Abstract
The gibbon genome exhibits extensive karyotypic diversity with an increased rate of chromosomal rearrangements during evolution. In an effort to understand the mechanistic origin and implications of these rearrangement events, we sequenced 24 synteny breakpoint regions in the white-cheeked gibbon (Nomascus leucogenys, NLE) in the form of high-quality BAC insert sequences (4.2 Mbp). While there is a significant deficit of breakpoints in genes, we identified seven human gene structures involved in signaling pathways (DEPDC4, GNG10), phospholipid metabolism (ENPP5, PLSCR2), β-oxidation (ECH1), cellular structure and transport (HEATR4), and transcription (ZNF461), that have been disrupted in the NLE gibbon lineage. Notably, only three of these genes show the expected evolutionary signatures of pseudogenization. Sequence analysis of the breakpoints suggested both nonclassical nonhomologous end-joining (NHEJ) and replication-based mechanisms of rearrangement. A substantial number (11/24) of human–NLE gibbon breakpoints showed new insertions of gibbon-specific repeats and mosaic structures formed from disparate sequences including segmental duplications, LINE, SINE, and LTR elements. Analysis of these sites provides a model for a replication-dependent repair mechanism for double-strand breaks (DSBs) at rearrangement sites and insights into the structure and formation of primate segmental duplications at sites of genomic rearrangements during evolution.
Chromosomal evolution in primates has been investigated at several levels of resolution, including comparative chromosome banding (Yunis and Prakash 1982), gene mapping (Turleau et al. 1983), cross-species chromosomal painting (Jauch et al. 1992; Murphy et al. 2005), comparative genome hybridization painting (Carbone et al. 2006), and fluorescent in situ hybridization (FISH) (Wienberg 2005). In general, linkage groups, gene order, and function have remained relatively unchanged since the common catarrhine primate ancestor (Haig 1999). Recent studies have not only identified the role of segmental duplications in disease and evolution but have also supported a nonrandom “fragile-breakage” model for chromosomal rearrangements (Armengol et al. 2003; Pevzner and Tesler 2003; Bailey et al. 2004). Overall, ∼40% of chromosomal rearrangements are associated with segmental duplications in mammals (Bailey and Eichler 2006). Segmental duplications are also a major impetus for the evolution of novel genes and gene functions by duplication and domain accretion (Eichler 2001; Samonte and Eichler 2002). However, in certain primate lineages, the position of chromosomal breaks and the evolutionary rate of rearrangements follow unpredictable patterns (O'Brien and Stanyon 1999) and the role of segmental duplications is not well established.
Gibbons, extant genera among the hominoids, show both anatomical and behavioral specializations. Compared with other apes, gibbons are small, slender, and agile, exhibit no sexual dimorphism, and have very long arms adapted for a spectacular arm swinging locomotion called “brachiation” (Clutton-Brock et al. 1977; Gebo 1996; Plavcan 2001; Usherwood and Bertram 2003). Gibbons have loud vocalizations and live in small monogamous families composed of a mated pair and offspring (Harcourt et al. 1981; Plavcan 2001; Dooley and Judge 2007). In contrast to other apes, which show limited chromosomal variation, gibbons (family Hylobatidae) exhibit rapid chromosomal evolution with a diverse karyotypic pattern among different species and subspecies (O'Brien and Stanyon 1999; Muller et al. 2003). Humans and gibbons are estimated to have separated from their common hominoid ancestor between 15 and 20 million years ago (mya) (Goodman 1999), and, subsequently, waves of synteny block rearrangements in the common gibbon ancestor (Hylobatidae) gave rise to four distinct gibbon genera with varying chromosomal numbers (Jauch et al. 1992; Muller et al. 2003). Furthermore, 84 of the 107 synteny breaks in gibbons, relative to humans, are specific to the gibbon lineage, inherited from the common gibbon ancestor, while the remainder (23/107) occurred in the common hominoid ancestor (Roberto et al. 2007). Interestingly, 14 of the 84 gibbon synteny breaks are specific to the white-cheeked gibbon (Nomascus leucogenys, NLE), suggesting increased chromosomal rearrangement in that gibbon lineage (Muller et al. 2003).
The orthologous chromosomal blocks between human and NLE gibbon were recently mapped by two studies using bacterial artificial chromosome (BAC) end sequencing or array painting and confirmed by FISH (Carbone et al. 2006; Roberto et al. 2007). The average breakpoint resolutions of these two studies were ∼80 kbp and 200 kbp, respectively (Carbone et al. 2006; Roberto et al. 2007). At this level of resolution, molecular mechanisms causing synteny breaks were not clear; however, segmental duplications were estimated to be associated with 46% of the rearrangements (Carbone et al. 2006). Although the potential for disruption of several genes in the vicinity of the breaks was suggested, the effect of the breaks on the gene structures, per se, was not well defined at this resolution. Previously, sequencing of a subset of gibbon BAC clones revealed segmental duplications or interspersed repeats at the breakpoints, although a detailed analysis of these regions was not presented (Carbone et al. 2006). While karyotypic variations are implicated for anatomical and phenotypic differences between hominoid species (Ferguson-Smith and Trifonov 2007), a high-resolution comparative genomics approach is imperative to identify the underlying causative molecular event.
We performed a sequence-based assessment of human and white-cheeked gibbon synteny breaks (1) to determine the sequence architecture and genomic characteristics predisposing to synteny breaks and chromosomal instability in gibbons, and (2) to determine the extent of gene rearrangements, correlating these with signatures of molecular evolution. Since regions of chromosomal rearrangement are frequently enriched in complex repetitive structures that are sometimes difficult to resolve by whole-genome sequence assembly, we targeted large-insert gibbon BAC clones for complete high-quality sequence analysis. Our analysis has characterized a subset of human–gibbon breakpoints at the sequence level, provided insight into the mechanism of rearrangement, and identified genes that potentially contribute to the evolution of the gibbons.
Results
Sequence resolution of human–gibbon breakpoints
We previously mapped the position of gibbon rearrangements orthologous to human chromosomes (HSA) by BAC-end sequence mapping and FISH (Fig. 1A; Roberto et al. 2007). Based on the BAC-end sequencing data and FISH-derived framework of human and NLE gibbon maps, we selected 24 gibbon BACs that span the syntenic breaks on the human genome for complete insert sequencing (see Methods). Our target set included eight intrachromosomal and 16 interchromosomal gibbon rearrangements with respect to the human genome (Table 1). We purposefully biased against regions associated with segmental duplications (SDs) due to the inherent difficulties in resolving breakpoints within duplicated regions, ambiguity associated with experimentally validating these events by FISH, and difficulties in obtaining large-insert clones. As such, we anticipated that we would enrich for rearrangement events mediated by nonhomologous end-joining (NHEJ) as opposed to nonallelic homologous recombination (NAHR). Each of the 24 BACs was sequenced (generating ∼4.2 Mbp of finished, high-quality NLE genomic sequence) and aligned to the human genome sequence assembly (Build 35; Fig. 1B). The NLE gibbon synteny blocks mapped unambiguously to orthologous regions on human chromosomes, consistent with the experimental FISH results (Table 1; Fig. 1A,B).
