Abstract
In mammals, genome-wide chromatin maps and immunofluorescence studies show that broad domains of repressive histone modifications are present on pericentromeric and telomeric repeats and on the inactive X chromosome. However, only a few autosomal loci such as silent Hox gene clusters have been shown to lie in broad domains of repressive histone modifications. Here we present a ChIP-chip analysis of the repressive H3K27me3 histone modification along chr 17 in mouse embryonic fibroblast cells using an algorithm named broad local enrichments (BLOCs), which allows the identification of broad regions of histone modifications. Our results, confirmed by BLOC analysis of a whole genome ChIP-seq data set, show that the majority of H3K27me3 modifications form BLOCs rather than focal peaks. H3K27me3 BLOCs modify silent genes of all types, plus flanking intergenic regions and their distribution indicates a negative correlation between H3K27me3 and transcription. However, we also found that some nontranscribed gene-poor regions lack H3K27me3. We therefore performed a low-resolution analysis of whole mouse chr 17, which revealed that H3K27me3 is enriched in mega-base-pair-sized domains that are also enriched for genes, short interspersed elements (SINEs) and active histone modifications. These genic H3K27me3 domains alternate with similar-sized gene-poor domains. These are deficient in active histone modifications, as well as H3K27me3, but are enriched for long interspersed elements (LINEs) and long-terminal repeat (LTR) transposons and H3K9me3 and H4K20me3. Thus, an autosome can be seen to contain alternating chromatin bands that predominantly separate genes from one retrotransposon class, which could offer unique domains for the specific regulation of genes or the silencing of autonomous retrotransposons.
Post-translational modifications on histone tails either reflect or directly influence the transcriptional status of genes and are classified as active, when they correlate with expressed genes, or, as repressive, when they correlate with silent genes. Chromatin immunoprecipitation (ChIP) using histone modification antibodies and tiling array analysis (ChIP-chip) or new generation sequencing technology (ChIP-seq), has been used to profile histone modifications of mouse and human chromosomes (Bernstein et al. 2007; Schones and Zhao 2008). Together these analyses show that active histone modifications such as H3K4 methylation and histone acetylation are enriched on expressed genes over short focal regions at promoters and nonpromoter putative gene-regulatory regions (Heintzman et al. 2007). However, these active marks can also be found at silent gene promoters in undifferentiated embryonic stem (ES) cells and in T cells, and, active transcription has been found to correlate with additional modifications such as H3K36 tri-methylation (me3), that spreads through the transcribed gene body (Bernstein et al. 2006b; Roh et al. 2006; Barski et al. 2007). Repressive histone modifications, such as H3K9me3, H4K20me3, and H3K27me3 have been associated in many cells types with gene silencing or heterochromatin formation (Martens et al. 2005; Boyer et al. 2006; Regha et al. 2007; Wutz 2007). In contrast to active histone modifications that are restricted to gene regulatory elements or the transcribed gene body, repressive histone modifications have also been shown to cover much larger regions, such as silent gene clusters, pericentromeric and telomeric repeats or mega-base-pair domains on the inactive X chromosome (Chadwick and Willard 2004; Schotta et al. 2004; Squazzo et al. 2006).
Repressive H3K27me3 modifications have attracted particular attention as they have been shown to repress developmentally important genes and are thought to maintain stem cell pluripotency (Boyer et al. 2006). However, while some studies have shown that Polycomb repressor complex 2 (PRC2) that catalyzes H3K27me3, is required for ES cell differentiation (Pasini et al. 2007), other studies have shown that ES cells retain pluripotency in the absence of functional PRC2 (Chamberlain et al. 2008). A role for H3K27me3 in repressing developmentally important genes is, however, supported by genome-wide mapping in combination with functional studies. Thus, H3K27me3 or PRC2 have been identified on key ES cell developmental regulatory genes, on genes showing lineage-specific activation and on highly conserved noncoding elements (Azuara et al. 2006; Bernstein et al. 2006a; Bracken et al. 2006; Lee et al. 2006; Squazzo et al. 2006). These studies focused on gene promoters or genomic regions containing key developmental genes and demonstrated that H3K27me3 mainly forms focal peaks of enrichment on CG-rich silent gene promoters. However, in some regions notably the four mammalian Hox gene clusters, both PRC2 and H3K27me3 covered broad domains from 10 kb up to 140 kb, which spanned entire genes or gene clusters (Ringrose 2007). Four studies have also mapped H3K27me3 modifications across the whole mouse or human genome (Barski et al. 2007; Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007). These studies came to generally similar conclusions: that H3K27me3 largely formed focal modifications at silent gene promoters with a few exceptions showing modifications of broad domains containing gene clusters. In contrast, genome-wide profiles of the Drosophila genome show that H3K27me3 covers large genomic domains containing silent genes that included genic and intergenic regions (Schwartz et al. 2006; Beisel et al. 2007).
Mammalian chromosomes are known to be longitudinally organized in a manner that can be visualized as a banding pattern in stained metaphase chromosomes, which shows that dark Giemsa (or G-bands) alternate with light reverse-Giemsa (or R-bands), along the chromosome length (Craig and Bickmore 1993). Cytological studies based on DNA probe hybridization have shown that genes and repeats are differently distributed between dark and light bands. Dark bands are enriched for autonomous long interspersed elements (LINEs) retrotransposons, while light bands are enriched for genes and nonautonomous short interspersed elements (SINEs) retrotransposons (Boyle et al. 1990). These banding patterns are suggested to reflect differences in chromatin structure with dark G-bands containing more condensed late-replicating chromatin. However, a detailed comparison of chromatin composition between dark and light bands has not yet been made.
Here we used ChIP-chip to profile active and repressive histone modifications on mouse chr 17 in a differentiated cell type, to examine the relationship between histone modifications, silent and expressed genes and interspersed repeats, along the chromosomal length. Our results using an algorithm that identified broad regions of histone modifications named broad local enrichments (BLOCs), were confirmed by unsupervised segmentation of continuous genomic data by hidden Markov models (Day et al. 2007) and by ChIP-seq analysis, and show that while active, H3K4me and acetylation modifications form peaks—H3K27me3 forms BLOCs with an average size of 43 kb that overlap silent genes and intergenic regions. Furthermore, we show that H3K27me3 is not randomly distributed along the chromosome length instead it is enriched in regions of high gene and SINE density and depleted in regions of high LINE/long-terminal repeat (LTR) and low gene density. We also show that regions of high LINE/LTR density are depleted of active histone modifications, but are enriched for repressive H3K9me3 and H4K20me3 histone modifications. Together this shows that specific enriched histone modifications distinguish gene-poor/LINE–LTR-rich chromosomal domains from gene-rich/SINE-rich domains, which have previously been shown to correlate, respectively, with dark and light Giemsa bands in metaphase chromosomes. These two types of chromosome histone domains may provide unique compartments for the specific regulation of gene expression or the silencing of autonomous retrotransposons.
Results
Active histone modifications form peaks, but H3K27me3 forms BLOCs
The distribution of active histone modifications H3K4me2, H3K4me3 and H3K9Ac and the repressive H3K27me3 modification were mapped across nonrepeat regions of mouse chr 17 in two independent mouse embryonic fibroblast (MEF) cell lines MEFB1 and MEFF (Regha et al. 2007; Supplemental Table 1) using a custom chr 17 oligonucleotide NimbleGen tiling array chip described in Methods (Fig. 1). Focal sites of enrichment (peaks) were identified by ChIPOTle (Buck et al. 2005) using 1.5 kb sliding windows and peaks within 0.5 kb were fused into one peak. The number of active histone modification peaks was similar in both MEF cell lines (data not shown). Combining the data from both the MEFF and the MEFB1 cell line (Fig. 2A), the most abundant active histone modification was H3K4me2 (758 peaks), followed by H3K9Ac (425 peaks) and H3K4me3 (275 peaks). H3K27me3 was initially analyzed using ChIPOTle and 476 variably sized peaks of H3K27me3 were identified with an average width of 2.2 kb (±1.5 kb). However, upon visual inspection it was evident that H3K27me3 peaks were contained in broadly enriched large genomic regions (Fig. 1). The ChIPOTle program is designed to identify peaks and it detected irregular enrichments and depletions within these broadly enriched regions as peaks. We therefore developed a new algorithm (BLOCs) to identify broad regions of H3K27me3 enrichment that identified 257 and 344 H3K27me3 BLOCs along chr 17 in MEFB1 and MEFF, respectively. 91% of ChIPOTle identified H3K27me3 peaks lie within BLOCs.
