Abstract
Alkali (Li+, Na+, K+, Rb+, and Cs+) and halide (F−, Cl−, Br−, and I−) ions play an important role in many biological phenomena, roles that range from stabilization of biomolecular structure, to influence on biomolecular dynamics, to key physiological influence on homeostasis and signaling. To properly model ionic interaction and stability in atomistic simulations of biomolecular structure, dynamics, folding, catalysis, and function, an accurate model or representation of the monovalent ions is critically necessary. A good model needs to simultaneously reproduce many properties of ions, including their structure, dynamics, solvation, and moreover both the interactions of these ions with each other in the crystal and in solution and the interactions of ions with other molecules. At present, the best force fields for biomolecules employ a simple additive, nonpolarizable, and pairwise potential for atomic interaction. In this work, we describe our efforts to build better models of the monovalent ions within the pairwise Coulombic and 6-12 Lennard-Jones framework, where the models are tuned to balance crystal and solution properties in Ewald simulations with specific choices of well-known water models. Although it has been clearly demonstrated that truly accurate treatments of ions will require inclusion of nonadditivity and polarizability (particularly with the anions) and ultimately even a quantum mechanical treatment, our goal was to simply push the limits of the additive treatments to see if a balanced model could be created. The applied methodology is general and can be extended to other ions and to polarizable force-field models. Our starting point centered on observations from long simulations of biomolecules in salt solution with the AMBER force fields where salt crystals formed well below their solubility limit. The likely cause of the artifact in the AMBER parameters relates to the naive mixing of the Smith and Dang chloride parameters with AMBER-adapted Åqvist cation parameters. To provide a more appropriate balance, we reoptimized the parameters of the Lennard-Jones potential for the ions and specific choices of water models. To validate and optimize the parameters, we calculated hydration free energies of the solvated ions and also lattice energies (LE) and lattice constants (LC) of alkali halide salt crystals. This is the first effort that systematically scans across the Lennard-Jones space (well depth and radius) while balancing ion properties like LE and LC across all pair combinations of the alkali ions and halide ions. The optimization across the entire monovalent series avoids systematic deviations. The ion parameters developed, optimized, and characterized were targeted for use with some of the most commonly used rigid and nonpolarizable water models, specifically TIP3P, TIP4PEW, and SPC/E. In addition to well reproducing the solution and crystal properties, the new ion parameters well reproduce binding energies of the ions to water and the radii of the first hydration shells.
I. Introduction
Salt and solvation are fundamental to chemistry, biology, and life. Monovalent salts such as Na+, K+, and Cl− are critically important in regulating the homeostasis and electric potentials of cells, and monovalent ions serve as important building blocks of biomolecular structure by stabilizing proteins, lipids, and nucleic acids through both specific and nonspecific interactions.1–6 Water and ions also modulate biomolecular stability, dynamics, and folding,7–17 and ions are important mediators of catalytic activity.18–23 To properly model biological phenomena at the atomic level, an accurate treatment of the ions and solvent is crucial. This is nontrivial, because the interactions of salt with salt, water with salt, and water with water are subtle and represent a balance of large electrostatic and dipole interactions, small additive van der Waals interactions, and large changes in entropy, among other interactions. At low-to-moderate salt concentrations and also at physiological salt concentrations, crystals of salt should dissolve in water. As will be discussed in greater detail below, this is not always the case in simulation because of misbalances in the commonly applied potentials. These misbalances can lead to erroneous conclusions about a given ion’s interaction with a biomolecule. Compounding the problem is the considerable disagreement among many of the calculated and measured values, such as ΔGhydration, coordination numbers, and the first peak of the radial distribution functions (RDFs) from the many different sets of ion parameters available (see Table 1).(24)
Table 1. Summary of Previously Reported Ion Parameters for MD Simulation.
| source | target properties | combining rule | force-field type | boundary | ions | water model |
|---|---|---|---|---|---|---|
| this work | Eion−water, dion−water, ΔGhydration, lattice energy, lattice spacing | Lorentz−Berthelot | nonpolarizable | PBC/Ewald | Li+, Na+, K+, Rb+, Cs+, F−, Cl−, Br−, I− | TIP3P, TIP4PEW, SPC/E |
| Jensen and Jorgensen(75) | ΔGhydration, water RDF | geometric | nonpolarizable | SBC | Li+, Na+, K+, Rb+, Cs+, F−, Cl−, Br−, I−, NH4+ | TIP4P |
| Åqvist(63) | ΔGhydration | geometric | nonpolarizable | SBC | Li+, Na+, K+, Rb+, Cs+, Mg2+, Ca2+, Sr2+, Ba2+ | SPC, TIP3P |
| Lamoureux and Roux(95) | Eion−water, dion−water, ΔG hydration | Lorentz−Berthelot | polarizable | SBC | Li+, Na+, K+, Rb+, Cs+, F−, Cl−, Br−, I− | SWM4-DP |
| Beglov and Roux(96) | ΔGhydration, water RDF | Lorentz−Berthelot | nonpolarizable | SBC | Na+, K+ | TIP3P |
| Roux(97) | ΔGhydration | Lorentz−Berthelot | nonpolarizable | SBC | Cl− | TIP3P |
| Smith and Dang(94) | Eion−water, water RDF | Lorentz−Berthelot | polarizable | PBC/Ewald | Na+, Cl− | RPOL |
| Dang(98) | Eion−water, water RDF | Lorentz−Berthelot | polarizable | PBC/Ewald | Li+, F− | POL1 |
| Dang(99) | Eion−water, water RDF | Lorentz−Berthelot | nonpolarizable | PBC/Ewald | Cs+ | SPC/E |
| Dang(64) | Eion−water, water RDF | Lorentz−Berthelot | nonpolarizable | PBC/Ewald | Na+, K+, Rb+, Cl− | SPC/E |
| Dang and Garrett(100) | Eion−water, water RDF | Lorentz−Berthelot | polarizable/nonpolarizable | PBC/(Ewald?) | I− | SPC/E, RPOL |
| Alejandre and Hansen(49) | hydrogen-bond strength | Lorentz−Berthelot | nonpolarizable | PBC/Ewald | Na+, Cl− | SPC/E |
| Lopes and Padua(101) | Born−Huggins−Mayer form (Fumi/Tosi data) | geometric | nonpolarizable | PBC/Ewald | Br−, Cl− | N/A |
| Lenart et al.(102) | Huggins−Mayer potental (Fumi/Tosi data) | N/A | N/A | Implicit water | Na+, K+, Cl− | N/A |
| Teleman and Ahlstrom(103) | Kirkwood−Slater formula | N/A | nonpolarizable | N/A | Ca2+ | SPC |
| Weerasinghe and Smith(104) | Kirkwood−Buff theory | Lorentz−Berthelot (w/ exception) | nonpolarizable | PBC/Ewald | Na+, Cl− | SPC(/E), TIP3P |
| Straatsma and Berendsen(105) | HF-SCF energy | N/A | nonpolarizable | PBC | Na+, K+, F− | SPC |
| Sremaniak et al.(106) | Heinzinger | unknown | nonpolarizable /polarizable | SBC | Br− | POL1, SPC/E |
| Peslherbe et al.(107) | HF-SCF energy | N/A | nonpolarizable /polarizable | N/A | I− | TIP4P, polarizable OPCS |
Issues and Artifacts in Simulations with Common Ion Potentials
The lack of consistency among the various models is a significant cause for concern because these models are routinely applied in biomolecular simulations to probe the influence ions exert on biomolecular structure and dynamics,25–44 to understand the process of crystallization/dissolution,45–49 or to better understand ion pairing.50–52 Hints of a potential problem with the ion parameters distributed with the standard AMBER ff94,(53) ff98,(54) and ff9955,56 (AMBER ff9X) force fields emerged in our simulations of A-DNA—B-DNA conformational transitions of nucleic acids with various added salt concentrations, where A-DNA was not stable at high salt, as expected.57–60 As more people applied these force fields, higher salt concentrations were studied, and simulations were routinely pushed beyond ∼10 ns timescales, the larger community started to become aware of issues with crystallization, specifically the anomalous formation of salt crystals well below the saturation limit.41,42,48,61,62
The AMBER ff9X force fields use the AMBER-adapted Åqvist parameters(63) for the cations and the Cl− parameters of Dang.(64) These were chosen on an ad hoc basis more than a decade ago and were never systematically assessed or validated. The lack of balance between the ion−ion interactions leads to spontaneous crystallization of NaCl or CsCl at concentrations greater than 1 M and KCl crystals at concentrations greater than ∼200 mM. Because the solubility of these salts is considerably higher than that in water (>∼5 molality), crystallization at low molarities caused us significant concern. Despite widespread acknowledgment of the problem, these negative findings—which are typically more difficult to publish—have not been widely discussed in the literature. Instead, discussion has been relegated to public forums such as the AMBER e-mail reflector, at various conferences, and through the ABC collaborations.65,66 In spite of this, the anomalous crystallization has been used as a rationale for the observation in MD simulation that Na+ ions condense around DNA better than K+,(41) simulations performed in an attempt to rationalize the inconclusive experimental findings.14,67,68 As an example of the anomalous crystallization in the published literature, see Figure 2 of Savelyev and co-workers,(42) Figure 1 of Chen and Pappu,(62) or Figure 1 from our previous work.(61) The misbalance in the ion parameters could significantly alter the interpretations of recent simulation experiments investigating the influence of ions on nucleic acid37,43,44 and lipid(69) structure and dynamics. Moreover, although recent MD simulations suggest that ion pairing is much more common than expected,(52) loosely consistent with interpretations of the results seen in conductivity experiments,(70) clearly, the observed crystallization at low molar concentrations is unrealistic. Yet, what is the proper balance between the crystal and the solution phase? Moreover, how much ion pairing should be observed and on what time scales?
Finding the Proper Balance between the Crystal Phase and the Solution Phase
How well can a simple and nonpolarizable Lennard-Jones (LJ) ion model perform? We attempt to address this question through the development of a new and self-consistent set of monovalent ion parameters across the whole series of Li+, Na+, K+, Rb+, Cs+, F−, Cl−, Br−, and I−. Our intent from the outset was to develop parameters that provide the proper balance between the calculated properties of both crystal and solution phases. To develop the parameters consistently and correctly requires choosing a common representation—for example, Ewald electrostatics with specific choices of common water models. This is necessary because the results depend critically on the choice of simulation conditions. To be consistent with the majority of simulation conditions employed in simulations of biomolecules, we initially focused on developing parameters consistent with each of three different and commonly used water models (TIP3P,(71) SPC/E,(72) and TIP4PEW(73)) in application with periodic boundary conditions (PBC) with a complete treatment of the long-ranged electrostatics(74) and pairwise additive potential, that is, nonpolarizable PBC simulations with an Ewald potential and spherical truncation of van der Waals interactions in the 8−10 Å range with inclusion of a continuum or homogeneous density correction for the long-range van der Waals energies and virials. In addition to choosing the appropriate simulation conditions, a correct set of parameters requires maintaining proper size consistency across the monovalent ion series and careful parametrization to both solution and crystal phase experiment. We believe that the systematic parametrization described herein provides the optimal way at present, given current experimental knowledge, to develop consistent ion potentials for use in simulations with the commonly employed biomolecule force fields.
Although the present research is limited to standard alkali and halide monovalent ions with three different water models, this approach can be readily extended to other mono- or multi-valent ions and other water models and simulation conditions, and ultimately, it can be extended to polarizable or higher-order treatments of the ion−water interaction. The experimental points of reference for the parametrization target are hydration free energies of the ions in solution (ΔGhydration), ion−water energies, and also the lattice constants (LC) and lattice energies (LE) of the salt crystals. This contrasts with much of the previous work (see Table 1) which focuses either solely on the hydration free energy, or combinations of ΔGhydration, first peaks of the relevant ion−water RDFs, and/or ion−water energetics in comparison with ab initio calculations. The results are validated by comparison to known first peaks of the RDFs and gas-phase ion−water and ion−water cluster energies.
Applying Standard Pairwise Additive, Nonpolarizable Intermolecular Potentials
Within the context of the majority of biomolecular simulation codes and force fields at present, a very simple intermolecular potential is applied. This includes a Coulombic potential with fixed charges on the atoms to represent the electrostatics and a LJ potential (see eq 1) that represents core-repulsion with an inverse r12 term and dispersion−attraction with an inverse r6 term, where r is the distance between particles. The LJ potential defines the energy Uij with two parameters: the well depth of the potential (εij) and the radius (Rmin,ij) at the minimum of the potential energy as a function of the distance between two particles i and j (rij).
With this simple pairwise additive (or nonpolarizable) force-field treatment for the alkali and halide ions, there are rather few parameters to manipulate, or degrees of freedom to adjust, while simultaneously retaining computational efficiency. Among the parameters available, the ion charge and mass are predetermined. The only parameters left to adjust are the interatomic dispersion−attraction and repulsive potentials, or Rmin,ij and εij, for each pair of distinct atom types.
The Results Depend Critically on the Details of the Simulation Protocol and Model
The total LJ potential is a pairwise sum over all pair interactions; as such, its value will be dependent on the choice of cutoff and cutoff method, and the LJ potential will be influenced indirectly by the choice of methods to treat the electrostatics and the choice of solvent model. In this regard, parameters generated with solvent boundary potentials(63) (SBC) or for use with truncated cutoffs(75) may not be applicable to periodic boundary simulations applying an Ewald potential treatment of the electrostatics.(76) Additionally, an ion model parametrized to work with one particular water model may not be transferable to another water model.24,51,75 Thus, when considering ion models, users need to worry about the combining rules, cutoff treatment, water model, and treatment of the electrostatics. Ideally, parameters will be transferable to different models and treatments; in practice, this is not always the case. Unlike a bond vibration, which is local and highly transferable, the nonbonded potential is highly sensitive to the details of the simulation protocol. This is most notable with the varied set of ion parameters in Table 1; changes in the water model, among similar models such as TIP3P,(71) SPC, SPC/E,(72) TIP4P,71,77 and TIP4PEW,(73) also affect the results but to less of an extent. However, these small differences are enough to alter interaction with protein side chains(78) and will alter the salt solubility. A more serious anomaly, as shown in the Supporting Information (Table S26), is the TIP5PEW(79) results for ΔGhydration of the ions which are significantly different than those obtained with the other common water models.
An additional issue that relates to simulation protocol is the combining rules. Because the LJ potential is a sum over all the atoms, in order to calculate the total energy, the coefficients for every possible pair of each of the different atom types are required. The Rmin and ε for two different atom types are usually mixed and calculated as the average of the parameters of the two individual atomic parameters. How this mixing is done depends on the choice of the combining rule. The AMBER,80,81 CHARMM,(82) and NAMD(83) programs, by default, apply the Lorentz−Berthelot84–87 combining rules which use the arithmetic mean for the combined Rmin and the geometric mean for the combined ε. Other programs, such as BOSS(88) and GROMOS,(89) use geometric mean combining rules. The combining rules are equivalent for mixing atoms with the same parameters, that is, for a homodiatomic molecule; the differences come when mixing atoms with different types (i.e., where each interacting atom has different Rmin values). Because the combining rules are different between different parametrizations and codes, care needs to be taken to use the correct mixing rule. The incorrect choice can lead to drastically different results.62,90–93 In AMBER, parameters that were developed with geometric mean combining rules, such as those of Åqvist,(63) are typically adapted to Lorentz−Berthelot/additive mixing rules by altering the water−ion mixed parameters to match the additive mixing that would have been calculated with the geometric mean. This retains the correct balance between water−ion interactions at the expense of the ion−ion interaction. Alternatively, as the premixed parameters are stored within the runtime parameter/topology file, these values can be manually adjusted by hand to reproduce the correct mixing; this is also possible in CHARMM/NAMD by using the NBFIX code to alter the mixed parameters.
Mapping the ΔGhydration = f(Rmin, ε) hypersurface
Our starting point in the development of the new set of consistent ion parameters across the entire monovalent ion series was to map out the complete set of minima and well-depth pairs, that is, the Rmin and ε of eq 1 that reproduce a chosen (experimental) ΔGhydration. This mapping was performed by assuming a simple pairwise additive Ewald and LJ intermolecular potential with a specific choice of a water model. By mapping the complete hypersurface ΔGhydration = f (Rmin, ε) for various water models, we could rationally choose the appropriate well depth (ε) for a given Rmin. This contrasts with previous work where the choice of ε has been somewhat ad hoc and poorly described.63,75,94 In addition, our mapping with different water models allowed a more detailed understanding of the influence of the water model (and simulation conditions) on the hypersurface. After this hypersurface was mapped, it was possible for a given choice of Rmin—a Rmin value chosen to well reproduce target experimental values—to determine the matching ε for a given ΔGhydration. Picking the appropriate target values, specifically the experimental ΔGhydration and Rmin, proved to be a challenging task.
Why Develop New Ion Parameters?
As discussed, the current AMBER ion parameters represent a mix of Åqvist cation and Dang chloride ion parameters and are misbalanced. This misbalance leads to enhanced ion pairing at low salt concentrations and crystallization above ∼1 M, depending on the choice of salt. However, given the large set of ion parameters that are already available (Table 1), it is important to argue why a new set of parameters is needed. It is also important to put this new work into the context of the existing parameter sets. Toward this end, concurrent to the development of the new set of monovalent ion parameters, we also examined many of the existing ion parameter sets. As discussed below, these studies and concurrent literature analysis helped us rationalize why a new parameter set was necessary and affirmed our contention that the pure Åqvist or pure Smith−Dang−Garrett ion parameters are not optimal.
Note that in this work, we have focused only on the commonly applied LJ potentials. The reason for this is their wide use, use that spans the bulk of the biomolecular simulation codes and force fields in common use. Beyond LJ potentials, more complex potentials for the ions have also been applied in a variety of studies of melting, such as using the Huggins−Mayer potential(108) parametrized(109) to fit Fumi−Tosi data.110,111 This model includes an exponential repulsive term and a −r−8 term for dipole−quadrupole attraction in addition to the standard −r−6 induced-dipole attraction.101,102 These potentials have not been widely applied in simulations of biomolecules, although the procedure that we employed herein could be used to help optimize parameters of this type.
