Abstract
Chromosomal aberrations have been thought to be random events. However, recent findings introduce a new paradigm in which certain DNA segments have the potential to adopt unusual conformations that lead to genomic instability and nonrandom chromosomal rearrangement. One of the best-studied examples is the palindromic AT-rich repeat (PATRR), which induces recurrent constitutional translocations in humans. Here, we established a plasmid-based model that promotes frequent intermolecular rearrangements between two PATRRs in HEK293 cells. In this model system, the proportion of PATRR plasmid that extrudes a cruciform structure correlates to the levels of rearrangement. Our data suggest that PATRR-mediated translocations are attributable to unusual DNA conformations that confer a common pathway for chromosomal rearrangements in humans.
Chromosomal aberrations, including translocations or deletions, have been thought to be essentially random events. Although most of the events arise in intergenic regions resulting in harmless consequences, disruption of important genes occasionally leads to cancer or genetic disease. Analysis of chromosomal aberrations observed in such disorders reveals that a subset of translocations or deletions arises in a nonrandom and recurrent fashion, indicating evidence of DNA segments that are susceptible to breakage in human chromosomes. Recently, formation of non-B DNA structures has been increasingly recognized as a source of genomic instability leading to these nonrandom chromosomal aberrations. In follicular lymphomas, the breakpoints of the disease-causing translocation cluster within a 150-bp genomic region of the BCL2 gene, which potentially forms a triplex DNA structure (Raghavan et al. 2004, 2005). PKD1, a causative gene for polycystic kidney disease, has a 2.5-kb poly(purine·pyrimidine) tract within an intron that can form a variety of non-B DNA structures and cause gross deletions and translocations (Bacolla et al. 2001, 2004).
We previously identified unique sequences, palindromic AT-rich repeats (PATRRs), on chromosomes 11 and 22 by analysis of the breakpoint of a recurrent constitutional chromosomal translocation, t(11;22)(q23;q11). Translocation carriers manifest no clinical symptoms but often come to attention subsequent to the birth of chromosomally unbalanced offspring. The breakpoints of most translocation carriers are slightly different from one another, but are concentrated close to the center of the palindromes (Kurahashi et al. 2000; Edelmann et al. 2001). t(11;22) is also detectable as de novo translocations in sperm from normal healthy males at frequencies of 10−4 to 10−5, but not in other mitotic cells (Kurahashi and Emanuel 2001). Similar PATRR sequences have been found at the breakpoints of t(17;22)(q11;q11) (Kehrer-Sawatzki et al. 1997; Kurahashi et al. 2003), t(4;22)(q35;q11) (Nimmakayalu et al. 2003), t(1;22)(p21.2;q11) (Gotter et al. 2004), and t(8;22)(q24.13;q11.21) (Gotter et al. 2007). It is therefore accepted that PATRRs can cause recurrent and nonrecurrent chromosomal translocations in humans.
Palindromic sequences, or inverted repeats, have been known to be unstable and represent hotspots for deletion or recombination in bacteria, yeast, and mammals (Gordenin et al. 1993; Leach 1994; Collick et al. 1996; Akgün et al. 1997). This genetic instability has generally been related to DNA replication: Slow replication was observed in an inverted-repeat sequence in Escherichia coli (Leach 1994), and inverted repeats lead to deletions or chromosomal rearrangements more frequently in yeast that are deficient in DNA polymerase activity (Ruskin and Fink 1993; Lemoine et al. 2005). Slow progression of the replication fork increases the chance of forming secondary structures at long tracts of single-stranded DNA in the lagging-strand template. Such secondary structures may be an obstacle to fork progression or a target for nucleolytic attack, thus permitting DNA breakage leading to deletion or recombination (Leach 1994).
However, PATRR-mediated genomic instability leading to structural rearrangement in humans is likely to be independent of replication. This is because the translocation could only be detected in sperm and not in other somatic cells (Kurahashi and Emanuel 2001). Furthermore, the frequency of de novo translocations does not appear to be influenced by increasing age (Kato et al. 2007). Recently, another hypothesis for palindrome instability in eukaryotic cells has emerged. Alu repeats artificially inserted in an inverted orientation in the yeast genome undergo double-strand breaks (DSBs) and enter a break–fusion cycle resulting in dicentric chromosomes (Lobachev et al. 2002; Narayanan et al. 2006). On the basis of the finding that the end of the DSB has a closed hairpin, it has been proposed that the break might be caused by a cruciform-specific resolution activity similar to Holliday junction resolvase, which generally is thought to act on intermediates produced through homologous recombination in the repair of DSBs or stalled replication.
