Abstract
Structural modification of peptidoglycan (PG) is one of the mechanisms that pathogenic bacteria use to evade the host innate immune system. For the noninvasive human gastric pathogen Helicobacter pylori, PG delivery to the host cells is one trigger of the immune response. H. pylori HP310 was markedly up-expressed upon cell exposure to oxidative stress. However, disruption of HP310 did not produce a phenotype distinguishable from the parent, including oxidative stress resistance characteristics. HP310 shows very weak homology to a known gene pgdA encoding PG deacetylase in Streptococcous pneumoniae. PGs from wild type H. pylori and the HP310 mutant were purified and analyzed by matrix-assisted laser desorption ionization time-of-flight and high pressure liquid chromatography. The parent strain PG is partially deacetylated, whereas several major PG-deacetylated muropeptides are absent or significantly reduced in the HP310 mutant. PG deacetylase activity was directly demonstrated by use of pure PG and HP310 protein by measuring the release of acetic acid. The Gram-negative bacterium H. pylori is highly resistant to lysozyme (up to 50 mg/ml), but the HP310 mutant is less resistant to lysozyme compared with the parent strain. Complementation of an hp310 strain with the wild type gene restored lysozyme resistance. The purified PG from the mutant is more susceptible to lysozyme (0.3 mg/ml) digestion than the wild type PG. The PG deacetylation appears to confer lysozyme resistance to escape immune detection. HP310 is representative of a new subfamily of bacterial PG deacetylases.
Helicobacter pylori, a pathogenic bacterium infecting over 50% of humans, is the etiological agent for gastritis, peptic ulcer, and gastric cancer (1). H. pylori is highly adapted to its ecologic niche, the human gastric mucosa. The pathogenesis of H. pylori relies on its persistence in surviving a harsh environment, including acidity, peristalsis, and attack by phagocyte cells and their released reactive oxygen species (ROS)2 (2). H. pylori has a unique array of features that permit entry into the mucus, attachment to epithelial cells, evasion of the immune response, and as a result, persistent colonization and transmission. Numerous virulence factors in H. pylori have been extensively studied, including urease, flagella, BabA adhesin, the vacuolating cytotoxin (VacA), and the cag pathogenicity island (cag-PAI).
Peptidoglycan (PG) was found in recent years to be an important factor involved in virulence by pathogenic bacteria (3). PG is one of the main protective barriers in the bacterial cell wall. PG consists of glycan strands made of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), which are cross-linked by short peptide chains. In mammalian cells, a group of pattern recognition molecules (Nod1 and Nod2) can sense bacterial PG degradation products (muropeptides), initiating innate immune responses (4, 5). For example, human Nod1 (Toll-like receptor) specifically detects a unique (GlcNAc-MurNAc) tripeptide motif found in Gram-negative bacterial PG, resulting in activation of the transcription factor NF-κB pathway (6). The induction of interleukin-8 production by gastric epithelial cells (the first line of defense against invasive pathogens) was shown to be dependent on the Nod1 pathway (7). H. pylori, as a noninvasive pathogen, can inject its PG into the host epithelial cells by a bacterial type IV secretion system encoded by the cag pathogenicity island (7). Most recently it was shown that PG modification, specifically N-deacetylation, is a highly efficient mechanism used by pathogenic bacteria to evade innate host defenses (8). Although PG deacetylation was suggested to play such a role in H. pylori (8), the PG structure (the deacetylation pattern) and the gene/enzyme responsible for PG deacetylation have not been described in H. pylori.
During the process of colonizing the host, H. pylori induces a strong inflammatory response, generating large amounts of ROS. Increased levels of ROS in the gastric mucosa have been measured in H. pylori-infected patients (9, 10). Exposure of gastric epithelial cells to H. pylori resulted in an inflammatory reaction with production of ROS and nitric oxide (11). Recent studies further demonstrated that increased levels of ROS are generated in H. pylori-infected gastric epithelial cells and that this may be one mechanism leading to apoptosis associated with infection (12). H. pylori survives these oxidative stress conditions via a battery of diverse activities (detoxification and repair), and it persistently colonizes the gastric mucosa (13). The oxidative stress-combating proteins that have been identified include superoxide dismutase (SodB) (14), catalase (KatA) (15), alkylhydroperoxide reductase (AhpC) (16), the neutrophil-activating protein (NapA) (17), NADPH quinone reductase (MdaB) (18), methoinine sulfoxide reductase (Msr) (19), and several DNA repair enzymes (20–22). Disruption of any of these genes resulted in a significantly reduced ability to colonize the host. Among these oxidative stress-combating proteins, KatA, AhpC, and NapA are abundantly expressed in H. pylori (23).
While studying the oxidative stress induction of potential virulence factors in H. pylori, we observed a hypothetical protein (HP310) that was greatly up-expressed upon increased oxygen tension exposure. By characterizing an HP310 mutant of H. pylori, the purified PGs, and the purified HP310 protein, we demonstrated that HP310 is a PG deacetylase. H. pylori is highly resistant to lysozyme; deacetylation of its PG contributes to this resistance.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions—H. pylori strain 43504 was used as the wild type. Cultures were grown microaerobically at 37 °C in CO2 incubators under controlled oxygen concentration as indicated throughout the text. Brucella agar (Difco) was used in preparing the plates supplemented with 10% defibrinated sheep blood (Gibson Laboratories, Inc.). Kanamycin and chloramphenicol antibiotics were used in concentration of 50 μg/ml. Escherichia coli DH5α strains were grown in Luria-Bertani agar or broth supplemented with 100 μg/ml ampicillin, 30 μg/ml chloramphenicol, or 50 μg/ml kanamycin. Rosetta strains were used for overexpression of HP310.
Cloning of HP310 and Construction of an hp310:kan Mutant—Forward primer 5′ AAACGAAGTGGCACTGCAAC 3′ and reverse primer 5′ CACCGCATAATCCATCTCAC 3′ were designed based on the published genome sequences and used for PCR amplification of HP310. The 827-bp-long PCR fragment was cloned into pGEMT vector (Promega). The php310: kan construct was created via insertion of a kanamycin resistance cassette into the BamHI site within the HP310 fragment and confirmed by restriction analysis. This plasmid was used for natural transformation of H. pylori, and the hp310:kan mutant strain was obtained by incubating cells on blood agar plates with kanamycin at 5% O2.
Complementation of HP310—A 210-bp DNA fragment containing the H. pylori ureA promoter was cloned into pET21a vector digested with BglII and NdeI. The resulting plasmid was then cut with XhoI and NdeI and ligated with the hp310 gene fragment flanked by digestion sites XhoI and NdeI. The plasmid pET21-pUreA-HP310 was then digested with SphI, blunted, and digested again with XhoI. The 1400-bp-long fragment containing the PureA promoter with the hp310 gene was then ligated into a pEU39Cm vector (pre-digested with SmaI and XhoI). The ligation product was transformed into E. coli DH5α by selection for ampicillin resistance. The resulting recombinant plasmid pEU39Cm-pUreA-HP310 was confirmed by restriction digestion analysis. The plasmid pEU39 contains a 2.04-kb fragment of H. pylori genomic DNA covering the open reading frame HP0405. Previously, our laboratory showed that disruption of HP0405 in H. pylori had no obvious phenotype. In plasmid pEU39Cm-pUreA-HP310, HP0405 was disrupted into two pieces flanking the Cm-pUreA-HP310. When this plasmid was used to transform H. pylori hp310:kan mutant (selection for Cmr Kanr), the intact hp310 gene (driven by ureA promoter) and the Cm cassette were inserted into the genome at HP0405 locus. This produced a merodiploid strain, hp310:kan-hp310+-Cm, which contains the original interrupted hp310 and an intact copy of hp310 at an unrelated site.