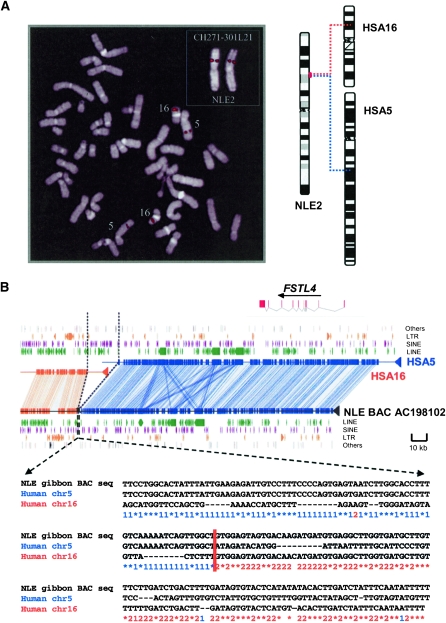
Figure 1.
(A) Identification of gibbon BAC clones at the breakpoint of synteny. All BAC clones were experimentally validated by FISH as described previously (Roberto et al. 2007). In this example, a gibbon BAC clone spanning the breakpoint shows a single signal on gibbon chromosome 2 (NLE 2), but FISH mapped to human shows two signals on chromosomes 5 and 16, identifying an interchromosomal rearrangement (as represented by the chromosomal ideogram). (B) Sequence architecture at human–NLE gibbon synteny breaks. (Top panel) Miropeats analysis of the gibbon BAC, CH271-301L21 (AC198102), when compared with segments of human chromosome 5 (132461336–132644892, blue) and chromosome 16 (73369800–73421145, orange). Representative repeat elements, LINE, SINEs, LTRs, segmental duplications, and genes mapping to the synteny blocks with arrows denoting transcriptional orientation are also shown based on human genome annotation. (Bottom panel) Three-way ClustalW alignment between human and NLE gibbon sequences at the breakpoint with 1 (blue) denoting a sequence identity with the human chromosome 5 segment and 2 (orange) indicating sequence identity with the human chromosome 16 segment. The figure shows a class I breakpoint where the human breakpoints abut precisely at the point of fusion on the gibbon chromosome.
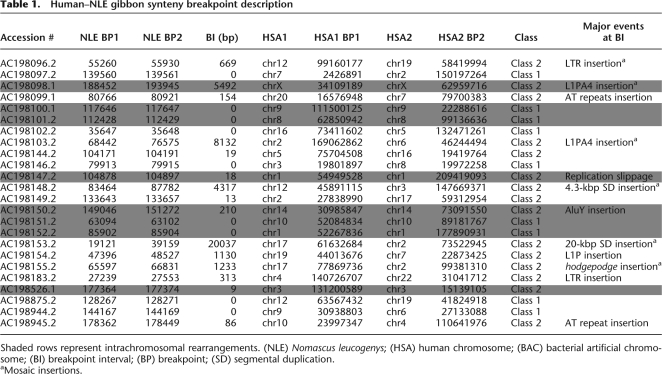
Table 1.
Human–NLE gibbon synteny breakpoint description
Shaded rows represent intrachromosomal rearrangements. (NLE) Nomascus leucogenys; (HSA) human chromosome; (BAC) bacterial artificial chromosome; (BI) breakpoint interval; (BP) breakpoint; (SD) segmental duplication.
aMosaic insertions.
Breakpoint analysis
We compared orthologous human and gibbon genomic sequences using a modified miropeats analysis (Parsons 1995) and a multiple sequence alignment analysis (ClustalW) (Higgins et al. 1996) to precisely identify the breakpoint or breakpoint interval for each event (see Methods). We manually curated all multiple sequence alignments and, due to the sequence heterogeneity and complexity of several breakpoints, we inspected regions flanking each of the breakpoints for orthology based on the analysis of high-quality alignments. The repeat content of both gibbon BACs and human orthologous regions was annotated using RepeatMasker (http://repeatmasker.org) and DupMasker (Jiang et al. 2008) (Supplemental Tables 1, 2). In addition, we examined the gibbon BAC sequences for the presence of lineage-specific gibbon duplications by identifying regions of excess read depth from available gibbon whole-genome shotgun (WGS) sequence data (Bailey et al. 2002).
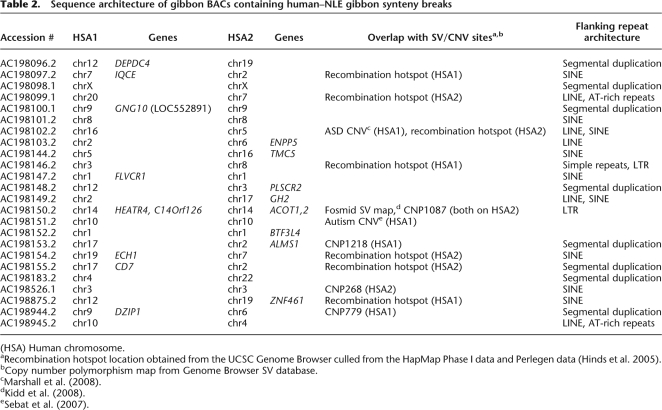
Table 2.
Sequence architecture of gibbon BACs containing human–NLE gibbon synteny breaks
(HSA) Human chromosome.
aRecombination hotspot location obtained from the UCSC Genome Browser culled from the HapMap Phase I data and Perlegen data (Hinds et al. 2005).
bCopy number polymorphism map from Genome Browser SV database.
cMarshall et al. (2008).
eSebat et al. (2007).
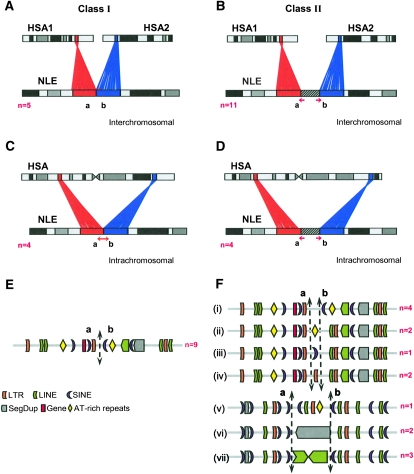
A comparison of human and gibbon breakpoints revealed two distinct classes: class I (n = 9), where the two syntenic regions precisely abut the breakpoint, and class II (n = 15), where the breakpoint could only be assigned to a sequence interval (termed breakpoint interval) (Fig. 2; Supplemental File 1). Class II breakpoints typically included additional sequences, ranging in length from 9 bp to 20 kbp, that did not map to either human orthologous chromosomal region (Table 1; Supplemental File 1). Nine class II breakpoints contained intervals ranging between 9 bp and 669 bp that also included insertions of AT-rich repeats, LTR (Supplemental Fig. 1), and AluY repeat elements, and one breakpoint interval contained insertion sequences generated by a replication slippage event (Table 1; Supplemental File 1). The 669-bp insertion formed a “mosaic” structure consisting of a series of three LTR5 elements and L1 repeats interspersed with nonrepeat sequences (Supplemental Fig. 1). We found no significant difference in the distribution of class I and class II events (Supplemental Table 3) when considering rearrangement events that occurred early within the gibbon phylogeny or, more recently, within the Nomascus lineage (Misceo et al. 2008).
Figure 2.
Class I and class II breakpoints. The schematic shows the types of rearrangements identified by high-resolution sequence analysis: Class I and class II breakpoints causing inter- (A,B) or intrachromosomal (C,D) rearrangements are shown. Based on the sequence context, the number (n) of different human–gibbon breakpoints identified from both categories (E,F) are also shown. Note that the class II breakpoints contain: (i) nonrepeat sequences, (ii) AT-rich repeats, (iii) SINEs (AluY element), (iv) LTR insertions, (v) a “hodgepodge” of repeats, (vi) segmental duplications, and (vii) LINE-1 elements. The diagram is not to scale.
