Abstract
Determining the underlying logic that governs the networks of gene expression in higher eukaryotes is an important task in the post-genome era. Sequence-specific transcription factors (TFs) that can read the genetic regulatory information and proteins that interpret the information provided by CpG methylation are crucial components of the system that controls the transcription of protein-coding genes by RNA polymerase II. We have previously described Stable Isotope Labeling by Amino acids in Cell culture (SILAC) for the quantitative comparison of proteomes and the determination of protein–protein interactions. Here, we report a generic and scalable strategy to uncover such DNA protein interactions by SILAC that uses a fast and simple one-step affinity capture of TFs from crude nuclear extracts. Employing mutated or nonmethylated control oligonucleotides, specific TFs binding to their wild-type or methyl-CpG bait are distinguished from the vast excess of copurifying background proteins by their peptide isotope ratios that are determined by mass spectrometry. Our proof of principle screen identifies several proteins that have not been previously reported to be present on the fully methylated CpG island upstream of the human metastasis associated 1 family, member 2 gene promoter. The approach is robust, sensitive, and specific and offers the potential for high-throughput determination of TF binding profiles.
The interactions between transcription factors (TFs) and their DNA binding sites are an integral part of gene regulatory networks and represent the key interface between the proteome and genome of an organism. These sequence-specific factors exert their effects through dynamic interactions with a plethora of protein complexes that modify and remodel chromatin, change the subnuclear localization of target genes, and regulate the promoter recruitment, activity, and processivity of the transcriptional machinery (for review, see Kadonaga 2004; Remenyi et al. 2004). Besides sequence-specific binding, a certain class of TFs interacts with so-called CpG islands that consist of clustered arrays of the dinucleotide sequence CG in a methylation (5-methyl cytosine)-dependent manner (Ohlsson and Kanduri 2002). These CpG islands are found in the proximal promoter regions of almost half of the genes in the human genome (Ohlsson and Kanduri 2002) and can be methylated in a tissue-specific manner or upon transformation to malignancy (Robertson 2005).
Thus, the determination and characterization of TF binding sites throughout the whole human genome is pivotal to our understanding of how genes are differentially expressed. While much progress has been made in the high-throughput identification of potential binding sites for a given protein by both the microarray chip-based readout of chromatin immunoprecipitation assays (ChIP-chip) and protein binding microarrays (Mukherjee et al. 2004; Warren et al. 2006), a scalable complementary technique that—in an unbiased way—reveals proteins binding in a sequence-specific manner to a given site is presently not available. Traditional methods for the unbiased identification of sequence-specific nucleic acid binding proteins employ a combination of several steps of classical chromatography followed by a final affinity purification step that uses their cognate recognition sequence as a ligand (Kadonaga 2004).
The classical approach is laborious and requires monitoring the purification process by functional assays (electrophoretic mobility shift assay [EMSA], DNA footprinting, in vitro transcription) and is thus impractical on a proteomic scale. Routine high-throughput identification of sequence-specific DNA binding factors is mainly hampered by their low abundance, the degeneration of their binding sites, and the competition by unspecific binding of positively charged nuclear proteins to the negatively charged phosphate backbone of DNA.
In contrast, computer predictions of TF DNA sequence binding specificities are fast and simple but have certain limitations (for review, see Bulyk 2003). First, they are based on experimental data derived from the published literature and may therefore not be sufficiently comprehensive and sensitive or may be subject to sampling biases. Second, they do not take into account the context dependency of TF binding and the effects of interactions between base pair positions in the binding sequence. Third, they are relatively poor predictors of quantitative binding to variant DNA motifs (Udalova et al. 2002; Tompa et al. 2005). Lastly, they cannot predict which isoforms or which polypeptides of a TF protein family are binding to a given element (Saccani et al. 2003).
Recent breakthroughs in quantitative protein mass spectrometry (for review, see Ong and Mann 2005) are providing us with the tools that will enable us to tackle many of the obstacles mentioned above. In a tour de force analyzing 53 ion exchange chromatography fractions of a tryptic digest by mass spectrometry in combination with their isotope coded affinity tag (ICAT) technology (Himeda et al. (2004) demonstrated that it is indeed possible to identify a sequence-specific factor by quantitative proteomics.
We have previously described Stable Isotope Labeling by Amino acids in Cell culture (SILAC) for the quantitative encoding of proteomes (Ong et al. 2002). Following a one-step affinity purification from SILAC-encoded extracts, specific binders can be directly distinguished by the isotope ratios of their tryptic peptides as determined by mass spectrometry provided that a specificity control can be designed (Schulze and Mann 2004; Schulze et al. 2005). Here we use immobilized oligonucleotides harboring functional DNA elements for the TFs AP2 and ESRRA (also known as ERRalpha) in their wild-type and control (point mutated) form to prove that we are able to uncover such interactions by SILAC. Investigating a CpG island of the human metastasis associated 1 family, member 2 (MTA2) gene, we further demonstrate that a similar approach can be used to identify proteins preferentially binding to methylated CpG sites. Apart from ZBTB33 (also known as Kaiso), which was known to bind the methylated MTA2 CpG island, our screen resulted in the identification of several known as well as unknown methyl-CpG-dependent binding partners. The DNA protein interaction screen proved sensitive, robust, fast, and specific and could in principle become a standard technique to investigate functional cis-elements on DNA.
Results
A generic strategy and proof of principle for the determination of DNA–protein interactions
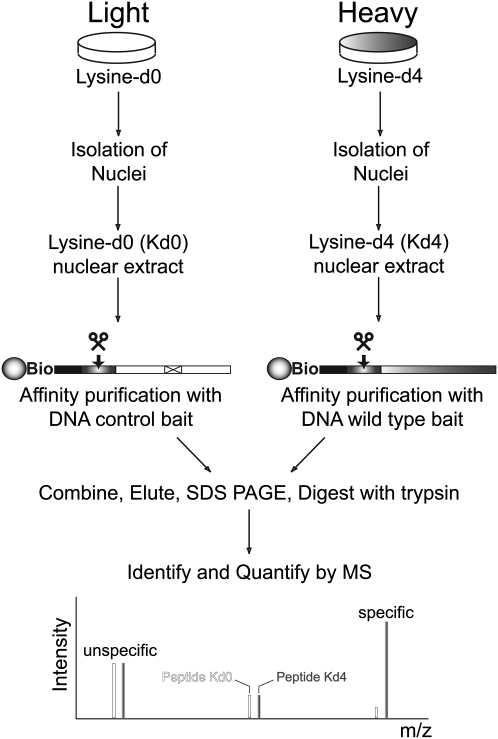
Modern mass spectrometric technology has become very powerful at determining the components of complex protein mixtures (for review, see Cox and Mann 2007; Cravatt et al. 2007). We have previously used this technology to determine peptide–protein interactions in the following way. Two cell populations are metabolically encoded through a stable isotope variant of an essential amino acid. In this approach, which we have termed SILAC, the peptides originating from the two cell populations can be distinguished because they have different masses. Pull-downs with a bait from the lysine-d4 encoded cell lysate and with a control from the nonencoded cell lysates are mixed and analyzed together. In this way, background proteins binding nonspecifically to the beads used in the pull-downs will be present in a one-to-one ratio in the two SILAC forms. Binders specific to the bait will have a quantitative ratio between the two forms. Here we develop the same principle into an assay for DNA–protein interactions (Fig. 1).