Protein Gel Electrophoresis and Protein Identification by N-terminal Sequencing—Bacteria were harvested from the plates and resuspended in PBS containing 20 mm sodium phosphate and 150 mm NaCl (pH 8.0). After washing once with the buffer, cells were resuspended in the same buffer and were broken by two passages through a French pressure cell at 138,000 kPa (SLM instruments, Inc.). The cell lysates were obtained by centrifugation (8,000 rpm for 10 min), and the supernatant was transferred to a clean tube. The protein concentration of cell extract was determined by Bradford protein assay (Bio-Rad). Seven micrograms of cell extract was mixed with the SDS buffer and incubated at 90 °C for 5 min. Proteins were then separated on the 12.5% SDS-polyacrylamide gel by electrophoresis for 1.5 h at 100 V.
The protein band of interest, which was resolved in SDS-PAGE, was subjected to N-terminal sequencing. The proteins on the gel were transferred onto a polyvinylidene difluoride membrane (24), and the band of interest was excised and sent for N-terminal sequencing (Protein Sequencing Lab, Georgia State University).
Anti-HP310 Antibody and Immunoblotting—Two peptides, each consisting of 15 amino acids, were synthesized by Antagene, Inc. (Sunnyvale, CA): peptide 1, IHPDVSARPQVLLMH; peptide 2, EILVAYGVDIDAVAG. The sequences of the two peptides are specific for HP310 in H. pylori. They are identical in the published genome sequences from three different strains but have no homology with any other known PG deacetylases. Polyclonal antibody was produced in rabbits against the mixture of the two synthesized and conjugated HP310-specific peptides by Antagene, Inc., using their conventional antiserum production protocols.
Crude cell extracts of H. pylori WT or hp310:kan mutant grown under 2% O2 or 12% O2 conditions were subjected to SDS-PAGE. The proteins were then electrotransferred onto a nitrocellulose membrane. The membrane was then incubated with anti-HP310 antiserum (1:500 dilution) followed by incubation with secondary goat anti-rabbit-IgG conjugated with alkaline phosphatase (1:1000 dilution; Bio-Rad).
Overexpression/Purification of His6-tagged H. pylori HP310 Protein—Primers (reverse, 5′ CACGACTCGAGTTTTTTTCTAGGGTT 3′; forward, 5′ CGCCGGCATATGGCAAAAGAAATTTT 3′) were designed to amplify a fragment of HP310 with flanking restriction sites of XhoI and NdeI. A 900-bp-long PCR product was digested with XhoI and NdeI and cloned into pET21a vector (Novagen). The resulting plasmid pET21a-HP310-His6 was confirmed by restriction digestion analysis and then transformed into E. coli Rosetta cells. The cells were grown at 37 °C to an A600 of 0.5 in 500 ml of LB medium with 100 μg/ml ampicillin and 50 μg/ml kanamycin. The cells were then induced by 1 mm isopropyl β-d-thiogalactopyranoside and incubated with shaking for another 4 h. The cells were then harvested by centrifugation (4,000 × g, 20 min, 4 °C). Cells were washed with 50 mm NaH2PO4 (pH 8), 300 mm NaCl, and 10 mm imidazole and resuspended in the same buffer. Cells were lysed by three passages through the French pressure cell and centrifuged (10,000 × g, 20 min) to collect the supernatant. The supernatant was centrifuged again at 45,000 × g for 80 min. The collected supernatant was applied on the nickel-nitrilotriacetic acid affinity column (Qiagen) and washed with lysis buffer (containing 10 mm imidazole) and collected until A280 reached the base line. The proteins were then washed with buffer containing 20 mm imidazole until A280 reached base line again. The protein was eluted (250 mm imidazole) and collected until A280 reached a base line. Fractions were then analyzed by gel electrophoresis.
Paper Disk Assay for Sensitivity—Sensitivity of the wild type versus mutant to different oxidative stress generators (30% hydrogen peroxide, up to 20 mm paraquat, 10, 20, 50% of t-butyl hydroperoxide, and up to 50% cumene hydroperoxide), dyes (up to 500 mm benzyl viologen, 10 mg/ml gentian violet, 11 mg/ml crystal violet), and detergents (100% Triton X-100, 100% Tween 20, 100% SDS) was measured by using sterile filter paper disks saturated with each agent. The saturated disks were placed onto the BA plate inoculated with H. pylori strain mutant and wild type as described previously (16). Plates were then incubated for 2 days, and the zones of inhibited growth were measured.
Measurement of Deacetylase Activity by Determination of Acetic Acid Release—To measure deacetylase activity, 50 mm HEPES buffer (pH 7) was used as described (25). Purified HP310 protein at various concentrations was added with 1 mg of peptidoglycan derived from the hp310:kan mutant strain in a total volume of 1 ml. After incubation at 37 °C for 4 h, the sample was centrifuged, and the supernatant was collected to be assayed for acetic acid release. The release was determined with the acetic acid kit (R-biopharm). The final calculations were made by using the formula provided in the kit.
Isolation and Preparation of Peptidoglycan—H. pylori cells were harvested and washed with ice-cold 20 mm sodium acetate (pH 5), centrifuged at 8,000 × g for 30 min at 4 °C, and then resuspended in 10 ml of the same buffer. The suspension was added dropwise to 10 ml of boiling 4% SDS buffered with 20 mm sodium acetate (pH 5) and boiled for 30 min (the reaction was carried out in a way to allow constant stirring to prevent spilling of the solution out of the hot flask). The samples were then cooled to room temperature overnight, and SDS-insoluble material was collected by centrifugation at 65,000 rpm for 1 h. The pellet was washed five times with warm water and resuspended in 5 ml of 100 mm Tris-HCl (pH 7.5) and 10 mm NaCl. 10 μg/ml DNase and 50 μg/ml of RNase (Sigma) were added and incubated at 37 °C with shaking for 2 h. 50 μg/ml proteinase K (Invitrogen) was added to the reaction and incubated at 37 °C with shaking overnight. The SDS-insoluble material was re-extracted by boiling in 1% SDS for 15 min and collected/washed by centrifugation four times with warm water. The peptidoglycan pellet was resuspended in distilled water and lyophilized.
Preparation of Muropeptides—Lyophilized peptidoglycan was suspended in 200 μl of 20 mm sodium phosphate buffer (pH 4.8), using sonication in a bath-type sonicator for 3 h at 40 °C to assist resuspension. Following sonication, the suspensions were digested with 50 μg/50 μl of muramidase (lysozyme, EC 3.2.1.17, from chicken egg white, Sigma L-7001, 40,800 units/mg solid) 18 h at 37 °C. The enzyme incubations were placed in a 100 °C bath for 3 min, cooled, then centrifuged (14,000 × g 3 min) to remove insoluble material (contaminants and any undigested peptidoglycan). Supernatants, containing the digested peptidoglycan, were mixed with an equal volume of 0.5 m borate buffer (pH 8.0) containing freshly dissolved sodium borodeuteride (10 mg/ml) and treated for 1 h at room temperature to reduce the free reducing ends of the muropeptide oligosaccharide moiety. Excess borodeuteride was destroyed, and the pH was adjusted to 4.0 by adding 20% phosphoric acid (∼50 μl). The reactions were then filtered (0.2-μm nylon spin filters) prior to HPLC analysis.