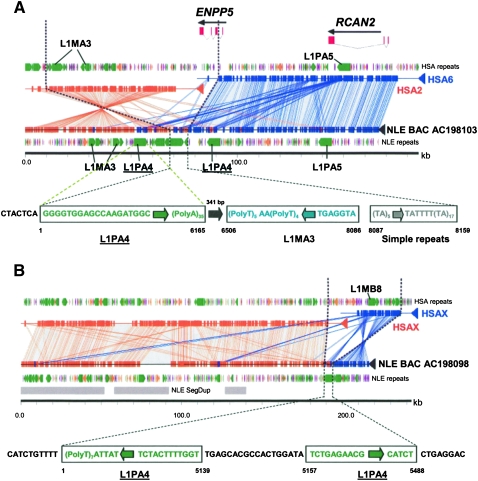
Six breakpoints contained larger insertion sequences ranging from >1 kbp up to 20 kbp in length (Table 1; Supplemental File 1). Three of these corresponded to LINE elements (one case with an L1P insertion [1.1 kbp] and two cases with L1PA4 elements [8 kbp and 5.5 kbp]) (Fig. 3A,B; Supplemental File 1). Of note, one of these breakpoints contained three L1PA4 elements arranged in tandem in gibbons but was absent in the corresponding syntenic region in humans (Fig. 3A). While the 1.1-kbp interval consisted of a single L1P element, the 8-kbp and 5-kbp intervals both consisted of a combination of L1PA4, L1MA3, simple repeats, or nonrepeat sequences (Fig. 3A,B; Supplemental File 1). No target site duplications (TSDs) were associated with these elements (Supplemental Table 4), suggesting an endonuclease-independent retrotransposition process (Morrish et al. 2007).
Figure 3.
L1PA4 repeat insertions at the breakpoints. Human–gibbon pairwise alignment by miropeats is shown. The NLE gibbon-specific segmental duplications are also remarkable. LINE-1 elements, L1PA4 (green block arrows), and L1MA3 (dark green arrows) in the vicinity of human–gibbon synteny breaks are shown. There are three L1PA4 elements (underlined) in panel A and one in panel B. Note that the L1PA4 elements are specific to the NLE gibbon chromosomal segment. (Black dotted lines) Extent of breakpoint intervals and sequence structure of each repeat. The directions of the arrows denote orientation of the LINEs, and the numbers denote the sequence extent.
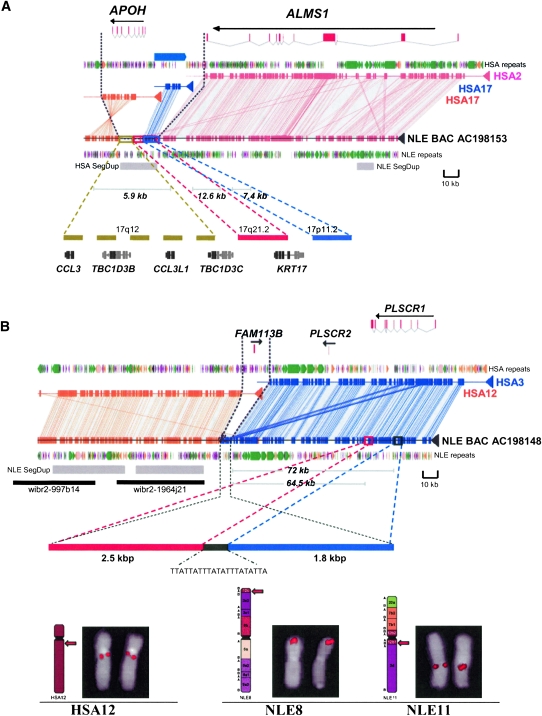
Although we biased our initial selection against segmental duplications, we found that one-third (8/24) of the sequenced gibbon BACs contained segmental duplications flanking the breakpoint intervals, ∼58% (135/234 kbp) of which occurred specifically within the gibbon lineage (Supplemental Table 5). We identified two breakpoint intervals that were themselves novel gibbon SDs (20 kbp and 4.3 kbp in length) (Fig. 4A,B) and spanned the breakpoint interval. Both SDs were also mosaic in their organization. For example, our sequence analysis of the 20-kbp SD showed that it mapped to multiple locations on human chromosome 17. It consisted of three major segments: a 5.9-kbp fragment, containing the gene structures for CCL3, CCL3L1, and a previously identified “core” duplicon (partial duplications of the TBC1D3B and TBC1D3C genes) on chr17q12 (Jiang et al. 2007; Sharp et al. 2008), a 12.6-kbp segment mapping to the KRT17 gene on chr17q21.2, and an overlapping 7.4-kbp segment that lacked genes (Fig. 4A). The second duplication at a gibbon breakpoint was smaller in size, a 4.3-kbp SD insertion. It shared high sequence identity (>95% identity, >1 kbp) to two sequences located 72 kbp and 64.5 kbp upstream of the translocation on chromosome 3 (Fig. 4B), possibly as a result of skipping of templates during replication (Fig. 4B; Lee et al. 2007; Smith et al. 2007; Payen et al. 2008). In both cases, the SDs mapped at the junctions of interchromosomal translocation fusion points (in gibbon) but were formed from template sequences located on only one of the two chromosomes involved in the translocation process.
Figure 4.
Segmental duplication insertions at the breakpoints. Alignments between the NLE BAC sequences and human chromosomes are shown. These breakpoints belong to class II category. (A) Note the insertion of an ∼20 kbp segmental duplication (gray box) at the breakpoint. The sequence interval maps to several regions on human chromosome 17, some of which are depicted (solid colored bars). The length of each insertion segment, encompassing gene structures, and karyotypic mapping location are also shown. Gene fragments that do not map to the breakpoint sequences are shown in gray. (B) Insertion of a 4.3-kbp sequence at the breakpoint. Please note that the NLE gibbon BAC is in the reverse orientation. A schematic depicting the arrangement of a 4.3-kbp sequence block at the breakpoint derived from ∼2.5-kbp and 1.8-kbp blocks located ∼72 kbp and 64.5 kbp upstream, respectively, are also shown. The location of human fosmid probes (black bar), wibr2-1964j21 (chr12: 45810892–45850262) and wibr2-997b14 (chr12: 45855081–45893396), used to map the NLE-specific segmental duplication, is also shown. (Bottom panel) Representative comparative FISH signal pattern on human (HSA) and gibbon (NLE) chromosomes using a human fosmid (wibr2-1964j21) probe mapping to segmental duplications ∼8 kbp downstream from the breakpoint (see Roberto et al. [2007] for FISH methods). Both the fosmids showed signals on NLE8 (12c) and NLE11 (12b1), displaying the presence of duplications on both translocated chromosomes. Syntenic blocks between human and gibbon chromosomes are reported diagrammatically on the left side of NLE chromosomes, according to Roberto et al. (2007).
The final class II breakpoint carried a 1.2-kbp insertion that was a “hodgepodge” of LINE, SINE, and LTR elements (Table 1; Supplemental Fig. 2; Supplemental File 1). BLAST analysis showed this breakpoint interval sequence did not map en bloc to either the human or the macaque genome, indicating that this particular constellation of sequence elements formed within the gibbon lineage. Similarly, our sequence analysis showed the divergence estimates of the LINE insertions and both SD insertions to be consistent with events that had occurred specifically within the gibbon lineage (Supplemental Tables 2, 3). Irrespective of their mechanism of origin, these data argue that human–gibbon synteny breaks are particularly receptive for the accumulation of additional retrotransposons and segmental duplications.