Existing Ion Parameters
The Åqvist monovalent ion parameters are some of the most widely used set of LJ parameters. They were parametrized to reproduce early experimental ion hydration free energies(112) and first peaks of the ion−water RDFs.113,114 Their agreement with the early experimental results is excellent; however, since the parameters were published, the experimental results have improved considerably.95,115–118 An issue with the Åqvist parametrization is that the procedure for determining Rmin and ε was not well-specified and appears somewhat arbitrary. As this work demonstrates, there are many different combinations of Rmin and ε that can reproduce the same hydration free energy. An additional issue with these parameters is the boundary model, specifically the surface constraint all-atom solvent (SCAS) model.(119) This surrounds the ion with spherical clusters of water; however, the surface potential of water was not included when calculating the hydration free energies. Subsequent studies by Darden and co-workers point out that the Åqvist parameters underestimate the ΔGhydration when a fully periodic Ewald treatment of the solvation is applied compared to calculations with a solvent boundary potential equivalent to that of Åqvist.(76) Interestingly, as Åqvist targeted higher ΔGhydration values than those that are currently accepted today, the Ewald numbers are fortuitously closer to experiment than are those obtained with the SCAS solvation model. Despite this, the Åqvist ions crystallize too easily, both with AMBER-adapted and pure geometric mean combining rules.41,42,48,61,62 Although less crystallization is seen when pure geometric mean combining rules with the Åqvist Na+ and Smith and Dang Cl− ion parameters are applied, as shown by Chen and Pappu,(62) our simulation results suggest that the crystal is overstabilized compared to simulations with other ion parameters, including our new set. MD simulations were performed for ∼250 ns on both 1 and 4 M solutions by using geometric mean combining rules consistent with Chen and Pappu(62) and protocols as described in the section. Although crystal formation was not readily observed, clear solvent separated, and small crystal-like domains were evident (see Figure S10 in the Supporting Information). The exact simulations were with 838 TIP3P waters in truncated octahedron periodic cells with either 19 or 73 of each ion (leading to 1.08 and 4.22 M solutions, respectively), where the initial ion positions were randomized. Moreover, simulation of a solvated crystal at ∼1.26 M (including 4501 TIP3P water molecules and 108 Na+ and 108 Cl− built initially into a cubic crystal and the conditions above) shows only minimal melting in simulations on the ∼75 ns time scale.
The Smith−Dang−Garrett parameters, in our experience, appear reasonably well-balanced for NaCl and KCl; however, nonpolarizable parameters for many of the other larger or smaller monovalent ions are not available. Also, the sampling of the (Rmin, ε) space was limited because ε was, without discussion in the publications, somehow fixed to ε = 0.1 kcal/mol. The target values for validation were the gas-phase binding energies of the ions to water.64,94,98,99 Because this approach focuses on the water−ion interaction, it comes at the expense of ion−ion interactions. In our studies of NaCl and KCl salts, very limited ion pairing (essentially none) is evident, and there is no crystallization, even at very high salt concentrations. Solvated salt crystals quickly dissolve in water with these potentials. For example, a ∼4 M solvated crystal (838 TIP3P waters, 73 of each ion) dissolves completely within ∼10 ns of MD simulation, although at ∼6 M (838 TIP3P waters, 100 of each ion), a partial ∼4 × 4 × 4 is crystal is still evident at ∼367 ns.
Similarly, the Beglov and Roux parameters are available only for K+, Na+, and Cl−, and these ions tend to crystallize below their solubility limit (but less so than the Åqvist ions). These parameters were parametrized to reproduce the ion−water RDFs and hydration free energies; however, the Rmin and ε values were chosen rather arbitrarily, and the details of the optimization procedure were not clearly stated. Note that although these parameters tend to crystallize below their solubility limit, at concentrations below 2 M, no crystallization artifacts were detected. At 2 M salt concentrations, in simulations on the ∼200 ns time scale of both NaCl and KCl, microcrystals are clearly evident (at a level greater than that observed in Figure S10b in the Supporting Information, particularly for KCl).
Finally, although the Jensen and Jorgensen parameters map out the entire monovalent ion series, they are targeted for solvent boundary simulations or PBC simulations without Ewald and geometric mean combining rules. As the results will likely be considerably different with an Ewald treatment of the electrostatics24,76 or with Lorentz−Berthelot combining rules,62,90–93 we argue that these parameters will not be compatible with many of the common biomolecular force fields, such as those from AMBER(81) and CHARMM.(120)Despite strong concerns raised by Jorgensen and others about possible artifacts from Ewald simulation, specifically those related to the applied periodicity and the surface boundary conditions in the limit of the summation, we believe that Ewald treatments are the most appropriate, particularly for highly charged polyelectrolytes such as nucleic acids. These limitations of the Ewald potential deserve more careful scrutiny and are discussed in the next section. To optimize their ion parameters, Jensen and Jorgensen focused on the interactions of the ions with water including the ion−water RDFs and the ion hydration free energies. Similar to other work, the ε values of the ions were fixed to specific values in an ad hoc manner.
How Significant Are the Artifacts from an Ewald Treatment of the Electrostatics?
The most commonly discussed artifact of an Ewald treatment relates to the imposition of true periodicity; the obvious worry is that the periodic images will inhibit motion and perturb the structure. Two simple model systems clearly display the artifact. The first is two ions at half a periodic box length separation. If the periodicity is not imposed, the ions will be attracted or repulsed depending on the sign of the charges. However, in a truly periodic system, intuition suggests that effectively, there will be no net force on the ions because of the interaction with the periodic images. The second model system involves the free rotation of a dipole in a periodic box; clearly, the periodic images of the dipole will tend to perturb the free rotation compared to that of an isolated dipole. The significance of these artifacts was reinforced by a paper, that was later retracted, that claimed that Ewald methods inhibit translational motion.(121) Notwithstanding, for systems with a sufficiently high dielectric, such as in water, periodicity does not seem to drastically perturb ion interactions122–124 nor inhibit rotation of dipoles.(125) Moreover, despite considerable investigation of the implications of true periodicity, the structure and motional artifacts appear small.59,60,126–128 Where large artifacts are predicted from continuum calculations,(129) in practice, during solvated MD simulation, such structural perturbation is not observed.(130) Additionally, the effect of true periodicity appears small even in lower dielectric environments, such as membranes, where perturbation of the structure and dynamics is observed to far less of an extent than what is seen with truncated or smoothed cutoffs or incorrect representation of long-range electrostatics.131–133
A second perceived shortcoming of Ewald simulations relates to the boundary conditions in the limit of the summation and the convergence of the Ewald sum.(134) Notably, the commonly employed Ewald construction turns a conditionally convergent series—a series the value in the limit of which depends on how the summation is performed—into a sum of two absolutely converging series (the direct and reciprocal sums). The conditionally convergent slop falls into an additional term that is related to the net-dipole of the unit cell.(134) If the boundary around the infinite crystal in the summation limit is assumed to have an infinite dielectric constant, this term disappears. These are the so-called tin-foil boundary conditions employed by most of the biomolecular simulation packages. Alternative boundary conditions in the summation limit have been investigated by a few groups to assess if the results are drastically altered and to determine whether the infinite crystal should be surrounded by a dielectric of 1, a dielectric closer to the dielectric of the system, or an infinite dielectric. Although the long-range dipole correlations can be minimized by incorporating boundary conditions closer to the simulation dielectric,135–138 this approach has not been widely adopted by the community. The reasons for this likely center on the distinct lack of significant periodicity or boundary artifacts elucidated in the many simulations applying an Ewald potential performed to date, including those with different treatment of the infinite boundary (for example, comparing tin-foil to dielectric of 1 simulations).
A third and related perceived shortcoming of Ewald simulations relates to energy divergence and the fictitious net-neutralizing plasma that implicitly adds a uniform counterion density to prevent the energy of net-charged systems from diverging. Although it is intuitively expected that the Ewald energies will diverge for a system with a net charge, in practice, this does not occur. The reason this does not occur is often attributed to the addition of an artificial net-neutralizing plasma; however, in practice, no special code or term is added to handle systems with a net charge differently. The origin of the plasma is in the standard Ewald construction that omits the zeroth-order term in the reciprocal space contribution. Regardless and although the energies should in principle diverge for a periodic system with a net charge, the forces do not, and hence, the dynamics are proper. In practice, essentially all Ewald implementations omit this zeroth-order term, and therefore, these Ewald implementations work just fine for a system with a net charge. Additionally, some of the programs have experimented with added code that smears the net charge (by subtracting the total charge divided by the number of atoms from each atom); however, this is a more drastic approximation, particularly for small systems. That systems with a net charge can be properly represented is demonstrated by free-energy simulations where the charge state is being changed. In fact, a large series of ionic free-energy studies demonstrate that even in very small systems, such as an ion with only 64 waters in the periodic unit cell, good estimates of the charging free energy can be obtained.76,137,139–145 These free-energy studies on rather small hydrated ion systems also suggest that the imposition of true periodicity does not lead to serious artifact in simulations.
II. Computational Methods and Data Sources
In this work, we have developed a set of monovalent ion parameters across the series of alkali cations and halide anions that are directly applicable to simulations in explicit solvent (TIP3P, TIP4PEW, or SPC/E) with Ewald treatments of the electrostatics. Alkali metal ions and halide ions investigated include Li+, Na+, K+, Rb+, Cs+, F−, Cl−, Br−, and I−. A significant challenge was choosing the appropriate target values to guide the parametrization.
Solution Phase Properties of the Ions: Free Energies of Ionic Hydration
Because it is practically impossible to separately measure the independent contributions of the cations and anions in experiment, choices among the existing estimates of the ion hydration free energies need to be made. Numerous groups have reported experimental single-ion hydration free-energy values each based on different approaches to decompose the neutral pair free energies (or other estimates) into single-ion components.95,115–118,146–152 These approaches and the underlying assumptions are very nicely summarized by Grossfield et al.(118) The extrathermodynamic approaches are based on simplifying assumptions about reference salts or proton solvation.The other class of investigations are based on either Born equation/radii estimations(146) or cluster-pair approximations that couple experimental free energies of small ion−water clusters with estimates of the free energy of hydration for a proton.115,149 None of these approaches can be fully validated or assessed because we lack the means to verify the choices experimentally. Moreover, each approach has its own set of limitations; however, recent work provides compelling evidence of the reliability and generality of the cluster-pair approximation approach.152,153 Effectively, the free energies of ionic solvation mostly agree among these methods, with the exception of an approximately constant offset in the absolute values. As discussed by Lamoureux and Roux,(95) the hydration free energies fall into two groups with a constant ∼12−15 kcal/mol difference in the overall free energies between the two groups (with the cations ∼12−15 kcal/mol more negative and the anions ∼12−15 kcal/mol less negative). Although a complete consensus regarding the differences has not emerged, it has been suggested that the difference comes from the presence or the absence of a water surface potential associated with the vacuum/liquid interface.95,118,151 Essentially, the hydration free energies from the cluster-pair approximation include this surface potential,95,115,149,152 whereas some of the other estimates do not.116,117,148 As this offset is largely determined by the estimation of the hydration free energy of the proton, the relative free energies among the monovalent cations or among the monovalent anions are mostly consistent across approaches. For example, excluding the choice of surface potential, on the basis of a similar summation of the proton and hydroxide free energies, the hydration free energies of Schmid et al.(117) are consistent within 0.5 kcal/mol with those of Tissandier et al.,(115) Zhan and Dixon,(154) and Grossfield et al.(118) Comparing the recent values from Kelly et al.,(152) the difference from Schmid is larger because of a different interpretation of the proton and hydroxide solvation free energies (∼3.7 kcal/mol); however, if a −14.6 kcal/mol offset is applied to the Schmid values, the rms deviation from the Kelly et al. values is only 0.3 kcal/mol. An outlier set is the Marcus values,116,148 because these values are based on an alternative summation model for the proton and hydroxide free energies.(118) Effectively, if considerations about the exact value of the surface potential (or offsets between the cation and anion free energy values) are omitted, only two sets of values are found, either those consistent with Schmid et al.(117) or those of Marcus.(116)
If we take as a given that these are the two sets to target for hydration free energies (ΔGhydration), the next question centers on the value of this surface or phase potential and whether it needs to be explicitly included in the estimation of the free energies of the isolated ions. Clearly, for a finite system, such as the spherical solvent boundary potential(96) or SCAS model,(63) there is a clear water/vacuum interface, and this extra offset needs to be included.(95) However, in an infinitely periodic representation, such as Ewald with tin-foil boundary conditions, there is no clear vacuum/water interface. Effectively, within the context of an Ewald treatment as used here, it is most consistent to target directly the Schmid et al.(117) and Marcus(116) values because these values do not include this vacuum/water surface/interface potential. Also, consistent with Schmid and standard practice with computer simulations, our reference state is chosen to be a 1 M gas-phase standard state. To compare with experimental estimates that transfer ions from the gas to the liquid phase with standard states of 1 atm and 1 M, respectively, 1.9 kcal/mol should be added to the single-ion hydration free energies.(118)
On the basis of the arguments listed above, we limited our scope to investigation of the hydration free energies (ΔGhydration) from Schmid et al.(117) and those of Marcus.(116) These are both relatively recent, and the numbers from both sources share common themes, most notably that the differences of hydration free energy of any two cations or anions are in agreement. These results also do not include a water/vacuum interface potential, consistent with our Ewald estimation of the free energy of ionic hydration. The main difference between the two sources lies in the hydration free energies of the anions. The hydration free energies of the Schmid anions are, on average, ∼8 kcal/mol more negative than those of the Marcus anions. As discussed in greater details in the section, our final ion parametrizations are based on the Schmid values because these led to a more consistent set of parameters across all of the experimental data considered, specifically the LC and LE. This is consistent with findings by other groups.(118)
Crystal-Phase Properties of the Ions: LE and LC
Experimentally, every combination of the alkali ions and the halide ions can form salt crystals, although each salt has a different solubility in water. The crystal structures of most of alkali halides are of the NaCl type, which is a simple cubic structure. All the alkali−halide crystals are of this type, except for CsCl, CsBr, and CsI, which have the structure of a body-centered cubic lattice (or CsCl type).
There are two important properties of the salt crystals which are amenable to experimental measurement: the LE and the LC. The LE is defined as the loss in the total lattice potential energy when the crystal structures transform into the gas phase. These values can be calculated via two independent approaches. The first is via the theoretical methods applied by Huggins155–157 and Kapustinskii.(158) The alternative means is to calculate the LE by using a traditional Born−Fajans−Haber cycle,159–161 which combines together various empirical results. Both the theoretical and empirical (or experimental) values are listed in the CRC Handbook.(162) They are referred to as LE (theory) and LE (expt) and are shown here in Table 2. The LE (expt) is on average ∼4.8 kcal/mol larger than LE (theory). The LCs (Table 3) on the other hand, fall into a very narrow range of values and are available from many sources.108,163,164 The values listed in the first column of Table 1.2 from Sirdeshmukh et al.(164) were used in this research. The LC is the length of the edge of the (ideal) unit crystals. Because all the alkali halide crystals are isometric, LC can be easily converted into interionic distance.
Table 2. LE (kcal/mol) of Crystals of Alkaline Halide Salts as Obtained from the CRC Handbook,(162) Including both the Theoretical LEs Estimated from the Kapustinskii Equation (Top Half) and the Empirical/Experimental LEs Calculated from a Born−Fajans−Haber Cycle (Bottom Half).
| Li | Na | K | Rb | Cs | |
|---|---|---|---|---|---|
| Theory | |||||
| F | 246.2 | 217.5 | 193.1 | 185.0 | 177.8 |
| Cl | 199.3 | 183.8 | 167.5 | 162.5 | 157.0 |
| Br | 188.3 | 175.0 | 160.4 | 155.6 | 151.1 |
| I | 174.5 | 163.0 | 151.1 | 147.5 | 143.4 |
| Experiment | |||||
| F | 250.7 | 222.3 | 198.1 | 190.0 | 181.4 |
| Cl | 206.5 | 188.8 | 172.1 | 166.1 | 160.1 |
| Br | 196.0 | 180.2 | 165.2 | 159.7 | 154.6 |
| I | 182.6 | 168.5 | 155.4 | 151.1 | 146.5 |
Table 3. LC (Angstroms) of Crystals of Alkaline Halide Salts (Top Half) and Interionic Distance (Angstroms) Calculated from the LC at Room Temperature (Bottom Half)a.
| Li | Na | K | Rb | Cs | |
|---|---|---|---|---|---|
| LC | |||||
| F | 4.027 | 4.634 | 5.347 | 5.652 | 6.014 |
| Cl | 5.140 | 5.640 | 6.293 | 6.581 | 4.123 |
| Br | 5.501 | 5.977 | 6.600 | 6.889 | 4.295 |
| I | 6.023 | 6.473 | 7.066 | 7.342 | 4.568 |
| Distance | |||||
| F | 2.014 | 2.317 | 2.674 | 2.826 | 3.007 |
| Cl | 2.570 | 2.820 | 3.147 | 3.291 | 3.571 |
| Br | 2.751 | 2.989 | 3.300 | 3.445 | 3.720 |
| I | 3.011 | 3.236 | 3.533 | 3.671 | 3.956 |
The data were obtained from the first column of Table 1.2 from Sirdeshmukh et al.(164)
Simulation Conditions
All of the simulations were performed by using the sander and PMEMD modules of AMBER9.80,81 Unless otherwise mentioned, the nonbonded cutoff distance was 9 Å (with an additional 2 Å buffer for the list of pair interactions with an automatically triggered pair-list update when atoms have moved more than 1 Å between updates and also application a continuum/homogeneous density correction for the long-range van der Waals energies and virials). In cases where the edge of the box was smaller than 22 Å, the pair-list cutoff was set to half of the periodic box size. All of the MD and free energy simulations applied the particle mesh Ewald method74,165 to properly treat the long-range electrostatic interactions with the three different water models. As discussed in the Supporting Information, the results are not terribly sensitive to the van der Waals cutoff or whether a long-range van der Waals correction is applied (see Table S25 in the Supporting Information).
Note that throughout the manuscript, published parameters are referred to by the authors names and were obtained from the following references: Jensen and Jorgensen,(75) Smith, Dang, and Garrett (Li+,(98) Na+,(94) K+,(64) Rb+,(64) Cs+,(99) F−,(98) Cl−,(64) I−(100)), Beglov and Roux,96,97 and Åqvist.(63) Parameters that are normally run with geometric mean combining rules were adapted to work with the AMBER (Lorentz−Berthelot) combining rules. Specifically, for the Jensen and Jorgensen and Åqvist parameters, the van der Waals radii (Rmin) were back calculated to reproduce the water−ion mixing with TIP4P water for the Jensen and Jorgensen and TIP3P water for the Åqvist parameters to match what would be obtained by using geometric mean combining rules. Because σ (in Å) values were reported by Jensen and Jorgensen, the Rmin for each ion was obtained from the relationship that follows, where 1.7699 Å is the Rmin/2 value of TIP4P water:
The Jensen and Jorgensen parameters used were as follows (ion, Rmin/2 (Å), ε (kcal/mol)): (Li+, 1.606984, 0.0005), (Na+, 2.251455, 0.0005), (K+, 2.762419, 0.0005), (Rb+, 2.947136, 0.0005), (Cs+, 3.193406, 0.0005), (F−, 1.711269, 0.71), (Cl−, 2.226677, 0.71), (Br−, 2.353895, 0.71), and (I−, 2.601774, 0.71).