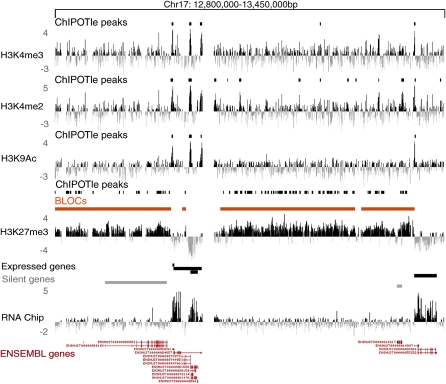
Figure 1.
H3K27me3 forms BLOCs covering silent genes and intergenic regions. A screen shot from the UCSC genome browser showing the distribution of histone modifications across a 650 kb region of mouse chr 17 (UCSC Mouse [mm6], March 2005) in mouse embryo fibroblasts (MEFs). Profiles for three active histone modifications (H3K4me3, H3K4me2, and H3K9Ac) and the repressive H3K27me3 are displayed. Active histone modifications form peaks detected by ChIPOTle (Buck et al. 2005) (short black bars), while H3K27me3 spreads over large regions (orange bars) called BLOCs (broad local enrichments) that are also detected as dense clusters of ChIPOTle peaks. Expression was determined by hybridizing polyA RNA to the tiling array (RNA chip track). Black and gray bars above the RNA chip track represent, respectively, expressed and silent genes (Supplemental Fig. 4). Positions of genes are shown by Ensembl predictions. Y-axis: log2 ChIP/input ratio or cDNA/input. X-axis: 50 bp oligonucleotides from repeat-masked sequence included on the NimbleGen custom mouse chr 17 tiling array.
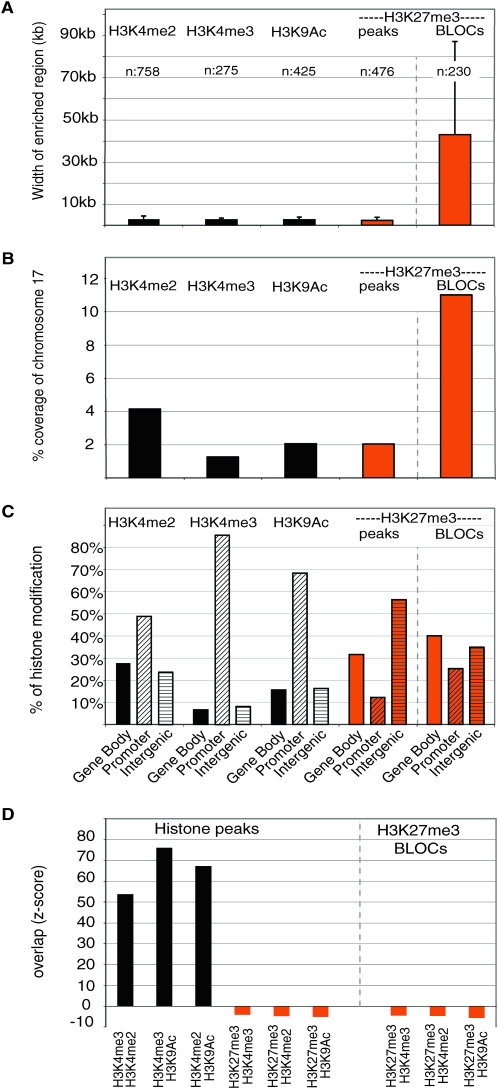
Figure 2.
Overview of active and repressive marks on mouse chr 17 in MEFs. The analysis of modifications forming peaks (H3K4me2, H3K4me3, and H3K9Ac) was based on the sum of all chr 17 regions with a probe density of at least eight oligonucleotides per 1500 bp. The analysis of modifications forming BLOCs (H3K27me3) was based on the whole mouse chr 17 tiling array. Data from two independent MEF cell lines (MEFB1 and MEFF) were merged for all analyses (Supplemental Table 1). (A) Chromosome-wide analysis showing the average width of genomic regions enriched by four histone modifications on chr 17, as analyzed by ChIPOTle (all modifications) or the BLOCs algorithm (H3K27me3). Only regions found to be enriched in all technical and biological replicates (Supplemental Table 1) were used in this analysis. (B) The percentage of chr 17 covered by four histone modifications analyzed by ChIPOTle peaks (all modifications) or the BLOCs algorithm (H3K27me3). (C) The percentage of ChIPOTle peaks (all modifications) or BLOCs (H3K27me3) located at the gene body excluding the promoter (solid bar), promoters (diagonally striped bar), and intergenic regions (horizontally striped bar). (D) The significance of overlap between genomic regions enriched for different histone modifications was calculated by identifying Z-scores for all possible pairs of ChIPOTle peaks (all modifications) and BLOCs (H3K27me3) (details in Methods). A high Z-score identifies an overlap that occurs more often than expected compared to a randomized data set.
All three active histone modifications formed typical peak shapes with an average width of 2.4–2.9 kb, while H3K27me3 BLOCs had an average size of 43 kb with a high variation (Fig. 2A). 22 H3K27me3 BLOCs exceeded 100 kb and the maximum BLOC was 337 kb. Peaks of active histone modifications covered only a small portion of the total chromosome (2%–4%), while H3K27me3 BLOCs covered 11% of chr 17 (Fig. 2B). Active histone modification peaks were more often on genes with a stronger bias toward promoters than intergenic regions, most notable for H3K4me3 and H3K9Ac (Fig. 2C). H3K4me3 had the strongest association with genes, followed by H3K9Ac and H3K4me2. In contrast, H3K27me3 BLOCs (as well as H3K27me3 ChIPOTle peaks) did not specifically mark promoters, but were more equally distributed over genes and intergenic regions (Fig. 2C).
Silent and expressed genes cluster close to H3K27me3 BLOCs
Visual inspection of H3K27me3 BLOCs across chr 17 indicated that although they contain silent genes they are often closely flanked by expressed genes (Fig. 1) Supplemental Figures 1–3 show different screen shots showing respectively that H3K27me3 negatively correlates with transcription, that H3K27me3 BLOCs mark intergenic regions bounded on both sides by a transcribed region lacking H3K27me3 and last, that a H3K27me3 BLOC extends over a silent gene and intergenic region up to the next expressed gene.
We therefore determined how H3K27me3 BLOCs and active histone peaks are spatially distributed across whole chr 17 in relation to expressed and silent genes. Gene expression status was determined by cDNA hybridization to the same tiling array used for ChIP-chip (see Methods). An RNA chip analysis was performed independently on MEFB1 and MEFF and both cell lines gave similar results (Supplemental Fig. 4; data not shown). This analysis showed that H3K4me2, H3K4me3, and H3K9Ac were most often associated with expressed genes. On average 89% of expressed genes were marked by H3K4me2, 64% by H3K4me3, and 73% by H3K9Ac (data not shown). A small number of silent genes also contained active histone modification peaks, 26% with H3K4me2, 6% with H3K4me3, and 1% with H3K9Ac (data not shown). These three active histone modifications were frequently located on the same genomic regions, but were rarely found to associate with H3K27me3, whether analyzed as ChIPOTle peaks or as BLOCs (Fig. 2D). Figure 3A shows the combined H3K27me3 log2 ChIP/input ratios relative to the transcription start site (TSS), gene length and flanking regions for all expressed genes on mouse chr 17 (Fig. 3A, top), all silent genes analyzed together (Fig. 3A, middle), and silent genes lying in BLOCs (Fig. 3A, bottom). The analysis shows that expressed genes are depleted for H3K27me3 for the gene length, but not for the immediate flanking sequences possibly indicating a negative correlation between transcription and H3K27me3. Silent genes lying within BLOCs show a significant H3K27me3 enrichment over the gene body, but not specifically over the promoter, compared to expressed genes (P < 10−4). The same enrichment is detectable at a reduced level if all silent genes are analyzed together. The comparison between these two groups of silent genes indicates a population of silent genes that lack H3K27me3.