Figure 1.
Principle of the DNA–protein interaction screen. Proteomes are metabolically labeled with 2H4-lysine (Lysine-d4) to allow discrimination based on differences in peptide mass (4 Da). Biotinylated (Bio) DNA molecules bearing functional elements (e.g., potential TF binding sites or methylated CpGs) are synthesized and immobilized on streptavidin magnetic beads. Control columns are designed to lack the functional element of interest (e.g., point mutations in cis-elements or nonmethylated CpGs). Nuclear extracts from unlabeled (Lysine-d0) and labeled cells are prepared and subjected to DNA affinity chromatography, followed by the release of DNA–protein complexes by restriction enzyme digestion. After in-gel digestion with trypsin, differentially labeled forms of lysine containing tryptic peptides are detected by mass spectrometry (MS). Peptides originating from proteins specifically recognizing functional elements will have a larger peak intensity of the lysine-d4 (Kd4) form. Unspecific interaction partners will have 1:1 ratios between both isotopic forms.
Because the pull-down experiment is performed from nuclear extracts and TFs are typically of low abundance, we decided to grow the cells in suspension and to encode them with 2H4-lysine (lysine-d4) as the SILAC amino acid, because deuterated amino acids are more economical than 13C or 15N substituted amino acids. We routinely prepared nuclear extracts from spinner cultures of unlabeled and 2H4-lysine-labeled HeLa S3 cells. Known ratio-mixing experiments demonstrated virtual identity of different extracts in terms of relative protein abundance as determined by measuring the isotope ratios of lysine-containing peptides from the 100 most abundant proteins by nanoLC-MS (data not shown).
As a proof of principle we decided to investigate proteins binding to the DNA sequence GCCCGGGGC, a known binding site for the TF AP2 that interacts as a homodimer with this palindromic DNA sequence motif (Egener et al. 2005). Indeed, SELEX experiments have determined that it is an optimal binding sequence (Mohibullah et al. 1999). We designed the DNA bait in the following way (Fig. 2A). A double-stranded DNA segment harboring the above-mentioned optimal AP2 binding site was biotinylated at the 5′-end of the sense strand for coupling to streptavidin beads. The sequence length of 40 bp was chosen as a compromise between minimizing nonspecific binding (requiring short sequences) and given ‘native context’ to the binding site (requiring long sequences). This sequence length is suitable for automated oligodeoxynucleotide synthesis and sufficient for metazoan DNA binding factors, which usually target 5 to 12 base pairs (Wingender et al. 2001). Furthermore, we designed the bait to contain a restriction site (6 bp) and a spacer to the 5′-end to enable elution of bound proteins by restriction endonuclease digestion, which is very efficient (Supplemental Fig. S1).
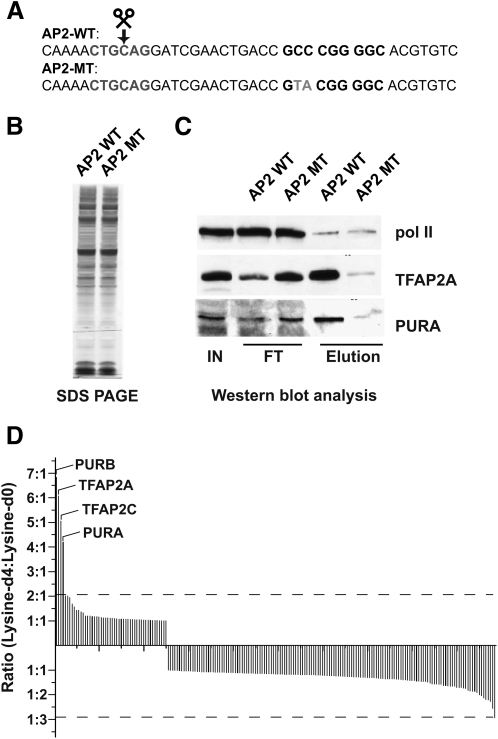
Figure 2.
Properties of the DNA–protein interaction screen exemplified by a functional element harboring a binding site for the TF AP2. (A) The sequence of the DNA baits used for the experiment. The AP2 binding site is shown in boldface type, whereas the point mutations designed to disrupt DNA binding are highlighted in gray. (B–D) SILAC is essential for the identification of specific binders. (B) Specific interaction partners cannot be identified by one-dimensional SDS-PAGE and silver staining of the eluted material from the AP2 wild-type (WT) and mutant (MT) columns. (C) Western blot analysis of selected specific interaction partners recapitulates results from the SILAC analysis. Equal amounts (10% of total) of input (IN) and flow-through (FT) as well as eluted (Elution) material (50% of total) from the AP2 wild-type (WT) and mutant (MT) columns, respectively, were analyzed by immunoblotting with antibodies directed against TFAP2A, the POLR2A subunit of RNA polymerase II, and PURA. POLR2A serves as a control for equalized total protein amounts and unspecific binding. (D) SILAC analysis discriminates specific from unspecific binders. Proteins containing peptides with lysine-d4 to lysine-d0 ratios of greater than 3:1 are considered to be specific.
We also synthesized a control bait, designed to abolish binding of specific but not of nonspecific binders to the sequence. As shown in Figure 2A, this was accomplished with two point mutations introduced in the AP2 binding site (GTACGGGGC), which have previously been shown to abrogate binding of AP2 in a band-shift assay (Mohibullah et al. 1999). The SDS-PAGE pattern of proteins binding to the wild-type and control bait, respectively, appeared essentially identical (Fig. 2B). Therefore, it was impossible to identify any wild-type-specific polypeptide band just based on visually comparing the protein composition. This demonstrates the necessity of a quantitative proteomics approach to uncover such proteins.
