacking’ mature cortical cells activated during lateral root development. Molecular Plant–Microbe Interactions. 2000;13:170–182. doi: 10.1094/MPMI.2000.13.2.170. [DOI] [PubMed] [Google Scholar]
- Miksicek RJ. Commonly occurring plant flavonoids have estrogenic activity. Molecular Pharmacology. 1993;44:37–43. [PubMed] [Google Scholar]
- Mo Y, Nagel C, Taylor LP. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proceedings of the National Academy of Sciences, USA. 1992;89:7213–7217. doi: 10.1073/pnas.89.15.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends in Plant Science. 1998;3:212–217. [Google Scholar]
- Mol JNM, Robbins MP, Dixon RA, Veltkamp E. Spontaneous and enzymic rearrangement of naringenin chalcone to flavanone. Phytochemistry. 1985;24:2267–2269. [Google Scholar]
- Morris AC, Djordjevic MA. The Rhizobium leguminosarum biovar trifolii ANU794 induces novel developmental responses on the subterranean clover cultivar Woogenellup. Molecular Plant–Microbe Interactions. 2006;19:471–479. doi: 10.1094/MPMI-19-0471. [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. The Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- Olivia M, Dunand C. Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytologist. 2007;176:37–43. doi: 10.1111/j.1469-8137.2007.02184.x. [DOI] [PubMed] [Google Scholar]
- O'Prey J, Brown J, Fleming J, Harrison PR. Effects of dietary flavonoids on major signal transduction pathways in human epithelial cells. Biochemical Pharmacology. 2003;66:2075–2088. doi: 10.1016/j.bcp.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiology. 2001;126:536–548. doi: 10.1104/pp.126.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. Flavonoids and auxin transport: modulators or regulators? Trends in Plant Science. 2007;12:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Burbulis IE, Winkel-Shirley B. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Molecular Biology. 1999;40:45–54. doi: 10.1023/a:1026414301100. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Shirley BW. Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Plant Physiology. 1996;111:339–345. doi: 10.1104/pp.111.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul J-M, Kerhoas L, Caboche M, Lepiniec L. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. The Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond JW, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG. Flavones induce expression of nodulation genes in Rhizobium. Nature. 1986;323:632–635. [Google Scholar]
- Ringli C, Biogler L, Kuhn BM, Leiber R-M, Diet A, Santelia D, Frey B, Pollmann S, Klein M. The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. The Plant Cell. 2008;20:1470–1481. doi: 10.1105/tpc.107.053249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasser M, Lu G-H, Hahlbrock K, Weisshaar B. A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes and Development. 2002;16:138–149. doi: 10.1101/gad.212702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelia D, Henrichs S, Vincenzetti V, et al. Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. Journal of Biological Chemistry. 2008;283:31218–31226. doi: 10.1074/jbc.M710122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. The Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biological Chemistry. 2000;381:749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- Sheahan JJ. Sinapate esters provide greater UV-B attenuation than flavonoids in Arabidopsis thaliana (Brassicaceae) American Journal of Botany. 1996;83:679–686. [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlates with increased constitutive auxin signalling. PLoS Genetics. 2006;2:1898–1911. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. Sugar-induced signal transduction in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Grotewold E. Flavonoids as developmental regulators. Current Opinion in Plant Biology. 2005;8:317–323. doi: 10.1016/j.pbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. Conditional male fertility in chalcone synthase-deficient petunia. Journal of Heredity. 1992;83:11–17. [Google Scholar]
- Thompson MV, Holbrook NM. Root–gel interactions and the root waving behavior of Arabidopsis. Plant Physiology. 2004;135:1822–1837. doi: 10.1104/pp.104.040881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biology. 2005;7:581–591. doi: 10.1055/s-2005-873009. [DOI] [PubMed] [Google Scholar]
- Vartanian N, Marcotte L, Giraudat J. Drought rhizogenesis in Arabidopsis thaliana. Plant Physiology. 1994;104:761–767. doi: 10.1104/pp.104.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, et al. Plant ABC proteins: a unified nomenclature and updated inventory. Trends in Plant Science. 2008;13:151–159. doi: 10.1016/j.tplants.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. The Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosythesis phenotypes. Proceedings of the National Academy of Sciences, USA. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]