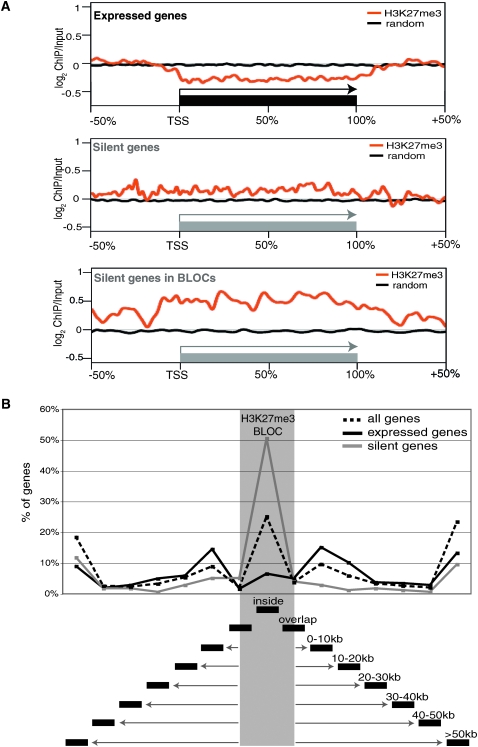
Figure 3.
Silent and expressed genes cluster close to H3K27me3 BLOCs. (A) The combined log2 ChIP/input ratios from one MEFF chr 17 ChIP-chip replicate are shown for all expressed genes (top) all silent genes (middle), and silent genes in BLOCs (bottom) relative to the transcription start site (TSS), gene length, and flanking regions. All positions are relative and the length of the gene (black box: expressed/gray box: silent) is defined as 100% and the flanking regions are ±50% of the gene length. Orange line: continuous log2 ChIP/input values, black line: randomized data set. (B) The graph indicates the distance of all genes (dotted line) or of expressed (black line) or silent (gray line) genes relative to the closest H3K27me3 BLOC (gray shaded area) in the MEFF cell line. Distances were calculated separately for each gene and then combined into distance bins (black boxes underneath). The percentage of genes in each bin is indicated on the y-axis. Silent genes are inside and expressed genes are outside, but close to H3K27me3 BLOCs.
Since the analysis of expressed genes in Figure 3A indicated a negative correlation between transcription and H3K27me3, we plotted the distribution of silent and expressed genes relative to H3K27me3 BLOCs for MEFF (Fig. 3B) and MEFB1 (Supplemental Fig. 5A). The distance of each gene to the closest H3K27me3 BLOC was calculated for the whole of chr 17 using three groups of genes: all genes, expressed genes and silent genes and grouped into 10 kb distance bins. “All genes” showed a characteristic three-peak pattern—enriched within H3K27me3 BLOCs, but also in the 0–10 kb flanking distance bins. Silent genes are enriched within H3K27me3 BLOCs, but not in the flanking distance bins. In contrast, the opposite pattern is seen for expressed genes, which are rarely found within BLOCs, but are enriched in the 0–10 kb flanking distance bins. This analysis, which shows that silent genes lie inside H3K27me3 BLOCs, while active genes immediately flank BLOCs, also indicates a negative correlation between H3K27me3 and transcription.
Validation of H3K27me3 ChIP-chip BLOCs
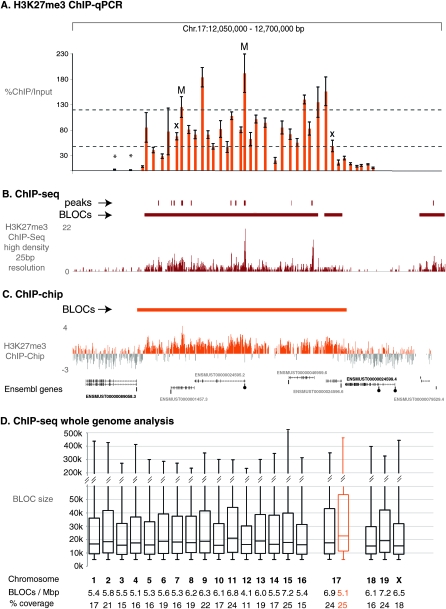
We performed two tests on the H3K27me3 ChIP-chip data. First, we determined the statistical significance of the difference of the H3K27me3 enrichment within BLOCs and the remaining chip, which shows the signal within BLOCs is significantly higher (P < 10−4) (Supplemental Fig. 6). Second, we performed an analysis of whole chr 17 H3K27me3 ChIP-chip profiles and expression identified by RNA chip using hidden Markov modeling (HMM) and Viterbi segmentation (Day et al. 2007), which confirmed that H3K27me3 and transcription show a negative correlation and that H3K27me3 forms BLOCs (Supplemental Fig. 7). Previous descriptions of H3K27me3 profiles across whole mouse and human genomes, based on ChIP-chip or on ChIP-seq analyses, have mostly shown that H3K27me3 forms focal modifications at silent gene promoters with a few exceptions showing modifications of broad domains containing gene clusters (Barski et al. 2007; Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007). In order to test if the identification of H3K27me3 BLOCs was a consequence of the low dynamic range of tiling arrays, we selected one large 365.4 kb BLOC on mouse chr 17 and performed a scanning qPCR assay throughout the BLOC, using primers spaced at ∼10 kb intervals and the same MEFF ChIP material used for the ChIP-chip analysis (Fig. 4A). The results show that significant H3K27me3 enrichment is found only inside the ChIP-chip identified BLOC, P = 10−4 for qPCR bars 4–31 (Fig. 4A, numbered left to right), compared with the 10 flanking primers. The scanning qPCR assay shows that levels of H3K27me3 enrichment are not uniform throughout the BLOC and six enriched peaks (defined as signals > mean + 1 standard deviation of all signals) could be seen. The boundaries of the enriched domain detected by qPCR coincide with ends of expressed genes (Supplemental Fig. 4), in a similar manner as shown above for the identified ChIP-chip H3K27me3 BLOCs.
Figure 4.
qPCR and ChIP-seq validation of H3K27me3 ChIP-chip BLOCs. (A) Scanning qPCR of one MEFF H3K27me3 ChIP-chip BLOC spanning 365.4 kb in a 650 kb region on mouse chr 17 (12.05–12.70 Mb, UCSC Mouse [mm8], February 2006) with 38 primers (orange bars, error bars indicate variation in three technical replicates) spaced ∼10 kb. Y-axis: %ChIP/input. Mock IP samples were lower than 10% of input with two exceptions (marked with X). Asterisk: low relative qPCR value. M: qPCR assay located in H3K27me3 peaks previously identified in this region by ChIP-seq (Mikkelsen et al. 2007). Lower dotted line indicates the cutoff for significant signals, upper dotted line indicates cutoff for enriched peaks. Primer sequences, qPCR assay details and Ct values are shown in Supplemental Table 2. (B) ChIP-seq of the H3K27me3 ChIP sample assayed by qPCR in A (details in Methods). The ChIP-seq sequence-tag abundance is displayed as 25 bp densities (high density). ChIP-seq BLOCs (horizontal dark red bar) in this region as well as significantly enriched regions (vertical dark red bars) are shown above the ChIP-seq track. Ensembl genes (expressed: black font, silent: gray font, see Supplemental Fig.4) are shown underneath. (C) H3K27me3 ChIP-chip profile (orange peaks) for the region analyzed in (A,B). The orange bar marks the BLOC identified in this region. Genes in this region (black font: expressed, gray font: silent, see Supplemental Fig. 4) are indicated with CG-poor promoters indicated underneath by a bar and CG-rich promoters indicated by a bar plus circle. (D) ChIP-seq BLOCs are identified on all mouse chromosomes in one MEFF data set. Box and whisker plots illustrate the size distribution for each chromosome. The number of BLOCs per Mb and % chromosome coverage by BLOCs is shown below. The ChIP-chip (orange) and ChIP-seq (black) BLOCs from one MEFF data set across chr 17 correlate well, as 82.3% of 1 kb windows show the same H3K27me3 BLOC state (BLOC or no BLOC). Note that MEFF cells are XO (data not shown) and thus have one active X chromosome.