Lysine-d4 encoded nuclear extract from 2.5 × 108 cells was then incubated with the oligonucleotide containing the AP2 binding site whereas the point mutated control DNA was exposed to the nonlabeled extract. After mild washing, beads from the two experiments were combined, and the baits and bound proteins were liberated by restriction enzyme digest and the eluted material was separated by one-dimensional SDS-PAGE. The entire gel lane was cut into six pieces, which were trypsin digested and analyzed by liquid chromatography tandem mass spectrometry as described before (Ong et al. 2004). More than 250 proteins were identified in this experiment, but quantitative analysis of the SILAC peptide pairs showed that almost all of them were present in approximately equal amounts (Fig. 2D). This means that they bound equally to both baits or to the beads. As expected we identified TFAP2A (AP2-alpha) as a specific interaction partner (ratio 6.75 between bait and control). Interestingly, in addition to the expected binding partner, the Pur proteins PURA (ratio 4.22) and PURB (ratio 6.73), which can either function as transcriptional activators or repressors, were also found to interact specifically with the wild-type bait (Fig. 2C; Supplemental Table S1). The AP2 family consists of five members, TFAP2A (alpha), TFAP2B (beta), TFAP2C (gamma), TFAP2D (delta), and TFAP2E (epsilon) (Tummala et al. 2003). Alignment of the peptides identifying AP2 showed that we had sequenced peptides common to all five proteins as well as unique peptides proving the presence of TFAP2A and TFAP2C (Supplemental Table S1; Supplemental Fig. S2). In the case of PURA and PURB, which are closely related proteins encoded by two different genes, we sequenced peptides specific for each, so we conclude that both were present. An antibody specific for PURA was obtained and confirmed presence of this protein (Fig. 2C). Both PURA and PURB can bind as homo- or heterodimers in a rather sequence-independent manner to double-stranded DNA that is characterized by a high degree of polypurine–polypyrimidine asymmetry (Bergemann et al. 1992).
Pur proteins have been shown to recognize the structure formed by GGN repeats (Knapp et al. 2006). Such a repeat is present on the antisense strand (GGCGGT) and overlaps with the AP2 consensus site. The first GGN repeat is disrupted by two nucleotide changes (to TAC) in the AP2 mutant site (Fig. 2A), which may explain the reduced binding of PURA and PURB to the mutant DNA and emphasizes our ability to identify structure-dependent associations in the screen. Figure 2D shows the ratios for all proteins identified in this experiment. As can be seen in the figure, there is a gap between the nonspecific and specific binders, which have ratios between 1.0 and 2.0, and the specific proteins, which have a ratio of greater than 3.0 in this experiment. However, this represents an empirically derived threshold because the AP2 proteins were known binders and the PURA protein was shown to bind to our column. As discussed elsewhere (MacCoss et al. 2003; Ong et al. 2003; McClatchy et al. 2007), in general the level of significance of a measured ratio depends on the experiment and within an experiment depends on the signal-to-noise ratio of peptides and the peptide ratio variance of the protein being quantified. Therefore, to objectively measure the degree of specificity of binding to a functional DNA element the statistical significance of the relative protein changes has to be determined. Hence, in the following experiments that mainly deal with screening for unknown interaction partners we employed our newly developed MaxQuant software (Cox and Mann 2007, 2008), which calculates the significance of a ratio being separated from the main distribution as a function of protein intensity.
Determination of TFs binding to bioinformatically predicted DNA cis-elements
Many bioinformatics and functional genomics experiments result in the prediction of functional DNA sequences, which may recruit specific effector proteins. In principle, our approach should be well suited to determine such candidate proteins.
To test this with a specific example, we synthesized a bait for a putative DNA binding factor from a recent experiment from Mootha et al. (2004). In that experiment, the transcriptional coactivator PPARGC1A (also known as PGC1-alpha) was overexpressed, leading to the differential expression of mRNAs measured by a microarray experiment. Regulated mRNAs were identified and corresponding genes assessed for possible binding motifs for TFs. Of the three motifs found we picked one that had been hypothesized to be a binding site for the orphan nuclear receptor (NR) ESRRA (ERR-alpha). That site had indirectly been confirmed by a luciferase reporter assay upon cotranfection of ESRRA- and PPARGC1A-expressing plasmids.
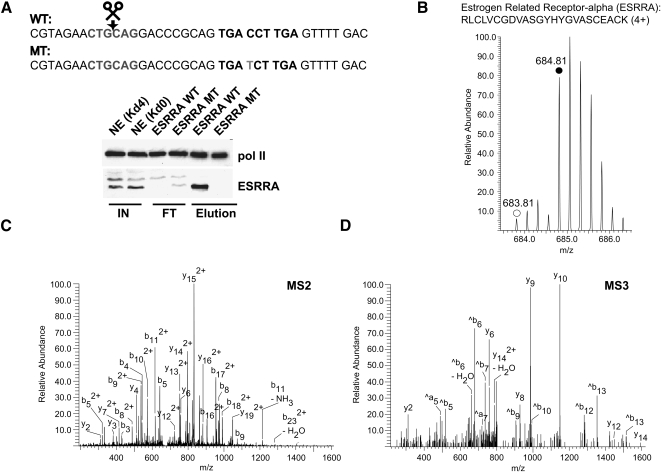
After synthesis of the DNA sequence—a 26-bp sequence from the promoter region of the human ESRRA gene that contains a predicted autoregulatory binding motif for ESRRA itself (Mootha et al. 2004)—and a single point mutant control in the middle of the motif (see Fig. 3A) we performed the SILAC binding experiment as before but analyzed the data by the MaxQuant software. To keep the number of false positive-specific binders as small as possible, stringent criteria were applied. Therefore, we only accept proteins that have been quantified by at least two peptides showing a ratio that is more than three standard deviations (P < 0.0012) apart from the center of the distribution of all proteins. Out of a total of 703 identified proteins, we sequenced 18 unique peptides of ESRRA and quantified its highly significant ratio (P = 1.09 × 10−49) between bait and control as roughly nine times (9.02), which correlates well with data obtained by immunoblot analysis (Fig. 3A). The MS, MS/MS, and MS3 spectra of one of the peptides employed for quantification are shown in Figure 3B–D. This confirms in vitro binding of the protein to this bioinformatically determined motif. Although expected, this confirmation is important because it is well known that the sequence context of a putative TF binding site can influence the ability of the protein to actually bind to it (Hoglund and Kohlbacher 2004). Notably, ESRRA does not interact with consensus binding sites present on the P450 promoter (Johnston et al. 1997).