In this study, we have investigated the mechanism of PATRR-mediated translocations in human cells using a plasmid-based model with PATRRs derived from human chromosomes 11 (PATRR11) and 22 (PATRR22). We observed that the cruciform structure is the cause of DNA breakage that leads to chromosomal rearrangement, not the palindromic sequence per se. Our data not only prove that cruciform DNA structures provoke genomic rearrangement in vivo, but also support the existence of such non-B form DNA in living cells.
Results
PATRRs induce translocation-like rearrangements in cultured human cells
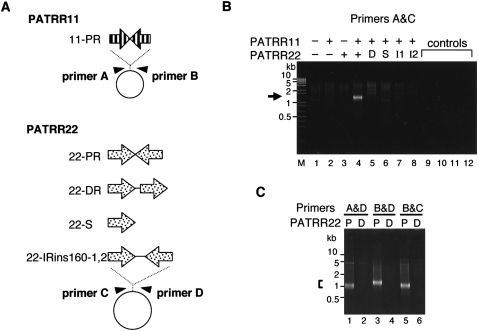
To investigate whether PATRRs could induce translocation-like rearrangements in human cultured cells, we transfected two plasmids, one with the PATRR11 and the other with the PATRR22 sequence (11-PR and 22-PR) (Fig. 1A), into HEK293 cells and attempted to detect illegitimate joining between them. After 24 h of transfection, total cellular DNA was isolated. PCR with primers designed such that one was flanking the 11-PR and the other was flanking the 22-PR produced PCR products in the DNA obtained by simultaneous transfection of 11-PR and 22-PR (Fig. 1B). Simple mixture of the two plasmids without transfection did not yield such PCR products, excluding the possibility of a PCR artifact. The size of the product was consistent with that expected from the translocation junction of the t(11;22) in humans, which consists of the two PATRRs joined together at the center of the palindromes accompanied by occasional small deletions. On the other hand, no prominent PCR product was detected when modified versions of the 22-PR plasmid were substituted, such as those with a direct repeat (22-DR), a single repeat unit (22-S), or an inverted repeat with a 0.16-kb spacer at the center of the palindrome (22-IRins160). To confirm these results, we also used three other pairs of primers so as to amplify all possible junctions between PATRR11 and PATRR22 (Fig. 1C). All primer pairs exhibited the rearrangements between 11-PR and 22-PR, but not between 11-PR and 22-DR, indicating that all four possible fusions occurred between PATRR11 and PATRR22. From these results, it appears that the two PATRRs indeed mediate a translocation-like rearrangement in human cultured cells similar to the constitutional t(11;22) rearrangement in humans. These results also indicate that the palindromic configuration, and not the nature of the single repeat unit constituting the PATRR, is essential for the rearrangement.
Figure 1.
PCR detection of PATRR-mediated translocation-like rearrangements in HEK293 cells. (A) PATRR11 (11-PR, 445 bp) and PATRR22 (22-PR, 597 bp) were inserted into pUC19 and pBR322, respectively. Three modified versions of PATRR22—direct repeat (22-DR), single repeat unit (22-S), and inverted repeats with 156-bp spacer (22-IRins160-1 and 2)—were tested. Arrowheads indicate PCR primers for detection. (B) PCR specific for the translocation-like rearrangement. Plasmids (+) with or (−) without the PATRR were introduced simultaneously into HEK293 cells, and the joined molecules were detected by PCR. (Lanes 1–4, arrows) PCR products ∼1.2 kb in size originating from rearranged molecules were detected only when both of the PATRR-bearing plasmids were co-transfected. (Lanes 5–8) No such prominent PCR product was obtained from the modified versions of the PATRR22, (D) 22-DR, (S) 22-S, or (I1 and I2) 22-IRins160s. Smears and faint bands may be PCR artifacts possibly produced by nonspecifically degraded plasmids. DNA samples derived from (lane 9) cells without transfection, (lane 10) cells transfected with plasmids without addition of the transfection reagent, (lane 11) a simple mixture of two PATRR-bearing plasmids, and (lane 12) water without DNA were used as templates as negative controls. (Lane M) A 1-kb plus DNA ladder. (C) The rearrangement was also detected by PCR with three other combinations of primer pairs when the 11-PR plasmid was co-transfected with (P) 22-PR, but not with (D) 22-DR.