We next analyzed the same MEFF chip material by whole genome sequencing (ChIP-seq, see Methods for details) and the results are similar to the scanning qPCR assay and show a broad enriched domain with oscillating peaks coincident with the ChIP-chip identified H3K27me3 BLOC. In Figure 4B a high-resolution (25 base pair [bp]) map of sequence tag density is shown and the peaks in ChIP-seq data were analyzed using a high and low cutoff (Supplemental Fig. 9). Only the low cutoff identified all six qPCR peaks from the scanning qPCR assay shown in Figure 4A. Supplemental Figure 8 shows screenshots of H3K27me3 ChIP-seq densities for the same regions in Figure 1 and Supplemental Figures 1–3 showing correspondence between BLOCs identified by ChIP-seq and ChIP-chip. Together, these experiments verify the existence of H3K27me3 BLOCs along mouse chr 17 and additionally show that a small number of enriched peaks are found within a BLOC. We then used the whole genome ChIP-seq data set to test if H3K27me3 BLOCs are a feature of the whole mouse genome. Figure 4D shows that H3K27me3 BLOCs are found on all mouse chromosomes with a mean size of 31.4 kb and a percentage chromosome coverage ranging from 11% to 26%. A genome-wide distance plot analysis of known genes (mm8) relative to the H3K27me3 ChIP-seq-BLOCs shows the same three-peak pattern as ChIP-chip BLOCs on chr 17, indicating a similar spatial distribution of H3K27me3 BLOCs across the whole genome (Supplemental Fig. 5B).
As an additional validation test for the H3K27me3 ChIP-chip BLOCs identified here, we compared our data with published H3K27me3 profiles from the same mouse region obtained by ChIP-seq of MEF cells (Mikkelsen et al. 2007). A distance distribution analysis (Supplemental Fig. 5C) performed on the Mikkelsen et al. (2007) data shows that H3K27me3 ChIP-seq peaks are located inside silent genes and intergenic regions, but not in expressed genes, similar to the analysis of H3K27me3 BLOCs shown in Figure 3B. The Mikkelsen et al. (2007) ChIP-seq data set identified 275 H3K27me3 peaks on chr 17 of which 79% lie inside ChIP-chip BLOCs identified here (Supplemental Fig. 9). Finally, we also compared our H3K27me3 ChIP-chip BLOCs with human ES cell ChIP-chip data (Pan et al. 2007). Notably, an orthologous human region shows conservation of most H3K27me3 BLOCs identified in the mouse region (Supplemental Fig. 9). Together, this shows that a generally similar H3K27me3 organization can be found in different data sets.
Histone modifications identify two types of chromosome domain on chr 17 that correlate with gene and repeat density
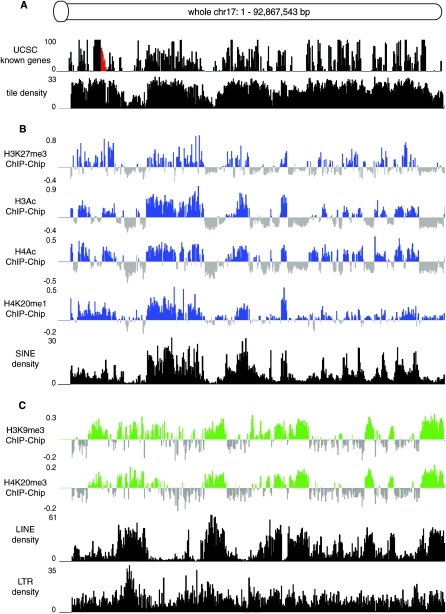
While the H3K27me3 data analysis indicated a negative correlation between transcription and H3K27me3 when the analysis focused on genes and intergenic regions, visual inspection of 5 Mb windows across whole chr 17 identified nontranscribed regions that lacked H3K27me3. We tested if H3K27me3 preferentially modifies genes in specific chromosomal domains regions by generating histone modification and gene and repeat density profiles at a low resolution across the whole of chr 17. Figure 5 shows log2 ChIP/input ratios in 200 kb nonoverlapping windows across the whole 93 Mb chr 17. All histone modification replicates were used for this analysis (Supplemental Table 1) and average signals from each 200 kb window were calculated (Methods) and aligned using the UCSC genome browser. Figure 5A shows that the gene density (calculated as the percent sequence coverage in nonoverlapping 200 kb windows from known genes (http://genome.ucsc.org assembly Mar05), is not uniform across mouse chr 17. The chip oligo probe tiling density was similarly calculated. Next the repeat density in these 200 kb nonoverlapping windows was calculated separately for three different interspersed repeat types: LINEs and LTRs (long autonomous retrotransposons) and SINEs (nonautonomous short <300 bp retrotransposons that co-opt the LINE transposition machinery) (Kazazian 2004). The tile density is reduced in LINE/LTR-rich regions compared to SINE-rich region, which is a consequence of the large size of LINE/LTR repeats compared to SINEs when the tiling array is designed from repeat-masked sequence. Figure 5B shows that H3K27me3, H3Ac, H4Ac, and H4K20me1 all identify the same gene-rich domains that correlate with SINE density, but not with LINE or LTR density. Figure 5C shows that the spaces between these gene-rich domains are enriched for the repressive histone modifications H3K9me3 and H4K20me3 and also for LINE and LTR repeats.
Figure 5.
Histone modifications identify two types of chromosome bands that correlate with gene and repeat density. (A) Gene density (http://genome.ucsc.edu) and probe tiling density (repeat-masked, see Methods for details) on the whole chr 17 tiling array chip, were analyzed in 200 kb nonoverlapping windows. The red area marks the imprinted Igf2r cluster (shown in Fig. 4) that contains three small H3K9me3/H4K20me3 peaks and lacks H3K27me3 (Regha et al. 2007). (B) Cumulative ChIP-chip profiles (log2 ChIP/Input hybridization signal means from MEFF and MEFB1 cells, Supplemental Table 1) for H3K27me3, H3Ac, H4Ac, H4K20me1 (blue bars), and SINE density (black bars). Each vertical bar indicates an average signal from 200 kb nonoverlapping windows. (C) Cumulative ChIP-chip profiles for H3K9me3 and H4K20me3 (green bars) and LINEs and LTRs (black bars). Details as in (B).
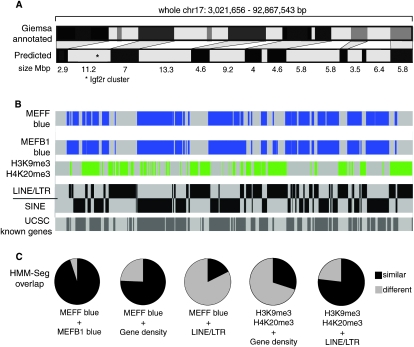
The profiles obtained for these two histone domain types were next quantitatively analyzed with the HMM-Seg program (Day et al. 2007) to identify enriched domains along chr 17 (Fig. 6). Figure 6A (top) shows the annotated metaphase Giemsa banding pattern (obtained from http://genome.ucsc.edu and based on size estimations of Giemsa stained metaphase chromosomes). Figure 6A (bottom) shows a banding pattern predicted from the histone domains identified here in nonsynchronized MEF cells, using the HMM-Seg patterns shown in Figure 6B (see Methods). Figure 6B shows enriched domains for H3K27me3, H3Ac, H4Ac, and H4K20me1 that are pooled and displayed separately for MEFF and MEFB1 along the length of chr 17 to show the concordance of these patterns between these two MEF cell lines (Fig. 6B, blue boxes). Enriched domains along the length of chr 17 for H3K9me3 and H4K20me3 were pooled for MEFF and MEFB1 (Fig. 6B, green boxes). Enriched domains for LINEs/LTRs (Fig. 6B, black boxes above line), SINEs (black boxes below line), and gene density (gray boxes), are shown as separate tracks. Figure 6, B and C, show that blue colored domains (H3K27me3, H3Ac, H4Ac, and H4K20me1, 95% of all windows showed the same segmentation state) correlate with gene density (77% of windows showing the same segmentation state). The segmentation of SINE, LINE, and LTR repeats revealed that LINEs and LTRs formed segments that were interspersed with SINE segments. SINEs (83% of windows with same segmentation state), but not LINE/LTRs (17% of windows show the same segmentation status) positively correlated with “blue” histone modifications. The H3K9me3 and H4K20me3 repressive histone modifications also formed segments (Fig. 6B, green boxes) that positively correlate with LINE/LTR repeat segmentation (77% windows show the same segmentation state), and negatively correlate with gene density (31% windows show the same segmentation state) (Fig. 6C). Together, the HMM-Seg analysis of the ChIP-chip profiles of active and repressive histone modifications show that mouse chr 17 in MEFs is organized into two alternating domain types. Type 1 is gene-rich and correlates with active histone modifications, repressive H3K27me3 modifications and SINE repeats. Type 2 is gene-poor and correlates with repressive H3K9me3 and H4K20me3 modifications and LINEs and LTR repeats.