Values of A and B were reported in Åqvist’s publication, and Rmin (Å) and ε (kcal/mol) are related to these values according to the following relationship, where 1.768 Å, 762.89 kcal1/2 mol−1/2 Å6, and 24.39 kcal1/2 mol−1/2 Å3 are Rmin/2, A, and B of TIP3P water.
The Åqvist parameters used in this work as follows (ion, Rmin/2 (Å), ε (kcal/mol)): (Li+, 1.137, 0.0183), (Na+, 1.868, 0.00277), (K+, 2.658, 0.000328), (Rb+, 2.956, 0.00017), and (Cs+, 3.395, 0.0000806).
Hydration Free Energy Calculations
A single ion was solvated in a cubic periodic box with either TIP3P,(71) SPC/E,(72) or TIP4PEW73,77 water molecules. The system was minimized with 1000 cycles by steepest descent and equilibrated in two steps, specifically 40 ps of MD at constant volume and another 40 ps of MD at constant 1 atm pressure. Weak coupling of temperature and pressure was performed with a coupling time of 1 ps.(166) The temperature was maintained at 298K. The number of grids for the reciprocal PME calculation was 2 per Ångstrom but not less than 30 in each dimension, and a cubic B-spline interpolation was used.(74) Tin-foil boundary conditions in the Ewald sum were applied, and net charge was not artificially altered through the addition of explicit counterions or through smearing the average net charge per atom over the whole system. Although the energies in principle will diverge with such a setup, the forces do not. Effectively, the system is neutralized by a fictitious net-neutralizing background; in practice, no additional term or code is included because the neutralization is implicit to the omission of the zeroth-order term in the reciprocal part of the Ewald sum when tin-foil boundary conditions are applied.(134)
To calculate the hydration free energies of the ions, a two-stage thermodynamic integration (TI) approach167,168 was used. In the first stage (charge neutralization), the charge of the ion was slowly neutralized in water, and in the second stage (disappearing), the van der Waals potentials were slowly removed. The TI simulations were performed by using the sander program in AMBER9.(81) The Hamiltonian mixing rule chosen followed equation 2, and the value λ in the equation indicates the mixed thermodynamic state between unperturbed (λ = 0) and perturbed (λ = 1) states. The k in the equation that modifies the mixing rule was set to 1 in the charge-neutralization stage and to 4 in the disappearing stage, consistent with recent work.(169)V0 and V1 refer to the unperturbed and perturbed Hamiltonians. For the nine-window charge-neutralization runs, λ values were set to be 0.00000, 0.11270, 0.24180, 0.37090, 0.50000, 0.62910, 0.75820, 0.88729, and 1.00000. For three-window charge-neutralization TI runs, they were set to be 0.11270, 0.50000, and 0.88729. For four-window charge-neutralization runs, a λ of 0.75820 was added to the previous list. For the nine-window disappearing runs, λ values were set to be same as those of nine-window charge-neutralization runs except that 1.00000 was switched to 0.94365. For the three-window disappearing runs, the windows were set to be 0.11270, 0.50000, and 0.88729. A λ of 0.94365 was added for four-window disappearing runs. The MD for each TI window was carried out for 600 ps, and the average derivative of Hamiltonian was calculated by using the data collected from the last 500 ps. The coupling constant for maintaining constant temperature and pressure was set to 10 ps in these runs. The derivative of the mixed Hamiltonian (V(λ)) with respect to λ was integrated to obtain the free energy difference between the two states. Initially, the derivatives were fit into either a cubic spline or a quadratic equation as a function of λ, and the functions were integrated analytically. As the calculated values were actually the dehydration free energies, the sign was flipped to obtain the hydration free energies.
Mapping out the ΔGhydration = f(Rmin, ε) Hypersurface
Because the potential energies and the hydration free energies of the ions do not depend on the mass of the ion, the free energy surfaces were mapped by assuming the masses of K+ and Cl−. Additionally, because the ion charges are fixed to +1 or −1, the cations (Li+, Na+, K+, Rb+, and Cs+) and the anions (F−, Cl−, Br−, and I−) were grouped separately. The only remaining degrees of freedom are the choice of water model and the ion’s van der Waals radii (Rmin) and well depths (ε). Three separate water models (TIP3P,(71) SPC/E,(72) or TIP4PEW73,77) were investigated, separately, for both the cation and anion ΔGhydration = f(Rmin, ε) hypersurfaces, for a total of six sets of simulations generating six hypersurfaces.
For each set of the simulations, the ε values of the LJ potential were sampled in the range of 0.00001−1 kcal/mol. The energy was expressed in a logarithmic scale, and the ε values were chosen to be evenly distributed across the entire scale. The Rmin values were chosen in the range of 1.2−6.4 Å for the cations and 3.2−9.0 Å for the anions, sampling at 0.2 Å intervals. Over the entire range of Rmin and ε values sampled, the free energies of hydration were calculated, as discussed previously. To map each hypersurface—specifically to fit a function that maps ΔGhydration = f(Rmin, ε)—the calculated hydration free energies (ΔG) were fit to a bicubic equation of either Rmin and εn or 1/Rmin and εn, where n is a constant to be fit with 16 coefficients for each of the terms in the equation. As a variable of the bicubic plane, εn was chosen instead of ε because it significantly reduced the deviation. The choice of the bicubic equation was made empirically. In general, although a higher-order bipolynomial plane could represent the data with lower deviations, these fits tended to generate more wiggles and larger excursions from the true values. On the other hand, a lower-order plane was found to be insufficient because of higher deviations and poorer fits to the data. Note that in the bicubic plane representation chosen, each variable has one inversion point and can represent two convexities. The bicubic plane fit proved to be robust with overall deviations in an acceptable range as is discussed in greater detail in the section.
In order to discover the 17 unknowns (the 16 coefficients and exponent n), n was assumed to be either positive or negative with optimal values confined in the range of −1.0 to 1.0. To sample different values of n, n was thoroughly scanned throughout this range, and at each fixed n value, the other 16 coefficients were determined through a least-squares fit. The parameters at the rms minimum were chosen as the best-fit parameters. All of the fitting was performed with a custom-designed Perl script. The final values of the fit are provided in the Supporting Information, Table S24.
MD Simulations of Infinitely Periodic Salt Crystals
Cubic CsCl-type crystals (CsCl, CsBr, and CsI) and NaCl-type crystals (all of the other ion pair combinations) were each built in a square periodic lattice with the appropriate packing. The CsCl-type crystals had 1024 explicit atoms, and the NaCl-type crystals had 1000 explicit atoms in the periodic lattice. NTP simulation was performed at constant 1 atm pressure at 300K (with weak-coupling times of 1 ps(166)). Each system was subjected to MD for 50 ps, and the last 30 ps were used to calculate the LC. Crystal stability was confirmed by measuring 12−14 different distances between adjacent cations and anions in the center of the crystals. If the standard deviation of these measurements was larger than 0.4 Å, the crystal was considered to be an unstable crystal. Because the number of the atoms in the periodic box is specified and the geometry of the crystals is known, the average LC of the crystals can be obtained from the average volume of the system. Specifically, LC = (length of the edge of unit periodic box)/(the number of unit crystals per axis) = (volume1/3)/(the number of unit crystals per axis). The optimal distance between a cation and an anion is calculated by multiplying either 1/2 (for NaCl-type crystal) or √3/2 (for CsCl-type crystal).
To calculate the LE, perfect cubic crystals made of either 1000 atoms (NaCl type) or 1024 atoms (CsCl type) were built according to a specified LC. As the LC increases from zero, the potential energies decrease and eventually pass through a minimum. This minimum potential energy was taken as the LE. Both the LC and the LE were fit into a bicubic spline with respect to the Rmin of a cation and the Rmin of an anion. To verify the absence of any size dependency on the calculated energies, LE were also calculated with a double-sized crystal (in each direction or ∼8000 atoms), and the change in potential energy of the system was found to be less than 0.1 kcal/mol.
Self-Consistent Fitting of the Ion Radii (Rmin) across the Monovalent Series
By using the ΔGhydration = f(Rmin, ε) relationship, given the choice of a specific hydration free energy for each ion, the ε values are determined by the choice of Rmin. Given this, the next stage is to determine the set of ion radii (the Rmin values of the anions and cations) across the entire monovalent ion series. The cation−anion distance (which is determined by the LC) and the LE of a crystal were interpolated by using the bicubic spline fitting method described previously; this used the Perl/PDL module wrapping the GNU Scientific Library.(170) To find the optimized Rmin values, eq 3 was minimized. In eq 3, F is the overall deviation and is unit-less because multiplication by the weighting factors was chosen to give unit-less numbers. δdis is the deviation from the literature value of the cation−anion distance, δLE is the deviation from the literature value of LE, Wdis is the weight on cation−anion distance, and WLE is the weight on LE. To minimize this equation, an arbitrary Rmin was assigned to each ion at the beginning of an optimization round. Then, the Rmin of one of the ions was changed by a random step size increment of 0.001 Å, and the new Rmin was determined. This process was repeated until F no longer decreased. Next, the Rmin of the other ions were changed, one by one in the same manner, until the lowest F was found. At this point, the random step size increment was reduced by half, and the same process was iterated until the random step size increment dropped below 10−12 Å.
During this process, restraints were applied to prevent ions from crossing the boundaries into the range where the crystals became unstable (as described in the previous section). To apply the restraints, the outermost boundary of the stable crystal was simplified as a quadratic curve, and Rmin was not allowed to change when crossing the boundary (between the shaded and white regions of Figure 5).
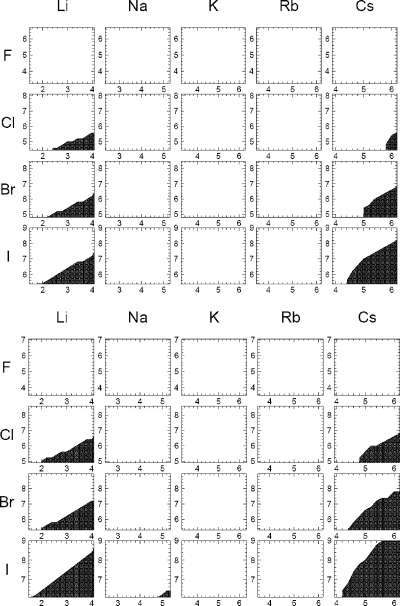
Figure 5.
Crystal instability (shaded) as a function of the van der Waals radii (Rmin) for each of the ion pairs. Some of the model crystals did not maintain their crystal structure during short MD simulation. This depends on the Rmin of each of the ion components scanned at 0.2 Å intervals, shown on the x-axis for the cations and the y-axis for the anions (in Å). For each Rmin pair, the corresponding ε values were chosen on the basis of the ΔG = f(Rmin, ε) hypersurfaces mapped (see Figures 3 and 4). The shaded areas indicate the regions where the crystals were unstable. Note that the resolution of the plot (at 0.2 Å steps in Rmin) should be considered accordingly. The regions of unstable crystal varied depending on water model and target hydration free energies. The top plots are with the parameters optimized to target the Schmid hydration free energy values, and the bottom plots target the Marcus values, both in TIP3P water. The figures for the TIP4PEW and SPC/E water models are shown in the Supporting Information.
Calculating the First Peak of the Ion−Water RDF
A single ion was solvated in a periodic box of water (TIP3P, 448 water molecules; TIP4PEW, 436 water molecules; or SPC/E, 439 water molecules), and the system was minimized for 1000 cycles by steepest descent. MD simulation was then performed at 298K for 10 ps at constant volume, followed by 30 ps at constant pressure (1 atm). After this initial equilibration, MD simulation was performed for 2 ns with trajectory snapshots saved every 0.5 ps. The radial density profile of the oxygen atoms of the water molecules as a function of the distance from the ion was measured at 0.01 Å intervals. After acquiring the density profile, the densities around the peak of the first water shell within the range of ±0.1 Å were fit into a quadratic equation with respect to the ion−oxygen distance. Then, the profiles around the vertex of the fit quadratic equation within the range of ±0.1 Å were fit into another quadratic equation. This last step was repeated until the ion−oxygen distance of the vertex of the fit quadratic equation did not change. This final distance was considered to be the radius of the first water shell.
Calculating the Binding Energy with Water Molecules
Models of single ions interacting with one or more water molecules were built and minimized in vacuo. Because the potential energy of either a single isolated ion or a single isolated water molecule (with the rigid water models used) is zero, the potential energy of the combined system is equivalent to the binding energy. For the cations and the anions with a single water molecule, C2v structures and Cs structures, respectively, represent the global minimum structures. In addition to these basic geometries, various other ion−water structures were built (see Figures 1 and 2). In the figures, the indices include two numbers (e.g., x + y) which indicate the number of water molecules in the first and second hydration shells, respectively. Some of these indices have the letter h next to them, which refers to a halfway or mixed interaction of the water molecules with both the first and the second hydration shells. Unfortunately, because the default minimization algorithms in AMBER are unstable, dated, and limited, the sander program could not accurately minimize the structures with fixed water bond distances (i.e., SHAKE(171)). Therefore, the energy minimization was performed with a home-built Perl script validated to match the AMBER energies.
Figure 1.
Molecular graphic visualizations of the optimized structures of alkali cation interactions with various numbers of water molecules. The cation is shown in blue, and the waters are shown in red (oxygen) and white (hydrogens). The molecular graphics are drawn with perspective.
Figure 2.
Molecular graphic visualizations of the optimized structures of halide anion interactions with various numbers of water molecules. The anion is shown in cyan, and the waters are shown in red (oxygen) and white (hydrogens). The molecular graphics are drawn with perspective.
III. Results and Discussion
Calculating the Hydration Free Energies of Monovalent Cation and Anion Models
A very large series of TI runs were necessary to scan the (Rmin, ε) free energy surface. In order to optimize the conditions and to minimize the computational burden, a series of calculations were performed with different sizes of the periodic box, different numbers of windows, and different integration methods for the free energy derivatives. For the tests, the TI calculations were performed by using the Åqvist Na+ and K+ and Dang Cl− parameters.(99) Tables S1−S3 in the Supporting Information show the calculated hydration free energies for these test calculations. For the charging free energy calculations, because the rms deviation for the less computationally demanding three-window TI run was only 0.07 kcal/mol from the inherently more accurate nine-window runs, the three-window protocol was applied. For disappearing of the van der Waals radii, the four-window protocol was sufficient to keep the rms deviation below 0.06 kcal/mol, compared to eight-window cubic spline integration. Together, the three-window charging and four-window disappearing protocols, with an overall deviation of 0.10 kcal/mol compared to the results of the nine-window cubic spline calculation, were applied in the large-scale sampling of the (Rmin, ε) space. Consistent with intuition and published results,140,143 as the number of water molecules in the system increases, the free energies converge. Because the values do not change significantly beyond 400 waters, 456 water molecules were used in the cation TI calculations, and 445 water molecules were used in the anions TI calculations; these smaller system sizes and less accurate integration of the free energies provide a small trade off in accuracy compared to significantly larger computational costs required for larger boxes and more free energy windows. Additionally, the results are not overly sensitive to the choice of van der Waals cutoff in the 8−10 Å range or whether a continuum energy and virial correction for the long-range van der Waals was included (see Supporting Information, Table S25).
To map out the ΔGhydration = f(Rmin, ε) surface, the total number of TI calculations performed was 276 for the cations (alkali ions) in TIP3P water, 298 for the anions (halide ions) in TIP3P water, 256 for the cations in TIP4PEW water, 240 for the anions in TIP4PEW water, 260 for the cations in SPC/E water, and 228 for the anions in SPC/E water. The different counts relate to differences in the crystal stability with each water model. The data obtained was fit into the bicubic equations as described in the section. The rms deviations of the hydration free energies after the fit to the calculated data were 0.27 kcal/mol for the cations in TIP3P water, 0.26 kcal/mol for the anions in TIP3P water, 0.28 kcal/mol for the cations in TIP4PEW water, 0.31 kcal/mol for anions in TIP4PEW water, 0.23 kcal/mol for cations in SPC/E, and 0.27 kcal/mol for anions in SPC/E. The fits are valid under the following conditions:
0.00001 kcal/mol < ε < 1 kcal/mol
1.2 Å < Rmin (cation/TIP3P) < 6.2 Å
3.2 Å < Rmin (anion/TIP3P) < 9.0 Å
1.2 Å < Rmin (cation/TIP4PEW) < 6.2 Å
3.4 Å < Rmin (anion/TIP4PEW) < 9.0 Å
1.2 Å < Rmin (cation/SPC/E) < 6.4 Å
3.2 Å < Rmin (anion/ SPC/E) < 9.0 Å
−114 kcal/mol < ΔGhydration (cation) < −59 kcal/mol
−120 kcal/mol < ΔGhydration (anion) < −65 kcal/mol
Correlating Rmin and ε
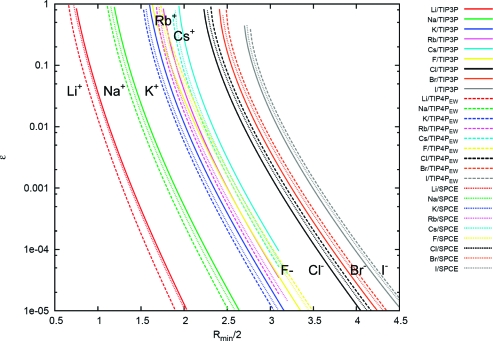
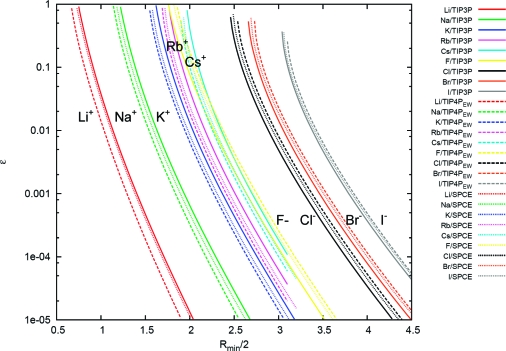
Careful observation of the surface mapping the hydration free energies to Rmin and ε shows that the hydration free energy increases as Rmin decreases and ε decreases. However, exceptions are found when ε is high (around 1 kcal/mol or higher). This occurs mainly with the anions. Likely, similar exceptions will be found with the cations but over a range of ε higher than what was sampled in our calculations. When this occurs, the hydration free energy starts to decrease as ε decreases. This effect is due to the contributions from the van der Waals-disappearing stage where the free energies become negative, implying that the charge-free (or inert) ions become soluble (rather than hydrophobic). This behavior destroys the 1:1 mapping of Rmin and ε because multiple values of ε are possible in that range that reproduce a given hydration free energy. To avoid this, these unrealistic regions of the ion potential should be excluded. Fortunately, over most of the range of Rmin and ε, the hydration free energies followed the pattern described initially, and it was possible to provide the 1:1 mapping between (Rmin, ε) and ΔG. This behavior, where ε decreases monotonically as Rmin increases, is shown in Figures 3–4. As the results show, when the unrealistic regions are omitted, ε is completely dependent on Rmin for a given hydration free energy. Notwithstanding shifts in the curves depending on the choice of water model, the shapes of the correlation curves for a given ion were very consistent across all the water models. Overall and weakly dependent on Rmin, there is approximately a 4 kcal/mol discrepancy between the hydration free energies calculated with the TIP3P and with the TIP4PEW water models. Essentially, cations are more soluble in TIP3P water than in TIP4PEW water, whereas anions are more soluble in TIP4PEW water. The SPC/E water model led to hydration free energies between the TIP3P and TIP4PEW values. These shifts very likely will also occur with other water models and suggest the impossibility of generating a simple set of ion LJ parameters that are truly water-model independent (consistent with recent work of Jensen).(78) Note also, as shown in the Supporting Information, that the TIP5P results differ considerably and more drastically than the three water models shown here.