Analysis of Lysozyme Digestion Products of PG by Mass Spectrometry—Portions of the reaction mixtures resulting from lysozyme digestion were reduced with borodeuteride as described above, and then desalted using a Superdex-Peptide HR 10/30 FPLC column (Amersham Biosciences) eluted with 50 mm ammonium acetate (pH 6.0), collecting 0.4-ml fractions. The eluant was monitored at 206 nm, and fractions containing peptides and muropeptides (eluting prior to the salt peak) were combined, concentrated by rotary evaporation, and analyzed by MALDI-TOF mass spectrometry. Individual muropeptide peaks obtained by HPLC elution were also isolated, desalted, and prepared for MALDI analysis using the Superdex column. MALDI mass spectrometry was performed on a Voyager-DE TOF spectrometer (Applied Biosystems, Boston) in the positive and negative modes, using a matrix of 100 mm 2,5-dihydroxybenzoic acid in 90% methanol. The instrument was operated at an accelerating voltage of 25 kV with extraction delay time of 150 ns. Samples were desorbed with a nitrogen laser (λ = 337 nm), and the detector sensitivity was 1000 mV full scale. Mass spectra were recorded over an m/z range of 500–20,000; spectra are the summation of 200 acquisitions. Maltooligosaccharides, standard peptides, and muramyldipeptide (N-acetylmuramic acid-l-alanyl-d-isoglutamine, Sigma A-9519) were used as calibration standards.
Chemical N-Acetylation of Peptidoglycan—Portions of the peptidoglycan preparations were resuspended by sonication as described above and subjected to N-acetylation, using saturated NaHCO3 and 5% acetic anhydride essentially as described (26). The N-acetylated peptidoglycan was recovered by centrifugation (14,000 × g, 3 min) and then subjected to lysozyme digestion, borodeuteride reduction, workup, and HPLC analysis as described above.
Cell Survival Assay for Assessing Lysozyme Sensitivity—H. pylori strains were grown on BA plates to late log-phase, and the cells were suspended in PBS at a concentration of ∼109 cells/ml. Upon addition of lysozyme (final concentration 50 mg/ml), the cell suspensions were incubated at 37 °C under 2% O2 condition with occasional shaking. Samples were then removed at various time points (0, 2, 4, and 6 h), serially diluted, and spread onto BA plates. Colony counts are recorded after 4 days of incubation in a microaerobic atmosphere (2% O2) at 37 °C.
Turbidometric Assay of Peptidoglycan—The susceptibility of the isolated peptidoglycan to lysozyme was analyzed with a turbidometric assay (27, 28). The peptidoglycan of the wild type and mutant strains was sonicated and diluted to 0.5 mg ml–1 in 80 mm PBS (pH 6.4). After addition of 0.3 mg/ml lysozyme, the absorbance at 600 nm was monitored every hour.
RESULTS
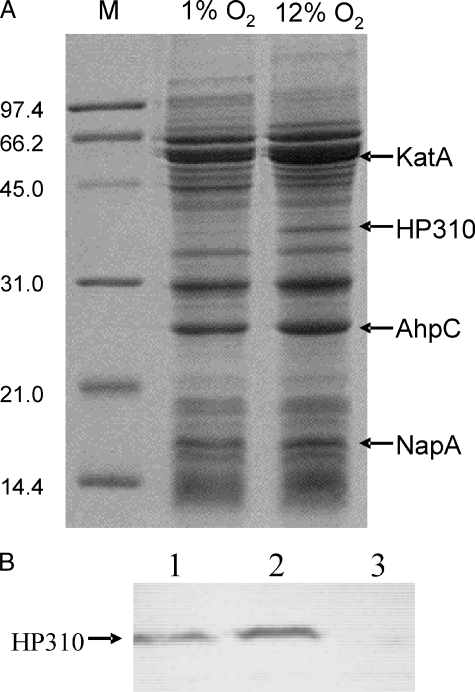
Oxidative Stress Induces Overexpression of a Hypothetical Protein (HP310) in H. pylori—The optimal O2 tension for the growth of H. pylori is 5–7% O2. Cells cultured at higher O2 tension may generate ROS and die (29). To overcome oxidative stress, the cells may modify their protein expression. We investigated the protein profiles of H. pylori (strain 43504) grown at 12% O2 versus at 1% O2. The major oxidative stress-related proteins, such as catalase (KatA), alkyl-hydroperoxide reductase (AhpC), and the neutrophil-activating protein (NapA), are abundantly expressed in H. pylori, and they can be easily identified by one-dimensional SDS-PAGE (23). As shown in Fig. 1, growth at 12% O2 did not change the expression of these proteins significantly. Intriguingly, the protein profile revealed a significant up-expression of an unknown protein of ∼38 kDa (Fig. 1). To identify the up-expressed protein, the gel containing the induced band was transferred onto a polyvinylidene difluoride membrane. The band was excised and subjected to N-terminal sequencing. The amino acid sequence (AKEILVAY) revealed its identity to a conserved hypothetical protein HP310 (33.6 kDa) from the published genome sequence (H. pylori strain 26695).
FIGURE 1.
SDS-PAGE of H. pylori proteins and Western blot analysis for HP310. A, wild type strain 43504 was grown at 1% O2 and 12% O2 conditions, and total protein extracts were subjected to analysis on a SDS-12.5% polyacrylamide gel. The arrows on the right mark several identifiable proteins (KatA, AhpC, and NapA) that are related to oxidative stress and a protein (HP310) that was up-expressed at 12% O2 condition. The M lane shows protein standards ranging from 14.4 to 97.4 kDa. B, 7 μg of crude cell extracts were resolved on a SDS-12.5% polyacrylamide gel followed by transfer onto a nitrocellulose membrane, and HP310 protein was detected using anti-HP310 antibody. Lane 1, WT cells grown at 2% O2; lane 2, WT cells grown at 12% O2; lane 3, hp310:kan mutant cells grown at 12% O2.
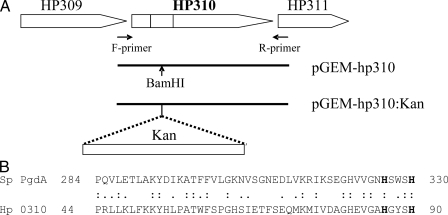
The sequence homology search revealed that HP310 contains a domain sequence similar to that of polysaccharide deacetylase (pfam01522). These proteins belong to the carbohydrate esterase family 4 (CE4), which also includes chitin deacetylase, acetylxylan esterase, chitooligosaccharide deacetylase, and PG deacetylase (30). The true homologues of HP310 (60–90% identity over the full length of protein) are present in the published genome sequences of many pathogenic bacteria, such as Campylobacter coli, Pseudomonas syringae, Yersinia pestis, Rhodococcus sp. RHA1, Corynebacterium efficiens, Rhodobacter sphaeroides, and Mycobacterium smegmatis; however, in none of them has this protein been studied. One member of the bacterial polysaccharide deacetylase family proteins that has been well characterized is the peptidoglycan deacetylase (PgdA) from Steptococcus pneumoniae (26). A small region (47 amino acids) at the N terminus of HP310 shows 36% identity with the catalytic C-terminal domain of PgdA protein (Fig. 2). PgdA protein was shown to be the primary enzyme responsible for deacetylation of the hexosamine residues of the peptidoglycan in S. pneumoniae (26).