Figure 6.
Chromosome band features analyzed by hidden Markov modeling with Viterbi segmentations. (A, top) Giemsa annotated, shows the whole chr 17 Giemsa banding pattern obtained from http://genome.ucsc.edu that is derived from measurement of Giemsa bands relative to whole chromosome length. (Bottom) Predicted, shows the whole chr 17 banding pattern predicted from the enriched histone domains shown in (B) that is based on a nonsynchronized, mainly interphase cell population. Gray shading indicates the predicted relationship between the two binding patterns. Light bands: gene-rich domains enriched for H3K27me3, H3Ac, H4Ac, H4K20me1, and SINEs. Dark bands: gene-poor domains enriched for H3K9me3, H4K20me3, and LINEs/LTRs. Domains sizes larger than 2.5 Mbp are shown and an asterisk marks the imprinted Igf2r cluster analyzed in Figure 4. (B) Hidden Markov modeling with Viterbi segmentations (HMM-Seg) for H3K27me3, H3Ac, H4Ac and H4K20me1 ChIP-chip hybridizations shown as blue boxes, separately for MEFF and MEFB1 cells. Note that all four histone modifications identify the same domains in two independent MEF cell lines. HMM-Seg for H3K9me3 and H3K20me3 is shown as green boxes using the combined MEFF and MEFB1 data. HMM-Seg is shown separately for LINEs + LTRs and for SINEs (black boxes). Note that areas of LINE and LTR density are distinct from areas of SINE density. HMM-Seg for gene density is shown underneath as dark gray boxes. (C) Quantitative analysis comparing the similarity of the above sets of HMM-Seg analyses. Pie charts show the amount of overlap of two groups of segments (similar: black) and the amount of nonoverlap (different: gray). Blue: H3K27me3, H3Ac, H4Ac, and H4K20me1.
A similar analysis of the Mikkelsen et al. (2007) ChIP-seq data set derived from formaldehyde-fixed chromatin, did not identify two alternating domain types on mouse chr 17 in MEF cells (see comment in Supplemental Fig. 10 legend; data not shown). However, two alternating domain types were seen on human chr 18 in human T cells using ChIP-seq data (Barski et al. 2007) that was derived from native ChIP, similarly to the data obtained here. Supplemental Figure 10 shows that on human chr 18 the repressive H3K27me3 and the active H3K4me3 marks form enriched domains that alternate with H3K9me3 enriched domains and the HMM-Seg algorithm also identifies these alternating domains. While repeat and gene density patterns on human chr 18 were not as clearly mutually exclusive as found on mouse chr 17, the H3K9me3 domains show a similar tendency to correlate with gene-poor and SINE-poor regions (Supplemental Fig. 10).
Discussion
Here we present an analysis of active and repressive histone modifications and their relationship to gene expression as well as to gene and retrotransposon density, along the length of mouse chr 17 in MEF differentiated cells. We show, using ChIP-chip and ChIP-seq data, that repressive H3K27me3 modifications are found as broad localized regions that we call BLOCs, which are also a feature of all mouse chromosomes. H3K27me3 BLOCs on chr 17 cover all types of silent genes as well as intergenic regions. Expressed genes are excluded from BLOCs, but are found in the immediate flanking regions. We expanded this gene-centered 100 bp high-resolution analysis of H3K27me3 to a chromosome-wide low-resolution analysis of the averaged H3K27me3 distribution over 200 kb windows. This lower resolution analysis showed that H3K27me3 is enriched in gene-rich/SINE-rich domains and depleted in gene-poor/LINE-rich domains. We further show that two active histone modifications (H3Ac, H4Ac) as well as H4K20me1 (whose role in gene expression is not yet clear), show the same chromosome domain enrichment as H3K27me3. In contrast, repressive H3K9me3 and H4K20me3 modifications show the opposite pattern and are enriched in gene-poor/LINE-rich chromosome domains. As metaphase chromosomes have been shown to contain alternating light R-bands and dark G-bands that are, respectively, gene-rich/SINE-rich and gene-poor/LINE-rich (Boyle et al. 1990), these results indicate that R- and G-bands are also associated with specific combinations of histone modification in nonsynchronized cell populations that principally contain interphase chromosomes.
Active histone modifications form peaks, but H3K27me3 forms BLOCs
The histone modification profiles identified here show characteristic features in terms of size of modified region, total chromosome coverage and position relative to genes and gene activity. Peaks of active histone modification had an average width of 2.6 kb and covered 2%–4% of mouse chr 17, with H3K4me2 being two- to threefold more abundant than H3K9Ac and H3K4me3. In contrast, repressive H3K27me3 modifications were found in broad localized regions named BLOCs that had an average size of 43 kb and covered 11% of mouse chr 17. These modifications also differed in position relative to genes and intergenic regions with active histone modifications mostly found on genes with a bias toward promoters. However, repressive H3K27me3 BLOCs were equally distributed over genes and intergenic regions and did not specifically mark promoters.
The ChIP-chip profiles described here for H3K4me2, H3K4me3, and H3K9Ac are in agreement with those obtained using ChIP-chip or ChIP-seq from different mammalian cell types, which have generally shown that these active histone modifications are enriched over short focal regions at promoters and nonpromoter putative gene-regulatory regions (Bernstein et al. 2007; Heintzman et al. 2007; Schones and Zhao 2008). In contrast, the ChIP-chip profiles described here for H3K27me3 show differences with previous results in terms of the types of genes modified and the size and position of the modified region. Our interest in H3K27me3 arose from a study of an imprinted gene cluster in MEFs where we noted that flanking silent nonimprinted genes, which lack clear developmental roles, were modified by broad regions of H3K27me3 (Regha et al. 2007). Previous results mainly from ES cells identified an association between H3K27me3 and silent developmental regulatory genes (Bernstein et al. 2006a; Lee et al. 2006). While an association with silent lineage-specific genes was also noted in differentiated cell types (Squazzo et al. 2006), the results presented here differ, as they show a general association between H3K27me3 and silent genes that lie in gene-rich/SINE-rich chromosomal domains in MEF differentiated cells. Thus, while H3K27me3 has been shown to play a role in repressing genes that promote ES cell differentiation (Boyer et al. 2006; Pasini et al. 2007), our results indicate that H3K27me3 may also play a general repressive role in gene regulation in differentiated cells.
The ChIP-chip profiles presented here show that H3K27me3 modifications mostly form BLOCs, not peaks. Previous studies that mainly focused on gene promoters identified punctate H3K27me3 patterns and a small number of broad or “blanket” H3K27me3 patterns covering silent gene clusters (Ringrose 2007). Only a few studies have generated continuous H3K27me3 profiles across large chromosomal regions in the mammalian genome, however, these also identified the majority of H3K27me3 enrichment as narrow peaks on silent gene promoters, although broad H3K27me3 enrichment was seen over silent Hox gene clusters and sometimes in the gene body of silent genes (Squazzo et al. 2006; Barski et al. 2007; Mikkelsen et al. 2007; Pan et al. 2007; Zhao et al. 2007). Despite these differences with published maps, there are four arguments that indicate the H3K27me3 BLOCs identified here accurately describe its profile across mouse chr 17 in MEFs. First, the specificity of the H3K27me3 antibody used here has been fully demonstrated (Peters et al. 2003; Perez-Burgos et al. 2004; Schwartz et al. 2006). Second, the native ChIP-chip technique used here produces profiles of active histone modification profiles in agreement with previous results (Bernstein et al. 2007; Heintzman et al. 2007; Schones and Zhao 2008). Third, the estimated chromosomal coverage of H3K27me3 BLOCs of 11%–26% in MEFs, is in agreement with quantitative mass spectrometry estimates that 10%–20% of histones are modified in ES cells by H3K27me3 (Peters et al. 2003). Lastly, we show that two alternative techniques (qPCR and ChIP-seq) identify H3K27me3 BLOCs similar to those seen in ChIP-chip analysis.