Figure 3.
Correlations between Rmin and ε for a given free energy of hydration by using the Schmid values. Shown are the correlations in Rmin and ε from the ΔGhydration = f(Rmin, ε) hypersurface at a given value of the hydration free energies. The chosen values are those from Schmid and co-workers (see Table 10).(117) Points on the same line have equivalent hydration free energies. The solid lines are for TIP3P water, the dashed lines are for TIP4PEW water, and the dotted lines are for SPC/E water; each ion is represented by a different color. The units for the x and y axes are Angstroms and kcal/mol, respectively.
Figure 4.
Correlations between Rmin and ε for a given free energy of hydration by using the Marcus values. Shown are the correlations in Rmin and ε from the ΔGhydration = f(Rmin, ε) hypersurface at a given value of the hydration free energies. The chosen values are those from Marcus (see Table 10).(116) Points on the same line have equivalent hydration free energies, and each ion is denoted by a different color. The solid lines are for TIP3P water, the dashed lines are for TIP4PEW water, and the dotted lines are for SPC/E water. The units for the x and y axes are Angstroms and kcal/mol, respectively.
Calculation of LC and LE
A rather wide range of parameters could be chosen for the alkali−halide crystals that correlate the specific Rmin values with the properties of the individual cations and anions. In the past, this has involved fixing ε to particular specific values followed by choosing Rmin to satisfy first peaks of RDFs, ion−water energies, or experimental measurements of the free energy of hydration. However, as the previous section shows, there are many combinations of Rmin and ε that reproduce a given ΔGhydration. In order to choose the appropriate set, we attempted to fit not only the solution properties but also the crystal properties. Every pair combination of the alkali and halide ions can crystallize, and many different properties of these crystals are experimentally known. By using this information, we explored whether we could self-consistently determine the Rmin values for each of the respective ions. With a specific choice of Rmin and the experimental ΔGhydration, ε is determined.
To do this, we initially investigated two very important properties of the crystals, specifically the LC, or equivalently the cation−anion distances in crystals, and the LE. Although both are distinct properties of the crystal, they are obtained from fairly distinct experimental methods. Because both are directly related to the ionicity of the ions in water, they provide independent measures of the crystal. Moreover, the values of these properties are in good agreement. To calculate LC and LE for interpolation, crystals every 0.2 Å of Rmin for both the cations and anions were built, and the LC and LE were calculated. These values were then fit into bicubic spline surfaces with respect to Rmin for each of the ions. Acceptable ranges of Rmin were limited by the results of the calculation of hydration free energies described above. However, the Rmin of some anions were also further limited by another factor: below a certain range of Rmin, the target hydration free energy of an ion was unachievable as illustrated in Figures 3 and 4.
Stability of the Crystals
Some of the crystals that were model-built to calculate the LC were not stable in MD simulation. In these cases, the crystals rapidly collapsed, usually immediately, as the MD was started. The distortion was readily evident and distinct from the behavior observed with stable crystals. The distortion tended to occur when there was a significant difference in the size of the two counterions, for example, with lithium iodide (with the NaCl-type crystals), or when the crystals were of the CsCl type. The size boundaries for the stable crystals were very distinctive, and any crystals beyond this boundary were unstable. This is shown for the TIP3P water model derived ion parameters in Figure 5. Because crystals located in the unstable regions cannot maintain their natural crystal form, these regions were avoided. Figures showing the crystal instability for the other water models are displayed in the Supporting Information.
Balancing the Crystal LC and LE
In order to determine the proper choice of a van der Waals radius (Rmin) for a given ΔGhydration, the calculated LC and LE of the different crystals across the monovalent ion series were used. Although ideal ion models should ultimately be able to reproduce both properties with low deviations, the simplified nonbonded model employed here has clear limitations. Specifically, the ion models cannot simultaneously reproduce both the LC and LE with low deviation. To better understand the balance, a series of weighting factors on the interionic distances (which are a surrogate property of the LC) and LE were investigated. Weighting ratios on the interionic distance and LE were applied in the range from 0:1 to 300:1. The first number in the ratio denotes the weight on interionic distance in units of Å−1, and the second number is the weight on the LE in units of kcal−1 mol. At each ratio across the range, the self-consistent fitting was repeated more than 50 times with random initial Rmin values of the ions. A set of local minimum solutions was consistently obtained at each ratio; however, some of these solutions had obvious problems. Specifically, (1) the size consistency of Rmin for the monovalent series was not always maintained (meaning that ions known to be larger than others became smaller than the others, both in the cation and the anion series), (2) the solutions fell right on the border of the stable and unstable crystal regions, suggesting that the solutions were forced to the stable regions, and (3) the Rmin of some of the ions reached beyond either their minimum or maximum value, values that are considerably out of the range of expectations. When these poor solutions were discarded, only a few reasonable solutions were found at each ratio.
In general, the F (or fit) in eq 3 was always larger (by ∼30% or worse) when the LE (theory) values were used instead of the LE (expt) values (see Table 2). This suggests that the LE (theory) values are less consistent. On the basis of this observation, we used the LE (expt) values in subsequent fits. At this stage, we also had concerns regarding the Rmin values for the larger ions, specifically Cs+ and I−. With these large, soft, and polarizable ions, it was clear that the simple nonpolarizable LJ model would be stretched to its breaking point. A concern was that the large ions, when interacting with solvent, would lack sensitivity with respect to small changes in ion size. This could limit the accuracy during parametrization of the large ions. To better understand the size dependence and the interaction of the ions with water, the binding energies and the ion−oxygen distances of the individual ions with a single water molecule (specifically 1 + 0(C2v) structures for the cations and 1 + 0(Cs) and 1 + 0(C2v) structures for the anions) were measured at their minimized structures. The deviations from the references of ab initio calculated values (see Tables 6, 7, and Tables S6−S23 in the Supporting Information) are plotted in Figures 6 and S7 in the Supporting Information. Overall, the plots show some expected trends but also some surprises. Looking at the rms (energy), greater deviation in the cation energy is seen with the SPC/E water compared to TIP3P or TIP4PEW. This is consistent with the parametrizations because SPC/E gives up accurate water−water interaction energy in order to provide better diffusive properties, whereas the TIP3P model gets the water−water interaction energy correct at the expense of diffusion which is ∼2 times faster than that in experiment. TIP4PEW, with its extra point, is able to reproduce both the water−water interaction energy and diffusion constant.
Table 6. Comparison of the TIP3P Single Water−Ion Binding Distances for Various Ion Parameter Setsa.
| structures | thisresearch | Jensen andJorgensen | Smith−Dang−Garrett | Beglovand Roux | Åqvist | referencevalues | |
|---|---|---|---|---|---|---|---|
| Li+ | 1 + 0(C2v) | 1.90 | 1.96 | 1.91 | 1.95 | 1.86172,173 | |
| Na+ | 1 + 0(C2v) | 2.29 | 2.41 | 2.29 | 2.22 | 2.32 | 2.23(174) |
| K+ | 1 + 0(C2v) | 2.66 | 2.77 | 2.73 | 2.62 | 2.64 | 2.64(175) |
| Rb+ | 1 + 0(C2v) | 2.82 | 2.90 | 2.83 | 2.76 | 2.80(176) | |
| Cs+ | 1 + 0(C2v) | 3.00 | 3.08 | 3.00 | 2.96 | 2.96(177) | |
| F- | 1 + 0(Cs) | 2.56 | 2.73 | 2.64 | 2.45178,179 | ||
| 1 + 0(C2v) | 2.68 | 2.82 | 2.74 | 2.61(178) | |||
| Cl- | 1 + 0(Cs) | 3.09 | 3.26 | 3.21 | 3.08 | 3.12178,180 | |
| 1 + 0(C2v) | 3.15 | 3.30 | 3.26 | 3.14 | 3.20(178) | ||
| Br- | 1 + 0(Cs) | 3.26 | 3.38 | 3.32(178) | |||
| 1 + 0(C2v) | 3.30 | 3.42 | 3.35(178) | ||||
| I- | 1 + 0(Cs) | 3.48 | 3.64 | 3.62 | 3.61178,181 | ||
| 1 + 0(C2v) | 3.51 | 3.65 | 3.64 | 3.61178,181 | |||
| rms | 0.06 | 0.14 | 0.08 | 0.03 | 0.06 |
Shown are the single water oxygen−ion distances for various geometries (as shown in Figures 1 and 2) for distinct ion parameter sets with TIP3P water. The reference values are averages of one or more ab initio calculations. The distances have units of Å. The levels of QM theory applied for each reference cited are as follows: ref (172), HF/6−31++G(d,p); ref (173), MP2/aug-cc-pVDZ; ref (174), HF/TZ2P; ref (175), MP2/TZ2P; ref (176), MP2/aVDZ; ref (177),;MP2/aVTZ ref (178), MP2/6−311++G**; ref (179), MP2/aug-cc-pVDZ; ref (180), MP2/aug-cc-pVDZ; ref (181), MP2/aug-cc-pVDZ+diff
Table 7. Comparison of the TIP4PEW Single Water−Ion Binding Distances for Various Ion Parameter Setsa.
| structures | thisresearch | Jensen andJorgensen | Smith−Dang−Garrett | Beglovand Roux | Åqvist | referencevalues | |
|---|---|---|---|---|---|---|---|
| Li+ | 1 + 0(C2v) | 1.86 | 1.98 | 1.93 | 1.97 | 1.86172,173 | |
| Na+ | 1 + 0(C2v) | 2.25 | 2.43 | 2.31 | 2.24 | 2.34 | 2.23(174) |
| K+ | 1 + 0(C2v) | 2.63 | 2.79 | 2.76 | 2.65 | 2.66 | 2.64(175) |
| Rb+ | 1 + 0(C2v) | 2.78 | 2.92 | 2.85 | 2.78 | 2.80(176) | |
| Cs+ | 1 + 0(C2v) | 2.93 | 3.10 | 3.03 | 2.98 | 2.96(177) | |
| F- | 1 + 0(Cs) | 2.63 | 2.72 | 2.62 | 2.45178,179 | ||
| 1 + 0(C2v) | 2.80 | 2.86 | 2.77 | 2.61(178) | |||
| Cl- | 1 + 0(Cs) | 3.13 | 3.24 | 3.19 | 3.06 | 3.12178,180 | |
| 1 + 0(C2v) | 3.23 | 3.33 | 3.29 | 3.17 | 3.20(178) | ||
| Br- | 1 + 0(Cs) | 3.29 | 3.37 | 3.32(178) | |||
| 1 + 0(C2v) | 3.38 | 3.45 | 3.35(178) | ||||
| I- | 1 + 0(Cs) | 3.51 | 3.63 | 3.61 | 3.61178,181 | ||
| 1 + 0(C2v) | 3.58 | 3.68 | 3.67 | 3.61178,181 | |||
| rms | 0.07 | 0.15 | 0.09 | 0.03 | 0.07 |
Shown are the single water oxygen−ion distances for various geometries (as shown in Figures 1 and 2) for distinct ion parameter sets with TIP4PEW water. The reference values are averages of one or more ab initio calculations. The distances have units of Å. The levels of QM theory applied for each reference cited are as denoted in the footnote of Table 6.
Figure 6.
rms deviations of ion−water binding energies and ion−oxygen distances. The rms deviations of single water binding energy (left, y axis) and ion−oxygen distance (right, y axis) for the Cs+ and I− ions as a function of ion size (x axis in Å) with each of the water models and both the Schmid and Marcus hydration free energy target values are shown. The deviations for the energies and distances are reported in kcal/mol and Angtroms, respectively. The energy and distance are shown as solid and dotted lines, respectively, the data for Cs+ is shown in black, and the data for I− is shown in red. The composite data is shown as a single plot in Figure S7 in the Supporting Information.
The plots also display a profound sensitivity to the choice of water model and also the experimental hydration free energies used in the fit. For the cations, the results are in general agreement and consistent. The lowest rms for the binding energies occurs when Rmin/2 equals around 2 Å for Cs+, and this trend is consistent with the rms deviations of both the energy and the distance. Because the Schmid and Marcus hydration free energies of the cations are very similar (see Table 10), agreement for the plots of the rms deviations for the cations is expected and observed. However, the results for the anions are distinctly different. Whereas the lowest rms (energy) and rms (distance) for I− fall into a range (Rmin/2 ≈ 2.8 Å) similar to that of the Schmid hydration free energies, independently of the water models, the minimal rms shifts with the Marcus hydration free energies to put the Rmin/2, or van der Waals radii, above 4−4.5 Å. This is unreasonably large. Moreover, with the TIP3P water model, the model fit to the Marcus free energies appears to have a large discrepancy between the optimal Rmin for the energy compared to that for the distance. To prevent parametrization that leads to large ions, the Rmin of Cs+ was restrained to be less than 5.0 Å. With this restraint, no reasonable solution was found for the Marcus hydration free energies with the TIP4PEW water model at any weight ratio. This lack of consistency between the fit of energies and distances with the Marcus model, most strikingly with the better performing TIP4PEW water model, led us to adopt Schmid hydration energies for the final optimization of the Rmin values. In addition, the Rmin of I− was restrained to be less than 7 Å (which, according to Figure 6, is reasonable with the Schmid hydration free energies).
Table 10. Dehydration Free Energies (−ΔGhydration in kcal/mol) of the Alkali and Halide Ions with Sets of Available Ion Parameters in the Three Different Water Modelsa.
| thisresearch | Jensen andJorgensen | Smith−Dang−Garrett | Beglovand Roux | Åqvist | Schmid(117) | Marcus(116) | |
|---|---|---|---|---|---|---|---|
| TIP3P | |||||||
| Li+ | 113.7 | 106.9 | 114.7 | 109.5 | 113.8 | 113.5 | |
| Na+ | 88.7 | 78.8 | 89.4 | 92.2 | 84.2 | 88.7 | 87.2 |
| K+ | 70.7 | 61.5 | 66.9 | 71.3 | 67.0 | 71.2 | 70.5 |
| Rb+ | 65.7 | 56.4 | 63.5 | 61.4 | 66.0 | 65.7 | |
| Cs+ | 60.6 | 50.2 | 57.2 | 53.9 | 60.5 | 59.8 | |
| F- | 119.7 | 115.8 | 118.9 | 119.7 | 111.1 | ||
| Cl- | 89.6 | 89.2 | 85.9 | 92.3 | 89.1 | 81.3 | |
| Br- | 82.9 | 85.0 | 82.7 | 75.3 | |||
| I- | 74.0 | 77.0 | 70.8 | 74.3 | 65.7 | ||
| rms | 0.3 | 7.2 | 2.7 | 2.8 | 4.9 | ||
| TIP4PEW | |||||||
| Li+ | 113.7 | 101.7 | 109.0 | 103.9 | 113.8 | 113.5 | |
| Na+ | 89.0 | 74.7 | 85.5 | 87.7 | 80.1 | 88.7 | 87.2 |
| K+ | 70.7 | 58.1 | 63.7 | 67.9 | 63.6 | 71.2 | 70.5 |
| Rb+ | 65.6 | 53.3 | 60.1 | 58.6 | 66.0 | 65.7 | |
| Cs+ | 60.1 | 47.5 | 54.5 | 50.8 | 60.5 | 59.8 | |
| F- | 119.8 | 122.4 | 126.4 | 119.7 | 111.1 | ||
| Cl- | 89.2 | 93.0 | 89.1 | 96.5 | 89.1 | 81.3 | |
| Br- | 82.8 | 87.6 | 82.7 | 75.3 | |||
| I- | 74.5 | 79.4 | 72.6 | 74.3 | 65.7 | ||
| rms | 0.3 | 10.1 | 5.1 | 4.7 | 8.7 | ||
| SPC/E | |||||||
| Li+ | 113.3 | 105.6 | 113.7 | 108.1 | 113.8 | 113.5 | |
| Na+ | 88.4 | 76.4 | 87.2 | 90.4 | 82.2 | 88.7 | 87.2 |
| K+ | 71.0 | 59.5 | 65.3 | 69.3 | 64.8 | 71.2 | 70.5 |
| Rb+ | 65.6 | 54.5 | 61.6 | 59.4 | 66.0 | 65.7 | |
| Cs+ | 60.5 | 48.6 | 55.6 | 51.8 | 60.5 | 59.8 | |
| F- | 119.8 | 119.6 | 123.8 | 119.7 | 111.1 | ||
| Cl- | 89.3 | 91.0 | 87.8 | 95.1 | 89.1 | 81.3 | |
| Br- | 82.7 | 85.8 | 0.0 | 82.7 | 75.3 | ||
| I- | 74.4 | 77.5 | 71.8 | 74.3 | 65.7 | ||
| rms | 0.3 | 8.5 | 3.6 | 3.8 | 6.9 | ||
The dehydration free energies were calculated by using a two-stage thermodynamics integration disappearing the charges followed by the van der Waals in the various water models using a particle mesh Ewald treatment of the electrostatics as described in greater detail in the . The rms shows the deviations from Schmid’s dehydration free energies.
To understand the range of the acceptable solutions as a function of the weight ratios, the weight ratios were scanned across the full range from 0:1 to 300:1; in each case, the weight for the distance was adjusted, and the weight for LE was fixed at 1 kcal−1mol (i.e., 100:1, 150:1, 200:1, and so forth). Shown in Table 4 are the ranges of acceptable solutions. The acceptable ranges are subject to the criteria that the crystal is not in unstable regions (Figures 5 and S1−S6 in the Supporting Information), that size consistency is maintained, and that the ions do not get too large (as discussed). Detailed reasons why the ranges of the ratios were bounded are as follows: (a) the lower boundaries of the TIP3P and SPC/E derived models were limited because Rmin of I− was larger than 7 Å right below the range, and (b) the upper boundary of the SPC/E and the boundaries of TIP4PEW models hit the unstable boundary of the LiI crystals. Note that the ranges of acceptable solutions vary considerably depending on the choice of water model. The TIP3P-compatible ions have acceptable solutions in the range of 107:1−300:1 (possibly even up to 1:0), and the ranges for the other water models were 50.30:1−50.36:1 for the TIP4PEW-compatible ions and 12:1−42:1 for the SPC/E-compatible ions. These results are shown in Table 4 for the TIP3P-compatible ions and in the Supporting Information for TIP4PEW (Table S4) and SPC/E (Table S5).