Figure 3.
The orphan nuclear receptor ESRRA (ERRalpha) exhibits specific binding to its bioinformatically predicted binding sequence. (A) Western blot analysis (same conditions as in Fig. 2C) confirms the specific interaction of ESRRA as revealed by the SILAC experiment (B–D) (Table 1). Sequences of the DNA baits employed are depicted. The predicted ESRRA binding site derived from an autoregulatory motif of the ESRRA gene (Mootha et al. 2004) is shown in boldface type whereas the point mutation for the control column is highlighted in gray. (B) Representative peptide mass spectrum demonstrating the specific binding of ESRRA. The MS spectrum of a quadruply charged labeled (monoisotopic peak marked with solid circle) and unlabeled (monoisotopic peak marked with open circle) tryptic ESRRA peptide acquired in the ICR cell of the LTQ-FT (0.3 ppm mass deviation after recalibration) is shown. (C) MS/MS (MS2) fragmentation spectrum that identifies the peptide shown in B. The precursor ion (m/z 684.81) was fragmented in the linear ion trap (LTQ part) of the mass spectrometer to obtain sequence information. (D) MS/MS/MS (MS3) spectrum of the y 15(2+) ion present in the MS2 experiment (C) further confirming the identification of the peptide shown in B.
Another NR, NR2F6 (v-erb A related gene 2), also bound to the same DNA sequence with a significant ratio (P = 3.56 × 10−23) of 4.93 (Table 1). We detected and quantified (ratio 4.15) one peptide mapping to the NR protein NR2C1 (testicular receptor 2) but excluded it from our list because of our stringent criteria. ESRRA and NR2F6 bind to the estrogen receptor superfamily of hormone response elements on DNA that usually consists of two direct or inverted repeats of the hexamer motif (half-site) TGACCT separated by spacer nucleotides of variable length. Promiscuous (but sequence context-dependent) binding of orphan NRs as monomers to NR half-sites or extended half-sites (e.g., imperfect direct repeats) is well known (Wilson et al. 1993; Giguere et al. 1995), so it is not surprising that we found specific binding of two receptors to this motif (see Discussion). We also quantified eight proteins that are significantly (P < 0.0012) but only moderately specific (ratios between 1.5 and 2.4) for the wild-type promoter element (Supplemental Table S2).
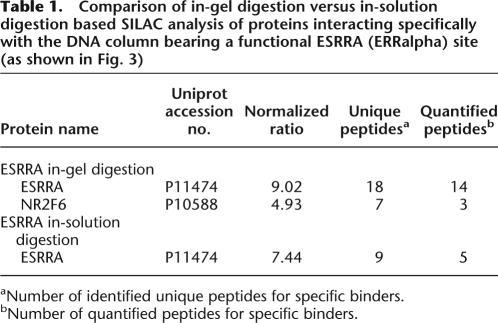
Table 1.
Comparison of in-gel digestion versus in-solution digestion based SILAC analysis of proteins interacting specifically with the DNA column bearing a functional ESRRA (ERRalpha) site (as shown in Fig. 3)
aNumber of identified unique peptides for specific binders.
bNumber of quantified peptides for specific binders.
Feasibility of scale-up
We next investigated whether the method could be streamlined as required to use it in large-scale experiments. In the work described above, protein eluates were separated by SDS-PAGE, excised, and in-gel digested. This serves to simplify the complex protein mixture and to lower the dynamic range requirements of the experiment. However, the gel step also results in an increase of analysis time as several bands are analyzed in succession and makes the procedure less automatable as in-gel protein digestion requires a more complicated protocol than in-solution digest.
We therefore repeated the experiment with the ESRRA motif using the in-solution digest instead of the gel. In this experiment, a total of 197 proteins were identified, including ESRRA sequenced with nine peptides. The protein ratio was 7.44, which is highly significant (P = 4.27 × 10−27) and very similar to the in-gel experiment (Table 1). We conclude that despite the great complexity of the proteins bound to DNA targets a single in-solution analysis and therefore automation is feasible. Furthermore, in this experiment, only lysine was labeled, whereas labeling both arginine and lysine results in better quantification as virtually all peptides can be quantified (Schulze et al. 2005). Furthermore, we did not yet use “exclusion lists,” which are lists of peptide masses of the most abundant background proteins, to exclude them from sequencing. This allows preferential sequencing of low-abundance peptides but was not possible because of software limitations on our mass spectrometer. If both steps are included, in-solution digests should perform similarly to the more laborious in-gel procedure, as we have already shown in the case of peptide pull-downs.
Identification of binding partners to modified DNA
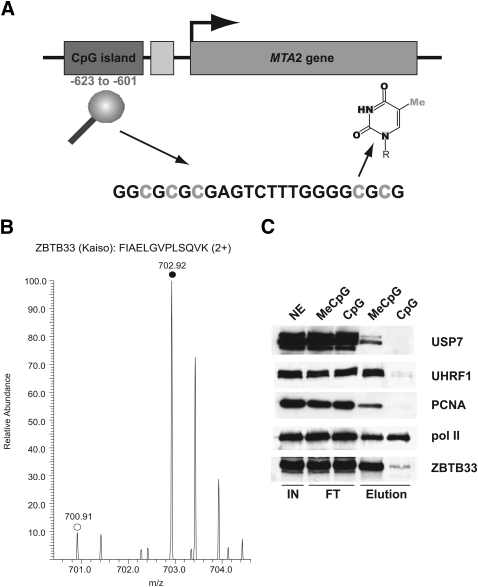
DNA modifications can also be important determinants for protein binding. For example, damaged DNA recruits DNA repair enzymes, and DNA methylation, in addition to its roles in epigenetics (Klose and Bird 2006), can directly influence TF binding to a given DNA sequence (Perini et al. 2005). In particular, it is well known that cancerous cells frequently hypermethylate promoter regions of tumor suppressor genes leading to their down-regulation (for review, see Robertson 2005). Conversely, MeCpG can also be a precondition for TF binding and transcriptional activation (Ego et al. 2005). To study methylation-dependent DNA binding of TFs, we selected the CpG island −623 to −601 upstream of the transcription start site of the MTA2 gene. A 41-bp bait containing this island in the fully methylated and nonmethylated form was synthesized as bait and control, respectively (Fig. 4A). We chose this region because it is an endogenously methylated site and because one binding protein, ZBTB33 (previously known as Kaiso), had already been determined in vitro and by ChIP (Yoon et al. 2003).