It is possible that identification of H3K27me3 modification patterns is influenced by the chosen peak-finding threshold that can affect the ability to detect peaks or BLOCs. In the example shown in Supplemental Figure 9, we also apply the BLOC algorithm to the ChIP-chip data from human ES cells previously characterized as mostly containing H3K27me3 peaks (Pan et al. 2007), which identifies H3K27me3 BLOCs that are conserved between the mouse and human genome. The BLOC algorithm failed to identify broad modified regions in a published ChIP-seq data set from mouse MEF cells (Mikkelsen et al. 2007), however 79% of H3K27me3 peaks indentified in this data set coincide with the BLOCs identified here. Notably, while the H3K27me3 ChIP-seq data generated here show oscillating peaks of enrichment throughout the BLOC, only 10%–15% of identified ChIP-seq peaks correlate with promoters (Supplemental Fig. 9; data not shown). This is in contrast to published ChIP-seq data (Mikkelsen et al. 2007), which show that 41% of observed H3K27me3 peaks correlate with promoters. While we currently have no explanation for this difference, our analysis of the published H3K27me3 ChIP-seq peaks (Mikkelsen et al. 2007) does show that they maintain the same spatial correlation relative to expressed and silent genes and intergenic regions as observed for H3K27me3 BLOCs.
Silent and expressed genes cluster close to H3K27me3 BLOCs
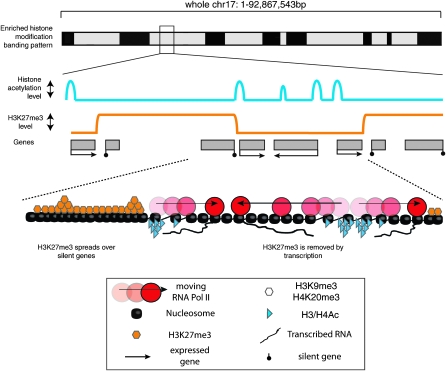
We used distance plots to show that silent genes lie within H3K27me3 BLOCs, which is in agreement with previous results that identify this modification as repressive (Boyer et al. 2006; Pasini et al. 2007). Notably, expressed genes were preferentially located in regions immediately flanking H3K27me3 BLOCs. This produced a characteristic three-peak pattern when expressed and silent genes on chr 17 were analyzed together, that is also reproduced in a whole genome ChIP-seq data set. As visual inspection of the data identified many incidents where H3K27me3 BLOCs appeared to be abruptly terminated by active transcription (Fig. 1; Supplemental Figs. 1–3), we suggest this indicates that active transcription may limit or erase H3K27me3 BLOCs. In support of this it has recently been shown in Drosophila that UTX, an H3K27me3 demethylase, co-localizes with the elongating form of RNA polymerase II (Smith et al. 2008). In the model shown in Figure 7, sites of initial H3K27me3 deposition are proposed to show the highest modification levels. This modification would then spread along the chromosome in an unknown manner, until it is excluded or erased by active transcription. This model is supported by observations in Drosophila, which show that initial targeting of H3K27me3 catalytic enzymes is directed to PREs (polycomb response elements) in genes and intergenic regions, while H3K27me3 forms broad domains including the entire transcription unit and regulatory regions (Schwartz et al. 2006). Similarly, in mammalian cells a nucleation model that leads to spreading of H3K27me3 has also been suggested from mapping sites of SUZ12 binding (Squazzo et al. 2006).
Figure 7.
Summary of H3K27me3 distribution along an autosomal chromosome. (Top) Predicted Giemsa banding pattern in metaphase chr 17 predicted from the data obtained in Figure 6 that is based on a nonsynchronized, mainly interphase cell population. (Middle) Schematic showing enlarged detail of the histone modification profile in regions with a high gene and SINE density that are enriched for repressive H3K27me3 and multiple active histone modifications. At a 100 bp high resolution level, repressive H3K27me3 and active H3Ac/H4Ac histone modifications are mutually exclusive. Expressed genes (black boxes) are characterized by active histone modifications at promoters (blue line) or by H3K36me3 throughout the gene body (not shown). In contrast, silent genes (gray boxes) and nontranscribed intergenic regions are covered by large BLOCs of H3K27me3 (orange line). BLOCs have sharp boundaries that are immediately flanked by transcripts from expressed genes. (Bottom) Enlarged detail showing sites of initial H3K27me3 deposition with the highest levels of H3K27me3 (piled orange hexagons). H3K27me3 could then spread in an unknown manner, along the chromosome, but is excluded or erased by RNA Pol II (red circles) or blocked by boundary elements marked by active histone modifications (blue triangles).
Histone modifications identify two types of chromosome bands that correlate with gene and repeat density
The analysis of H3K27me3 BLOCs relative to silent and expressed genes indicates it is induced by lack of active transcription rather than sequence-specific features. However, this interpretation is contradicted by the presence of gene-poor nontranscribed genomic regions that lack H3K27me3, which prompted us to examine the relationship between histone modifications and sequence features, such as genes and retrotransposon repeats along mouse chr 17. Our results show that histone modifications identify two types of alternating chromosomal domains that correlate with gene density and different types of retrotransposon elements. Type 1 is gene-rich and correlates with active H3Ac and H4Ac histone modifications, H4K20me1 (whose role in transcription is not clear) (Karachentsev et al. 2005; Papp and Muller 2006), repressive H3K27me3 modifications and nonautonomous SINE retrotransposons. Type 2 is gene poor and correlates with repressive H3K9me3 and H4K20me3 modifications and autonomous LINES and LTR retrotransposons. As previous studies of metaphase chromosomes (Boyle et al. 1990; Craig and Bickmore 1993) have shown that gene-rich/SINE-rich regions are lightly stained by Giemsa (known as light R-bands), while gene-poor/LINE-rich regions are darkly stained by Giemsa (known as dark G-bands), this indicates that specific patterns of histone modifications may discriminate Giemsa bands in nonsynchronized cell populations principally composed of interphase cells. The tiling array used in this ChIP-chip study was repeat masked, thus the obtained histone profiles do not directly result from hybridization to repeats themselves, but from hybridization to single copy sequences flanking the repeats. However, LTR retrotransposons have been shown to be enriched for H3K9me3 and/or H4K20me3 (Martens et al. 2005; Mikkelsen et al. 2007), thus the enrichment on flanking single copy regions may reflect retrotransposon epigenetic modifications.
A connection between histone modifications and chromosome domains has previously been made in human T cells, which similarly showed that dark G-bands correlate with H3K4 methylation depletion and H3K9me3 enrichment, while light R-bands correlate with H3K4 methylation enrichment and reduced H3K9me3 (Barski et al. 2007). Our low-resolution analysis of this data set supports this observation by showing that alternating domains of H3K4me3 (plus H3K27me3) and H3K9me3 occur along human chr 18 and that the H3K9me3 domains correlate with dark staining Giemsa bands (Supplemental Fig. 10). Mutual exclusive distribution of H3K27me3 and H3K9me3 based on ChIP-chip data has also been demonstrated in several mammalian cell lines (Squazzo et al. 2006). A published analysis of ENCODE regions (Thurman et al. 2007) using the same computational approach that we applied in Figure 6, also identified two types of domains. One domain termed “active” was gene and SINE rich and also enriched for active histone modifications, as we show here for Type 1 domains. The other domain termed “repressed” was gene poor and LINE/LTR rich, but in contrast to our results, was enriched for H3K27me3. This discrepancy may result from the analysis not including H3K9/H4K20 trimethylation and because ENCODE regions comprise 1% of the human genome from 100 dispersed regions, instead of a continuous chromosomal region. Support for the existence of two types of repressive chromatin similar to that described here comes from two other studies. The first study demonstrated that human alpha- and beta-globin genes are silenced by different mechanisms; alpha-globin that lies in a gene-rich domain was repressed by H3K27me3, while beta-globin that lies in a gene-poor domain was not (Garrick et al. 2008). In a second study, immunofluorescence of human metaphase chromosomes was used to identify two nonoverlapping chromatin states spread over mega-base-pair-sized domains, one defined by H3K27me3 and the other by H3K9me3 + HP1 + H4K20me3 (Chadwick, 2007). This study focused on the inactive X chromosome and concluded that alternating H3K27me3/H3K9me3 banding patterns reflect the specific organization of the inactive X chromosome into a heterochromatin state. Our results do not disagree with this conclusion that concerned the inactive X chromosome at metaphase, instead we show that a typical autosome is also organized into alternate H3K27me3 and H3K9me3 domains in nonsynchronized principally interphase, cell populations.