Table 4. Range of Acceptable Solutions at Different Ratios of Weights on Interionic Distance and LE on the Ion Radii Optimization (WLE = 1)a.
| Wdis = 107 | Wdis = 150 | Wdis = 200 | Wdis = 250 | Wdis = 300 | |
|---|---|---|---|---|---|
| Rmin/2 | |||||
| Li+ | 0.886 | 0.986 | 1.079 | 1.078 | 1.078 |
| Na+ | 1.325 | 1.361 | 1.379 | 1.384 | 1.387 |
| K+ | 1.687 | 1.702 | 1.709 | 1.712 | 1.713 |
| Rb+ | 1.803 | 1.811 | 1.815 | 1.817 | 1.818 |
| Cs+ | 1.968 | 1.974 | 1.978 | 1.979 | 1.980 |
| F- | 2.377 | 2.346 | 2.259 | 2.180 | 2.144 |
| Cl- | 2.614 | 2.526 | 2.500 | 2.498 | 2.497 |
| Br- | 2.657 | 2.616 | 2.602 | 2.602 | 2.601 |
| I- | 2.876 | 2.862 | 2.856 | 2.855 | 2.855 |
| rms (Distance) | |||||
| 0.028 | 0.022 | 0.018 | 0.018 | 0.017 | |
| rms (LE) | |||||
| 3.28 | 3.97 | 4.49 | 4.62 | 4.68 | |
By using eq 3, the radii of the ions (Rmin) were optimized at various ratios of the respective weights on the inter-ionic distance (Wdis) and LE (WLE). Displayed are the different Rmin/2 values optimized for the Schmid hydration free energy target values with TIP3P water. Acceptable solutions were those that led to stable crystals (as per Figure 5), maintained size consistency across the monovalent ion series, and had Cs+ < 2.5 Å and I− < 3.5 Å. The units of the weights are kcal−1mol (Wdis) and Å−1 (WLE). The same data for the TIP4PEW and SPC/E models are shown in the Supporting Information, Tables S4 and S5.
Improving the Fit by Fitting to QM-Derived Binding Energies and Distances of the Ions to a Single Water Molecule
Up to this point, the experimental data considered for the optimization of the ion radii (Rmin) included hydration free energies (of Schmid), LC, and experimental LE. This was done separately for each of the three water models investigated. In an effort to improve and refine the fitting, we now include ion−water binding energies and distances of each ion to single water molecules in comparison with high-level QM data. For this fit, the optimized structures chosen were the C2v geometries for the cations and the Cs and C2v geometries for the anions (as shown in Figures 1 and 2). The overall rms deviations of the energy and distance from the reference values (shown in Table 6) were calculated from the deviations of the individual ions and are shown in Figure 7 for TIP3P and in Figures S8 and S9 in the Supporting Information for TIP4PEW and SPC/E. The plots of the rms deviation show that the variations of both the deviations in energy and distance (shown on the y axis) are very small within the full range of acceptable weight ratios (as per Tables 4 and S4 and S5 in the Supporting Information). With the exception of the optimization with TIP4PEW where the minima in the deviations of both distance and energy coincided at a weight ratio of 50.36:1, distinct minima for the rms deviations of distance and energy occurred at different weight ratios (for example, at a weight ratio of 146:1 for the energy and 195:1 for the distance with TIP3P water). In these cases, we compromised by balancing both properties to choose an intermediate weight ratio. For TIP3P, we ultimately chose a weight ratio of 171:1, and for SPC/E, we chose a weight ratio of 27:1.
Figure 7.
rms deviations of single water—ion binding energies and distances. The rms deviations are plotted as a function of the weight on interionic distance (Å−1) with a fixed weight on the LE (1 kcal−1mol) over the range of acceptable weight ratios (as per Table 4). The units are kcal/mol for the energy and Angstrom for the distance. TIP3P ions have the minimal rms in energy at a weight ratio of 146:1, whereas the minimal rms in distance occurs at 195:1. TIP4PEW ions have the minimal rms deviation for both the energy and the distance at a weight ratio of 50.36:1. SPC/E is somewhat anomalous because the rms of the energy keeps increasing as the rms of the distance decreases. With the SPC/E model, because the minima are not colocalized, the weight ratio was taken at the middle point of both curves or at 27:1. Plots of the rms deviations for TIP4PEW and SPC/E are shown in the Supporting Information.
Performance of the Optimized LJ Parameters for the Alkali and Halide Ions: Multiple Water−Ion Binding Energies
In addition to single water−ion binding energies and distances (Tables 6–8), binding energies of the ions with multiple water molecules were also measured with the final set of optimized parameters (by using the parameters from Table 5 to give the results shown in Table 9). In general, with the exception of fluorine, the deviations of the monohydrates are relatively small. However, as the number of surrounding water molecules increases, the deviations become larger, particularly with the smaller ions, such as lithium. Although the deviations with the small ions are relatively larger than those seen with the larger ions, such deviations are also evident with all of the nonadditive pairwise nonpolarizable potentials, particularly for fluorine, suggesting that for the small ions, the LJ potential is suboptimal. These results also show a dependence on the choice of water model. Interestingly, with the ions parametrized with SPC/E water, the deviations of the monohydrates are generally larger than the deviations seen with the other water models. Investigations of the influence of the geometry on the results suggest that the different H−O−H angle of SPC/E water molecule is the responsible factor. As the bond distances and angles of the water models are fixed, perfect accuracy in the microscopic hydration structures is not expected; in fact, it is surprising that such a simple model of the ion−water interaction works as well as it does. Given these inaccuracies, the optimization considered only the binding energy and the binding distance of the ions with single water molecules when choosing the final ratio of weights on LE and interionic distances.
Table 8. Comparison of the SPC/E Single Water−Ion Binding Distances for Various Ion Parameter Setsa.
| structures | thisresearch | Jensen andJorgensen | Smith−Dang−Garrett | Beglovand Roux | Åqvist | reference values | |
|---|---|---|---|---|---|---|---|
| Li+ | 1 + 0(C2v) | 1.92 | 1.96 | 1.91 | 2.22 | 1.95 | 1.86172,173 |
| Na+ | 1 + 0(C2v) | 2.30 | 2.41 | 2.29 | 2.63 | 2.32 | 2.23(174) |
| K+ | 1 + 0(C2v) | 2.66 | 2.77 | 2.74 | 2.64 | 2.64(175) | |
| Rb+ | 1 + 0(C2v) | 2.79 | 2.90 | 2.83 | 2.76 | 2.80(176) | |
| Cs+ | 1 + 0(C2v) | 2.85 | 3.08 | 3.01 | 2.96 | 2.96(177) | |
| F- | 1 + 0(Cs) | 2.61 | 2.72 | 2.62 | 2.45178,179 | ||
| 1 + 0(C2v) | 2.78 | 2.86 | 2.78 | 2.61(178) | |||
| Cl- | 1 + 0(Cs) | 3.10 | 3.25 | 3.20 | 3.07 | 3.12178,180 | |
| 1 + 0(C2v) | 3.21 | 3.33 | 3.29 | 3.17 | 3.20(178) | ||
| Br- | 1 + 0(Cs) | 3.26 | 3.38 | 3.32(178) | |||
| 1 + 0(C2v) | 3.35 | 3.45 | 3.35(178) | ||||
| I- | 1 + 0(Cs) | 3.49 | 3.63 | 3.62 | 3.61178,181 | ||
| 1 + 0(C2v) | 3.56 | 3.68 | 3.67 | 3.61178,181 | |||
| rms | 0.08 | 0.14 | 0.08 | 0.03 | 0.06 |
Shown are the single water oxygen−ion distances for various geometries (as shown in Figures 1 and 2) for distinct ion parameter sets with SPC/E water. The reference values are averages of one or more ab initio calculations. The distances have units of Å. The levels of QM theory applied for each reference cited are as denoted in the legend to Table 6.
Table 5. Final Optimized LJ Parameters for the Alkali and Halide Ions with Different Water Modelsa.
| TIP3P |
TIP4PEW |
SPC/E |
||||
|---|---|---|---|---|---|---|
| Rmin/2 (Å) | ε (kcal/mol) | Rmin/2 (Å) | ε (kcal/mol) | Rmin/2 (Å) | ε (kcal/mol) | |
| Li+ | 1.025 | 0.027 9896 | 0.808 | 0.10 39884 | 0.791 | 0.33 67344 |
| Na+ | 1.369 | 0.087 4393 | 1.226 | 0.16 84375 | 1.212 | 0.35 26418 |
| K+ | 1.705 | 0.19 36829 | 1.590 | 0.27 94651 | 1.593 | 0.42 97054 |
| Rb+ | 1.813 | 0.32 78219 | 1.709 | 0.43 31494 | 1.737 | 0.44 51036 |
| Cs+ | 1.976 | 0.40 65394 | 1.888 | 0.39 44318 | 2.021 | 0.089 8565 |
| F- | 2.303 | 0.0033 640 | 2.538 | 0.0015 752 | 2.257 | 0.0074 005 |
| Cl- | 2.513 | 0.035 5910 | 2.760 | 0.011 6615 | 2.711 | 0.012 7850 |
| Br- | 2.608 | 0.058 6554 | 2.768 | 0.030 3773 | 2.751 | 0.026 9586 |
| I- | 2.860 | 0.053 6816 | 2.952 | 0.041 7082 | 2.919 | 0.042 7845 |
The ion radii (Rmin) were optimized on the basis of the Schmid hydration free energies, LC and LE (expt) from Tables 2 and 3, and single water−ion binding energies and distances were optimized on the basis of Tables 6–8. The weight ratios for the interionic distance and LE (expt) chosen were 171:1 for the TIP3P water model, 50.36:1 for the TIP4PEW water model, and 27:1 for the SPC/E water model. The well depth (ε) was determined from the bicubic fits of ΔGhydration = f(Rmin, ε).
Table 9. Multiple Water (TIP3P)−Ion Binding Energies for the Various Ion Parameters Compared to Ab Initio Valuesa.
| n | structure | this research | Jensen and Jorgensen | Smith−Dang−Garrett | Beglov and Roux | Åqvist | QM | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li+ | |||||||||||||
| 1 | 1 + 0(C2v) | −33.85 | (0.6) | −31.55 | (2.9) | −34.07 | (0.4) | −32.22 | (2.3) | −34.5172,173,182 | |||
| 2 | 1 + 1(Cs) | −48.98 | (3.0) | −46.45 | (5.5) | −49.19 | (2.8) | −47.16 | (4.8) | −52.0173,182 | |||
| 2 + 0(D2d) | −65.45 | (−1.5) | −61.03 | (2.9) | −65.90 | (−1.9) | −62.33 | (1.6) | −64.0172,173,182 | ||||
| 3 | 2 + 1(C2v) | −79.39 | (0.9) | −74.95 | (5.4) | −79.86 | (0.4) | −76.27 | (4.0) | −80.3173,182 | |||
| 3 + 0(D3) | −92.40 | (−5.5) | −86.36 | (0.6) | −93.13 | (−6.2) | −88.22 | (−1.3) | −86.9172,173,182 | ||||
| 4 | 3 + 1(C2v) | −106.74 | (−3.5) | −100.63 | (2.6) | −107.48 | (−4.3) | −102.51 | (0.7) | −103.2173,182 | |||
| 4 + 0(S4) | −114.10 | (−10.2) | −107.11 | (−3.2) | −115.14 | (−11.3) | −109.39 | (−5.5) | −103.9172,173,182 | ||||
| 5 | 4 + 1(C2) | −127.88 | (−9.6) | −120.83 | (−2.5) | −128.94 | (−10.6) | −123.13 | (−4.8) | −118.3172,173,182 | |||
| 6 | 4 + 2(Cs) | −139.08 | (−9.2) | −131.99 | (−2.1) | −140.15 | (−10.3) | −134.31 | (−4.5) | −129.8172,182 | |||
| 4 + 2(D2d) | −141.06 | (−10.8) | −134.00 | (−3.7) | −142.13 | (−11.8) | −136.32 | (−6.0) | −130.3(172) | ||||
| Na+ | |||||||||||||
| 1 | 1 + 0(C2v) | −24.29 | (0.0) | −21.62 | (2.7) | −24.43 | (−0.1) | −25.53 | (−1.2) | −23.24 | (1.1) | −24.3174,182 | |
| 2 | 1 + 1(Cs) | −38.10 | (0.8) | −35.05 | (3.9) | −38.25 | (0.7) | −39.54 | (−0.6) | −36.93 | (2.0) | −38.9(182) | |
| 2 + 0(D2d) | −47.22 | (−1.4) | −42.05 | (3.8) | −47.49 | (−1.7) | −49.59 | (−3.8) | −45.16 | (0.6) | −45.8174,182 | ||
| 3 | 2 + 1(C2v) | −60.95 | (−0.4) | −55.61 | (5.0) | −61.23 | (−0.6) | −63.38 | (−2.8) | −58.83 | (1.8) | −60.6(182) | |
| 3 + 0(D3) | −67.51 | (−3.4) | −60.18 | (4.0) | −67.90 | (−3.8) | −70.78 | (−6.6) | −64.55 | (−0.4) | −64.2174,182 | ||
| 4 | 3 + 1(C2v) | −81.50 | (−3.5) | −73.99 | (4.0) | −81.91 | (−3.9) | −84.84 | (−6.8) | −78.48 | (−0.5) | −78.0(182) | |
| 4 + 0(S4) | −85.07 | (−6.3) | −75.95 | (2.9) | −85.58 | (−6.8) | −88.99 | (−10.2) | −81.33 | (−2.5) | −78.8174,182 | ||
| 5 | 5 + 0(C2v) | −98.04 | (−6.3) | −88.00 | (3.7) | −98.63 | (−6.9) | −101.96 | (−10.3) | −93.80 | (−2.1) | −91.7(174) | |
| 4 + 1(C2) | −98.60 | (−6.1) | −89.35 | (3.2) | −99.12 | (−6.6) | −102.57 | (−10.1) | −94.81 | (−2.3) | −92.5174,182 | ||
| 6 | 4 + 2(D2d) | −111.87 | (−4.3) | −102.54 | (5.1) | −112.39 | (−4.8) | −115.83 | (−8.2) | −108.03 | (−0.4) | −107.6(174) | |
| K+ | |||||||||||||
| 1 | 1 + 0(C2v) | −18.51 | (0.1) | −16.61 | (2.0) | −17.53 | (1.1) | −18.87 | (−0.3) | −18.15 | (0.4) | −18.6175,182 | |
| 2 | 1 + 1(Cs) | −31.27 | (0.7) | −29.08 | (2.9) | −30.11 | (1.8) | −31.72 | (0.2) | −30.93 | (1.0) | −31.9175,182 | |
| 2 + 0(D2d) | −36.11 | (−0.8) | −32.41 | (2.9) | −34.21 | (1.1) | −36.79 | (−1.5) | −35.37 | (−0.1) | −35.3175,182 | ||
| 3 | 2 + 1(C2v) | −49.42 | (0.8) | −45.52 | (4.7) | −47.42 | (2.8) | −50.13 | (0.1) | −48.65 | (1.6) | −50.2175,182 | |
| 3 + 0(D3) | −51.96 | (−1.8) | −46.63 | (3.6) | −49.26 | (0.9) | −52.89 | (−2.7) | −50.80 | (−0.6) | −50.2175,182 | ||
| 4 | 4 + 0(C4) | −60.92 | (2.3) | −55.59 | (7.6) | −58.42 | (4.8) | −67.13 | (−3.9) | −64.40 | (−1.2) | −63.2(182) | |
| 3 + 1(C2v) | −65.49 | (−1.1) | −59.95 | (4.5) | −62.68 | (1.7) | −66.46 | (−2.1) | −64.30 | (0.1) | −64.4175,182 | ||
| 5 | 5 + 0(C2v) | −77.42 | (−3.6) | −69.48 | (4.3) | −73.54 | (0.3) | −78.57 | (−4.8) | −75.28 | (−1.5) | −73.8(182) | |
| 4 + 1(C2) | −79.23 | (−2.6) | −72.26 | (4.4) | −75.76 | (0.9) | −80.36 | (−3.7) | −77.57 | (−0.9) | −76.7175,182 | ||
| 5 + 0(C2) | −76.44 | (1.4) | −69.82 | (8.0) | −73.35 | (4.4) | −77.20 | (0.6) | −74.17 | (3.6) | −77.8175,182 | ||
| 3 + 2(Cs) | −76.10 | (1.8) | −70.41 | (7.5) | −73.22 | (4.7) | −77.09 | (0.8) | −74.89 | (3.0) | −77.9(175) | ||
| 6 | 4 + 2(Cs) | −90.32 | (−0.8) | −83.23 | (6.3) | −86.79 | (2.7) | −91.45 | (−2.0) | −88.62 | (0.9) | −89.5(175) | |
| 4 + 2(D2d) | −92.31 | (−1.3) | −85.19 | (5.8) | −88.77 | (2.2) | −93.45 | (−2.5) | −90.60 | (0.4) | −91.0(175) | ||
| 4 + 2(C2) | −89.24 | (5.2) | −82.80 | (11.6) | −86.19 | (8.2) | −90.03 | (4.4) | −87.21 | (7.2) | −94.4(175) | ||
| Rb+ | |||||||||||||
| 1 | 1 + 0(C2v) | −16.77 | (−0.8) | −15.20 | (0.7) | −16.44 | (−0.5) | −16.65 | (−0.7) | −15.9176,182 | |||
| 2 | 1 + 1(Cs) | −29.17 | (−0.8) | −27.36 | (1.0) | −28.80 | (−0.4) | −29.13 | (−0.7) | −28.4176,182 | |||
| 2 + 0(D2d) | −32.78 | (−2.7) | −29.69 | (0.4) | −32.12 | (−2.0) | −32.48 | (−2.4) | −30.1176,182 | ||||