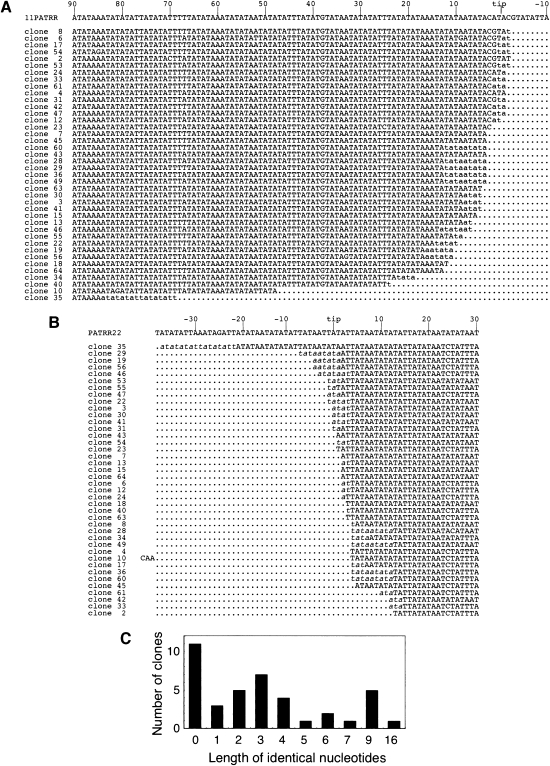
To determine the junction sequence, rearrangement-specific PCR products were isolated and cloned. The sizes of the PCR products seemed to be heterogeneous (Fig. 1B), suggesting that individual products represent different breakpoints from one another. The breakpoints are mainly located near the center of both PATRRs, and some of the clones show small deletions at the junction (Fig. 2A,B). Thirty of the 40 junctions manifested only four or fewer base pairs of nucleotide of microhomology between PATRR11 and PATRR22, and 11 with no overlap (Fig. 2C). In human t(11;22)s, since >75% of the junction sequences possess <5-bp microhomologies, the two PATRRs have been presumed to join via nonhomologous end-joining (NHEJ) (Kurahashi et al. 2007). These results suggest that both t(11;22) in humans and the rearrangement reaction in this plasmid model are likely to be processed through the same pathway: Both PATRRs are cleaved at the palindrome's center, and the DNA ends are then joined through NHEJ to produce fused molecules. The data appear to fit the previously proposed “center-break mechanism” of palindrome resolution in mammals (Cunningham et al. 2003).
Figure 2.
Junction sequences of translocation-like rearrangements in the plasmid-based system. (A) Sequences of the PATRR11 side of the junction. The first line is the sequence of the proximal arm of PATRR11. Sequences from a total of 40 rearrangement-specific PCR products derived from primers A and D in Figure 1A were aligned. In some cases, the exact origin of some nucleotides at the junctions could not be determined (lowercase) since the nucleotides are identical between the PATRR11 and the PATRR22. (B) Sequences of the PATRR22 side of the rearranged molecules. Junction sequences identical between PATRR11 and PATRR22 are shown in italic. The 3 nt of clone 10 is an insertion from unknown origin at the junction. (C) The number of identical nucleotides between PATRR11 and PATRR22 at the junctions.
PATRR-mediated rearrangement depends on secondary structure
To quantitatively estimate the incidence of this PATRR-mediated rearrangement, we established a fluorescence reporter system. Two plasmids were constructed: One has a promoter and a splice donor sequence derived from pCAGGS (Niwa et al. 1991) and the PATRR11 in this order (P-SD-11), and the other has the PATRR22 followed by the splice acceptor sequence of pCAGGS and the coding sequence of the GFP gene (22-SA-GFP) (Fig. 3A). If both PATRRs on the plasmids are cut and fused to one another, the rearrangement will enable splicing to occur between the splicing signals. The GFP gene will be under the control of the promoter 25% of the time, and we should detect such an event by green fluorescence. In fact, after simultaneous transfection of both plasmids, fused molecules that were detected by PCR after 3 h increased until 24 h post-transfection (Fig. 3A). At 48 h, GFP-positive cells comprised 11.0% of the total. Transfection of either the PATRR11 or the PATRR22 plasmids alone was negative for the presence of the PCR product (0.02% and 0.9%) (Fig. 3B,C). Furthermore, when the PATRR11-deleted plasmid (P-SD-11del) was introduced with the intact PATRR22 plasmid, the percentage of positive cells decreased to 3.0%, which is regarded as background for this experiment.