The findings in this study refine our understanding of the distribution of histone modifications in two ways. First we show by high-resolution ChIP profiling that H3K27me3 is not restricted to the promoter regions of silent genes, but instead generally marks broad localized regions that include silent genes and intergenic regions. Second, we use a low-resolution analysis to show that large chromosomal domains on an autosome are alternately enriched for distinctive histone modifications that correlate with gene density and different retrotransposon elements. While these low-resolution histone-banding patterns do not exclude the existence of smaller domains within them that may show contrary patterns, they do indicate the possibility that chromosomes are subdivided into domains that largely regulate genes and domains that largely silence autonomous retrotransposons. These two regions may impose different epigenetic constraints, e.g., genes lying close to LINE/LTR-rich bands may be more affected by epigenetic mechanisms normally directed toward transposon silencing, as has been well demonstrated in the plant genome (Weil and Martienssen 2008). An improved understanding of genes and their relative position to specific enriched histone domains may also give new insights into genes showing epigenetic dysregulation in development and disease.
Methods
Chromatin immunoprecipitation (ChIP), microarray design, and hybridizations
Native ChIP, T7 in vitro amplification, RNA-chip, and the MEFF and MEFB1 cell lines were described previously (Regha et al. 2007). Antibodies, cells, and replicates are listed in Supplemental Table 1. The custom mouse chr 17 NimbleGen tiling array was designed by identifying 50 bp windows containing at least 18 unique 17-mers with a maximum distance of 15 bp (Thomas Jenuwein and the GEN-AU bioinformatics team, pers. comm.). This identified 390,000/50-mers giving a resolution of ∼100 bp from single copy sequences (program is available on request) from mouse chr 17: 3021,656 to 92,867,543 (UCSC [mm6], March 2005, Build 34). chip hybridizations and scanning were performed by NimbleGen Systems Iceland and involved two technical replicates (with a dye swap) for 2 MEF cell lines (MEFB1 and MEFF) for most histone modification. The data were Tukey bi-weight normalized before analysis.
Identification of focal regions enriched in active histone modifications and H3K27me3 from ChIP-chip data
Using an implementation of ChIPOTle (Buck et al. 2005), a P-value was calculated for each 1500 bp window with at least 8 probes and assigned to bins of size 1 × 10−5. The cutoff for selection of enriched windows was determined by generating a null distribution through permutation of the signals (400 times) and assigning the resulting P-values to bins. The bin where the ratio of summed averaged randomized counts/summed real counts was still smaller than the chosen false discovery rate (1 × 10−7) was the upper limit for the P-value. A second selection step removed windows where the intensity of less than one-third of the probes was below the sum of the mean and the standard deviation of all the experimental probes. Peaks were joined if they overlapped or were within 500 bp of each other.
Identification of H3K27me3 broad local enrichments (BLOCs)
The start of a BLOC is defined by 10–13 consecutive probes, where 10 probes show a positive log2 (ChIP/input) value. The BLOC end was defined by 6–8 probes where six probes show a negative log2 (ChIP/input) value. BLOCs with a median log2 (ChIP/input) value greater than 0.25× standard deviations above the median log2 (ChIP/input) value of all probes of the chip were used. BLOCs that were separated by less than 10 kb were merged and only BLOCs larger than 5 kb in length were used for data analyses. In the case where several H3K27me3 chip replicates were available for one cell line, BLOCs were identified separately for each replicate. The BLOCs were fused by taking the overlapping BLOC regions and excluding regions that did not overlap. ChIP-seq BLOC finding was performed on 25 bp fragment density maps (see ChIP-seq below) from one MEFF genome data set. The BLOC start was defined by 20–23 windows, where 20 windows show a fragment density >0 and the BLOC end was defined by 6–8 windows where six windows show a fragment density <0. All 25 bp windows with no fragment density were assigned the value −1. The BLOCs program is available at http://genauwiki.imp.ac.at.
Gene expression analysis
These were based on cDNA hybridization to the NimbleGen chr 17 array relative to genomic DNA (RNA-chip). Informative genes were identified in two steps. First, only genes with at least five tiling array probes in exons were considered informative (750 of the 1282 genes on chr 17). The median signal of probes within exons was calculated. Genes with a value above the median value of all probes with a positive log2 (ChIP/input) value on the array were classified as expressed, while genes with a value below the median were classified as silent. Second, only genes with a coverage of at least eight tiling array probes per 1500 bp over >70% of the gene locus were used for data analyses. In MEFB1 cells this identified 553 of the 1282 genes on chr 17 from one replicate. In MEFF cells we only counted genes with the same expression status in two replicates and analyzed 518 of the 1282 genes on chr 17.
Combined analysis of histone modifications and location in genes
Focal regions enriched for active histone modifications as defined by ChIPOTle were identified for each hybridization replicate of a respective histone modification. The enriched regions from all hybridizations (Supplemental Table 1) were pooled for each histone modification. Overlap between the enriched regions was determined and those common to all files were pooled and used for data analyses in Figure 2. To determine how much of the single copy portion of chr 17 was covered by focal active histone modifications, informative regions with a probe density of at least eight oligos per 1500 bp were analyzed by ChIPOTle. 89,832,667 bp of chr 17 was present on the array, but only 53,415,732 bp (60%) was informative for analysis. The repetitive part was underrepresented. The total length covered by each histone modification was summed and divided by 53,415,732 to determine the proportion of single copy regions covered. For each histone modification the size of each enriched region was determined and the average size was calculated by dividing the summed length by the total number of enriched regions. For H3K27me3 the same analysis was performed, but here the whole of chr 17 was taken as informative. Genes were Ensembl transcripts. Promoters were defined as 1 kb upstream and 1 kb downstream of the exon 1 start position (2 kb region). The remainder of the gene was defined as the gene body. The percentage of ChIPOTle (H3K4me2, H3K4me3, H3K9Ac, and H3K27me3) or BLOC (H3K27me3) enriched regions that overlapped with genes or intergenic regions were then calculated.
Z-score analysis
We calculated an estimate of whether the observed overlap between two enriched regions for histone modifications was significant by randomly reshuffling the enriched regions 10,000 times to generate an empirical null distribution of the total overlap lengths. Only regions with a probe density >8 probes/1500 bp were used for analysis. The observed total overlap length was expressed as a Z-score relative to this null distribution: for example a Z-score of 10.0 indicates the actual overlap length was 10 standard deviation units higher than the mean of the random overlap length distribution.
Distance distribution analysis
The distance of the midpoint of each gene to the borders of the closest H3K27me3 BLOC was calculated. The distances were combined into 10 kb distance bins and plotted using MS-Excel. H3K27me3 BLOCs located completely within genes were excluded from the analysis. Genes that were more than 50% overlapped by an H3K27me3 BLOC were put into the “inside” bin (genes in BLOCs).
Histone profiling relative to gene expression
For each probe, the distance to the transcriptional start site of the genes of each category (expressed, silent, genes in BLOCs, see above) was calculated and normalized to the length of the gene. The normalized log2 (ChIP/input) values of the probes were smoothened by fitting a cubic smoothing spline, with the relative distances as predictors. A randomization and eventual smoothing of all the log2 (ChIP/input) values from the array across the relative distances indicates the background level of the array.