To explore a possible common mechanism for synteny breaks, we further analyzed breakpoint regions for enriched sequence motifs (Supplemental File 1; see Methods). We identified short stretches of 2–6 bp of microhomology in 50% (12/24) of the breakpoint regions from both classes (Supplemental File 1), suggesting a nonclassical NHEJ mechanism for synteny breaks (Yan et al. 2007). Such microhomology motifs have, for example, been associated with template switching double-strand break (DSB) repair (Lee et al. 2007; Smith et al. 2007). Also, previously described sequence motifs associated with DSBs and recombination hotspots (Abeysinghe et al. 2003) were identified in the region flanking the breaks (Supplemental File 1). Finally, several orthologous NLE breakpoint regions in humans mapped within known regions of human copy number variation and structural variation (see Methods; Table 2; Supplemental Table 6).
Gene content analysis
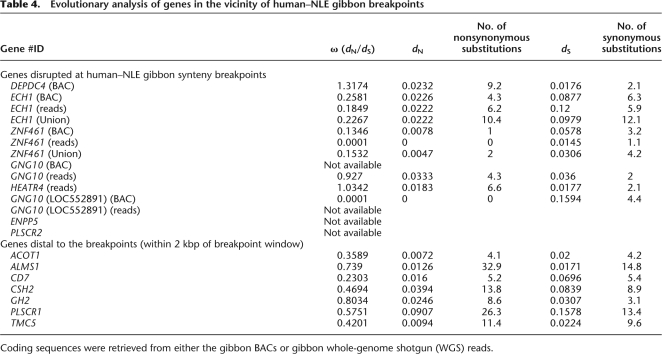
We identified seven human gene orthologs whose protein-coding sequences were disrupted by the rearrangement in gibbons (Table 3). These included genes involved in G-protein-coupled receptor signaling pathways (DEPDC4 and GNG10 (LOC552891), phospholipid metabolism including sphingomyelin hydrolysis (ENPP5) and transport (PLSCR2), peroxisomal β-oxidation (ECH1), cell structure organization (HEATR4), and ovarian and testicular functions (ZNF461 [also known as GIOT-1]) in humans (Mi et al. 2005). To test for the enrichment of genes at synteny breaks, we simulated a random distribution of breakpoints to the human genome assembly, excluding segmental duplications due to our initial bias in selecting against these regions for sequence analysis in the gibbon. The number of breakpoints mapping within human RefSeq coordinates was used to estimate an empirical P-value (n = 100 permutations). Compared with the random simulation (expected = 19, standard deviation = 3.5), the rate of gene disruption observed in 24 gibbon breakpoints was significantly lower (observed = 7), indicating that gibbon rearrangement breakpoints are biased against gene disruptions (P = 0.02) (see Methods; Supplemental Fig. 3; Supplemental Table 7).
Table 3.
Genes disrupted at human–NLE gibbon synteny breaks
Numbers in parentheses represent total exons.
aGNG10 (LOC552891) is an alternative splice variant of GNG10.
Interestingly, we found that 33% (8/24) of the BAC clones sequenced contained clusters of tandemly duplicated genes mapping within 50 kbp of the breakpoint, including the growth hormone cluster, KRAB-containing zinc finger genes (ZNF677, ZNF483, ZNF512, ZNF567, and ZNF382), vomeronasal type 1 receptors (VN1R2 and VN1R4), phospholipase scramblase (PLSCR1 and PLSCR2), and acyl-CoA thioesterases (ACOT1 and ACOT2) (Supplemental Figs. 4, 5; Supplemental Data). In some cases, paralogous genes (based on human gene annotation) were disrupted in gibbons (Supplemental Fig. 5A). For example, one breakpoint mapped to the 5′UTR of the somatotropin hormone, GH2, predicting a disruption of transcription due to uncoupling of the promoter from its coding sequence—an observation that was also reported by Carbone and colleagues (2006). Sequence analysis of the other gene family members, CSH1, CSH2, and CSHL1 within the gibbon, demonstrated numerous sequence variations, including obliteration of the start codon and point mutations in the sequence coding for the signal peptide domain of the proteins (Supplemental File 2). Similarly, the human paralogous gene, ACOT1, may be disrupted by the gibbon rearrangements, as SIM4 analysis predicted only the ACOT2 gene in gibbons (see Methods; Supplemental Fig. 5B).
We investigated whether the gibbon rearrangement events coincided with changes in the evolutionary pressure of genes mapping at the breakpoints or distal to the breakpoints. For this purpose, we performed a maximum-likelihood evolutionary analysis using Phylogenetic Analysis by Maximum Likelihood (PAML) to calculate dN/dS ratios (ω) (see Methods) (Yang 1997). First, we reconstituted a complete gibbon gene model based on the BAC sequence and the available gibbon whole-genome shotgun sequence (for the portion of the gene that was not represented within the BAC clone) (Table 4). Next, we created a multiple sequence alignment of the coding sequence from available genome sequence data and generated a phylogenetic gene tree with a minimum of five orthologous genes from various primate and mammalian lineages (Supplemental File 3). It should be noted that the latter approach in the case of duplicated genes is suboptimal as it is impossible to accurately distinguish paralogous genes from WGS read data. Thus, more rigorous tests of selection within the human and gibbon lineages are not possible until a high-quality sequence of all duplicated gene family members has been generated.
Table 4.
Evolutionary analysis of genes in the vicinity of human–NLE gibbon breakpoints
Coding sequences were retrieved from either the gibbon BACs or gibbon whole-genome shotgun (WGS) reads.
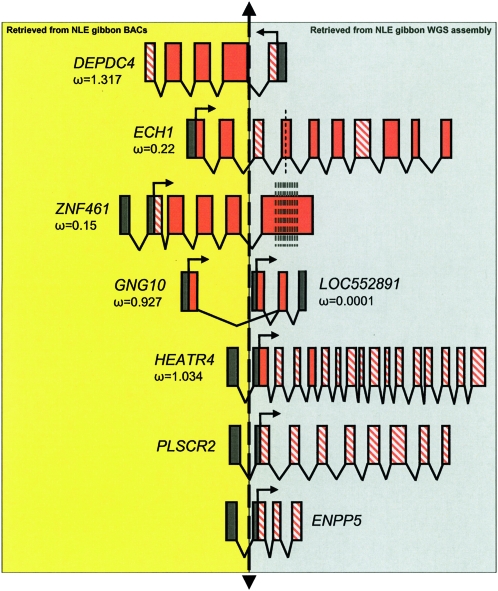
Three genes disrupted in the protein-coding sequences clearly showed a relaxation of selection pressure within the gibbon lineage; namely, DEPDC4 (ω = 1.31), HEATR4 (ω = 1.03), and GNG10 (ω = 0.927), consistent with pseudogenization as a result of the rearrangement (ω ∼21 for gibbon branch in the phylogeny; Table 4). Two additional gibbon gene models showed the presence of multiple nonsense mutations despite dN/dS ratios suggesting purifying selection (ω < 1) (Fig. 5; Table 4; Supplemental File 3); namely, ECH1 (ω = 0.25 and 0.18) and ZNF461 (ω = 0.13 and 0.0001). A comparison using a free codon-substitution model for neutral (ω = 1) or conserved (ω = 0.5) evolution in the gibbon branch for all analyzed genes suggested a significantly conserved evolution for ECH1, ZNF461, and GNG10 (LOC552891) (see Methods; Supplemental Table 7). Coding sequences for PLSCR2 and ENPP5 were not available (in the current gibbon WGS assembly) for evolutionary analysis. As expected, analysis of genes distal to the breakpoints demonstrated signatures of purifying selection (Table 4; Supplemental Table 8; Supplemental File 3).