Figure 3.
Fluorescence detection of the PATRR-mediated rearrangement. (A) Plasmids used for the detection system. When the two plasmids harboring PATRR11 (P-SD-11) and PATRR22 (22-SA-GFP) are rearranged within each PATRR region, the transcript from the fusion product splices out the junction sequence and (middle panel) expresses the downstream GFP gene product. As a positive control, an expression vector, which includes this expected rearranged fragment, was also produced. (Arrowheads) PCR with flanking primers detected the junction fragment of ∼620 bp as early as 3 h after transfection, and (lower panel, arrow) the amount of product increased until 24 h post-transfection. (B) Flow cytometry. The GFP signal was measured 48 h after transfection. P-SD-11 and 22-SA-GFP were transfected alone or simultaneously into the cells. (Area shown in black in the histograms, R2) GFP-positive cells. (C) Percentage of GFP-positive cells. Bars, SD (n = 3).
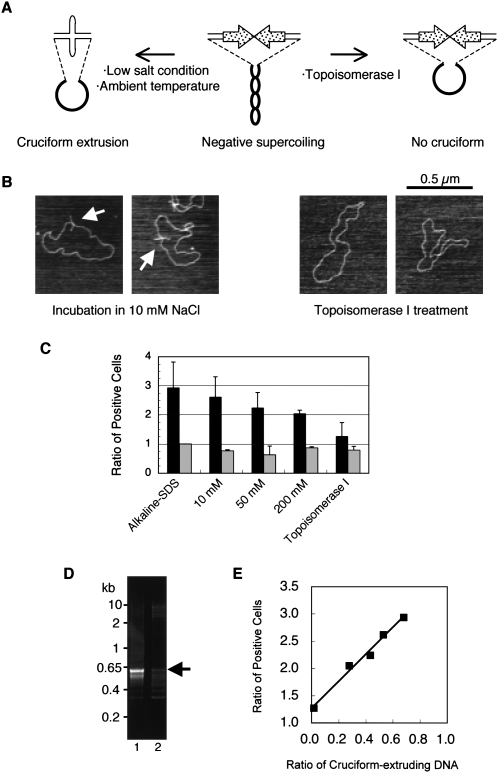
We hypothesized that the rearrangement mediated by the PATRRs is caused by DNA instability induced by the secondary structure adopted by the PATRRs. Palindromic sequences can form cruciform structures in the presence of negative torsion on the double-stranded DNA (Fig. 4A, left; Sinden 1994). To investigate the effect of secondary structure on the rearrangement, we attempted to prepare topoisomers of the PATRR plasmid with or without a cruciform configuration. Since the PATRR in the plasmid extrudes a cruciform during isolation from E. coli using the standard alkaline-SDS method, we applied a nondenaturing method using Triton X-100 that does not induce extrusion of the cruciform (Kurahashi et al. 2007) from the PATRR22-bearing plasmid. Plasmid DNA was treated with topoisomerase I to eliminate negative supercoiling in order to limit the potential for cruciform extrusion (Fig. 4A, right). After topoisomerase I treatment, the plasmid no longer extruded a cruciform, which was confirmed by atomic force microscopy (AFM) (Fig. 4B, right). When the PATRR22 plasmid treated to eliminate cruciform extrusion was transfected with the cruciform-extruding PATRR11 plasmid, GFP-positive cells were decreased almost to background levels (Fig. 4C) as compared to what was observed when the PATRR22 with the cruciform was used (P = 0.043, t-test). This substantial decrease of rearrangements was confirmed by semiquantitative PCR (Fig. 4D). Quantitative dot-blot hybridization indicated no difference in transfection efficiency between these topoisomers (data not shown). The results indicate that the PATRR-mediated rearrangement was induced only when the PATRRs extruded cruciform arms. It also suggests that after introduction into the cell, the plasmids with an intact cruciform were cut in a relatively short time before changes in superhelicity and assembly into nucleosomes.
Figure 4.