ChIP-seq
Sequencing libraries were obtained from 10 ng of ChIP DNA by adaptor ligation, gel purification and 18 cycles of PCR. Sequencing was carried out using the Illumina Genome Analyzer (GA) I system according to the manufacturer's protocol. 4468,089 uniquely alignable sequence tags were mapped to the mouse genome (NCBI build 36) using the GA-Pipeline V0.3. Tags passing the standard GA-Pipeline quality threshold and mapping uniquely with not more than two mismatches were used for data analyses. High density map: A theoretical fragment density for each 25 bp window was calculated. Uniquely aligned tags within 200 bp and oriented toward it, were counted as 1 for each 25 bp window and as 0.25 if they were within 200 bp and 300 bp. Low-density map: For each 200 bp window the uniquely aligned tags were counted. Peaks of significant enrichment were identified using the USeq toolkit (http://useq.sourceforge.net/): A sliding window of 1 kb was used to calculate smoothened window scores (ScanSeqs). For each window a Bonferroni corrected P-value was estimated using a global Poisson distribution and windows with a minimum score of 10 (low cut off) or 60 (high cut off) and a maximum distance of 500 bp were combined to enriched regions (EnrichedRegionMaker).
Statistical significance
P-values were calculated using an unpaired t-test on http://www.graphpad.com using the mean and the standard deviation.
Whole chromosome profiles
The average normalized log2 (ChIP/input) ratios of all probes within nonoverlapping 200 kb windows was calculated for each replicate (Supplemental Table 1) of each histone modification examined in MEFF and MEFB1 cell lines. The data of all replicates were combined by averaging the 200 kb windows. Repeat and gene densities (“known genes”) were calculated as percent sequence coverage of 200 kb nonoverlapping windows. The positions of SINE, LINE, LTR repeats, and known genes were obtained from the UCSC genome browser (http://genome.ucsc.edu).
HMM-Seg
Paired H3K27me3 and RNA chr 17 profiles were analyzed by HMM-Seg with described parameters, except that the data were “smoothened” over 15 kb in Supplemental Figure 7 (Day et al. 2007). The unmodified ChIP/input ratios were averaged over 1 kb nonoverlapping windows and these data sets were used as the HMM-Seg input. In Figure 6, single H3K27me3, H3Ac, H4Ac, and H4K20me1 profiles were analyzed separately for the MEFF and MEFB1 cell line by averaging the unmodified ChIP/input ratios over 1 kb nonoverlapping windows and the data “smoothed” over 200 kb. For H3K9me3 and H4K20me3 profiles in Figure 6, the average log2 (ChIP/input) ratios were calculated together for all available replicates (Supplemental Table 1). The repeat segmentation was obtained by calculating the LINE, SINE, and LTR repeat coverage for 1 kb nonoverlapping windows that were cumulatively analyzed. The gene segmentation was obtained by calculating the coverage of known genes for 1 kb nonoverlapping windows. The repeat and gene data sets were the same as for whole chromosome profile described above. The “predicted” banding pattern on chr 17 was estimated by defining a pattern that best fits the combined H3K27me3, H3Ac, H4Ac, and H4K20me1 profiles and the SINE profiles (forming the light staining bands) or the combined H3K9me3 and H4K20me3 profiles and the LINE/LTR profiles (forming the dark staining bands). Small gaps in the profile were neglected and the overall structure of the annotated Giemsa bands was retained where possible. The sizes of the predicted bands were calculated based on their coverage of chr.17.
Quantitative analysis of HMM-Seg overlap
HMM-Seg converts the whole chromosome into blocks of two states, 0 and 1. Two HMM-Seg results were compared by testing the whole chromosome in 1 kb nonoverlapping windows to determine if both segmentations showed the same state (0 or 1). The percentage of similarity was calculated by dividing the number of windows showing the same state by the total number of windows on chr 17. The result was visualized as pie charts using MS-Excel.
Real-time qPCR
Primers (Supplemental Table 2) were designed by PrimerExpress and qPCR performed with the ABI PRISM 7000 using MESA GREEN qPCR (Mastermix Plus for SYBR ASSAY- dTTP), with the primers under the following cycling conditions: 2 min 50°C, 10 min 95°C, 40 cycles of 15 s 95°C and 1 min 60°C. ChIP and Mock material were assayed undiluted, while input DNA was diluted 1:100. DNA quantification was made by the standard curve method using serial dilutions of input DNA. Relative quantification and statistics were performed as described in the manufacturer's protocol (Applied Biosystems).
Acknowledgments
We thank all members of the Barlow group and members of the GEN-AU Epigenetics project for support, Leonie Ringrose and Anton Wutz, for their comments on the paper. Project support was from GEN-AU Epigenetic Plasticity of the Mammalian Genome (GZ200.141/1-VI/2006), the EU-FW6 IP “HEROIC” (LSHG-CT-2005-018883), the NoE “The Epigenome” (LSHG-CT-2004-053433), and FWF SFB F17 Modulators of RNA Fate and Function (SFBF01718 B10).
Footnotes
[Supplemental material is available online at www.genome.org. The microarray and sequence data from this study have been submitted to Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE11389.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.080861.108.
References
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beisel C., Buness A., Roustan-Espinosa I.M., Koch B., Schmitt S., Haas S.A., Hild M., Katsuyama T., Paro R. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc. Natl. Acad. Sci. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Duncan E.M., Masui O., Gil J., Heard E., Allis C.D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Meissner A., Lander E.S. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Boyle A.L., Ballard S.G., Ward D.C. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. 1990;87:7757–7761. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.J., Nobel A.B., Lieb J.D. ChIPOTle: A user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 2005;6:R97. doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick B.P. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma. 2007;116:147–157. doi: 10.1007/s00412-006-0085-1. [DOI] [PubMed] [Google Scholar]
- Chadwick B.P., Willard H.F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S.J., Yee D., Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.M., Bickmore W.A. Chromosome bands—Flavours to savour. Bioessays. 1993;15:349–354. doi: 10.1002/bies.950150510. [DOI] [PubMed] [Google Scholar]
- Day N., Hemmaplardh A., Thurman R.E., Stamatoyannopoulos J.A., Noble W.S. Unsupervised segmentation of continuous genomic data. Bioinformatics. 2007;23:1424–1426. doi: 10.1093/bioinformatics/btm096. [DOI] [PubMed] [Google Scholar]
- Garrick D., De Gobbi M., Samara V., Rugless M., Holland M., Ayyub H., Lower K., Sloane-Stanley J., Gray N., Koch C., et al. The role of the polycomb complex in silencing α-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Karachentsev D., Sarma K., Reinberg D., Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes & Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H., Jr Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:552–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Papp B., Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes & Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Burgos L., Peters A.H., Opravil S., Kauer M., Mechtler K., Jenuwein T. Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 2004;376:234–254. doi: 10.1016/S0076-6879(03)76016-9. [DOI] [PubMed] [Google Scholar]
- Peters A.H., Kubicek S., Mechtler K., O'Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Regha K., Sloane M.A., Huang R., Pauler F.M., Warczok K.E., Melikant B., Radolf M., Martens J.H., Schotta G., Jenuwein T., et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L. Polycomb comes of age: Genome-wide profiling of target sites. Curr. Opin. Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Cui K., Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D.E., Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Kahn T.G., Nix D.A., Li X.Y., Bourgon R., Biggin M., Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster . Nat. Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Lee M.G., Winter B., Droz N.M., Eissenberg J.C., Shiekhattar R., Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol. Cell. Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo S.L., O'Geen H., Komashko V.M., Krig S.R., Jin V.X., Jang S.W., Margueron R., Reinberg D., Green R., Farnham P.J. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R.E., Day N., Noble W.S., Stamatoyannopoulos J.A. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917–927. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil C., Martienssen R. Epigenetic interactions between transposons and genes: Lessons from plants. Curr. Opin. Genet. Dev. 2008;18:188–192. doi: 10.1016/j.gde.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Wutz A. Xist function: Bridging chromatin and stem cells. Trends Genet. 2007;23:457–464. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A., et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]