| 3 | 3 + 0(D3) | −47.28 | (−4.1) | −42.79 | (0.4) | −46.30 | (−3.2) | −46.72 | (−3.6) | −43.1176,182 | |||
| 2 + 1(C2v) | −45.90 | (−1.0) | −42.63 | (2.3) | −45.20 | (−0.3) | −45.59 | (−0.7) | −44.9176,182 | ||||
| 4 | 4 + 0(D4) | −59.45 | (−6.2) | −53.74 | (−0.4) | −58.18 | (−4.9) | - | - | −53.3(176) | |||
| 4 + 0(S4) | −60.24 | (−5.8) | −54.47 | (0.0) | −58.96 | (−4.5) | −59.34 | (−4.9) | −54.5176,182 | ||||
| 3 + 1(C2v) | −60.60 | (−4.0) | −55.91 | (0.7) | −59.59 | (−3.0) | −60.04 | (−3.4) | −56.6(182) | ||||
| 4 + 0(C4) | −57.18 | (1.6) | −52.66 | (6.1) | −56.04 | (2.7) | −55.61 | (3.1) | −58.8176,182 | ||||
| 5 | 4 + 1(C2) | −73.29 | (−5.3) | −67.33 | (0.7) | −71.97 | (−4.0) | −72.36 | (−4.4) | −68.0(176) | |||
| 3 + 2(Cs) | −71.07 | (−0.3) | −66.25 | (4.6) | −70.03 | (0.8) | −70.50 | (0.3) | −70.8(176) | ||||
| 5 + 0(C5) | −71.44 | (0.1) | −66.16 | (5.3) | −70.07 | (1.4) | −69.31 | (2.2) | −71.5(176) | ||||
| 5 + 0(C2) | −71.71 | (0.2) | −65.93 | (6.0) | −70.28 | (1.6) | −69.84 | (2.1) | −71.9(176) | ||||
| 4 + 1 h(C1) | −70.83 | (3.0) | −65.76 | (8.0) | −69.57 | (4.2) | −69.22 | (4.6) | −73.8(176) | ||||
| 6 | 4 + 2(Cs) | −84.28 | (−3.6) | −78.19 | (2.5) | −82.93 | (−2.2) | −83.33 | (−2.6) | −80.7(176) | |||
| 4 + 2(D2d) | −86.24 | (−4.3) | −80.12 | (1.8) | −84.89 | (−3.0) | −85.29 | (−3.4) | −81.9(176) | ||||
| 5 + 1 h(C1) | −84.67 | (0.4) | −78.40 | (6.7) | −83.13 | (2.0) | −82.77 | (2.3) | −85.1(176) | ||||
| 4 + 2(C2) | −84.42 | (2.8) | −78.79 | (8.4) | −83.06 | (4.1) | −82.85 | (4.4) | −87.2(176) | ||||
| Cs+ | |||||||||||||
| 1 | 1 + 0(C2v) | −15.08 | (−1.0) | −13.57 | (0.5) | −14.70 | (−0.6) | −14.60 | (−0.5) | −14.1177,182 | |||
| 2 | 1 + 1(Cs) | −27.09 | (−1.6) | −25.34 | (0.2) | −26.67 | (−1.2) | −26.62 | (−1.1) | −25.5(182) | |||
| 2 + 0(D2d) | −29.50 | (−2.9) | −26.54 | (0.1) | −28.75 | (−2.2) | −28.52 | (−1.9) | −26.6(182) | ||||
| 3 | 3 + 0(D3) | −42.64 | (−4.4) | −38.33 | (−0.1) | −41.54 | (−3.3) | −41.12 | (−2.9) | −38.2(182) | |||
| 2 + 1(C2v) | −42.41 | (−1.6) | −39.25 | (1.6) | −41.62 | (−0.8) | −41.38 | (−0.6) | −40.8177,182 | ||||
| 4 | 4 + 0(S4) | −54.48 | (−6.1) | −48.90 | (−0.5) | −53.03 | (−4.6) | −52.38 | (−4.0) | −48.4(182) | |||
| 3 + 1(C2v) | −55.73 | (−4.4) | −51.20 | (0.1) | −54.58 | (−3.3) | −54.16 | (−2.9) | −51.3177,182 | ||||
| 4 + 0(C4) | −53.78 | (1.1) | −49.49 | (5.4) | −52.50 | (2.4) | −51.36 | (3.5) | −54.9177,182 | ||||
| F- | |||||||||||||
| 1 | 1 + 0(C2v) | −18.17 | (1.6) | −17.43 | (2.4) | −18.01 | (1.8) | −19.8(178) | |||||
| 1 + 0(Cs) | −21.23 | (5.2) | −19.31 | (7.1) | −20.49 | (5.9) | −26.4178,179 | ||||||
| 2 | 2 + 0(C2h) | −41.56 | (4.7) | −37.89 | (8.4) | −40.17 | (6.1) | −46.3(178) | |||||
| 2 + 0(C1) | −42.06 | (6.2) | −38.65 | (9.6) | −40.79 | (7.5) | −48.3(178) | ||||||
| 3 | 3 + 0(C3) | −62.32 | (3.1) | −57.84 | (7.6) | −60.70 | (4.7) | −65.5178,179 | |||||
| 4 | 4 + 0(C4h) | −79.37 | (−2.5) | −72.46 | (4.4) | −76.80 | (0.1) | −76.9(178) | |||||
| 3 + 1(Cs) | −77.61 | (0.7) | −72.20 | (6.1) | −75.64 | (2.7) | −78.3(178) | ||||||
| 4 + 0(C4) | −81.24 | (−2.9) | −75.85 | (2.4) | −79.35 | (−1.0) | −78.3(178) | ||||||
| 4 + 0(C1) | −80.11 | (−1.2) | −74.41 | (4.5) | −78.09 | (0.8) | −78.9(178) | ||||||
| Cl- | |||||||||||||
| 1 | 1 + 0(C2v) | −13.67 | (−0.6) | −13.18 | (−0.1) | −13.01 | (0.1) | −14.04 | (−0.9) | −13.1(178) | |||
| 1 + 0(Cs) | −14.26 | (0.1) | −13.49 | (0.8) | −13.39 | (0.9) | −14.66 | (−0.3) | −14.3178,180 | ||||
| 2 | 2 + 0(C2h) | −27.99 | (−1.0) | −26.51 | (0.5) | −26.29 | (0.7) | −28.79 | (−1.8) | −27.0(178) | |||
| 2 + 0(C1) | −29.13 | (−0.5) | −27.74 | (0.9) | −27.51 | (1.1) | −29.91 | (−1.3) | −28.6178,180 | ||||
| 3 | 3 + 0(C3h) | −40.81 | (−1.5) | −38.67 | (0.6) | −38.32 | (1.0) | −41.99 | (−2.7) | −39.3(178) | |||
| 2 + 1(Cs) | −42.37 | (−0.7) | −40.53 | (1.2) | −40.35 | (1.4) | −43.27 | (−1.6) | −41.7178,180 | ||||
| 3 + 0(C3) | −44.50 | (−1.0) | −42.74 | (0.7) | −42.29 | (1.2) | −45.64 | (−2.2) | −43.5178,180 | ||||
| 4 | 4 + 0(C4h) | −53.00 | (−4.0) | −50.10 | (−1.1) | −49.65 | (−0.6) | −54.58 | (−5.6) | −49.0(178) | |||
| 4 + 0(Ci) | −55.96 | (−4.1) | −53.52 | (−1.6) | −52.97 | (−1.1) | −57.47 | (−5.6) | −51.9(178) | ||||
| 3 + 1(Cs) | −56.31 | (−3.6) | −54.04 | (−1.3) | −53.60 | (−0.9) | −57.63 | (−4.9) | −52.7(178) | ||||
| 4 + 0(C4) | −59.16 | (−2.5) | −57.24 | (−0.6) | −56.53 | (0.1) | −60.62 | (−3.9) | −56.7178,180 | ||||
| Br- | |||||||||||||
| 1 | 1 + 0(C2v) | −12.60 | (−0.7) | −12.36 | (−0.5) | −11.9(178) | |||||||
| 1 + 0(Cs) | −12.91 | (−0.2) | −12.53 | (0.2) | −12.7(178) | ||||||||
| 2 | 2 + 0(C2h) | −25.34 | (−1.5) | −24.62 | (−0.8) | −23.8(178) | |||||||
| 2 + 0(C1) | −26.58 | (−0.8) | −25.90 | (−0.1) | −25.8(178) | ||||||||
| 3 | 3 + 0(C3h) | −36.91 | (−2.3) | −35.89 | (−1.3) | −34.6(178) | |||||||
| 2 + 1(Cs) | −39.24 | (−1.0) | −38.30 | (−0.1) | −38.2(178) | ||||||||
| 3 + 0(C3) | −40.99 | (−1.3) | −40.20 | (−0.5) | −39.7(178) | ||||||||
| 4 | 4 + 0(C4h) | −47.76 | (−4.3) | −46.36 | (−2.9) | −43.5(178) | |||||||
| 4 + 0(Ci) | −51.22 | (−4.2) | −50.09 | (−3.1) | −47.0(178) | ||||||||
| 3 + 1(Cs) | −52.04 | (−1.2) | −50.98 | (−0.2) | −50.8(178) | ||||||||
| 4 + 0(C4) | −54.93 | (−2.3) | −54.19 | (−1.6) | −52.6(178) | ||||||||
| I- | |||||||||||||
| 1 | 1 + 0(C2v) | −11.24 | (−1.1) | −10.95 | (−0.8) | −10.59 | (−0.4) | −10.2178,181 | |||||
| 1 + 0(Cs) | −11.34 | (−0.8) | −10.99 | (−0.5) | −10.62 | (−0.1) | −10.5178,181 | ||||||
| 2 | 2 + 0(C2h) | −22.27 | (−2.2) | −21.59 | (−1.5) | −20.87 | (−0.8) | −20.1(178) | |||||
| 2 + 0(C1) | −23.61 | (−1.3) | −22.95 | (−0.6) | −22.24 | (0.1) | −22.3178,181 | ||||||
| 3 | 3 + 0(C3h) | −32.39 | (−3.5) | −31.42 | (−2.5) | −30.33 | (−1.4) | −28.9178,181 | |||||
| 2 + 1(Cs) | −35.59 | (−2.1) | −34.69 | (−1.2) | −33.90 | (−0.4) | −33.5(178) | ||||||
| 3 + 0(C3) | −36.87 | (−0.9) | −36.12 | (−0.2) | −35.01 | (0.9) | −35.9178,181 | ||||||
| 4 | 4 + 0(C4h) | −41.68 | (−4.8) | −40.37 | (−3.5) | −38.91 | (−2.1) | −36.8178,181 | |||||
| 4 + 0(Ci) | −45.66 | (−4.6) | −44.54 | (−3.4) | −43.12 | (−2.0) | −41.1(178) | ||||||
| 3 + 1(Cs) | −47.06 | (−1.9) | −46.06 | (−0.9) | −44.80 | (0.4) | −45.2(178) | ||||||
| 4 + 0(C4) | −50.00 | (−1.4) | −49.34 | (−0.7) | −47.84 | (0.8) | −48.6178,181 | ||||||
Water−ion binding energies (kcal/mol), with deviations from the reference values shown in parenthesis, are reported for the various ion parameter sets. The structures were optimized as described in the section by using the geometries depicted in Figures 1 and 2. Empty values mean that the ion parameters for that ion were not defined in the particular parameter set, and a ‘-’ indicates that a minimized structure at that geometry was not found. Similar results with TIP4PEW and SPC/E are shown in the Supporting Information. The reference values are the average of various ab initio calculations (QM) from the following references: ref (172), MP2/6−31++G(d,p)//HF/6−31++G(d,p); ref (173), MP2/CBS for n = 1, MP2/aug-cc-pVQZ//MP2/aug-cc-pVTZ for 2 + 0(D2d), MP2/aug-cc-pVDZ for 1 + 1(Cs) and n = 3−6; ref (182), MP2/6−31+G*//RHF/6−31+G*; ref (174), MP2/TZ2P (50% BSSE corrected); ref (175), MP2/TZ2P (BSSE uncorrected); ref (176), MP2/aVDZ (BSSE uncorrected); ref (177), MP2/aVTZ (BSSE uncorrected); ref (178), MP2/6−311++G**; ref (179), MP2/aug-cc-pVDZ; ref (180), MP2/aug-cc-pVDZ (50% BSSE corrected); ref (181), MP2/aug-cc-pVDZ+diff.
Performance of the Optimized LJ Parameters for the Alkali and Halide Ions: Hydration Free Energies
Although the bicubic equation which fits Rmin and ε to ΔGhydration can accurately estimate the hydration free energies with low errors, the free energies of hydration of the optimized ion parameters were recalculated with the optimized parameters from Table 5 for confirmation. With all of the water models tested, the rms deviations were less than 0.3 kcal/mol for the ion parameters when comparing the calculated hydration free energies to the target values. As shown in Table 10, this error is significantly less than the error seen with all of the other ion parameter models tested. However, this is expected because the other parameters were optimized with different target values in mind. For example, the Jensen and Jorgensen’s ions are optimized for the TIP4P water model(77) (not the TIP4PEW model and not with a particle mesh Ewald treatment of the electrostatics), and their ions targeted Marcus’s hydration free energies.(116) The major difference is not due simply to the small differences between the TIP4P and TIP4PEW water models, because our fitting to Marcus’s hydration energies with the TIP4PEW water would be expected to be in reasonable agreement with their results. However, their rms deviation from Marcus’s hydration free energies are ∼12.3 kcal/mol (not shown in the table), which is considerably larger than what we expect. The boundary conditions are the primary difference; they used spherical boundary conditions, and this research employed PBC with an Ewald treatment of the long-range electrostatics. Interestingly, systematic deviations from Marcus’s hydration free energies were observed in Jensen and Jorgensen’s ions. For the cations, they were less negative by 12.3 kcal/mol, and for the anions, they were more negative by 12.3 kcal/mol in average. This exactly corresponds to the rms deviation. The surface potential should be compensated if spherical boundary condition is used instead of periodic boundary condition.76,165 Jensen and Jorgensen estimated the surface potential energy of TIP4P water as −3 kcal/mol for monovalently charged ions according to ref (183); however, another group estimated −0.63 Ve−1 (184) (i.e., −14.5 kcal/mol for monovalent ions). Interestingly, the difference of the two numbers is 11.5 kcal/mol, which is very close to the 12.3 kcal/mol difference. The discrepancies in the estimated values suggest that Jensen and Jorgensen’s estimation of the surface potential might be inappropriate, and therefore, their values might be suboptimal. To date, the Jensen and Jorgensen ion parameters are the only other complete set of alkali and halide ion parameters. Of the partial sets, the hydration free energies of Smith−Dang−Garrett’s ions are closer to those of Schmid than those of Marcus in TIP3P, TIP4PEW, and SPC/E water, but their rms deviations were all higher than those observed with the optimized parameters developed in this work. Beglov and Roux’s ions and Åqvist’s ions also exhibited relatively high rms deviations.
Performance of the Optimized LJ Parameters for the Alkali and Halide Ions: Interionic (Cation−Anion) Distances in the Crystals
The rms deviations of the calculated interionic distances were less than 0.1 Å (specifically, 0.02Å for TIP3P-compatible ions, 0.04 Å for TIP4PEW-compatible ions, and 0.08 Å for SPC/E-compatible ions, Table 11). Among the cations, the deviations of Li+ and Na+ were relatively larger. A clear anomaly is the deviation of Cs+ in SPC/E water. Among the anions, crystals containing F− and I− ions had rms deviations that were relatively larger than those of the cations. When comparing other parameter sets, the rms deviations of the Jensen and Jorgensen’s ions were 0.18 Å for all the ions, and their lithium−iodide crystal was unstable, such that the interionic distance was not measurable. Smith−Dang−Garrett’s ions lack bromide salts, and the overall rms deviations were 0.12 Å. Only NaCl and KCl could be combined with Beglov and Roux’s ions, and their deviations was 0.08 Å. When the rms deviations were compared by using only crystals that exist in each set, the newly optimized parameters showed lower deviations in each case.
Table 11. Interionic Distances of the Alkali−Halide Crystals with the Various Parameter Setsa.
| Li+ | Na+ | K+ | Rb+ | Cs+ | rms | |
|---|---|---|---|---|---|---|
| This Research (TIP3P-Compatible Ions) | ||||||
| F- | 2.06 | 2.37 | 2.69 | 2.83 | 2.98 | 0.04 |
| Cl- | 2.57 | 2.82 | 3.16 | 3.30 | 3.56 | 0.01 |
| Br- | 2.76 | 2.98 | 3.30 | 3.45 | 3.73 | 0.01 |
| I- | 3.01 | 3.20 | 3.51 | 3.66 | 3.96 | 0.02 |
| rms | 0.03 | 0.03 | 0.01 | 0.01 | 0.02 | 0.02 |
| This Research (TIP4PEW-Compatible Ions) | ||||||
| F- | 2.13 | 2.43 | 2.74 | 2.87 | 2.99 | 0.08 |
| Cl- | 2.60 | 2.85 | 3.17 | 3.31 | 3.55 | 0.03 |
| Br- | 2.78 | 3.00 | 3.31 | 3.45 | 3.71 | 0.02 |
| I- | 3.05 | 3.23 | 3.51 | 3.66 | 3.94 | 0.02 |
| rms | 0.07 | 0.06 | 0.03 | 0.02 | 0.02 | 0.04 |
| This Research (SPC/E-Compatible Ions) | ||||||
| F- | 2.16 | 2.45 | 2.75 | 2.87 | 2.90 | 0.11 |
| Cl- | 2.63 | 2.89 | 3.19 | 3.31 | 3.44 | 0.07 |
| Br- | 2.79 | 3.03 | 3.33 | 3.45 | 3.60 | 0.06 |
| I- | 3.05 | 3.25 | 3.54 | 3.66 | 3.85 | 0.05 |
| rms | 0.08 | 0.08 | 0.05 | 0.02 | 0.12 | 0.08 |
| Jensen and Jorgensen | ||||||
| F- | 2.19 | 2.58 | 2.94 | 3.08 | 3.27 | 0.25 |
| Cl- | 2.76 | 2.99 | 3.31 | 3.44 | 3.75 | 0.17 |
| Br- | 2.92 | 3.11 | 3.41 | 3.53 | 3.87 | 0.13 |
| I- | 3.36 | 3.61 | 3.73 | 4.12 | 0.11 | |
| rms | 0.18 | 0.18 | 0.17 | 0.16 | 0.19 | 0.18 |
| Smith−Dang−Garrett | ||||||
| F- | 2.07 | 2.38 | 2.80 | 2.89 | 3.06 | 0.08 |
| Cl- | 2.71 | 2.92 | 3.30 | 3.39 | 3.67 | 0.12 |
| Br- | ||||||
| I- | 3.20 | 3.35 | 3.68 | 3.76 | 4.10 | 0.14 |
| rms | 0.14 | 0.10 | 0.14 | 0.08 | 0.11 | 0.12 |
| Roux | ||||||
| Cl- | 2.73 | 3.08 | 0.08 | |||
| rms | 0.09 | 0.07 | 0.08 | |||
The rms indicates deviations from the literature values (Table 3). The units are Angstroms.