Translocation-like rearrangements induced by cruciform DNA. (A) Cruciform extrusion of the PATRR in plasmids with negative superhelicity. (Middle) The plasmid purified from E. coli is negatively supercoiled. In low salt conditions, a PATRR plasmid with negative superhelicity energetically favors cruciform extrusion forming intrastrand base-pairing at ambient temperature. (Left) The positive free energy of cruciform formation is offset by relaxation of the negative superhelical density. (Right) If the negative superhelicity is abrogated by topoisomerase I prior to cruciform extrusion, relaxed plasmid without extruded cruciform can be obtained. (B) AFM image of the 22-SA-GFP plasmid. Plasmid DNA purified by the Triton method was incubated in 10 mM NaCl (left) or treated with topoisomerase I (right) and observed by AFM. (Arrows) Cruciform extrusion of the PATRR plasmids. (C) The incidence of translocation-like rearrangements reflects the method of preparation for the PATRR plasmids. The numbers of GFP-positive cells were enumerated after co-transfection with various 22-SA-GFP plasmid topoisomers and cruciform-extruding P-SD-11. The plasmid topoisomers were prepared by the alkaline-SDS method and by the Triton method followed either by incubation in various NaCl concentrations or by topoisomerase I treatment. (Left bars, black) The results of 22-SA-GFP with the P-SD-11 plasmid; (right bars, gray) results of 22-SA-GFP with the P-SD-11del plasmid. (Vertical scale) The ratio of values relative to that obtained for the 22-SA-GFP isolated by the alkaline-SDS method with the P-SD-11del. Bars, SD (n = 3). (D) Rearrangement-specific PCR. Cruciform-extruding 22-SA-GFP obtained by the alkaline-SDS method (lane 1) or non-cruciform 22-SA-GFP obtained by the Triton method followed by topoisomerase I treatment (lane 2) were transfected with cruciform-extruding P-SD-11 for 48 h. In contrast to the strong band in the cruciform-positive sample (arrow), only a faint band was observed with non-cruciform DNA. (E) Proportional relation between cruciform extrusion and rearrangement frequency. The degrees of cruciform extrusion observed by AFM correlate with the ratio of GFP-positive cells (r = 0.992, P = 0.0008).
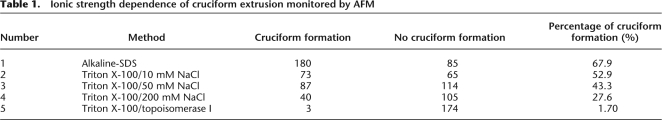
Next, DNA isolated by the Triton method was incubated in different ionic conditions ranging from 10 mM to 200 mM NaCl. Since isolated plasmid still has negative torsion, cruciform extrusion is facilitated in low ionic buffer (Fig. 4A; Kogo et al. 2007). After incubation of Triton-isolated PATRR22 plasmid in 10 mM NaCl, the plasmid exhibited cruciform extrusion, which was apparent in AFM images (Fig. 4B, left). When this PATRR22 plasmid was used for transfection experiments together with a cruciform-extruding PATRR11 plasmid, the rearrangement was observed at a level comparable to that of the PATRR22 plasmid purified by the alkaline-SDS method (Fig. 4C). On the other hand, incubation in 200 mM NaCl resulted in a decreased level of the rearrangement. The ionic strength–dependence of cruciform extrusion could be monitored by AFM (Table 1; Supplemental Fig. S1) or by measuring the extent of digestion of the plasmid with structure-specific nucleases, for example, T7 endonuclease I (Supplemental Fig. S2A,B). The level of rearrangement observed was proportional to the amount of cruciform plasmid (Fig. 4E; Supplemental Fig. S2C). Therefore, these results strongly suggest that PATRR-mediated rearrangement is not caused by the PATRR sequence itself, but rather by the unusual secondary structure—cruciform extrusion.
Table 1.
Ionic strength dependence of cruciform extrusion monitored by AFM
Discussion
Non-B DNA structure mediates chromosomal translocation
Using the plasmid-based system described in this study, we have demonstrated that palindrome-mediated rearrangement in humans is induced by cruciform extrusion of palindromic sequences at the breakpoint. To date, two lines of evidence point to involvement of cruciform DNA in the development of palindrome-mediated rearrangements. Regarding the recurrent constitutional t(11;22), differences in de novo translocation frequency in sperm is related to polymorphic sequence variation of the PATRR (Kato et al. 2006), and predisposition for cruciform structure formation of each PATRR is linked to translocation frequency (Kogo et al. 2007). These data indirectly but strongly suggest that the presence of cruciform DNA in human chromosomes is etiologic in causing this chromosomal aberration. Our current data directly show that only cruciform-forming PATRRs cause DNA rearrangement leading to translocations in human cells.