Figure 4.
Methyl-CpG specific binding partners can be discriminated by SILAC analysis. (A) Scheme of the CpG island in the upstream region of the human MTA2 gene that is located at position −623 to −601 relative to the transcription start site (arrowhead). The cytosine residues highlighted in gray were either fully methylated (MeCpG) or not methylated (CpG) in the SILAC experiment. The structure of 5-methyl cytosine is shown. (B) Representative peptide mass spectrum demonstrating the preferential binding of the methyl-CpG binding protein ZBTB33 (Kaiso), which was known to interact with the fully methylated MTA2 CpG island. The MS spectrum of a labeled (monoisotopic peak marked with filled circle) and unlabeled (monoisotopic peak marked with open circle) tryptic ZBTB33 peptide acquired is shown. (C) Western blot analysis (same conditions as in Fig. 2C) of selected specific interaction partners recapitulates results from the SILAC analysis (Table 2).
As shown in Figure 4B and Table 2, this experiment resulted in the identification and quantification of ZBTB33 with a ratio of 11.26 (P = 2.04 × 10−8) using two peptides. Of 328 quantified proteins, seven had significant fold changes between experiment and control, showing that they bind more tightly to the methylated than to the nonmethylated CpG island (Table 2).
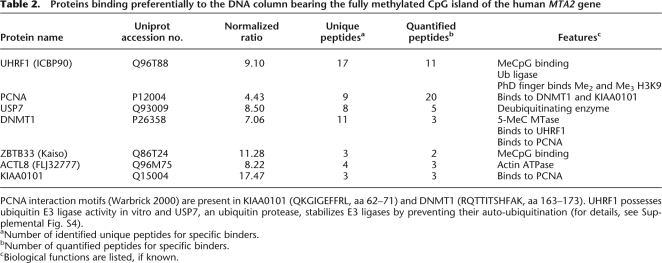
Table 2.
Proteins binding preferentially to the DNA column bearing the fully methylated CpG island of the human MTA2 gene
PCNA interaction motifs (Warbrick 2000) are present in KIAA0101 (QKGIGEFFRL, aa 62–71) and DNMT1 (RQTTITSHFAK, aa 163–173). UHRF1 possesses ubiquitin E3 ligase activity in vitro and USP7, an ubiquitin protease, stabilizes E3 ligases by preventing their auto-ubiquitination (for details, see Supplemental Fig. S4).
aNumber of identified unique peptides for specific binders.
bNumber of quantified peptides for specific binders.
cBiological functions are listed, if known.
Apart from ZBTB33, we found several putative and not previously described methyl-CpG-specific binding partners of the MTA2 CpG island, namely the SRAYDG domain containing protein UHRF1 (formerly known as ICBP90), the replication clamp protein PCNA, the PCNA interacting proteins DNA methyltransferase 1 (DNMT1) and KIAA0101/p15PAF, the ubiquitin-specific protease USP7, as well as the hypothetical protein ACTL8 (actin-like 8/FLJ32777) (Table 2; Supplemental Fig. S3). Antibodies available for ZBTB33, UHRF1, PCNA, and USP7 further confirmed specific binding of these factors to the methylated probe (Fig. 4C). Interestingly, we also identified another known DNMT1 interaction partner, the histone H3 K9 methyltransferase EHMT2 (also known as G9A) (Esteve et al. 2006), with two peptides and quantified one peptide with a ratio of 3.40 but excluded it because of our stringent criteria (see above and Supplemental Methods). ZBTB33 and UHRF1 have been shown to directly interact with methyl-CpG sites (Daniel et al. 2002; Unoki et al. 2004; Bostick et al. 2007), so we assume direct binding of these factors to the 5-methyl cytosine modified bait. The other proteins do not posses any known methyl-CpG binding domain but some of them (PCNA, DNMT1) have been identified as interaction partners of NP95, the mouse ortholog of UHRF1 (Sharif et al. 2007). Therefore, their specific binding to the methylated bait is most likely indirect (see Discussion and Supplemental Fig. S4).
Discussion
The ability to rapidly determine the protein binding partners of a DNA sequence of interest is becoming increasingly important given the current pace of genome sequencing, which accompanies the discovery of highly conserved noncoding sequences as well as novel potential TF binding sites in mammals (Bejerano et al. 2004; Cawley et al. 2004; Harbison et al. 2004; Xie et al. 2005). In this study we have outlined a strategy for unbiased identification of sequence and modification-specific DNA binding proteins using SILAC-based quantitative proteomics.
General aspects of the screen (specificity and sensitivity of the approach)
The screen uses custom-made oligonucleotides with a biotin linker and takes advantage of differential metabolic labeling of cell populations using SILAC. This allows the distinction of proteins by mass spectrometry. To identify specific candidate binding partners of a DNA element of interest, it is crucial to design appropriate control baits, in which the binding of specific interaction partners is severely diminished. In the case of modification-dependent DNA protein interactions the control bait will simply contain the identical DNA sequence in the unmodified form. For sequence context-dependent interactions the design of suitable controls can be achieved in several ways. If data correlating the effect of mutations with the activity of a putative cis-element are not available, two other possibilities can be pursued. First, phylogenetic footprinting analysis of the cis-element by multiple species alignment of synthenic regions (for review, see Wasserman and Sandelin 2004) will often reveal highly conserved nucleotides, which can be mutated in the control. Second, the use of the complete reverse sequence of the cis-element or part of it can be a suitable control (Travis et al. 1993), because it does not change the overall nucleotide composition of the double helix and conserves the GC/AT content of each DNA strand.
The fact that the vast majority of binding partners identified in our pilot screens either reproduce and confirm published data or can be placed in a functional biological context argues for a low false-positive detection rate of the technique, especially when the robust statistics of the MaxQuant software are applied (Graumann et al. 2007, 2008). However, as is the case for every scalable screening method we cannot exclude false-negative results. This is exemplified by the experiment we performed to detect MeCpG-dependent binders (Fig. 4; Table 2), which failed to identify the indirect interaction partner EHMT2 (G9A) (part of an UHRF1 complex) due to sensitivity issues in combination with our stringent statistical filtering (see Results). Furthermore, when working with a single cell line and with unstimulated cells we cannot find proteins that are not expressed in that cell line or need to be modified in a signal-dependent manner to bind DNA. In contrast to most previous work from our laboratory, the DNA protein interaction screen involves the preparation of nuclear extracts from the labeled and unlabeled cell population, respectively, which represents a second sample manipulation step after cell lyses. Therefore, standardized and reproducible conditions in cell growth and nuclear extract preparation are crucial for the success of the assay system.