Figure 5.
Gene disruptions at synteny breaks. The schematic shows the seven genes mapping to the breakpoints (dashed vertical arrow). One part of the gene is contained in the BAC (yellow region) and the other part is lost due to synteny break (gray region). Both gene parts were reconstructed either from the gibbon BAC sequences or contigs assembled from the gibbon WGS reads (see text). Coding exons (orange, completely retrieved sequences; stripes, missing sequences in gibbons) and noncoding exons (black) are depicted. (Black arrows) Transcriptional orientation. The dN/dS ratios (ω) and number of synonymous and nonsynonymous substitutions calculated for the available gene fragments (orange) are also shown. Vertical dashed lines on the exons indicate location of stop codons. The figure is not to scale.
Discussion
Gibbons are known to have a rapid rate of chromosomal evolution among the hominoids, mainly involving large-scale rearrangements and rapid karyotypic divergence (Muller et al. 2003; Carbone et al. 2006; Roberto et al. 2007). In contrast to human and great ape segmental duplications, where ∼70% of all large-scale evolutionary rearrangements map to regions of segmental duplication (Kehrer-Sawatzki and Cooper 2008), initial studies of the gibbon reported that only 46% of gibbon breakpoints mapped to sites of segmental duplication in the human lineage (Carbone et al. 2006). BAC sequence analysis of a smaller subset identified segmental duplications or interspersed repeats at most breakpoints; however, two clones in this initial study also showed evidence of “micro-rearrangements” containing disparate repeat sequences derived from various human chromosomal locations (Carbone et al. 2006). These initial data from Carbone and colleagues hinted at potential alternate mechanisms of rearrangements, although the number of sites and the extent of sequence analysis were limited. In this study, we expanded upon earlier work (Carbone et al. 2006; Roberto et al. 2007) to present single-base-pair resolution of 24 human–gibbon breakpoints of synteny within the context of 4.2 Mbp of high-quality gibbon BAC sequence.
The most striking finding was the presence of additional sequences for ∼40% of the gibbon sites of translocation, suggesting a more complex rearrangement mechanism than simply nonallelic homologous recombination or nonhomologous end joining. The largest (1–20 kbp) of these insertion sequences consisted of various classes of repetitive DNA including segmental duplications and L1 repeats. Detailed sequence analyses of these new insertions reveals two important features. First, we note that in the case of L1 elements, we observed no target-site duplications, suggesting that they did not originate as a result of typical endonuclease-mediated retrotransposition (Morrish et al. 2007). Second, in many cases the new insertion sequences are mosaic structures composed of disparate common repeats or duplicated sequences (Figs. 3, 4; Supplemental Fig. 2) that originate upstream of the rearrangement breakpoint.
At least two different mutational mechanisms are consistent with these observations. Since microhomology was observed in 50% of the human–NLE gibbon breaks (Supplemental Fig. 6), one possibility may be a microhomology-mediated end-joining (MMEJ) mechanism, recently reported as a nonclassical NHEJ mechanism for translocations in mammals (Yan et al. 2007). Sequence microhomology and site-specific recombinogenic sequences in the vicinity of the breakpoints have been associated with translocations in evolutionary rearrangements and cancer (Kehrer-Sawatzki et al. 2002; Abeysinghe et al. 2003; Wei et al. 2003). We identified sequence motifs (e.g., topoisomerase II and translin sites) consistent with DSB and repair mechanisms generating overhangs at several human–NLE gibbon breakpoints (Negrini et al. 1993; Kanoe et al. 1999; Wei et al. 2003). We propose that these overhangs may have been repaired by an “error-prone” mechanism, creating some of the smaller breakpoint intervals (Fig. 6A).
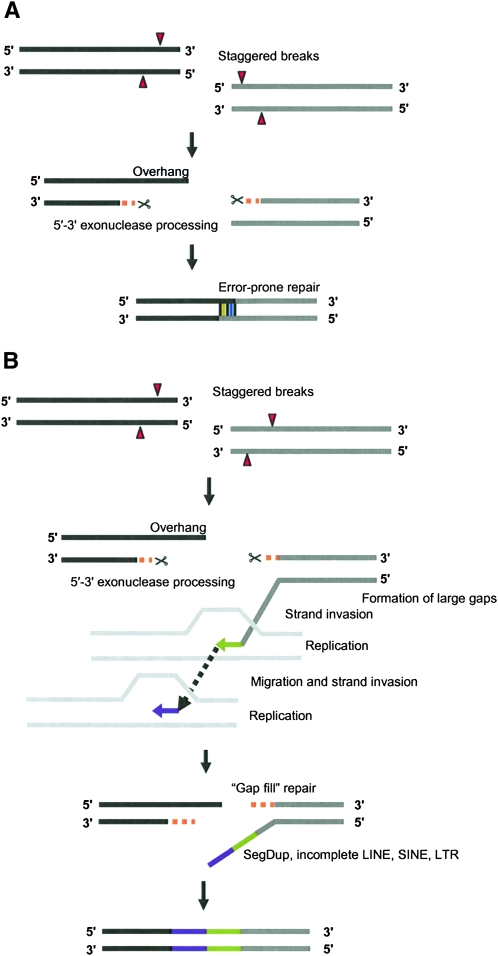
Figure 6.
Models for human–NLE gibbon rearrangements. (A) An error-prone repair mechanism for smaller breakpoint intervals (<20 bp). DNA strands from two chromosomes (black and gray bars) are shown. Staggered double-strand breaks are processed by 5′–3′ exonuclease, creating overhangs. These overhangs are filled by an error-prone repair mechanism, creating shorter insertions. (B) “Gap-fill” model for larger breakpoint intervals. Large gaps are generated by double-strand breaks (due to possible collapsed or stalled replication forks) at rearrangement sites. These staggered breaks are processed by exonucleases to generate long 3′ overhangs. Replication is initiated by strand invasion to repair the gap. However, likely due to low processivity of the replication-dependent repair process (McVey et al. 2004), only smaller-length sequence stretches are synthesized. Consequently, a series of strand invasion, replication, and uncoupling of the replication machinery is necessary to fill the large gap. Thus, a less-efficient replication-based repair process generates a mosaic of incomplete repetitive elements at the larger breakpoint intervals.
Both the microhomology and, more importantly, the mosaic architecture of the larger breakpoint intervals are also consistent with more recently proposed replication-based mechanisms such as FoSTeS (fork stalling template switching) (Lee et al. 2007) and MMIR (microhomology/microsatellite-induced replication) (Payen et al. 2008). Template switching as a result of multiple rounds of strand invasion from DSB sites generated by stalled or collapsed replication forks to ectopic sites could, in principle, explain some of the events we have observed (see “gap-fill model,” Fig. 6B) (McVey et al. 2004; Lee et al. 2007; Smith et al. 2007). Repeat-rich sequences frequently serve as preferred templates because of their tendency to interfere with replication fork progression, leading to the formation mosaic structures at the point of rearrangement (Figs. 3, 4, 6B; Supplemental Fig. 7; Kehrer-Sawatzki and Cooper 2008; Payen et al. 2008). A remarkable example was the presence of a 4.2-kbp gibbon-specific segmental duplication mapping precisely at the translocation fusion point between chromosomes 3 and 12. Sequence analysis revealed that this segmental duplication actually consisted of duplicatively transposed sequences mapping 72 kbp and 64.5 kbp further upstream of the point of fusion on chromosome 3.