Most of the examples of genomic instability originating from non-B DNAs have been explained by replication-dependent phenomena. Inverted-repeat sequences have been known to be unstable in bacteria and eukaryotic cells, particularly under conditions of slow fork progression caused by DNA polymerase deficiency (Ruskin and Fink 1993; Lemoine et al. 2005). Perturbation of the replication fork would facilitate hairpin formation at long tracts of single-stranded portions of the lagging-strand template. Similarly, some fragile sites in human chromosomes are comprised of highly AT-rich tandem repeats, suggesting the involvement of DNA secondary structure in genomic or chromosomal instability. They manifest late replication and are sensitive to additional replication delay by DNA synthesis inhibitors, which leads to the induction of fragility (Glover et al. 2005). Trinucleotide repeat expansions are generally believed to arise by formation of a secondary structure in the lagging strand of the replication fork (Mirkin 2007). In our present experiments, we used nonreplicating plasmid, and, in fact, there was no increased amount of plasmid DNA after introduction into the cell. Furthermore, the PATRR plasmids were cut within a short time, suggesting that the PATRR underwent no DNA replication during its cleavage. Indeed, the absence of evidence for de novo translocations in somatic cells (Kurahashi and Emanuel 2001) and the demonstration of age independence of translocation frequency (Kato et al. 2007) are features that are also inconsistent with involvement of DNA replication in the process. Thus, it is conceivable that PATRR-mediated rearrangements are generated by genomic instability via a replication-independent non-B DNA conformation.
The existence of non-B DNA structures in living cells has long been proposed and is still a controversial question. In addition to several previous reports about non-B structures in E. coli (McClellan et al. 1990; Dayn et al. 1991, 1992; Leach 1994) and cruciform conformation at DNA replication origins in mammalian cells (Zannis-Hadjopoulos et al. 1988), recent data have also highlighted the possibility that such non-B DNA conformation contributes to chromosomal instability in humans (Bacolla et al. 2004; Raghavan et al. 2004). Our present results provide physical evidence of cutting of cruciform DNA at the breakpoint of t(11;22) in our plasmid-based model system producing rearranged molecules similar to those observed in human t(11;22)s. In addition, the extreme AT richness of the PATRRs may support this hypothesis, since AT-rich inverted repeats, indeed, rapidly convert into DNA cruciforms even at low supercoiling levels (Greaves et al. 1985; Panyutin et al. 1985). This is in contrast to relatively GC-rich inverted repeats, for which cruciform extrusion is kinetically forbidden in physiological conditions (Courey and Wang 1983). These data strongly suggest that the PATRR sequence adopts a cruciform structure in living cells.
Non-B DNA structure and sperm-specific translocations
If this is the case, then, why are de novo t(11;22)s detected in sperm, but not in other mitotic cells in humans (Kurahashi and Emanuel 2001)? Our current data indicate that all of the enzymatic activities necessary for PATRR-mediated translocations may be available in cultured human cells. However, transfection of PATRR plasmids with canonical double-stranded DNA structure did not generate any translocations in our model. This suggests that what is lacking for translocation formation is the conformation of the cruciform. Likewise, such human cell lines did not produce any translocations derived from the endogenous PATRR11 and PATRR22. This implies that the reason PATRR-mediated translocations do not arise somatically is that the PATRRs do not extrude a cruciform conformation in the chromosomal context of somatic cells.