FIGURE 2.
A, organization of hp310 gene and surrounding genes in H. pylori genome and DNA constructs for generating the HP310 mutant. A fragment containing the hp310 gene was PCR-amplified using the F- and R-primer pair and cloned into pGEM-T to obtain pGEM-hp310. A kanamycin resistance cassette (Kan) was then inserted within the HP310 fragment at the BamHI site, and the plasmid pGEM-hp310:Kan was used for transformation of wild type H. pylori to create the hp310:kan mutant. B, amino acid sequence alignment. The N-terminal portion of HP310 (residues 44–90, equivalent to the region marked with gray bar in A) shows a limited homology to the C-terminal catalytic domain (residues 284–330) of S. pneumoniae PgdA (peptidoglycan deacetylase). The two catalytic histidine residues are in bold.
Construction and Primary Characterization of hp310:kan Mutant—To investigate the physiological role of HP310, we constructed an H. pylori mutant defective in HP310 by insertion of a kanamycin resistance cassette (Fig. 2A). The hp310: kan mutant strain was obtained after natural transformation by incubating cells on blood agar plates with kanamycin (50 μg/ml) at 5% O2. The mutant strain showed a similar growth phenotype to the wild type. The correct insertion of the kan cassette within the hp310 gene on the genome was verified by PCR analysis (not shown). Both the wild type and hp310:kan mutant strains can tolerate 12% O2 for growth; the growth rate was slightly reduced compared with that at the optimal growth condition (5% O2).
Western blotting was performed using polyclonal anti-HP310 antibody against the cell extracts of H. pylori WT or hp310:kan mutant grown under 2% O2 or 12% O2 condition (Fig. 1B). The assay detected the immuno-reactive bands migrating at the expected size of HP310 in the WT cells. The expression of HP310 in the cells grown at 12% O2 is about 3-fold higher compared with that in the cells grown at 2% O2 (based on the densitometric measurement of the bands). This result confirms the induction of HP310 by oxidative stress. The disruption of HP310 in the mutant strain was also confirmed by the same (Western blot) approach; no HP310 protein band was detected in the hp310:kan mutant cells.
As HP310 is induced by oxidative stress, we wanted to know if it is an oxidative stress-combating factor. Initially, we characterized the effect of oxygen on the survival of the hp310:kan mutant compared with the wild type. In this assay, H. pylori cell suspensions (in PBS at a concentration of ∼108 cells/ml) were incubated at 37 °C under normal atmospheric conditions (21% O2). Samples were then removed at various time points, serially diluted, and spread onto BA plates to determine the viability based on cell counts. With this assay, a similar air survival effect was found for both the WT and the HP310 mutant, with recovery of about 106 viable cells after 10 h of exposure to air (data not shown).
Further characterizations were done by disk inhibition assays to determine the sensitivity of the HP310 mutant to the oxidative stress-generating agents as well as to certain dyes and detergents (supplemental Table 1). Neither dyes (benzyl viologen, gentian violet, or crystal violet) nor the detergents (Triton, Tween 20, or SDS) were shown to have a more deleterious impact on the growth of the mutant compared with the wild type. Oxidative stress generators like t-butyl hydroperoxide, cumene hydroperoxide, and paraquat (but not H2O2), evoked an increase in the zone of growth inhibition for the mutant compared with the wild type. However, somewhat surprisingly, this increase was not as striking as observed previously for other oxidative stress resistance mutants (e.g. ahpC mutants (16)). The slight increase in sensitivity to organic peroxides prompted us to measure the sensitivity of the mutant to unsaturated fatty acid and the level of lipid peroxides within the mutant cells compared with the wild type; these assays were done as described previously (31). However, HP310 mutant cells showed a similar sensitivity to linolenic acid as the wild type strain, and they had a level of lipid peroxidation similar to the wild type (data not shown), suggesting that HP310 is not involved in lipid peroxidation prevention. From these results we conclude that the major physiological role of HP310 is not in combating oxidative stress agents, although its expression is induced by oxidative stress.
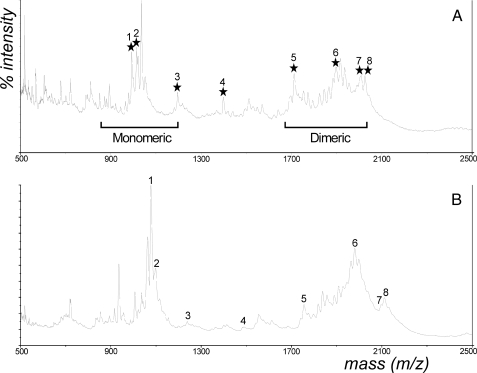
H. pylori Wild Type PG Is Partially N-Deacetylated—HP310 contains a sequence motif indicative of a possible polysaccharide deacetylase, and it displays a limited homology to the peptidoglycan deacetylase (PgdA) from S. pneumoniae. We examined whether HP310 has a function in PG deacetylation. For this purpose, we isolated and prepared PG from H. pylori. First, we analyzed the muropeptide composition of H. pylori WT PG directly by mass spectrometry. MALDI mass spectra for muropeptides were obtained both before and after chemical N-acetylation of the intact PG (Fig. 3, upper and lower panel, respectively). The major observed ions, tentative peptide assignments, and the mass shift because of N-acetylation are reported in Table 1. All mass spectra revealed two main families of digestion products, the monomeric and dimeric muropeptides, detected as their sodiated molecular ions. The monomeric muropeptides have the general formula: disaccharide-peptide, where the disaccharide consists of the borodeuteride reduced GlcNAc-MurNAc, and the peptide portion is typically a tetra- or pentapeptide. Dimeric muropeptides are of the general formula disaccharide-peptide-peptide-disaccharide. The observed molecular masses are similar, but not identical, to those derived from other Gram-negative bacterial PGs (32), reflecting variability in the peptide portion.
FIGURE 3.
Positive ion MALDI-TOF mass spectra of the muropeptide products derived from H. pylori wild type peptidoglycan. A, PG subjected directly to muramidase digestion. B, PG was chemically N-acetylated prior to muramidase digestion. Muropeptide peaks labeled by an asterisk correspond to N-deacetylated muropeptides that are shifted after chemical acetylation.
TABLE 1.
Positive ion MALDI-TOF MS analysis of H. pylori wild type muropeptides
|
Muropeptidea
|
Peak no.
|
Observed
ionb
|
||
|---|---|---|---|---|
| Before N-acetylation | After N-acetylation | Mass difference | ||
| 894 | ||||
| 965 | ||||
| 979 | ||||
| 1 | 993 | 1078 | 42 × 2 = 2 acetyl groups | |
| 2 | 1015 | 1098 | 42 × 2 = 2 acetyl groups | |
| 1021 | ||||
| Tetra-G-G | 1036 | |||
| 1052 | ||||
| 3 | 1196 | 1238 | 42 × 1 = 1 acetyl groups | |
| 4 | 1399 | 1483 | 42 × 2 = 2 acetyl groups | |
| 5 | 1713 | 1756 | 42 × 1 = 1 acetyl groups | |
| 6 | 1896 | 1980 | 42 × 2 = 2 acetyl groups | |
| 1917 | ||||
| Tetra-G-G-tetra | 1937 | |||
| 1955 | ||||
| 7 | 2009 | 2093 | 42 × 2 = 2 acetyl groups | |
| 8 | 2026 | 2111 | 42 × 2 = 2 acetyl groups | |
Tetra = disaccharide-tetrapeptide (Ala-Glu-Lys-Ala); disaccharide = GlcNAc-MurNAc, reduced with sodium borodeuteride; G = Gly; assignments represent the most likely fit based on typical amino acid components.