Performance of the Optimized LJ Parameters for the Alkali and Halide Ions: LE of the Crystals
Because the LE was weighted less in the self-consistent fitting with the TIP3P-compatible ions, as compared with the TIP4PEW and SPC/E compatible ions, the rms deviation of TIP3P-compatible ions (4.2 kcal/mol) was higher than that obtained with the TIP4PEW-compatible ions (2.0 kcal/mol) or SPC/E-compatible ions (1.9 kcal/mol). Nonetheless, the rms deviations are lower with the new parameters than is observed with any of the other ion sets listed in Table 12, where the overall rms deviations were >5 kcal/mol in each case. Likewise, the partial rms deviations measured with the only corresponding crystals provided by other groups were all lower than those of the crystals of the groups.
Table 12. LE of the Alkali−Halide Crystalsa.
| Li+ | Na+ | K+ | Rb+ | Cs+ | rms | |
|---|---|---|---|---|---|---|
| This Research (TIP3P) | ||||||
| F- | 261.3 | 228.1 | 202.2 | 193.4 | 184.9 | 6.1 |
| Cl- | 213.2 | 194.0 | 174.7 | 167.7 | 159.0 | 4.1 |
| Br- | 200.0 | 184.9 | 167.8 | 161.3 | 153.0 | 3.2 |
| I- | 183.8 | 172.4 | 157.9 | 152.2 | 144.0 | 2.5 |
| rms | 6.6 | 5.0 | 3.0 | 2.1 | 2.4 | 4.2 |
| This Research (TIP4PEW) | ||||||
| F- | 253.1 | 222.8 | 198.8 | 190.4 | 183.3 | 1.4 |
| Cl- | 209.8 | 191.3 | 173.2 | 166.3 | 158.1 | 2.1 |
| Br- | 197.6 | 183.2 | 167.0 | 160.6 | 152.4 | 2.0 |
| I- | 181.8 | 171.4 | 157.8 | 152.2 | 143.9 | 2.1 |
| rms | 2.2 | 2.4 | 1.7 | 0.8 | 2.2 | 2.0 |
| This Research (SPC/E) | ||||||
| F- | 251.5 | 222.5 | 199.1 | 191.7 | 188.0 | 3.1 |
| Cl- | 208.1 | 189.5 | 172.3 | 166.6 | 161.4 | 1.0 |
| Br- | 197.4 | 182.0 | 166.2 | 160.8 | 155.1 | 1.2 |
| I- | 182.1 | 170.5 | 157.1 | 152.3 | 145.9 | 1.4 |
| rms | 1.2 | 1.4 | 1.1 | 1.2 | 3.4 | 1.9 |
| Jensen and Jorgensen | ||||||
| F- | 255.8 | 213.8 | 185.8 | 177.3 | 167.3 | 11.1 |
| Cl- | 208.1 | 188.5 | 168.0 | 161.1 | 150.5 | 5.2 |
| Br- | 197.9 | 182.4 | 164.1 | 157.5 | 146.9 | 3.9 |
| I- | 180.4 | 170.7 | 156.5 | 150.9 | 139.6 | 3.4 |
| rms | 3.0 | 4.5 | 6.6 | 6.9 | 10.0 | 6.6 |
| Smith−Dang−Garrett | ||||||
| F- | 268.4 | 223.6 | 198.0 | 191.8 | 181.4 | 8.0 |
| Cl- | 205.2 | 189.5 | 167.9 | 163.6 | 153.8 | 3.6 |
| Br- | ||||||
| I- | 175.5 | 166.6 | 151.3 | 148.0 | 138.6 | 5.3 |
| rms | 11.0 | 1.4 | 3.4 | 2.5 | 5.8 | 5.9 |
| Roux | ||||||
| Cl- | 202.6 | 180.2 | 11.3 | |||
| rms | 13.8 | 8.1 | 11.3 | |||
The rms indicates deviations from the literature values (Bottom Half of Table 2).
Performance of the Optimized LJ parameters for the Alkali and Halide Ions: Radii of the First Water Shell
The radii of the first water shell around the ions are displayed in Table 13. Among the water models, the optimized parameters for TIP3P water displayed the lowest rms deviation (0.07 Å) compared to the reference values. Additionally, the optimized parameters show lower deviations than the Jensen and Jorgensen ions, independent of water model, despite the fact that the Jensen and Jorgensen ions were parametrized to fit these radii. On the other hand, the Smith−Dang−Garrett ions showed consistently lower deviation (0.05 Å); however, the differences are only 0.02−0.05 Å. The rms deviations of Beglov and Roux ions are intermediate; however when individual ions are compared, the deviations of the Beglov and Roux ions are larger. Because the ion−water distance varies depending on the choice of counterion, the literature values(185) are variable and less reliable than the ion−water binding structures discussed previously. Because of this, the radii of the first water shell were not considered in the optimization of the ion radii (Rmin). Although the radii of the first water shell varied by about 0.2 Å over the possible range of Rmin (data not shown), it is surprising to have such low deviations, even though the property was not considered in the optimization process.
Table 13. Radii of the First Hydration Shella.
| thisresearch | Jensen andJorgensen | Smith−Dang−Garrett | Beglov andRoux | Åqvist | Marcus(185) | |
|---|---|---|---|---|---|---|
| TIP3P | ||||||
| Li+ | 1.96 | 2.04 | 1.97 | 2.03 | 2.08 | |
| Na+ | 2.38 | 2.49 | 2.37 | 2.31 | 2.41 | 2.356 |
| K+ | 2.75 | 2.86 | 2.83 | 2.71 | 2.72 | 2.798 |
| Rb+ | 2.92 | 3.00 | 2.93 | 2.85 | 2.89 | |
| Cs+ | 3.11 | 3.19 | 3.11 | 3.05 | 3.139 | |
| F- | 2.63 | 2.80 | 2.70 | 2.63 | ||
| Cl- | 3.13 | 3.29 | 3.24 | 3.12 | 3.187 | |
| Br- | 3.29 | 3.41 | 3.373 | |||
| I- | 3.51 | 3.65 | 3.64 | 3.647 | ||
| rms | 0.07 | 0.09 | 0.05 | 0.07 | 0.06 | |
| TIP4PEW | ||||||
| Li+ | 1.92 | 2.08 | 2.00 | 2.07 | 2.08 | |
| Na+ | 2.35 | 2.51 | 2.39 | 2.33 | 2.43 | 2.356 |
| K+ | 2.72 | 2.88 | 2.85 | 2.73 | 2.75 | 2.798 |
| Rb+ | 2.87 | 3.02 | 2.94 | 2.88 | 2.89 | |
| Cs+ | 3.04 | 3.22 | 3.13 | 3.07 | 3.139 | |
| F- | 2.69 | 2.78 | 2.68 | 2.63 | ||
| Cl- | 3.16 | 3.26 | 3.23 | 3.10 | 3.187 | |
| Br- | 3.31 | 3.39 | 3.373 | |||
| I- | 3.52 | 3.63 | 3.61 | 3.647 | ||
| rms | 0.09 | 0.10 | 0.05 | 0.06 | 0.05 | |
| SPC/E | ||||||
| Li+ | 1.98 | 2.03 | 1.97 | 2.02 | 2.08 | |
| Na+ | 2.38 | 2.49 | 2.38 | 2.31 | 2.41 | 2.356 |
| K+ | 2.74 | 2.86 | 2.83 | 2.71 | 2.73 | 2.798 |
| Rb+ | 2.88 | 3.00 | 2.93 | 2.85 | 2.89 | |
| Cs+ | 2.96 | 3.20 | 3.12 | 3.07 | 3.139 | |
| F- | 2.68 | 2.78 | 2.69 | 2.63 | ||
| Cl- | 3.13 | 3.28 | 3.23 | 3.10 | 3.187 | |
| Br- | 3.28 | 3.39 | 3.373 | |||
| I- | 3.50 | 3.63 | 3.62 | 3.647 | ||
| rms | 0.10 | 0.09 | 0.05 | 0.07 | 0.06 | |
The rms deviations from the reference values were calculated with all the ions available. Marcus’s numbers are displayed as the reference values. The radii of the first hydration shell were calculated as described in the section and are reported in Å.
Is a Water-Model-Independent Approach to Ion-Radii Optimization Possible?
The hydration free energy of an ion is a very important property and one of the key values that is targeted in parameter optimization. Proper balance and accurate representation is critically necessary in biomolecular simulation because the interactions of ions with molecules are numerous and ions play key physiological and structural roles. A difficulty with the current approach employed is that we separately targeted optimization with specific choices of the water model. This was done because we noted that the results (such as free energies of ionic hydration, radii of the first hydration shell, etc.) depended on the choice of water model. As shown in Figures 3 and 4, the ΔGhyd = f(Rmin, ε) curve is different with different choices of the water model. Because there are many different water models available, parametrization for each becomes a chore. An alternative approach is to focus instead on optimizing the radii (Rmin) and well depths (ε) on the basis of the LC, LE, and other water-model-independent properties alone. To do this, Rmin and ε need to be optimized simultaneously, which leads to significant increases in the degrees of freedom that need to be sampled. If each degree of freedom requires a minimum of 10 data points, this leads to a total of 104 data points which is feasible. However, because the hydration free energies will be less accurately reproduced, we did not investigate this path.
Importance of Self-Consistent Ion Models across the Monovalent Ion Series
In the optimization of the ion radii, all of the ions were considered simultaneously, and the optimization occurred self-consistently across the whole series. This proves to be advantageous compared to the standard approach of fitting each ion parameter independently, one by one. Self-consistent strategies reduce the risk of systematic deviation. For example, the optimized parameters with conventional methods reproduced the properties that they targeted very well but did not show comparable accuracy for the other properties that were not fit. For example, the independent fits of KCl salts in the AMBER force fields clearly show that misbalance as the ions tend to crystallize below their solubility limit. Moreover, the self-consistent strategy spreads out the error across the series rather than fitting one ion better at the expense of another.
IV. Conclusion
Force fields for ions are dependent on the specific choice of the water model. Although changes in hydration free energies of the ions with different water models are small, across the whole series, this leads to accumulation of error and poor representation of the ion−ion and ion−water interactions. In this research, we attempted to use a simple additive pairwise force field by assuming Lorentz−Berthelot combining rules and to reoptimize the van der Waals radii (Rmin) and well depth (ε) for alkali and halide ions. The primary target property was the hydration free energy. When extracting the dependency of Rmin and ε, we further optimized the two parameters by using LE and interionic distances of the alkali−halide crystals. According to the weight on the LE and interionic distance, distinct Rmin were determined. The ratio of the weight was finally fixed to the ratio which also minimizes the binding energy and binding distance of an ion with a single water molecule. The final parameters showed good agreement with the reference values of water binding energies of multiple water molecules and reproduced well first hydration shell radii.
A crucial element of our approach was the self-consistent fitting of the parameters. So far, the previous approaches have employed ad hoc choices regarding what properties to target and the range of Rmin and ε sampled, and they have also tended to optimize the individual ion parameters independently. However, because the crystal properties such as the LE and LC are based on pairs of cations and anions, optimization should be performed simultaneously across the entire series. Fitting the properties in this way reduces serious deviations that might occur when targeting properties of the ions individually. Also, we targeted a mixture of two pairwise properties (LE and LC), which when the appropriate weights on the properties were chosen, reduced the risk of focusing the fit on one property. In summary, this work provides a new set of ion parameters for the alkali and halide ions, for use in nonpolarizable simulations with an Ewald treatment of the electrostatics and Lorentz−Berthelot combining rules, in simulations with three different popular water models. These parameters (as shown in Table 5) are likely not transferable to different (geometric mean) combining rules, electrostatic treatments that omit the long-range, polarizable models, or to different water models. However the procedure and approach used here could easily be extended to include these alternate simulation protocols.
Acknowledgments
Support from the NIH R01-GM079383-01 for the AMBER force-field consortium and computer time from NSF LRAC MCA01S027 on TeraGrid resources at NCSA, TACC, PSC and Indiana University and the University of Utah Center for High Performance Computing (including resources provided by the NIH 1S10RR17214-01 on the Arches metacluster) is greatly acknowledged.
Supporting Information Available
Supporting Information is available that includes a wealth of additional information including (a) tables of the dependence of the hydration free energy calculations on the simulation protocol, (b) pictorial representations of the crystal instability with different water models, (c) plots of the deviations of ion−water binding energies and distances with different ion models and target free energies, (d) influence of the weighting ratios with different water models, (e) multiple-water—ion binding energies with the different water models, and (f) fitting parameters for the ΔGhydration = f(Rmin, ε) hypersurface. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Manning G. S. Q. Rev. Biophys. 1978, 2, 159. [DOI] [PubMed] [Google Scholar]
- Sinibaldi F.; Howes B. D.; Smulevich G.; Ciaccio C.; Coletta M.; Santucci R. J. Biol. Inorg. Chem. 2003, 8, 663. [DOI] [PubMed] [Google Scholar]
- Auffinger P.; Bielecki L.; Westhof E. Structure 2004, 12, 379. [DOI] [PubMed] [Google Scholar]
- Klein D. J.; Moore P. B.; Steitz T. A. RNA 2004, 10, 1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sissi C.; Chemello A.; Noble C. G.; Maxwell A.; Palumbo M. J. Mol. Biol. 2005, 353, 1152. [DOI] [PubMed] [Google Scholar]
- Ke A.; Ding F.; Batchelor J. D.; Doudna J. A. Structure 2007, 15, 281. [DOI] [PubMed] [Google Scholar]
- Wolf B.; Hanlon S. Biochem. 1975, 14, 1661. [DOI] [PubMed] [Google Scholar]
- Tatulian S. A. Eur. J. Biochem. 1987, 170, 413. [DOI] [PubMed] [Google Scholar]
- Hardin C. C.; Henderson E.; Watson T.; Prosser J. K. Biochem. 1991, 30, 4460. [DOI] [PubMed] [Google Scholar]
- Clarke R. J.; Lupfert C. Biophys. J. 1999, 76, 2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D. E. RNA 2004, 10, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai G. Philos. Trans. R. Soc. London, Ser. B 2004, 359, 1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova I.; Gupta S.; Chance M. R.; Brenowitz M. J. Mol. Biol. 2004, 342, 1431. [DOI] [PubMed] [Google Scholar]
- Stellwagen E.; Dong Q.; Stellwagen N. C. Biopolymers 2005, 78, 62. [DOI] [PubMed] [Google Scholar]
- Sinibaldi F.; Piro M. C.; Coletta M.; Santucci R. FEBS J. 2006, 273, 5347. [DOI] [PubMed] [Google Scholar]
- Cano S.; Caravaca J. M.; Martin M.; Daban J. R. Eur. Biophys. J. 2006, 35, 495. [DOI] [PubMed] [Google Scholar]
- Vieregg J.; Cheng W.; Bustamante C.; Tinoco I. Jr J. Am. Chem. Soc. 2007, 129, 14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R.; Bass B. L. Annu. Rev. Biochem. 1986, 55, 599. [DOI] [PubMed] [Google Scholar]
- Dahm S. C.; Uhlenbeck O. C. Biochem. 1991, 30, 9464. [DOI] [PubMed] [Google Scholar]
- Murray J. B.; Seyhan A. A.; Walter N. G.; Burke J. M.; Scott W. G. Chem. Bio. 1998, 5, 587. [DOI] [PubMed] [Google Scholar]
- Di Cera E. J. Biol. Chem. 2006, 281, 1305. [DOI] [PubMed] [Google Scholar]
- Roychowdhury-Saha M.; Burke D. H. RNA 2007, 13, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz P. A.; Frey P. A. Biochem. 2007, 46, 7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra M.; Karttunen M. J. Comput. Chem. 2004, 25, 678. [DOI] [PubMed] [Google Scholar]
- Cheatham T. E. III; Kollman P. A. J. Am. Chem. Soc. 1997, 119, 4805. [Google Scholar]
- Young M. A.; Jayaram B.; Beveridge D. L. J. Am. Chem. Soc. 1997, 119, 59. [Google Scholar]
- Hamelberg D.; McFail-Isom L.; Williams L. D.; Wilson W. D. J. Am. Chem. Soc. 2001, 122, 10513. [DOI] [PubMed] [Google Scholar]
- Spackova N.; Berger I.; Sponer J. J. Am. Chem. Soc. 2001, 123, 3295. [DOI] [PubMed] [Google Scholar]
- Korolev N.; Lyubartsev A. P.; Laaksonen A.; Nordenskiold L. Biophys. J. 2002, 82, 2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev N.; Lyubartsev A. P.; Laaksonen A.; Nordenskiold L. Nucleic Acids Res. 2003, 31, 5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev N.; Lyubartsev A. P.; Laaksonen A.; Nordenskiold L. Biopolymers 2004, 73, 542. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P.; Monticelli L.; Tieleman D. P. Biophys. J. 2004, 86, 1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neamtu A.; Suciu D. Rom. J. Physiol. 2004, 41, 69. [PubMed] [Google Scholar]