Sufficient negative supercoiling is a prerequisite for the formation of certain non-B DNA structures in vitro, and possibly in vivo as well (Sinden 1994). However, the level of supercoiling that is sufficient to support cruciform extrusion may not be feasible in the chromosomal context of the living cell, because the torsional stress imposed on DNA by replication or transcription should normally be relieved by the activity of topoisomerases. Although several reports have described unrestrained supercoiling in the proximity of many genes (Ljungman and Hanawalt 1992; Jupe et al. 1993), possibly caused by localized superhelical tension that cannot be relieved sufficiently by topoisomerase activity (Wang and Dröge 1996), this argument would not apply to cruciform extrusion of the PATRR because there is no active gene in the vicinity of PATRR11 or 22. On the other hand, an excess of negative superhelicity may be accumulated temporarily in the DNA during chromatin compaction in the late stages of spermatogenesis. The successive transition of chromatin components from histones to protamines will cause dynamic changes of chromatin structure (Meistrich et al. 2003), since DNA dissociation from histones may involve accumulation of free negative supercoiling. Non-B DNA structures such as cruciforms might be induced by this accumulated negative superhelicity in the DNA. Germinal expansion of CAG repeats in the Huntington's disease gene arises in post-meiotic cells (Kovtun and McMurray 2001). This observation allows for the speculation that DNA strand breaks may be induced by non-B DNA conformation during the compaction of chromatin, and the elongation of trinucleotide repeats would be promoted by gap-synthesis repair of such DNA breakage. In addition, the in vitro analysis shows that the PATRRs form a cruciform structure more stably at a slightly lower temperature than body temperature (Kurahashi et al. 2004). The lower temperature of the testis might stabilize cruciform structures ready for DSB formation. If this hypothesis is true, meiotic cells from females may not be as susceptible to producing translocations mediated by palindromes. Thus, it may be worth examining the parental origin of de novo t(11;22) translocations in humans.
Methods
Construction of the plasmids
PATRR11 and 22 were amplified from genomic DNA derived from case 3 in our previous report (Kurahashi et al. 2007) using PATRR-optimized PCR conditions (Inagaki et al. 2005) with the primers described in Table 1, and cloned into plasmids with different backbones (GenBank/EMBL/DDBJ accession nos. AB334268 and AB334267). To make modified PATRR22s, the distal arm of PATRR22 was cut out with S1 nuclease (TAKARA BIO) and a restriction enzyme (22-S; AB334270) and then reinserted in the opposite direction (22-DR; AB334269). Alternatively, PATRR22 was cut with S1 nuclease at the center, and a 156-bp fragment derived from pBR322 was inserted at that position in both directions (22-IRins160-1 and 2; AB334271, AB334272).
To generate the fluorescent detection system, PATRR11 was cloned downstream from an enhancer–promoter and a splice donor site from pCAGGS (Niwa et al. 1991) with the aid of a chloramphenicol-resistance gene and the replication origin from pACYC184 (P-SD-11). PATRR22 was also cloned upstream of a splice acceptor site of pCAGGS and the EGFP reporter gene with the aid of pBR322 (22-SA-GFP). Thus the backbones of these constructs have minimal regions of homology. For a PATRR11-deleted version (P-SD-11del), P-SD-11 was cleaved with T7 endonuclease I (New England Biolabs), digested with S1 nuclease, and then self-ligated.
Preparation of plasmid DNAs
For plasmids with cruciforms extruded, the DNAs were prepared by a standard alkaline-SDS method using the Endofree Plasmid Maxi Kit (QIAGEN). For plasmids without cruciform extrusion, the bacteria were lysed with a solution containing 3% Triton X-100 (Kurahashi et al. 2004) followed by column purification using the same kit at 4°C. The column eluate was divided into aliquots followed by ethanol precipitation, and the resulting pellets were dried and stored at −80°C until use. Each pellet was dissolved immediately prior to use into pre-chilled 10 mM Tris-HCl and 1 mM EDTA (TE) supplemented with 200 mM NaCl and stored on ice. The DNAs were then diluted with TE to produce the final NaCl concentrations and incubated for 16 h at 25°C to extrude the cruciform. Alternatively, unextruded plasmid was treated with topoisomerase I (New England Biolabs) for 16 h at 25°C. Cruciform formation was evaluated by AFM as previously described (Kurahashi et al. 2004), and by quantitative assessment of the amount of DNA sensitive to T7 endonuclease I in the presence of 200 mM NaCl at 25°C. This was estimated by determining the intensity of the cleaved band using ImageJ software.
Detection of DNA rearrangement in HEK293 cells
A total of 3.2 μg of plasmid DNA was transfected into HEK293 cells in a 30 mm dish at 50%–80% confluency using Lipofectamine 2000 (Invitrogen). For flow cytometry, the cells were collected at 48 h and applied to a FACScan (BD Bioscience). Rearranged molecules were detected by semiquantitative PCR from 100 ng of DNA. The PCR conditions were 25 cycles of 10 sec at 98°C and 5 min at 60°C with the primers described in Supplemental Table S1.
Acknowledgments
We thank J. Miyazaki of Osaka University for providing pCAGGS. We thank E. Hosoba, K. Nagaoka, and T. Mori for technical assistance. We also thank H. Yamaguchi, J. Okada, N. Yamamoto, and M. Nakamura for helpful discussions. These studies were supported by a grant-in-aid for Scientific Research and the 21st Century COE program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.I. and H.K.). These studies were also supported in part by funds from CA39926 (NIH) and the Charles E.H. Upham chair in Pediatrics (B.S.E.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.079244.108.