Although we have clearly biased against homology-based events, such insertions of mosaic structures have not yet been described at sites of rearrangement between humans and the African great apes, most of which have now been characterized at the molecular level (Kehrer-Sawatzki and Cooper 2007, 2008). Do these results provide any insight into the apparent increased tempo of large-scale rearrangements in the gibbon lineages? There are a few important facts. First, computational analyses of the human genome based on percent sequence identity suggest a burst of segmental duplications in the African great-ape lineage when compared with other apes (Cheng et al. 2005; Bailey and Eichler 2006). Second, most large-scale chromosomal rearrangements in humans and African great apes are intrachromosomal as opposed to interchromosomal translocations (Kehrer-Sawatzki and Cooper 2007, 2008). Third, 65%–70% of all great ape chromosomal rearrangements were associated with large blocks of segmental duplication (Cheng et al. 2005; Kehrer-Sawatzki and Cooper 2007), although the number appears to be lower in gibbons (46%) (Carbone et al. 2006). One possibility may be that a paucity of segmental duplications in ancestral gibbon genomes channeled rearrangement pathways away from NAHR, favoring these alternate mechanisms (e.g., MMIR, FoSTeS, break-induced replication). We speculate that the overall “rate of rearrangement” is largely constant among all ape genomes but that fewer SDs drive fewer homology-mediated events and, consequently, nonhomology-based mechanisms contribute more significantly to large-scale chromosomal rearrangements in gibbons. Many SD-mediated events have occurred among great apes, but because of the predominance of interspersed duplication blocks within close proximity along a chromosome, a large number of these African-ape events are below the level of cytogenetic resolution and instead are observed as an abundance of smaller structural variant events (Feuk et al. 2005; Newman et al. 2005).
In this model, intrachromosomal segmental duplications essentially “resolve” larger chromosomal rearrangements in the African great ape/human genomes (Kehrer-Sawatzki and Cooper 2007). Moreover, given that NAHR events are often associated with breakpoint reuse (Bailey et al. 2004; Murphy et al. 2005; Zody et al. 2008), at a constant rearrangement rate, the great apes would show apparently fewer structural changes, due to recurrent rearrangements involving the same chromosomal segments. However, gibbons with fewer SDs would tend to have more distinct structural changes, although with the same effective number of events. In this regard, it is interesting that we previously noted no apparent increase in smaller rearrangements in gibbon despite the nearly fourfold increase in gross chromosomal rearrangement events when compared with the African great apes (Roberto et al. 2007). High-quality sequence of many more breakpoints within ape lineages will be necessary to fully address this model.
Although the precise mechanism(s) underlying these events is not yet understood, it is clear that segmental duplications are intimately associated with large-scale chromosomal rearrangements. Even when we bias against SD regions such as in this study, the association resurfaces. Bailey et al. (2004) proposed that the association between segmental duplications and large-scale genomic rearrangements is not entirely causative. In our study, eight breakpoints mapped within 100 kbp of a previously characterized segmental duplication. Since no homology was detected at corresponding chromosomal positions of the rearrangement (Supplemental Data; Table 2), we exclude the possibility of homology-mediated (or NAHR) events. In four cases (Supplemental Table 5; Supplemental File 4), we identified gibbon-specific segmental duplications mapping distal to (within 50 kbp) gibbon fusion breakpoints. One example is the gibbon-specific segmental duplication mapping ∼8 kbp downstream from the HSA3 and HSA12 translocation breakpoint (Fig. 4B). FISH analysis using human fosmid probes showed signals on both translocated chromosomes (Fig. 4B); however, no direct involvement of SD was evident in this chromosomal rearrangement due to the absence of its homologous counterpart on the other side (HSA3) of the breakpoint. Similarly, when we reanalyzed the 11 gibbon BACs at breakpoints reported by Carbone and colleagues (Carbone et al. 2006) using our analytical pipeline (Supplemental Tables 9, 10; Supplemental File 4), we identified at least five breakpoints that contain segmental duplications. None of these, however, show evidence of homologous sequence at both corresponding regions in the human genome arguing, once again, against nonallelic homologous recombination between ancestral segmental duplications.
These data clearly reinforce the strong association between segmental duplications and chromosomal rearrangements (O'Brien and Stanyon 1999; Armengol et al. 2003; Bailey et al. 2004) and imply that regions of rearrangement may, in fact, also be the source of new duplications (Kehrer-Sawatzki et al. 2002; Ranz et al. 2007). These data support an alternative model associating segmental duplication and rearrangements reinforcing that DSBs can generate segmental duplications (Koszul et al. 2004; Smith et al. 2007; Kim et al. 2008). Our model extends these observations to include both translocations as well as inversions. As mentioned, one possibility may be that the rearrangement regions could also serve as preferential templates for subsequent or concurrent strand invasion of other regions during replication-dependent repair, spawning de novo segmental duplications at other sites (Koszul et al. 2004). This view is further supported by our observation of the 20-kbp segmental duplication block mapping to a core duplicon on chromosome 17. Thus, regions of genome rearrangement may, in fact, promote the formation of segmental duplications at other regions of the genome, as opposed to these being the cause of evolutionary rearrangements.
From the genic perspective, our analysis supports the more general observation that structural variation occurs preferentially near or within duplicated genes (Locke et al. 2006; Redon et al. 2006; Kidd et al. 2008). The functional redundancy conferred by such duplicated genes might make these rearrangements more tolerable in an evolving species as opposed to disruptions of unique, single-copy genes. The growth hormone gene cluster, for example, is specific to the primate lineage and originated from a single ancestral GH gene by duplications. It comprises paralogous growth hormone genes (GH1, pituitary, and GH2, placental) and two chorionic somatomammotropin genes (CSH1 and CSH2) (Barsh et al. 1983). The CSH1 gene duplicated further to yield a chorionic somatomammotropin gene (CSHL1) that later became a pseudogene by inactivation (Misra-Press et al. 1994). Likewise, the ACOT gene cluster is variable in copy number between species. This protein family regulates intracellular levels of lipids by hydrolysis of acyl CoAs to free fatty acids and CoASH with localizations in the cytosol (ACOT1) and mitochondria (ACOT2). While the human ACOT cluster is composed of ACOT1, ACOT2, and ACOT4, the mouse cluster contains six paralogous genes (Acot1–Acot6). Similarly, the vomeronasal receptors have undergone a steady evolutionary decline from mouse to humans, with gradual inactivation of pheromone sensation genes, VN1R2 and VN1R4, since the divergence of the Old World monkeys and the hominoids, ∼23 mya (Zhang and Webb 2003). These examples highlight both the variability in copy number and functional diversity for these genes, making them preferred targets for large-scale rearrangement events. Recently, Dumas et al. identified a high rate of lineage-specific gene duplication in gibbons (Dumas et al. 2007). Our preliminary analysis of the gibbon genome does not support this observation. Among the segmental duplications that we identified at the breakpoints, we were unable to find any overlap between genes in our analysis and the ones identified by Dumas and colleagues.
Three genes disrupted by rearrangement in gibbon showed signatures of selection consistent with pseudogenization. While it is tempting to speculate that some of these gene losses may have contributed to morphological and behavioral specialization in the gibbon lineage, further functional characterization of the genes and their impact on biochemical pathways and developmental lineages will be required. Our analysis, however, provides some interesting candidates for further investigation (i.e., loss of the growth hormone genes associated with lack of sexual dimorphism in the gibbon). Interestingly, not all genes appear to be dead as a result of rearrangement. Our preliminary analysis of two genes, ECH1 and ZNF461, suggests a model of purifying selection. While the functional implications of these results are unclear, our results raise the intriguing possibility that a gene broken by a rearrangement event may not be doomed to pseudogenization, and the underlying coding sequences may be exapted for other functions in the organism.