Ions are present as the sodiated molecular species, [M + Na]+1, rounded to nearest whole mass.
It can be seen from the spectra and Table 1 that a majority of the muropeptides derived from the WT PG gain one or two acetyl group (42 mass unit increment) as a result of chemical N-acetylation pretreatment of the PG. In Fig. 3 and Table 1, we highlighted eight peaks representing the deacetylated muropeptides in the intact PG that became acetylated after chemical N-acetylation. The data indicated that H. pylori WT PG is partially N-deacetylated. The available sites for chemical N-acetylation may consist of endogenously N-deacetylated carbohydrate moieties (i.e. N-deacetylated GlcNAc and MurNAc residues) or possibly free amino groups residing on peptide moiety having suitable amino acid composition.
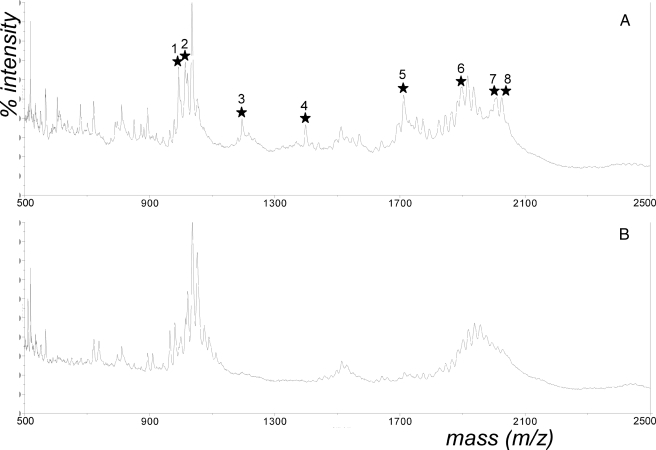
HP310 Encodes a Peptidoglycan Deacetylase—To examine whether HP310 encodes a PG deacetylase responsible for the observed deacetylation of the wild type PG, we analyzed the muropeptide composition of the PG from the hp310:kan mutant, compared with that of the WT PG (Fig. 4 and Table 2). The compounds previously shown to represent partly N-deacetylated muropeptides were either absent or significantly reduced in the mutant PG. For instance, the monomeric ions m/z 993 and 1015 (marked peaks 1 and 2) are abundant in the parent strain but are greatly diminished in the HP310 mutant (Fig. 4). These two parental muropeptides appear to gain two N-acetyl groups because of chemical N-acetylation (Table 1 and Fig. 3). These and other muropeptides that gain more than one N-acetyl group tend to be more abundant in the parental strain than the mutant. In addition, several of the parental muropeptide components, e.g. marked peaks 3 and 4 (monomeric ions m/z 1196 and 1399), are completely missing in the HP310 mutant.
FIGURE 4.
Comparison of MALDI-TOF mass spectra of the muropeptide products of PGs derived from H. pylori wild type strain (A) and the HP310:Kan mutant strain (B). Labeled muropeptide peaks corresponding to N-deacetylated muropeptides are the same as those in the Fig. 3A. Note that these peaks are missing or significantly reduced in the mutant PG.
TABLE 2.
Comparison of the muropeptides of H. pylori WT and HP310 mutant by MALDI-TOF analysis
|
Muropeptidea
|
Peak no.
|
Observed
ionb
|
|
|---|---|---|---|
| Parent strain | HP310:Kan | ||
| 894 | 894 | ||
| 965 | 965 | ||
| 979 | 981 | ||
| 1 | 993 | 993 | |
| 2 | 1015 | 1015 | |
| 1021 | 1022 | ||
| Tetra-G-G | 1036 | 1037 | |
| 1052 | 1053 | ||
| 3 | 1196 | ||
| 4 | 1399 | ||
| 5 | 1713 | ||
| 6 | 1896 | 1898 | |
| 1917 | 1920 | ||
| Tetra-G-G-tetra | 1937 | 1939 | |
| 1955 | 1957 | ||
| 7 | 2009 | ||
| 8 | 2026 | ||
Tetra = disaccharide-tetrapeptide (Ala-Glu-Lys-Ala); disaccharide = GlcNAc-MurNAc, reduced with sodium borodeuteride; G = Gly; assignments represent the most likely fit based on typical amino acid components.
Ions are present as the sodiated molecular species, [M + Na]+1, rounded to nearest whole mass.
In addition to N-acetylation content, other structural differences appear to exist in the muropeptides from the parent and mutant strains. The parent spectrum showed an additional higher mass ion family centered around 2026 mass units in the dimeric region that was absent from the mutant spectrum. In the parent strain, this secondary group of muropeptides also gained two N-acetyl groups, yielding the group around 2111 mass units (Fig. 3, peaks 7 and 8), and this latter ion group was also notably absent from the mutant spectrum (results not shown). The higher masses of these muropeptides (compared with the primary m/z 1940 grouping observed in both bacterial strains) suggests that there is an additional structural origin for their presence in the parent that is absent in the mutant. These muropeptides probably contain additional amino acid(s), or a somewhat different amino acid composition, than the muropeptides centered around 1940 mass units. This difference, in addition to N-deacetylation differences, could also contribute to altered peptidoglycan properties between these two bacterial strains.
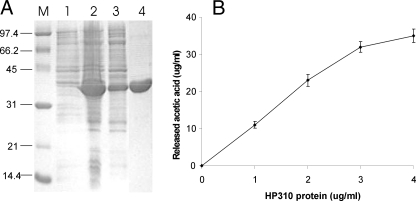
Comparative MALDI-TOF analysis of PG from the wild type and HP310 mutant indicated that HP310 has a role in PG N-deacetylation. To directly demonstrate such enzyme activity, we overexpressed H. pylori hp310 gene in E. coli and purified the His6-tagged H. pylori HP310 protein (Fig. 5A). We measured deacetylase activity of this protein with the PG isolated from the HP310 mutant as a substrate. 1 mg of the isolated PG was incubated with 0 and 1–4 μg of the purified HP310 protein for 4 h with shaking at 37 °C, and the concentration of the released acetic acid was determined (Fig. 5B). Under this assay condition, the specific activity was determined to be 2.7 μg/ml acetic acid released per h per μg of HP310 protein. Therefore, we conclude that HP310 protein is a PG deacetylase.
FIGURE 5.
Demonstration of deacetylase activity of HP310 protein. A, SDS-PAGE of the purified protein HP310-His6. His-tagged HP310 was overexpressed in E. coli in the presence of 1 mm isopropyl 1-thio-β-d-galactopyranoside. Lanes 1 and 2, proteins extracted from whole cells without and with isopropyl 1-thio-β-d-galactopyranoside induction. Lane 3, the purification intermediate (supernatant from a 45,000 × g centrifugation). Lane 4, the purified HP310-His6 protein. Lane M, protein standards. B, deacetylase activity of HP310 protein using H. pylori PG as a substrate. 1 mg of the isolated PG was incubated with various amounts of the purified HP310 protein (as indicated in x axis) for 4 h at 37 °C. The PG deacetylase activity is demonstrated as release of acetic acid during the reaction. The data are the means of three experiments with standard deviations.