In comparison to the EMSA technique, our solid phase approach is much less subjected to nonphysiological buffer conditions (ionic strength and pH), which occur during gel electrophoresis and can drastically disturb the equilibrium between free DNA and protein–DNA complexes (Sidorova et al. 2005). Thus, interactions revealed by our SILAC-based screen in vitro will most likely also have the potential to occur in vivo, where they are of course influenced by additional levels of regulation (chromatin structure and histone modifications).
Importantly, ChIP-chip studies have suggested that in vitro affinity of TFs for specific DNA sequences is often recapitulated in the relative occupancy of these regions in vivo (Horak et al. 2002; Harbison et al. 2004).
The energetics of protein–DNA interactions are dominated by interactions with the sugar–phosphate backbone of DNA; hence about two thirds of all contacts are not sequence specific (Hoglund and Kohlbacher 2004). Despite very large affinities for their target (dissociation constants between 10−8 to 10−12 M) the binding specificity of DNA binding proteins can therefore be often rather modest (as low as a factor 10). Apart from the ability to monitor all-or-nothing interactions (Schulze and Mann 2004), the SILAC methodology excels in its potential to accurately quantify relative differences in the range from two to 10 (Ong et al. 2003) and is hence well suited for our protein DNA interaction screen (see Tables 1, 2).
The amount of TFAP2A molecules per cell has been estimated to be around 200,000 (Egener et al. 2005), which means that we have used ∼500 pmol of TFAP2A as input in our pilot experiment (in the presence of 25 pmol of bait). Although drastically decreased (10 times) compared with related studies (Yaneva and Tempst 2003; Himeda et al. 2004), sample consumption is still significant and needs to be further optimized in the future. This will be achieved in several ways. The ratio of total protein input versus bait can be reduced by factor ten, which will not lead to a substantial decrease in the total amount of proteins bound to the bait. More importantly, the relative abundance of the bound factors will change only slightly or not at all. Because our LC tandem MS proteomic platform using the LTQ-FT is not so much limited by sample amount as by sample complexity, we do not expect any negative effect on our analysis (de Godoy et al. 2006). Double-labeling of the extracts with heavy lysine and arginine derivatives will roughly double the number of peptides that can be quantified and hence increase sensitivity. Moreover, several improvements in mass spectrometric analysis currently in development, such as the peptide exclusion lists mentioned above, should dramatically improve the sensitivity of peptide mixture analysis.
Determination of sequence-specific DNA protein interactions
Apart from validating the methodology, the results obtained with both the bait bearing a binding site for AP2 and the bait designed to confirm a direct interaction with ESRRA are functionally interesting. Our data clearly demonstrate that the point mutations in the AP2 site drastically reduce binding of PURA and PURB to the bait. Because the interaction of Pur proteins with DNA is mainly based on DNA structure (e.g., the propensity to form cruciform DNA) (Wortman et al. 2005), our observation suggests the ability to identify structure-dependent associations in our screen. At the same time, this example also demonstrates that we can monitor the specific binding of several candidate factors to our DNA columns. This is further corroborated by the observations obtained with the bait harboring an ESRRA binding motif present at the ESRRA gene promoter. Apart from confirming specific ESRRA binding we find that the NR NR2F6 is also able to bind this sequence element in vitro and might therefore play a role in regulating ESRRA gene expression. Indeed, it has been shown that NR2F6 can bind the same motif present upstream of the renin and luteinizing hormone receptor gene in vivo (Zhang and Dufau 2000; Liu et al. 2003). In both cases NR2F6 acts as a transcriptional repressor competing with binding of other NRs to the same motif in cis.
Thus, the DNA protein interaction screen is a powerful tool to conduct completely unbiased experiments that can follow up bioinformatic cis-element predictions. Unlike EMSA our screen is directly able to answer the question which proteins can potentially bind a sequence of interest. Therefore, changes in protein occupancy that are influenced by single nucleotide polymorphisms, disease-related nucleotide exchanges, or alterations in relative protein expression levels may be easily monitored.
Determination of DNA modification-dependent protein interactions
The experiment performed with the MTA2 promoter CpG island probe is to our knowledge the first example of a quantitative proteomics approach designed to identify factors that bind to DNA in a modification-dependent manner. Besides two known methyl-CpG interacting proteins, namely ZBTB33 and UHRF1, we identified two factors (PCNA, DNMT1) that are part of a recently described UHRF1 complex (Sharif et al. 2007), as well as several proteins (USP7, KIAA0101, ACTL8) that have so far not been directly implicated in the biology of DNA methylation (Fig. 4; Table 2). However, we have not found any protein belonging to the MBD (methyl-CpG binding domain) family (e.g., MECP2, MBD1 to MBD4), which have been discovered by means of binding to plasmid DNA-derived methylated DNA substrates in vitro (Boyes and Bird 1991). MBD3 interacts with DNA in a methylation-independent manner, and MBD4 is mainly linked to DNA repair processes (for review, see Klose and Bird 2006). The absence of MECP2, which is able to bind even a single MeCpG site, can simply be explained by the fact that this protein is not expressed in HeLa cells (Ng et al. 1999). A series of experiments has recently challenged the view that MBD proteins bind to methylated CpGs in a sequence- and context-independent manner (Ballestar et al. 2003; Fraga et al. 2003; Klose et al. 2005; Le Guezennec et al. 2006). MECP2, for instance, requires a run of four or more A/T base pairs adjacent to the methyl-CpG for efficient DNA binding in vitro and in cells (Klose et al. 2005). Although unlikely, the lack of comprehensive binding data for MBD family members makes it impossible for us to exclude that our assay system might be biased against detection of these proteins because of sensitivity issues or experimental conditions.
However, our results confirm (Fig. 4; Table 2) the specific binding of ZBTB33 to the methylated CpG island of the MTA2 gene in vitro (Yoon et al. 2003). Strikingly, the absence of ZBTB33 does not detectably alter the expression of MTA2 and other putative ZBTB33 target genes suggesting a redundant function (Prokhortchouk et al. 2006). UHRF1, being part of a multiprotein complex containing PCNA and DNMT1 (Supplemental Fig. S4), might represent such a redundancy factor. Hence, we conclude that our screening system can uncover both direct and indirect protein DNA interactions that are specific for modified DNA.