Methods
Gibbon BAC sequencing
Twenty-four bacterial artificial chromosomes (BACs) were chosen from the white-cheeked gibbon, Nomascus leucogenys/NLE, BAC library, CHORI-271, based on unambiguous signals with FISH (Roberto et al. 2007). The BACs were then subjected to whole-genome shotgun sequencing to at least sixfold sequence redundancy and assembled to completion at the Genome Sequencing Center, Washington University, St. Louis, Missouri.
The accession numbers of the BACs are as follows: AC198096.2, AC198097.2, AC198098.1, AC198099.1, AC198100.1, AC198101.2, AC198102.2, AC198103.2, AC198144.2, AC198146.2, AC198147.2, AC198148.2, AC198149.2, AC198150.2, AC198151.2, AC198152.2, AC198153.2, AC198154.2, AC198155.2, AC198526.1, AC198183.2, AC198944.2, AC198945.2, and AC198875.2 (Supplemental Data).
Sequence alignment and annotation
Gibbon BAC sequences were initially compared with human genome sequence using BLAST sequence similarity searches and miropeats (Altschul et al. 1990; Parsons 1995) to identify potential breakpoint intervals. Analysis for repeats on finished gibbon BAC sequences was performed using RepeatMasker, and segmental duplications (>94% identical, ≥10 kbp size) were detected using the whole-genome shotgun sequence detection (WSSD) strategy for gibbon (Bailey et al. 2002; Chen 2004). Human genomic coordinates corresponding to gibbon SDs (identified by WSSD mapped against the gibbon WGS clones) were intersected with human, chimp, orangutan, and macaque segmental duplication (T. Marques-Bonet and E.E. Eichler, unpubl.) to detect gibbon-specific SDs. Sequences homologous to known human SDs were detected on both syntenic human chromosomes and the gibbon BACs using DupMasker (Jiang et al. 2008). The sequences corresponding to syntenic regions on the human chromosomes and the gibbon BACs were aligned using ClustalW (Higgins et al. 1996). The exact sequence breaks in the alignments between gibbon and human sequences were identified as breakpoints or breakpoint intervals. To estimate the evolutionary age of various classes of repeats, sequence divergence from consensus repeat sequences was computed for each of the repeat elements mapping within and flanking the breakpoints.
Breakpoint analyses
Sequences around the breakpoints were compared with sequence motifs associated with DSBs, recombination, and chromosomal rearrangement, allowing for up to 2-bp mismatches. Sequences ±15 bp around the breaks were searched for previously reported 5–9-mer recombination hotspot sequences (Myers et al. 2005), topoisomerase consensus binding sites, topoIIv ([A/G]N[T/C]NNCNNG[T/C]NG[G/T]TN[T/C]N[T/C]) (Spitzner and Muller 1988), topoIId (GTN[T/A]A[C/T]ATTNATNN[A/G]) (Sander and Hsieh 1985), topoIIi ([T/C][T/C]CNTA[C/G][C/G]CC[T/G][T/C][T/C]TNNC) (Kas and Laemmli 1992), and translin recognition sites (ATGCAG and GCCC[A/T][G/C][G/C][A/T]) (Aoki et al. 1995) on both strands using C-program-based K-mer finder and BLAST (Altschul et al. 1990). A homology of >75% is considered a strong binding/cleavage site (Spitzner and Muller 1988). Sequence motifs identified in cancer-associated rearrangements were also compared with sequences near the human gibbon synteny breaks (Abeysinghe et al. 2003).
The significance of breakpoints within genes (human Refseq) and within human recombination hotspots was determined by simulation. Breakpoints were randomly distributed to the human genome assembly (Build 35), and the number of breakpoints mapping within human RefSeq coordinates and within human recombination hotspots (HapMap Phase II and Perlegen data [Hinds et al. 2005]) was used to estimate an empirical P-value (n = 100 permutations). For gene break simulation, segmental duplications were excluded from the human genome sequences duplications due to our initial bias in selecting against these regions for gibbon BAC sequence analysis.
Evolutionary gene analyses
To determine the gene structure, human cDNA sequences and gibbon BAC sequences were aligned using ClustalW. Exon–intron boundaries were determined using the SIM4 program (Higgins et al. 1996; Florea et al. 1998). Functional annotations for each of the genes were derived from www.pantherdb.org (Mi et al. 2005). The analysis of the evolution of the coding sequence was done by maximum likelihood using PAML (Yang 1997). The ratio dN/dS (ω), which compares the rate of nonsynonymous substitutions against the rate of synonymous substitutions, was used as a measure of evolutionary constraint. If a gene is under no selection (neutrality), it tends to have dN/dS close to 1 since the ratio of fixation of synonymous and nonsynonymous mutation will be the same. However, in a situation where the gene has a strong functional role, this ratio will tend to be <1 since the nonsynonymous mutation would tend to be removed from the population because of the disturbing effect on the functional protein. Finally, positive selection (adaptive evolution) acting continuously upon the gene generates a dN/dS ratio >1 as the new nonsynonymous substitutions acquired will be fixed more rapidly than the almost neutral synonymous substitutions.
To perform the evolutionary analysis on the coding sequences, we first retrieved the best orthologous sequences using the Ensembl predictions for as many eutherian species as possible (ranging from five to eight species using human, chimpanzee, orangutan, gibbon, macaque, lemur, mouse, and dog). A multiple-sequence alignment was then applied (using the translated amino acids as a unit for the alignments) and back-translating into DNA sequences. All the alignments were manually curated, and regions poorly aligned were removed (although this is a conservative measure against rapid evolution, we removed particular segments that were poorly aligned in more than one species). For the gibbon sequences containing stop codons, we used the longer translatable frame in order to study the amino acid evolution of the remaining part of the gene. We then used a codon-substitution branch model (CODEML) (Yang and Nielsen 2002). First, a free codon-substitution model (in which every branch of the tree is allowed to have different dN/dS) was applied to the accepted phylogeny for the species to estimate the evolutionary pressures at different times during the evolution of these genes. Then, in order to have a statistical significance to gibbon-specific estimations, different evolutionary situations were modeled and compared with the initial free model. Then, we compared a codon-substitution model for the branch leading to gibbons to a neutral evolution (ω = 1) or a conserved evolution (ω = 0.5) model. Likelihood ratio tests were performed using a χ2 distribution with as many degrees of freedom as differences of parameters in the model to estimate the significance of the comparison (Yang and Nielsen 2002).
Acknowledgments
We thank Drs. Can Alkan, Zhaoshi Jiang, and Ze Cheng for assistance with bioinformatics. We also thank Michelle O'Laughlin and Laura Courtney for help with BAC insert sequencing and Jeffrey Kidd for critical reading of the manuscript. This work was supported, in part, by an NIH grant HG002385 to E.E.E. T.M.-B. is supported by a Marie Curie fellowship. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.086041.108.