- Costanzo F.; Della Valle R. G.; Barone V. J. Phys. Chem. B 2005, 109, 23016. [DOI] [PubMed] [Google Scholar]
- Cheng Y.; Korolev N.; Nordenskiold L. Nucleic Acids Res. 2006, 34, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Xu Y.; Tang P. J. Phys. Chem. B 2006, 110, 12789. [DOI] [PubMed] [Google Scholar]
- Pal S.; Maiti P. K.; Bagchi B.; Hynes J. T. J. Phys. Chem. B 2006, 110, 26396. [DOI] [PubMed] [Google Scholar]
- Long H.; Kudlay A.; Schatz G. C. J. Phys. Chem. B 2006, 110, 2918. [DOI] [PubMed] [Google Scholar]
- Thomas A. S.; Elcock A. H. J. Am. Chem. Soc. 2006, 128, 7796. [DOI] [PubMed] [Google Scholar]
- Vrbka L.; Jungwirth P.; Bauduin P.; Touraud D.; Kunz W. J. Phys. Chem. B 2006, 110, 7036. [DOI] [PubMed] [Google Scholar]
- Savelyev A.; Papoian G. A. J. Am. Chem. Soc. 2006, 128, 14506. [DOI] [PubMed] [Google Scholar]
- Savelyev A.; Papoian G. A. J. Am. Chem. Soc. 2007, 129, 6060. [DOI] [PubMed] [Google Scholar]
- Cui S. T. Phys. Rev. Lett. 2007, 98, 138101. [DOI] [PubMed] [Google Scholar]
- Brovchenko I.; Krukau A.; Oleinikova A.; Mazur A. K. J. Am. Chem. Soc. 2008, 130, 121. [DOI] [PubMed] [Google Scholar]
- Degreve L.; da Silva F. L. B. J. Chem. Phys. 1999, 111, 5150. [Google Scholar]
- Degreve L.; Mazze F. M. Mol. Phys. 2003, 101, 1443. [Google Scholar]
- Yang Y.; Meng S.; Xu L. F.; Wang E. G. Phys. Rev. E 2005, 72, 012602. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Meng S. J. Chem. Phys. 2007, 126, 044708. [DOI] [PubMed] [Google Scholar]
- Alejandre J.; Hansen J.-P. Phys. Rev. E 2007, 76, 061505. [DOI] [PubMed] [Google Scholar]
- Degreve L.; da Silva F. L. B. J. Mol. Liq. 2000, 87, 217. [Google Scholar]
- Gavryushov S.; Linse P. J. Phys. Chem. B 2006, 110, 10878. [DOI] [PubMed] [Google Scholar]
- Chen A. A.; Pappu R. V. J. Phys. Chem. B 2007, 111, 6469. [DOI] [PubMed] [Google Scholar]
- Cornell W. D.; Cieplak P.; Bayly C. I.; Gould I. R.; Merz K. M.; Ferguson D. M.; Spellmeyer D. C.; Fox T.; Caldwell J. W.; Kollman P. A. J. Am. Chem. Soc. 1995, 117, 5179. [Google Scholar]
- Cheatham T. E. III; Cieplak P.; Kollman P. A. J. Biomol. Struct. Dyn. 1999, 16, 845. [DOI] [PubMed] [Google Scholar]
- Wang J.; Cieplak P.; Kollman P. A. J. Comput. Chem. 2000, 21, 1049. [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C.. Proteins 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T. E. III; Crowley M. F.; Fox T.; Kollman P. A. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatham T. E. III; Kollman P. A. Structure 1997, 5, 1297. [DOI] [PubMed] [Google Scholar]
- Cheatham T. E. III; Kollman P. A.. Molecular dynamics simulation of nucleic acids in solution: How sensitive are the results to small perturbations in the force field and environment. In Structure, motion, interactions and expression of biological macromolecules; Sarma M., Sarma R., Eds.; Adenine Press: Schenectady, NY, 1998; pp 99. [Google Scholar]
- Cheatham T. E. III; Young M. A. Biopolymers 2001, 56, 232. [DOI] [PubMed] [Google Scholar]
- Auffinger P.; Cheatham T. E. III; Vaiana A. C. J. Chem. Ther. Comp. 2007, 3, 1851. [DOI] [PubMed] [Google Scholar]
- Chen A. A.; Pappu R. V. J. Phys. Chem. B 2007, 111, 11884. [DOI] [PubMed] [Google Scholar]
- Aqvist J. J. Phys. Chem. 1990, 94, 8021. [Google Scholar]
- Dang L. X. J. Am. Chem. Soc. 1995, 117, 6954. [Google Scholar]
- Beveridge D. L.; Barreiro G.; Byun K. S.; Case D. A.; Cheatham T. E. III; Dixit S. B.; Giudice E.; Lankas F.; Lavery R.; Maddocks J. H.; Osman R.; Seibert E.; Sklenar H.; Stoll G.; Thayer K. M.; Varnai P.; Young M. A. Biophys. J. 2004, 87, 3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S. B.; Beveridge D. L.; Case D. A.; Cheatham T. E. III; Giudice E.; Lankas F.; Lavery R.; Maddocks J. H.; Osman R.; Sklenar H.; Thayer K. M.; Varnai P. Biophys. J. 2005, 89, 3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle B.; Denisov V. P. Biopolymers 1998, 48, 210. [DOI] [PubMed] [Google Scholar]
- Denisov V. P.; Halle B. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtovenko A. A.; Vattulainen I. J. Phys. Chem. B 2008, 112, 1953. [DOI] [PubMed] [Google Scholar]
- Fuoss R. M. Proc. Natl. Acad. Sci. U.S.A. 1980, 77, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. J. Chem. Phys. 1983, 79, 926. [Google Scholar]
- Berendsen H. J. C.; Grigera J. R.; Straatsma T. P. J. Phys. Chem. 1987, 91, 6269. [Google Scholar]
- Horn H. W.; Swope W. C.; Pitera J. W.; Madura J. D.; Dick T. J.; Hura G. L.; Head-Gordon T. J. Chem. Phys. 2004, 120, 9665. [DOI] [PubMed] [Google Scholar]
- Essmann U.; Perera L.; Berkowitz M. L.; Darden T.; Lee H.; Pedersen L. G. J. Chem. Phys. 1995, 103, 8577. [Google Scholar]
- Jensen K. P.; Jorgensen W. L. J. Chem. Ther. Comp. 2006, 2, 1499. [DOI] [PubMed] [Google Scholar]
- Darden T.; Pearlman D.; Pedersen L. G. J. Chem. Phys. 1998, 109, 10921. [Google Scholar]
- Jorgensen W. L.; Madura J. D. Mol. Phys. 1985, 56, 1381. [Google Scholar]
- Jensen K. P. J. Phys. Chem. B 2008, 112, 1820. [DOI] [PubMed] [Google Scholar]
- Rick S. W. J. Chem. Phys. 2004, 120, 6085. [DOI] [PubMed] [Google Scholar]
- Pearlman D. A.; Case D. A.; Caldwell J. W.; Ross W. S.; Cheatham T. E.; Debolt S.; Ferguson D.; Seibel G.; Kollman P. Comput. Phys. Commun. 1995, 91, 1. [Google Scholar]
- Case D. A.; Cheatham T. E. III; Darden T. A.; Gohlker H.; Luo R.; Merz K. M. Jr.; Onufriev A. V.; Simmerling C.; Wang B.; Woods R. J. Comput. Chem. 2005, 26, 1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. J. Comput. Chem. 1983, 4, 187. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. J. Comput. Chem. 2005, 26, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waals J. D. Z. Phys. Chem. 1890, 5, 133. [Google Scholar]
- Berthelot D. Compt. Rend. Acad. Sci. Paris 1889, 126, 1703. [Google Scholar]
- Lorentz H. A. Ann. Phys. 1881, 12, 127. [Google Scholar]
- Allen M. P.; Tildesley D. J.. Computer simulation of liquids; Oxford University Press: Oxford, 1987. [Google Scholar]
- Jorgensen W.BOSS, Version 3.6; Yale University: New Haven, 1995. [Google Scholar]
- van Gunsteren W. F.; Berendsen H. J. C.. Groningen molecular simulation (GROMOS) library manual; BIOMOS: Nijenborgh, Groningen, The Netherlands, 1987. [Google Scholar]
- Marrone T. J.; Merz K. M. J. Phys. Chem. 1993, 97, 6524. [Google Scholar]
- Aqvist J. J. Phys. Chem. 1994, 98, 8253. [Google Scholar]
- Marrone T. J.; Merz K. M. J. Phys. Chem. 1994, 98, 8256. [Google Scholar]
- Delhommelle J.; Millie P. Mol. Phys. 2001, 99, 619. [Google Scholar]
- Smith D. E.; Dang L. X. J. Chem. Phys. 1994, 100, 3757. [Google Scholar]
- Lamoureux G.; Roux B. J. Phys. Chem. B 2006, 110, 3308. [DOI] [PubMed] [Google Scholar]
- Beglov D.; Roux B. J. Chem. Phys. 1994, 100, 9050. [Google Scholar]
- Roux B. Biophys. J. 1996, 71, 3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L. X. J. Chem. Phys. 1992, 96, 6970. [Google Scholar]
- Dang L. X. Chem. Phys. Lett. 1994, 227, 211. [Google Scholar]
- Dang L. X.; Garrett B. C. J. Chem. Phys. 1993, 99, 2972. [Google Scholar]
- Lopes J. N. C.; Padua A. A. H. J. Phys. Chem. B 2006, 110, 19586. [DOI] [PubMed] [Google Scholar]
- Lenart P. J.; Jusufi A.; Panagiotopoulos A. Z. J. Chem. Phys. 2007, 126, 044509. [DOI] [PubMed] [Google Scholar]
- Teleman O.; Ahlstrom P. J. Am. Chem. Soc. 1986, 108, 4333. [Google Scholar]
- Weerasinghe S.; Smith P. E. J. Chem. Phys. 2003, 119, 11342. [DOI] [PubMed] [Google Scholar]
- Straatsma T. P.; Berendsen H. J. C. J. Chem. Phys. 1988, 89, 5876. [Google Scholar]
- Sremaniak L. S.; Perera L.; Berkowitz M. L. Chem. Phys. Lett. 1994, 218, 377. [Google Scholar]
- Peslherbe G. H.; Ladanyi B. M.; Hynes J. T. Chem. Phys. 2000, 258, 201. [Google Scholar]
- Huggins M. L.; Mayer J. E. J. Chem. Phys. 1933, 1, 643. [Google Scholar]
- Pettitt B. M.; Rossky P. J. J. Chem. Phys. 1986, 84, 5836. [Google Scholar]
- Fumi F. G.; Tosi M. P. J. Phys. Chem. Solids 1964, 25, 31. [Google Scholar]
- Tosi M. P.; Fumi F. G. J. Phys. Chem. Solids 1964, 25, 45. [Google Scholar]
- Burgess M. A.Metal ions in solution; Ellis Horwood: Chichester, England, 1978. [Google Scholar]
- Pauling L.The nature of the chemical bond; Cornell University Press: Ithaca, NY, 1960. [Google Scholar]
- Magini M. In Ions and molecules in solution; Tanaka N., Ohtaki H., Tamamushi R., Eds.; Elsevier: Amsterdam, 1983; Vol. 27; pp 470. [Google Scholar]
- Tissandier M. D.; Cowen K. A.; Feng W. Y.; Gundlach E.; Cohen M. H.; Earhart A. D.; Coe J. V.; Tuttle T. R. J. Phys. Chem. A 1998, 102, 7787. [Google Scholar]
- Marcus Y. Biophys. Chem. 1994, 51, 111. [Google Scholar]
- Schmid R.; Miah A. M.; Sapunov V. N. Phys. Chem. Chem. Phys. 2000, 2, 97. [Google Scholar]
- Grossfield A.; Ren P.; Ponder J. W. J. Am. Chem. Soc. 2003, 125, 15671. [DOI] [PubMed] [Google Scholar]
- King G.; Warshel A. J. Chem. Phys. 1989, 91, 3647. [Google Scholar]
- MacKerell A. D. Jr J. Comput. Chem. 2004, 25, 1584. [DOI] [PubMed] [Google Scholar]
- Teleman O.; Wallqvist A. Int. J. Quantum Chem. 1990, 24, 245. [Google Scholar]
- Hummer G.; Soumpasis D. M.; Neumann M. Mol. Phys. 1993, 81, 1155. [Google Scholar]
- Bader J. S.; Chandler D. J. Phys. Chem. 1992, 96, 6423. [Google Scholar]
- Khavrutskii I. V.; Dzubiella J.; McCammon J. A. J. Chem. Phys. 2008, 128, 044106. [DOI] [PubMed] [Google Scholar]
- Smith P. E.; Blatt H. D.; Pettitt B. M. J. Phys. Chem. 1997, 101B, 3886. [Google Scholar]
- Smith P. E.; Pettitt B. M. J. Chem. Phys. 1996, 105, 4289. [Google Scholar]
- Hunenberger P. H.; McCammon J. A. Biophys. Chem. 1999, 78, 69. [DOI] [PubMed] [Google Scholar]
- Hunenberger P. H.; McCammon J. A. J. Chem. Phys. 1999, 110, 1856. [Google Scholar]
- Weber W.; Hunenberger P. H.; McCammon J. A. J. Phys. Chem. B. 2000, 104, 3668. [Google Scholar]
- Villarreal M. A.; Montich G. G. J. Biomol. Struct. Dyn. 2005, 23, 135. [DOI] [PubMed] [Google Scholar]
- Feller S. E.; Pastor R. W.; Rojnuckarin A.; Bogusz S.; Brooks B. R. J. Phys. Chem. 1996, 100, 17011. [Google Scholar]
- Dufner H.; Kast S. M.; Brickmann J.; Schlenkrich M. J. Comput. Chem. 1997, 18, 660. [Google Scholar]
- de Vries A. H.; Chandrasekhar I.; van Gunsteren W. F.; Hunenberger P. H. J. Phys. Chem. B 2005, 109, 11643. [DOI] [PubMed] [Google Scholar]
- DeLeeuw S. W.; Perram J. M.; Smith E. R. Proc. R. Soc. London 1980, A373, 27. [Google Scholar]
- Roberts J. E.; Schnitker J. J. Chem. Phys. 1994, 101, 5024. [Google Scholar]
- Roberts J. E.; Schnitker J. J. Phys. Chem. 1995, 99, 1322. [Google Scholar]
- Hummer G.; Pratt L. R.; Garcia A. J. Phys. Chem. 1995, 99, 14188. [Google Scholar]
- Boresch S.; Steinhauser O. Ber. Bunsenges. Phys. Chem. 1997, 101, 1019. [Google Scholar]
- Figueirido F.; Del Buono G. S.; Levy R. M. J. Chem. Phys. 1995, 103, 6133. [Google Scholar]
- Hummer G.; Pratt L. R.; Garcia A. E. J. Phys. Chem. 1996, 100, 1206. [Google Scholar]
- Kalko S. G.; Sese G.; Padro J. A. J. Chem. Phys. 1996, 104, 9578. [Google Scholar]
- Hummer G.; Pratt L. R.; Garcia A. J. Chem. Phys. 1997, 107, 9275. [Google Scholar]
- Figueirido F.; Del Buono G. S.; Levy R. M. J. Phys. Chem. 1997, 101, 5622. [Google Scholar]
- Bogusz S.; Cheatham T. E. III; Brooks B. R. J. Chem. Phys. 1998, 108, 7070. [Google Scholar]
- Kastenholz M. A.; Hunenberger P. H. J. Chem. Phys. 2006, 124, 224501. [DOI] [PubMed] [Google Scholar]
- Latimer W. M.; Pitzer K. S.; Slansky C. M. J. Chem. Phys. 1939, 7, 108. [Google Scholar]
- Morris D. F. C. Electrochim. Acta 1982, 27, 1481. [Google Scholar]
- Marcus Y. J. Chem. Soc., Faraday Trans. 1 1986, 82, 233. [Google Scholar]
- Tuttle T. R.; Malaxos S.; Coe J. V. J. Phys. Chem. A 2002, 106, 925. [Google Scholar]
- Noyes R. M. J. Am. Chem. Soc. 1962, 84, 513. [Google Scholar]
- Asthagiri D.; Pratt L. R.; Ashbaugh H. S. J. Chem. Phys. 2003, 119, 2702. [Google Scholar]
- Kelly C. P.; Cramer C. J.; Truhlar D. G. J. Phys. Chem. B 2006, 110, 16066. [DOI] [PubMed] [Google Scholar]
- Kelly C. P.; Cramer C. J.; Truhlar D. G. J. Phys. Chem. B 2007, 111, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan C. G.; Dixon D. A. J. Phys. Chem. A 2002, 106, 9737. [Google Scholar]
- Cubicciotti D. J. Chem. Phys. 1959, 31, 1646. [Google Scholar]
- Huggins M. L. J. Chem. Phys. 1937, 5, 143. [Google Scholar]
- Huggins M. L. J. Chem. Phys. 1937, 15, 212. [Google Scholar]
- Kapustinskii A. F. Quart. Rev. Chem. Soc. 1956, 10, 283. [Google Scholar]
- Born M. V. Verh. Deutsch. Phys. Ges. 1919, 21, 679. [Google Scholar]
- Fajans K. Verh. Deutsch. Phys. Ges. 1919, 21, 714. [Google Scholar]
- Haber F. Verh. Deutsch. Phys. Ges. 1919, 21, 750. [Google Scholar]
- Lide D. R.Handbook of Chemistry and Physics, Internet Version, 87th ed.; Taylor and Francis: Boca Raton, 2007. [Google Scholar]
- Pauling L. J. Am. Chem. Soc. 1927, 49, 765. [Google Scholar]
- Sirdeshmukh D. B.; Sirdeshmukh L.; Subhadra K. G.. Alkali Halides. A Handbook of Physical Properties; Springer: Berlin, 2001; Vol. 49. [Google Scholar]
- Sagui C.; Darden T. A. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 155. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; van Gunsteren W. F.; DiNola A.; Haak J. R. J. Comp. Phys. 1984, 81, 3684. [Google Scholar]
- Kollman P. A. Chem. Rev. 1993, 93, 2395. [Google Scholar]
- Pearlman D. A. J. Phys. Chem. 1993, 98, 1487. [Google Scholar]
- Steinbrecher T.; Mobley D. L.; Case D. A. J. Chem. Phys. 2007, 127, 214108. [DOI] [PubMed] [Google Scholar]
- Galassi M.; Davies J.; Theiler J.; Gough B.; Jungman G.; Booth M.; Rossi F.. GNU Scientific Library Reference Manual, revised 2nd ed.; Network Theory Limited: Bristol, 2006. [Google Scholar]
- Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. J. Comp. Phys. 1977, 23, 327. [Google Scholar]
- Hashimoto K.; Kamimoto T. J. Am. Chem. Soc. 1998, 120, 3560. [Google Scholar]
- Feller D.; Glendening E. D.; Kendall R. A.; Peterson K. A. J. Chem. Phys. 1994, 100, 4981. [Google Scholar]
- Kim J.; Lee S.; Cho S. J.; Mhin B. J.; Kim K. S. J. Chem. Phys. 1995, 102, 839. [Google Scholar]
- Lee H. M.; Kim J.; Lee S.; Mhin B. J.; Kim K. S. J. Chem. Phys. 1999, 111, 3995. [Google Scholar]
- Park J.; Kolaski M.; Lee H. M.; Kim K. S. J. Chem. Phys. 2004, 121, 3108. [DOI] [PubMed] [Google Scholar]
- Kolaski M.; Lee H. M.; Choi Y. C.; Kim K. S.; Tarakeshwar P.; Miller D. J.; Lisy J. M. J. Chem. Phys. 2007, 126, 074301. [DOI] [PubMed] [Google Scholar]
- Kim J.; Lee H. M.; Suh S. B.; Majumdar D.; Kim K. S. J. Chem. Phys. 2000, 113, 5259. [Google Scholar]
- Xantheas S. S.; Dunning T. H. Jr J. Phys. Chem. 1994, 98, 13489. [Google Scholar]
- Xantheas S. S. J. Phys. Chem. 1996, 100, 9703. [Google Scholar]
- Lee H. M.; Kim K. S. J. Chem. Phys. 2001, 114, 4461. [Google Scholar]
- Glendening E. D.; Feller D. J. Phys. Chem. 1995, 99, 3060. [Google Scholar]
- Wilson M. A.; Pohorille A.; Pratt L. R. J. Chem. Phys. 1988, 88, 3281. [DOI] [PubMed] [Google Scholar]
- Brodskaya E. N.; Zakharov V. V. J. Chem. Phys. 1995, 102, 4595. [Google Scholar]
- Marcus Y. Chem. Rev. 1988, 88, 1475. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.