Perspectives in the context of genome-wide mapping of DNA protein interactions
The Tempst and the Aebersold groups have described related approaches to identify TFs by affinity purification procedures combined with mass spectrometry (Yaneva and Tempst 2003; Himeda et al. 2004). The study of Yaneva and Tempst (2003) requires prefractionation of nuclear extracts on a phosphocellulose column. In contrast, the quantitative proteomics approach by Himeda et al. (2004) used the ICAT technique (Gygi et al. 1999) to identify proteins binding to a functionally important enhancer element. Both methodologies have limitations that prevent their use in high-throughput investigations. Fractionation of nuclear extracts requires the parallel monitoring of the binding activities by EMSA. The ICAT approach requires chemical derivatization and subsequent clean-up steps by strong cation exchange chromatography before LC-MS analysis, increasing the complexity of analysis (see introductory section).
The strategy presented here has the potential to quantify almost all peptides present in a sample, because metabolic labeling of cells with “heavy” derivatives of lysine and arginine leads to complete encoding of tryptic peptides representing the total cellular proteome (Schulze et al. 2005). The screen is relatively simple to perform given high-sensitivity mass spectrometric equipment associated with advanced software. Streamlining the procedure should thus allow to perform it in large-scale formats. In general, it can be employed to detect the following type of interactions: (1) direct binding; (2) indirect binding; (3) partner binding, in which a protein exhibits a low-affinity interaction with DNA but specificity is achieved via binding to a nearby factor; and (4) modification-dependent binding regulated by signaling.
Therefore, the DNA protein interaction screen can serve as a follow-up experiment in approaches based on bioinformatics prediction of functional elements or discovery of disease-related nucleotide changes. In addition, it complements rather than competes with established genome-wide location analysis (“ChIP on chip” and “ChIP-seq” experiments). It is completely unbiased with respect to both the bait and the potential preys present in nuclear extracts and does not require any a priori knowledge about DNA binding specificities of the proteins involved. Whereas the DNA protein interaction screen reveals possible binders for a particular DNA sequence, the orthogonal ChIP-chip experiment delivers binding sites for a given TF. Currently, technical limitations in nucleotide resolution (in the range of several hundred base pairs) require extensive bioinformatic modeling of ChIP-chip data to define and locate the probable TF binding sites (Cawley et al. 2004; Harbison et al. 2004; Wei et al. 2006). Accordingly, there has been relatively little overlap between two sets of proposed TP53 (p53) binding sites in related studies (for review, see Holstege and Clevers 2006). Protein arrays and protein binding microarrays (PPMs) have so far been only successfully used in yeast (Hall et al. 2004; Mukherjee et al. 2004). Protein arrays cannot reveal cis-elements that are bound in a cooperative manner by two or more proteins. Like the protein array studies, PPM experiments require purification and subsequent labeling of proteins that cannot be performed at large scale in higher organisms. In conclusion, the SILAC-based DNA protein interaction screen will have an important and unique role in delivering highly validated candidate proteins representing potential in vivo binding partners of functional DNA elements.
Methods
Antibodies
The TFAP2A (AP2-alpha) and PCNA antibodies were purchased from Santa Cruz. The antibodies against PURA and ZBTB33 were a generous gift from E.M. Johnson (Mount Sinai School of Medicine, New York, NY) and A.B. Reynolds (Vanderbilt University, Nashville, TN), respectively. The ESRRA (ERRalpha) antibody was from Novus Biologicals, the UHRF1 (ICBP90) antibody from BD Biosciences, and the USP7 antibody (ab4080) from Abcam. The RNA polymerase II antibody directed against the C-terminal domain (CTD) of the POLR2A subunit was derived from hybridomas (WG16.1).
DNA affinity chromatography and quantitative proteomics
A detailed description can be found in the supplementary material. Briefly, HeLa-S3 cells were metabolically labeled with 2H4-lysine as the SILAC amino acid in RPMI medium followed by isolation and high salt extraction of nuclei (Dignam et al. 1983). Affinity purifications were performed with biotinylated double-stranded oligonucleotides (wild-type and control baits) that have been immobilized on streptavidin magnetic beads (Dynal MyOne, Invitrogen) at their maximum binding capacity of ∼200 pmol/mg. Routine analyses were carried out with nuclear extracts corresponding to 2.5 × 108 cells for both the 2H4-lysine-labeled and the unlabeled state in the presence of 0.4 mL of beads, respectively. Protein–DNA complexes were combined, eluted by restriction enzyme cleavage (PstI, NEB), and subjected to GeLC-MS analysis.
Data availability
Supplementary data accompanies this paper. Mass spectrometric raw data and MaxQuant evidence files have been uploaded to Tranche at http://tranche.proteomecommons.org.
Acknowledgments
We thank E.M. Johnson for anti-PURA and A.B. Reynolds for anti-ZBTB33 antibodies. CEBI (Center of Experimental Bioinformatics) is supported by a generous fund of the Danish National Research Foundation (Grundforskningfond). We thank members of CEBI and the Department of Proteomics and Signal Transduction at the Max-Planck-Institute for Biochemistry for constructive comments and discussion. We also thank Juergen Cox for developing the MaxQuant software and help in its application.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.081711.108.