References
- Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aoki K., Suzuki K., Sugano T., Tasaka T., Nakahara K., Kuge O., Omori A., Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- Armengol L., Pujana M.A., Cheung J., Scherer S.W., Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 2003;12:2201–2208. doi: 10.1093/hmg/ddg223. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Eichler E.E. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat. Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. http://genomebiology.com/2004/5/4/R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh G.S., Seeburg P.H., Gelinas R.E. The human growth hormone gene family: Structure and evolution of the chromosomal locus. Nucleic Acids Res. 1983;11:3939–3958. doi: 10.1093/nar/11.12.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L., Vessere G.M., ten Hallers B.F., Zhu B., Osoegawa K., Mootnick A., Kofler A., Wienberg J., Rogers J., Humphray S., et al. A high-resolution map of synteny disruptions in gibbon and human genomes. PLoS Genet. 2006;2:e223. doi: 10.1371/journal.pgen.0020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2004;4 doi: 10.1002/0471250953.bi0410s05. Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T.H., Harvey P.H., Rudder B. Sexual dimorphism, socionomic sex ratio and body weight in primates. Nature. 1977;269:797–800. doi: 10.1038/269797a0. [DOI] [PubMed] [Google Scholar]
- Dooley H., Judge D. Vocal responses of captive gibbon groups to a mate change in a pair of white-cheeked gibbons (Nomascus leucogenys) Folia Primatol. (Basel) 2007;78:228–239. doi: 10.1159/000102318. [DOI] [PubMed] [Google Scholar]
- Dumas L., Kim Y.H., Karimpour-Fard A., Cox M., Hopkins J., Pollack J.R., Sikela J.M. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17:1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E.E. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith M.A., Trifonov V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- Feuk L., MacDonald J.R., Tang T., Carson A.R., Li M., Rao G., Khaja R., Scherer S.W. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:e56. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L., Hartzell G., Zhang Z., Rubin G.M., Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebo D.L. Climbing, brachiation, and terrestrial quadrupedalism: Historical precursors of hominid bipedalism. Am. J. Phys. Anthropol. 1996;101:55–92. doi: 10.1002/(SICI)1096-8644(199609)101:1<55::AID-AJPA5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Goodman M. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. A brief history of human autosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999;354:1447–1470. doi: 10.1098/rstb.1999.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt A.H., Harvey P.H., Larson S.G., Short R.V. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Thompson J.D., Gibson T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Jauch A., Wienberg J., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Hubley R., Smit A., Eichler E.E. DupMasker: A tool for annotating primate segmental duplications. Genome Res. 2008;18:1362–1368. doi: 10.1101/gr.078477.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoe H., Nakayama T., Hosaka T., Murakami H., Yamamoto H., Nakashima Y., Tsuboyama T., Nakamura T., Ron D., Sasaki M.S., et al. Characteristics of genomic breakpoints in TLS-CHOP translocations in liposarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene. 1999;18:721–729. doi: 10.1038/sj.onc.1202364. [DOI] [PubMed] [Google Scholar]
- Kas E., Laemmli U.K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Cooper D.N. Structural divergence between the human and chimpanzee genomes. Hum. Genet. 2007;120:759–778. doi: 10.1007/s00439-006-0270-6. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Cooper D.N. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res. 2008;16:41–56. doi: 10.1007/s10577-007-1207-1. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Schreiner B., Tanzer S., Platzer M., Muller S., Hameister H. Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am. J. Hum. Genet. 2002;71:375–388. doi: 10.1086/341963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.M., Lam H.Y., Urban A.E., Korbel J.O., Chen X., Snyder M., Gerstein M.B. Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history. Genome Res. 2008;18:1865–1874. doi: 10.1101/gr.081422.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R., Caburet S., Dujon B., Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y., et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Adams M., Staeva-Vieira E., Sekelsky J.J. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila . Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D., Capozzi O., Roberto R., Dell'oglio M.P., Rocchi M., Stanyon R., Archidiacono N. Tracking the complex flow of chromosome rearrangements from the Hominoidea Ancestor to extant Hylobates and Nomascus Gibbons by high-resolution, punctuated synteny mapping. Genome Res. 2008;18:1530–1537. doi: 10.1101/gr.078295.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra-Press A., Cooke N.E., Liebhaber S.A. Complex alternative splicing partially inactivates the human chorionic somatomammotropin-like (hCS-L) gene. J. Biol. Chem. 1994;269:23220–23229. [PubMed] [Google Scholar]
- Morrish T.A., Garcia-Perez J.L., Stamato T.D., Taccioli G.E., Sekiguchi J., Moran J.V. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- Muller S., Hollatz M., Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum. Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Negrini M., Felix C.A., Martin C., Lange B.J., Nakamura T., Canaani E., Croce C.M. Potential topoisomerase II DNA-binding sites at the breakpoints of a t(9;11) chromosome translocation in acute myeloid leukemia. Cancer Res. 1993;53:4489–4492. [PubMed] [Google Scholar]
- Newman T.L., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien S.J., Stanyon R. Phylogenomics. Ancestral primate viewed. Nature. 1999;402:365–366. doi: 10.1038/46450. [DOI] [PubMed] [Google Scholar]
- Parsons J.D. Miropeats: Graphical DNA sequence comparisons. Comput. Appl. Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Payen C., Koszul R., Dujon B., Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl. Acad. Sci. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavcan J.M. Sexual dimorphism in primate evolution. Am. J. Phys. Anthropol. 2001;(Suppl. 33):25–53. doi: 10.1002/ajpa.10011.abs. [DOI] [PubMed] [Google Scholar]
- Ranz J.M., Maurin D., Chan Y.S., von Grotthuss M., Hillier L.W., Roote J., Ashburner M., Bergman C.M. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberto R., Capozzi O., Wilson R.K., Mardis E.R., Lomiento M., Tuzun E., Cheng Z., Mootnick A.R., Archidiacono N., Rocchi M., et al. Molecular refinement of gibbon genome rearrangements. Genome Res. 2007;17:249–257. doi: 10.1101/gr.6052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte R.V., Eichler E.E. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 2002;3:65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Sander M., Hsieh T.S. Drosophila topoisomerase II double-strand DNA cleavage: Analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J., et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.E., Llorente B., Symington L.S. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Spitzner J.R., Muller M.T. A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res. 1988;16:5533–5556. doi: 10.1093/nar/16.12.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turleau C., Creau-Goldberg N., Cochet C., de Grouchy J. Gene mapping of the gibbon. Its position in primate evolution. Hum. Genet. 1983;64:65–72. doi: 10.1007/BF00289482. [DOI] [PubMed] [Google Scholar]
- Usherwood J.R., Bertram J.E. Understanding brachiation: Insight from a collisional perspective. J. Exp. Biol. 2003;206:1631–1642. doi: 10.1242/jeb.00306. [DOI] [PubMed] [Google Scholar]
- Wei Y., Sun M., Nilsson G., Dwight T., Xie Y., Wang J., Hou Y., Larsson O., Larsson C., Zhu X. Characteristic sequence motifs located at the genomic breakpoints of the translocation t(X;18) in synovial sarcomas. Oncogene. 2003;22:2215–2222. doi: 10.1038/sj.onc.1206343. [DOI] [PubMed] [Google Scholar]
- Wienberg J. Fluorescence in situ hybridization to chromosomes as a tool to understand human and primate genome evolution. Cytogenet. Genome Res. 2005;108:139–160. doi: 10.1159/000080811. [DOI] [PubMed] [Google Scholar]
- Yan C.T., Boboila C., Souza E.K., Franco S., Hickernell T.R., Murphy M., Gumaste S., Geyer M., Zarrin A.A., Manis J.P., et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yunis J.J., Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- Zhang J., Webb D.M. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc. Natl. Acad. Sci. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zody M.C., Jiang Z., Fung H.C., Antonacci F., Hillier L.W., Cardone M.F., Graves T.A., Kidd J.M., Cheng Z., Abouelleil A., et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]