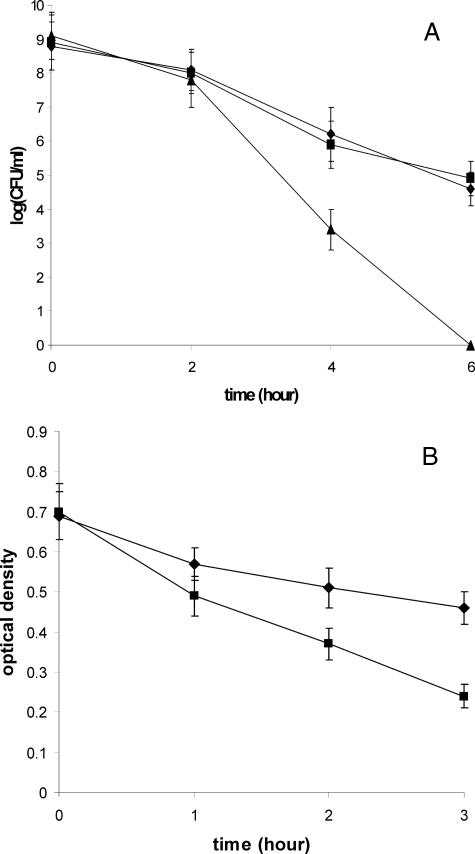
HP310 Confers Resistance to Lysozyme—Lysozyme is one of the first lines of the host defense against bacterial infections. Lysozyme is a muramidase that cleaves bacterial PG between the glycosidic β-1,4-linked MurNAc and GlcNAc. For several bacterial species, such as S. pneumoniae (26), Bacillus cereus (33), and Listeria monocytogenes (8), it was reported that N-deacetylation of PG confers resistance to lysozyme. In those studies, lysozyme at a concentration of less than 0.1 mg/ml was used for sensitivity tests. H. pylori, on the other hand, as a Gram-negative bacterium, has a different cell wall structure from those of Gram-positive ones. As expected, we found that H. pylori is highly resistant to lysozyme; it can tolerate 20 mg/ml lysozyme for growth (data not shown). To test lysozyme sensitivity of hp310:kan mutant compared with the wild type strain, we determined the cell survival curve after treatment with 50 mg/ml lysozyme (Fig. 6A). Two hours after the treatment, no significant difference was observed between the WT and the mutant. However, the number of viable mutant cells decreased more rapidly than the WT after that point. Six hours after the treatment, about 105 viable cells of the wild type strain survived, whereas the mutant cells were completely killed (no viable cells recovered). The results indicated that the HP310 mutant is considerably less resistant (i.e. more sensitive) to lysozyme compared with WT. However, this high concentration of lysozyme may not be physiologically relevant.
FIGURE 6.
Lysozyme sensitivity. A, cell survival curves of H. pylori strains (WT, diamond; hp310:kan mutant, triangle; the complemented strain, square) after treatment with 50 mg/ml lysozyme for the time indicated on x axis. B, absorbance at 600 nm of purified PG from H. pylori WT strain (diamond) or hp310:kan mutant (square) after treatment with 0.3 mg/ml lysozyme for the time indicated on x axis. The data are the means of three experiments with standard deviation as indicated.
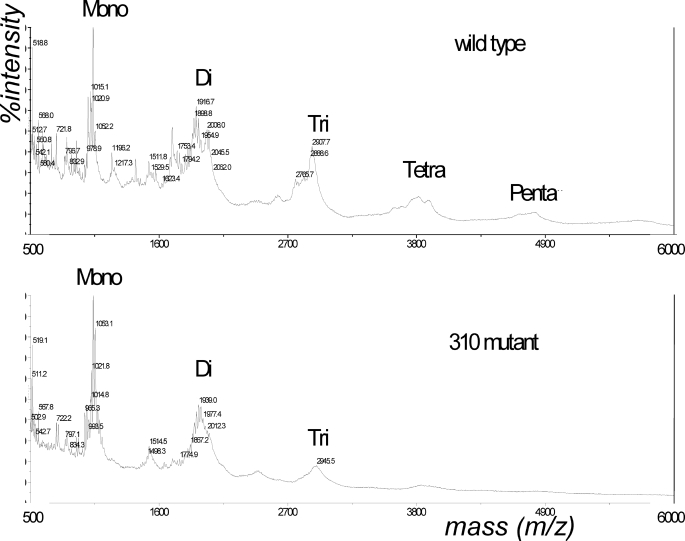
H. pylori delivers its PG into host cells (7) where the PG would be a direct target of the host-produced lysozyme. Thus, we compared the lysozyme sensitivity of the purified PG from WT and HP310 mutant cells (Fig. 6B). Upon treatment with 0.3 mg/ml lysozyme for 1–3 h, the purified PG from HP310 mutant was shown to be significantly more sensitive to lysozyme than the PG from WT. The sensitivity of PG to lysozyme digestion was also demonstrated by MALDI-TOF mass spectrometry analysis of lysozyme digestion products of PG. Upon digestion with 1 mg/ml lysozyme for 18 h, the HP310 mutant PG contained mainly monomeric and dimeric (with trace amount of trimer) muropeptides, whereas the WT PG contained additional multimeric muropeptides (tri-, tetra-, and pentamer) from the same treatment (Fig. 7). The abundance of higher oligomeric muropeptide products derived from the parent strain reflects the resistance of WT PG to lysozyme, undoubtedly arising from the extensive N-deacetylation and other structural differences.
FIGURE 7.
MALDI-TOF mass spectra showing the total array of muropeptides generated by lysozyme digestion of PGs. The PG derived from H. pylori wild type strain (upper panel) or the hp310:kan mutant strain (lower panel) was digested with 1 mg/ml lysozyme for 18 h.
Downstream of the hp310 gene on the H. pylori genome are hypothetical genes hp311 and hp312. To eliminate a possible polar effect of hp310:kan mutation on the downstream genes, we complemented the hp310 mutant strain by introducing a functional copy of the hp310 gene back into the hp310:kan mutant strain. The strain hp310:kan-hp310+-Cm contains a mutated hp310 gene at the original locus and an intact hp310 gene (driven by the ureA promoter) at an unrelated site (see “Experimental Procedures”). The lysozyme sensitivity of this new merodiploid strain was similar to the wild type strain (Fig. 6A).