References
- Ballestar E., Paz M.F., Valle L., Wei S., Fraga M.F., Espada J., Cigudosa J.C., Huang T.H., Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bergemann A.D., Ma Z.W., Johnson E.M. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol. 1992;12:5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Bulyk M.L. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cravatt B.F., Simon G.M., Yates J.R., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- Daniel J.M., Spring C.M., Crawford H.C., Reynolds A.B., Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy L.M., Olsen J.V., de Souza G.A., Li G., Mortensen P., Mann M. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egener T., Roulet E., Zehnder M., Bucher P., Mermod N. Proof of concept for microarray-based detection of DNA-binding oncogenes in cell extracts. Nucleic Acids Res. 2005;33:e79. doi: 10.1093/nar/gni1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ego T., Tanaka Y., Shimotohno K. Interaction of HTLV-1 Tax and methyl-CpG-binding domain 2 positively regulates the gene expression from the hypermethylated LTR. Oncogene. 2005;24:1914–1923. doi: 10.1038/sj.onc.1208394. [DOI] [PubMed] [Google Scholar]
- Esteve P.O., Chin H.G., Smallwood A., Feehery G.R., Gangisetty O., Karpf A.R., Carey M.F., Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes & Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Montoya G., Taysavang P., Wade P.A., Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V., McBroom L.D., Flock G. Determinants of target gene specificity for RORα1: Monomeric DNA binding by an orphan nuclear receptor. Mol. Cell. Biol. 1995;15:2517–2526. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann J., Hubner N.C., Kim J.B., Ko K., Moser M., Kumar C., Cox J., Schoeler H., Mann M. SILAC-labeling and proteome quantitation of mouse embryonic stem cells to a depth of 5111 proteins. Mol. Cell. Proteomics. 2007;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- Gygi S.P., Rist B., Gerber S.A., Turecek F., Gelb M.H., Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hall D.A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeda C.L., Ranish J.A., Angello J.C., Maire P., Aebersold R., Hauschka S.D. Quantitative proteomic identification of six4 as the trex-binding factor in the muscle creatine kinase enhancer. Mol. Cell. Biol. 2004;24:2132–2143. doi: 10.1128/MCB.24.5.2132-2143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund A., Kohlbacher O. From sequence to structure and back again: Approaches for predicting protein-DNA binding. Proteome Sci. 2004;2:3. doi: 10.1186/1477-5956-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Clevers H. Transcription factor target practice. Cell. 2006;124:21–23. doi: 10.1016/j.cell.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Horak C.E., Mahajan M.C., Luscombe N.M., Gerstein M., Weissman S.M., Snyder M. GATA-1 binding sites mapped in the beta-globin locus by using mammalian chIp-chip analysis. Proc. Natl. Acad. Sci. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.D., Liu X., Zuo F., Eisenbraun T.L., Wiley S.R., Kraus R.J., Mertz J.E. Estrogen-related receptor alpha 1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Mol. Endocrinol. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Sarraf S.A., Schmiedeberg L., McDermott S.M., Stancheva I., Bird A.P. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Knapp A.M., Ramsey J.E., Wang S.X., Godburn K.E., Strauch A.R., Kelm R.J., Jr Nucleoprotein interactions governing cell type-dependent repression of the mouse smooth muscle α-actin promoter by single-stranded DNA-binding proteins Purα and Purβ. J. Biol. Chem. 2006;281:7907–7918. doi: 10.1074/jbc.M509682200. [DOI] [PubMed] [Google Scholar]
- Le Guezennec X., Vermeulen M., Brinkman A.B., Hoeijmakers W.A., Cohen A., Lasonder E., Stunnenberg H.G. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang X., Sigmund C.D. Identification of a nuclear orphan receptor (Ear2) as a negative regulator of renin gene transcription. Circ. Res. 2003;92:1033–1040. doi: 10.1161/01.RES.0000071355.82009.43. [DOI] [PubMed] [Google Scholar]
- MacCoss M.J., Wu C.C., Liu H., Sadygov R., Yates J.R., III A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal. Chem. 2003;75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- McClatchy D.B., Liao L., Park S.K., Venable J.D., Yates J.R. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N., Donner A., Ippolito J.A., Williams T. SELEX and missing phosphate contact analyses reveal flexibility within the AP-2α protein: DNA binding complex. Nucleic Acids Res. 1999;27:2760–2769. doi: 10.1093/nar/27.13.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Zhang Y., Hendrich B., Johnson C.A., Turner B.M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Ohlsson R., Kanduri C. New twists on the epigenetics of CpG islands. Genome Res. 2002;12:525–526. doi: 10.1101/gr.18002. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Kratchmarova I., Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J. Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- Ong S.E., Mittler G., Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- Perini G., Diolaiti D., Porro A., Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl. Acad. Sci. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A., Sansom O., Selfridge J., Caballero I.M., Salozhin S., Aithozhina D., Cerchietti L., Meng F.G., Augenlicht L.H., Mariadason J.M., et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell. Biol. 2006;26:199–208. doi: 10.1128/MCB.26.1.199-208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A., Scholer H.R., Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Saccani S., Pantano S., Natoli G. Modulation of NF-κB activity by exchange of dimers. Mol. Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Schulze W.X., Mann M. A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 2004;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- Schulze W.X., Deng L., Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 2005 doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Sidorova N.Y., Muradymov S., Rau D.C. Trapping DNA-protein binding reactions with neutral osmolytes for the analysis by gel mobility shift and self-cleavage assays. Nucleic Acids Res. 2005;33:5145–5155. doi: 10.1093/nar/gki808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Travis A., Hagman J., Hwang L., Grosschedl R. Purification of early-B-cell factor and characterization of its DNA-binding specificity. Mol. Cell. Biol. 1993;13:3392–3400. doi: 10.1128/mcb.13.6.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala R., Romano R.A., Fuchs E., Sinha S. Molecular cloning and characterization of AP-2ɛ, a fifth member of the AP-2 family. Gene. 2003;321:93–102. doi: 10.1016/s0378-1119(03)00840-0. [DOI] [PubMed] [Google Scholar]
- Udalova I.A., Mott R., Field D., Kwiatkowski D. Quantitative prediction of NF-kappa B DNA-protein interactions. Proc. Natl. Acad. Sci. 2002;99:8167–8172. doi: 10.1073/pnas.102674699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M., Nishidate T., Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. BioEssays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Warren C.L., Kratochvil N.C., Hauschild K.E., Foister S., Brezinski M.L., Dervan P.B., Phillips G.N., Jr, Ansari A.Z. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wilson T.E., Fahrner T.J., Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol. Cell. Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E., Chen X., Fricke E., Geffers R., Hehl R., Liebich I., Krull M., Matys V., Michael H., Ohnhauser R., et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman M.J., Johnson E.M., Bergemann A.D. Mechanism of DNA binding and localized strand separation by Purα and comparison with Pur family member, Purβ. Biochim. Biophys. Acta. 2005;1743:64–78. doi: 10.1016/j.bbamcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M., Tempst P. Affinity capture of specific DNA-binding proteins for mass spectrometric identification. Anal. Chem. 2003;75:6437–6448. doi: 10.1021/ac034698l. [DOI] [PubMed] [Google Scholar]
- Yoon H.G., Chan D.W., Reynolds A.B., Qin J., Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Dufau M.L. Nuclear orphan receptors regulate transcription of the gene for the human luteinizing hormone receptor. J. Biol. Chem. 2000;275:2763–2770. doi: 10.1074/jbc.275.4.2763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary data accompanies this paper. Mass spectrometric raw data and MaxQuant evidence files have been uploaded to Tranche at http://tranche.proteomecommons.org.