References
- Akgün E., Zahn J., Baumes S., Brown G., Liang F., Romanienko P.J., Lewis S., Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A., Jaworski A., Connors T.D., Wells R.D. PKD1 unusual DNA conformations are recognized by nucleotide excision repair. J. Biol. Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collick A., Drew J., Penberth J., Bois P., Luckett J., Scaerou F., Jeffreys A., Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- Courey A.J., Wang J.C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983;33:817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Cunningham L.A., Coté A.G., Cam-Ozdemir C., Lewis S.M. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 2003;23:8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayn A., Malkhosyan S., Duzhy D., Lyamichev V., Panchenko Y., Mirkin S. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J. Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayn A., Malkhosyan S., Mirkin S.M. Transcriptionally driven cruciform formation in vivo. Nucleic Acids Res. 1992;20:5991–5997. doi: 10.1093/nar/20.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover T.W., Arlt M.F., Casper A.M., Durkin S.G. Mechanisms of common fragile site instability. Hum. Mol. Genet. 2005;14:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- Gordenin D.A., Lobachev K.S., Degtyareva N.P., Malkova A.L., Perkins E., Resnick M.A. Inverted DNA repeats: A source of eukaryotic genomic instability. Mol. Cell. Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L., Nimmakayalu M.A., Jalali G.R., Hacker A.M., Vorstman J., Duffy D.C., Medne L., Emanuel B.S. A palindrome-driven complex rearrangement of 22q11.2 and 8q24.1 elucidated using novel technologies. Genome Res. 2007;17:470–481. doi: 10.1101/gr.6130907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D.R., Patient R.K., Lilley D.M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J. Mol. Biol. 1985;185:461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Inagaki H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H. Palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved in primates. Hum. Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe E.R., Sinden R.R., Cartwright I.L. Stably maintained microdomain of localized unrestrained supercoiling at a Drosophila heat shock gene locus. EMBO J. 1993;12:1067–1075. doi: 10.1002/j.1460-2075.1993.tb05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Yamada K., Inagaki H., Kogo H., Ohye T., Emanuel B.S., Kurahashi H. Age has no effect on de novo constitutional t(11;22) translocation frequency in sperm. Fertil. Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Häussler J., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tümmers U., Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: Sequence analysis of the breakpoint regions. Hum. Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- Kogo H., Inagaki H., Ohye T., Kato T., Emanuel B.S., Kurahashi H. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun I.V., McMurray C.T. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;11:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T., Takata M., Toda T., Emanuel B.S. The constitutional t(17;22): Another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Hosoba E., Kato T., Ohye T., Kogo H., Emanuel B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R.F. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase: A model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Ljungman M., Hanawalt P.C. Localized torsional tension in the DNA of human cells. Proc. Natl. Acad. Sci. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- McClellan J.A., Boublíková P., Paleček E., Lilley D.M. Superhelical torsion in cellular DNA responds directly to environmental and genetic factors. Proc. Natl. Acad. Sci. 1990;87:8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M.L., Mohapatra B., Shirley C.R., Zhao M. Roles of transition nuclear protein in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Narayanan V., Mieczkowski P.A., Kim H.M., Petes T.D., Lobachev K.S. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Nimmakayalu M.A., Gotter A.L., Shaikh T.H., Emanuel B.S. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Panyutin I., Lyamichev V., Mirkin S. A structural transition in d(AT)n·d(AT)n inserts within superhelical DNA. J. Biomol. Struct. Dyn. 1985;2:1221–1234. doi: 10.1080/07391102.1985.10507634. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Chastain P., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J. Biol. Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Fink G.R. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics. 1993;133:43–56. doi: 10.1093/genetics/134.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.R. DNA structure and function. Academic Press; San Diego: 1994. [Google Scholar]
- Wang Z., Dröge P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996;15:581–589. [PMC free article] [PubMed] [Google Scholar]
- Zannis-Hadjopoulos M., Frappier L., Khoury M., Price G.B. Effect of anti-cruciform DNA monoclonal antibodies on DNA replication. EMBO J. 1988;7:1837–1844. doi: 10.1002/j.1460-2075.1988.tb03016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]