DISCUSSION
During studies on H. pylori adaptation to oxidative stress, we noticed that HP310 was greatly up-expressed when cells are exposed to an elevated oxygen condition. The induction of HP310 by oxidative stress was further confirmed by Western blot analysis. Thus, we expected that HP310 may be an oxidative stress-combating factor. However, the air survival assay revealed no difference between the HP310 mutant and the wild type strain. Although the HP310 mutant was slightly more sensitive to organic peroxides than the wild type strain, it showed a similar sensitivity to unsaturated fatty acid as the wild type and had a similar level of lipid peroxidation to the wild type. These results led us to conclude that the major physiological role of HP310 is not in combating oxidative stress. Still, as H. pylori encounters oxidative stress in the host, we hypothesize that oxidative stress serves as a signal for inducing HP310 expression in the host. How HP310 expression is regulated is among the many interesting questions that remain to be investigated. All three genes that may form an operon with hp310 (hp309, hp311, and hp312) are hypothetical genes, so that no insight into the oxidative stress response could be gleaned from the genome. Homologues of HP310 are present in the published genome sequences of many pathogenic bacteria, such as C. coli, P. syringae, Y. pestis, Rhodococcus sp. RHA1, C. efficiens, R. sphaeroides, and M. smegmatis. However, the genomic area surrounding the HP310 homologue seems to be different in each bacterium. Thus, the regulation of HP310 expression by oxidative stress may be unique in H. pylori. H. pylori lacks the oxidative stress response regulators OxyR and SoxR; and our preliminary results3 suggests that the HP310 expression is probably regulated at the post-transcription level through aconitase (AcnB). Further investigation is underway to study the induction of HP310 by oxidative stress.
Two observations strongly suggest that HP310 is a PG deacetylase. Inactivation of HP310 resulted in the absence or significantly reduced amount of deacetylated muropeptides in the bacterial PG; and a PG deacetylase activity was directly demonstrated by use of pure HP310 protein by measuring the release of acetic acid from pure PG. We did not perform a detailed structural analysis of H. pylori PG, but we showed that H. pylori WT PG is partially deacetylated by comparing the MALDI mass spectra for muropeptides obtained before and after chemical N-acetylation of the intact PG. The H. pylori wild type PG profile had molecular ions that correspond to muropeptides containing multiple sites of non-N-acetylation; many peaks were shifted by 2 × 42 mass units after the PG was chemically N-acetylated, indicating they gained two acetyl groups. As demonstrated in other bacteria, the primary location of PG N-deacetylation would be GlcNAc residues (8, 26, 32–34). The deacetylation could also be on MurNAc residues, hypothesized to be the function of a PG deacetylase PdaA in B. subtilis (25). Another site of chemical N-acetylation is likely on free amino groups carried on the peptide moiety of the muropeptides (e.g. lysine or DAP free amino groups). Nevertheless, we identified eight peaks representing the deacetylated muropeptides in the WT PG that were absent or greatly diminished in the HP310 mutant PG, indicating that HP310 is responsible for the PG deacetylation. The HP310 mutant cells still contain some deacetylated PG; the enzyme responsible for the residual deacetylase activity is currently unknown. In M. smegmatis, five genes homologous to HP310 are present in the genome, whereas no other HP310 homologue was found in the H. pylori genome.
The N-terminal portion of HP310 shows limited homology to the C-terminal catalytic domain of S. pneumoniae PgdA. S. pneumoniae PgdA is a representative member of CE4 esterases that was shown to be a metalloenzyme (35). The two catalytic histidine residues are conserved in HP310, but the metal ligand amino acid residues (Asp-275, Arg-364, Asp-391, and His-417) identified in S. pneumoniae PgdA (35) could not be found at the corresponding positions within HP310. Thus, HP310 likely uses a different mechanism in catalyzing deacetylation of PG. The function of the C-terminal part of HP310 that shows no homology to known proteins is unknown. It may be involved in substrate recognition and/or specificity or in interaction with other proteins. Although HP310 shows limited homology to S. pneumoniae PgdA, the true homologues of HP310 (60–90% identity over the full length of protein) are present in many pathogenic bacteria as mentioned above. These proteins form a new subfamily of bacterial polysaccharide deacetylases. H. pylori HP310 is the first protein in this subfamily that has been studied. By using the purified HP310 protein and determining the release of acetic acid, we demonstrated that PG is a substrate of this enzyme. Whether HP310 protein can deacetylate other polysaccharides (such as lipopolysaccharide) remains to be determined.
Lysozyme is a part of the first defense of host organisms against invasive bacteria. Bacteria have developed strategies to counteract the hydrolytic activity of lysozyme by modification of their PG, preventing the binding of lysozyme to the polysaccharide substrate. Two main types of PG modification conferring lysozyme resistance have been characterized for different species of bacteria as follows: O-acetylation occurring on the C-6 hydroxyl moiety of the MurNAc residues (27, 36) and N-deacetylation of the GlcNAc residues (8, 26, 33, 34). Most of these were studied in Gram-positive bacteria, and the genes responsible for these activities are oatA (for O-acetyltransferase) and pgdA (for PG N-deacetylase), respectively. For Gram-negative bacteria, so far only an O-acetyltransferase gene (pacA) was identified to be involved in lysozyme resistance (37). In Gram-negative bacteria, PG is protected from lysozyme because of the presence of an outer membrane. Thus, H. pylori was shown to be highly resistant to lysozyme (up to 50 mg/ml). The HP310 mutant is less resistant to lysozyme killing compared with the parent strain; but this high concentration of lysozyme is likely not physiologically relevant. As H. pylori is a noninvasive pathogen, these results suggest that cellular protection from lysozyme killing may not be a major role of PG deacetylation in H. pylori, unlike those proposed for invasive pathogens. Using the purified PG, we further demonstrated that the mutant PG is more susceptible to lysozyme (0.3 mg/ml) digestion than the wild type PG. This result indicated that PG deacetylation in H. pylori indeed confers lysozyme resistance to the PG; on the other hand another (possibly more important) determinant with such a role may exist in H. pylori, such as an O-acetyltransferase.
N-Deacetylation of PG in Streptococcus (38) and in Listeria (8) was shown to be a virulence factor. It was proposed that PG N-deacetylation functions as a double protection mechanism for Listeria against the innate immune defenses by escaping the action of lysozyme and providing a mechanism to evade TRL2 and the Nod proteins (8). PG is known to be the pathogen-associated molecular pattern recognized by the Nod proteins of the host innate immune system (5). The role of N-deacetylation of H. pylori PG in escaping the host innate immune system was suggested by showing that in vitro fully N-deacetylated H. pylori PG completely lost its ability to be sensed by both Nod1 and Nod2 compared with native H. pylori PG (8). Human Nod1 was shown to be the crucial intracellular sensor of bacterial PG degradation products in epithelial cells, resulting in activation of the transcription factor NF-κB pathway; and the Nod1-dependent proinflammatory pathway depends on the ability of bacterial pathogen to translocate PG to the host cells (6, 7). Indeed, a bacterial type IV secretion system encoded by H. pylori cag pathogenicity island was shown to be responsible for delivering PG into host cells (7). Interestingly, a unique diaminopimelate-containing GlcNAc-MurNAc tripeptide, but not any other muropeptides (bacterial PG degradation products), can be detected by human Nod1 to activate an immune response (6). Lysozyme and peptidases (e.g. dd-endopeptidase and dd-carboxypeptidase) in the host cells are required for the formation of this unique muropeptide. In this sense, H. pylori PG deacetylation resulting in lysozyme resistance would inhibit the degradation of PG upon delivery into host cells. Currently we are performing a series of in vivo assays using a mouse infection model to investigate the role of H. pylori PG deacetylation in evasion of host immune responses.
Supplementary Material
This work was supported by the University of Georgia Foundation and in part by Department of Energy Grant DE-FG02–93ER20097 (to Complex Carbohydrate Research Center). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; WT, wild type; PG, peptidoglycan; MurNAc, N-acetylmuramic acid; MDP, muramyldipeptide (N-acetylmuramic acid-l-alanyl-d-isoglutamine); MALDI-TOF, matrix-assisted laser desorption-ionization-time of flight; TMS, trimethylsilyl; HPLC, high pressure liquid chromatography.
G. Wang and R. J. Maier, unpublished data.
References
- 1.Dunn, B. E., Cohen, H., and Blaser, M. J. (1997) Clin. Microbiol. Rev. 10 720–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGee, D. J., and Mobley, H. L. (1999) Curr. Top. Microbiol. Immunol. 241 155–180 [DOI] [PubMed] [Google Scholar]
- 3.Boneca, I. G. (2005) Curr. Opin. Microbiol. 8 46–53 [DOI] [PubMed] [Google Scholar]
- 4.Strober, W., Murray, P. J., Kitani, A., and Watanabe, T. (2006) Nat. Rev. Immunol. 6 9–20 [DOI] [PubMed] [Google Scholar]
- 5.Meylan, E., Tschopp, J., and Karin, M. (2006) Nature 442 39–44 [DOI] [PubMed] [Google Scholar]
- 6.Girardin, S. E., Boneca, I. G., Carneiro, L. A., Antignac, A., Jehanno, M., Viala, J., Tedin, K., Taha, M. K., Labigne, A., Zahringer, U., Coyle, A. J., DiStefano, P. S., Bertin, J., Sansonetti, P. J., and Philpott, D. J. (2003) Science 300 1584–1587 [DOI] [PubMed] [Google Scholar]
- 7.Viala, J., Chaput, C., Boneca, I. G., Cardona, A., Girardin, S. E., Moran, A. P., Athman, R., Memet, S., Huerre, M. R., Coyle, A. J., DiStefano, P. S., Sansonetti, P. J., Labigne, A., Bertin, J., Philpott, D. J., and Ferrero, R. L. (2004) Nat. Immunol. 5 1166–1174 [DOI] [PubMed] [Google Scholar]
- 8.Boneca, I. G., Dussurget, O., Cabanes, D., Nahori, M. A., Sousa, S., Lecuit, M., Psylinakis, E., Bouriotis, V., Hugot, J. P., Giovannini, M., Coyle, A., Bertin, J., Namane, A., Rousselle, J. C., Cayet, N., Prevost, M. C., Balloy, V., Chignard, M., Philpott, D. J., Cossart, P., and Girardin, S. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baik, S. C., Youn, H. S., Chung, M. H., Lee, W. K., Cho, M. J., Ko, G. H., Park, C. K., Kasai, H., and Rhee, K. H. (1996) Cancer Res. 56 1279–1282 [PubMed] [Google Scholar]
- 10.Davies, G. R., Simmonds, N. J., Stevens, T. R., Sheaff, M. T., Banatvala, N., Laurenson, I. F., Blake, D. R., and Rampton, D. S. (1994) Gut 35 179–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nardone, G., Rocco, A., and Malfertheiner, P. (2004) Aliment. Pharmacol. Ther. 20 261–270 [DOI] [PubMed] [Google Scholar]
- 12.Ding, S. Z., Minohara, Y., Fan, X. J., Wang, J., Reyes, V. E., Patel, J., Dirden-Kramer, B., Boldogh, I., Ernst, P. B., and Crowe, S. E. (2007) Infect. Immun. 75 4030–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, G., Alamuri, P., and Maier, R. J. (2006) Mol. Microbiol. 61 847–860 [DOI] [PubMed] [Google Scholar]
- 14.Seyler, R. W., Jr., Olson, J. W., and Maier, R. J. (2001) Infect. Immun. 69 4034–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, A. G., Wilson, J. E., Danon, S. J., Dixon, M. F., Donegan, K., and Hazell, S. L. (2003) Microbiology 149 665–672 [DOI] [PubMed] [Google Scholar]
- 16.Olczak, A. A., Olson, J. W., and Maier, R. J. (2002) J. Bacteriol. 184 3186–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, G., Hong, Y., Olczak, A., Maier, S. E., and Maier, R. J. (2006) Infect. Immun. 74 6839–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, G., and Maier, R. J. (2004) Infect. Immun. 72 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alamuri, P., and Maier, R. J. (2004) Mol. Microbiol. 53 1397–1406 [DOI] [PubMed] [Google Scholar]
- 20.Loughlin, M. F., Barnard, F. M., Jenkins, D., Sharples, G. J., and Jenks, P. J. (2003) Infect. Immun. 71 2022–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, G., Alamuri, P., Humayun, M. Z., Taylor, D. E., and Maier, R. J. (2005) Mol. Microbiol. 58 166–176 [DOI] [PubMed] [Google Scholar]
- 22.O'Rourke, E. J., Chevalier, C., Pinto, A. V., Thiberge, J. M., Ielpi, L., Labigne, A., and Radicella, J. P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2789–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olczak, A. A., Wang, G., and Maier, R. J. (2005) Free Radic. Res. 39 1173–1182 [DOI] [PubMed] [Google Scholar]
- 24.Towbin, H., Staehelin, T., and Gordon, J. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukushima, T., Kitajima, T., and Sekiguchi, J. (2005) J. Bacteriol. 187 1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vollmer, W., and Tomasz, A. (2000) J. Biol. Chem. 275 20496–20501 [DOI] [PubMed] [Google Scholar]
- 27.Bera, A., Herbert, S., Jakob, A., Vollmer, W., and Gotz, F. (2005) Mol. Microbiol. 55 778–787 [DOI] [PubMed] [Google Scholar]
- 28.Hash, J. H. (1967) J. Bacteriol. 93 1201–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, A. M., Li, Q., Nagata, K., Tamura, T., Shimono, K., Sato, E. F., and Inoue, M. (2004) Free Radic. Biol. Med. 36 1126–1133 [DOI] [PubMed] [Google Scholar]
- 30.Caufrier, F., Martinou, A., Dupont, C., and Bouriotis, V. (2003) Carbohydr. Res. 338 687–692 [DOI] [PubMed] [Google Scholar]
- 31.Wang, G., Hong, Y., Johnson, M. K., and Maier, R. J. (2006) Biochim. Biophys. Acta 1760 1596–1603 [DOI] [PubMed] [Google Scholar]
- 32.Antignac, A., Rousselle, J. C., Namane, A., Labigne, A., Taha, M. K., and Boneca, I. G. (2003) J. Biol. Chem. 278 31521–31528 [DOI] [PubMed] [Google Scholar]
- 33.Psylinakis, E., Boneca, I. G., Mavromatis, K., Deli, A., Hayhurst, E., Foster, S. J., Varum, K. M., and Bouriotis, V. (2005) J. Biol. Chem. 280 30856–30863 [DOI] [PubMed] [Google Scholar]
- 34.Meyrand, M., Boughammoura, A., Courtin, P., Mezange, C., Guillot, A., and Chapot-Chartier, M. P. (2007) Microbiology 153 3275–3285 [DOI] [PubMed] [Google Scholar]
- 35.Blair, D. E., Schuttelkopf, A. W., MacRae, J. I., and van Aalten, D. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15429–15434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebert, L., Courtin, P., Torelli, R., Sanguinetti, M., Chapot-Chartier, M. P., Auffray, Y., and Benachour, A. (2007) Infect. Immun. 75 5390–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dillard, J. P., and Hackett, K. T. (2005) Infect. Immun. 73 5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vollmer, W., and Tomasz, A. (2002) Infect. Immun. 70 7176–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.