Abstract
Androgen receptor (AR) plays a critical role in development and maintenance of male reproductive functions and the etiology of prostate cancer. As a ligand-regulated transcription factor, identification and characterization of AR coregulators are essential for understanding the molecular mechanisms underlying its diverse biological functions. Here we reported the identification of a novel AR coactivator, deleted in breast cancer 1 (DBC1), through a biochemical approach. DBC1 interacts with AR in a ligand-stimulated manner and facilitates AR transcriptional activation in transfected cells as well as in Xenopus oocytes. In in vitro gel shift experiments, recombinant DBC1 drastically enhanced AR DNA-binding activity. Expression of DBC1 also enhanced the binding of AR to chromatinized template in vivo, whereas knockdown of DBC1 impaired the binding of AR to endogenous prostate-specific antigen (PSA) gene in the prostate cancer cell line LNCaP. Thus, our data identify DBC1 as a novel AR coactivator.
Androgen receptor (AR)2 mediates the diverse biological functions of androgens, including development and maintenance of male reproductive functions and the etiology of prostate cancer (1, 2). As a member of nuclear receptor superfamily (NR), AR is a modular protein, containing an N-terminal domain, a DNA binding domain, and a multifunctional C-terminal ligand-binding domain (LBD) (3). Like other NRs, transcriptional regulation by AR is believed to be mediated coordinately through accessory cofactors, termed coactivators or corepressors (4–6). The coactivators facilitate transcriptional activation through diverse mechanisms, including functioning as bridging factors between receptors and basal transcription machinery to enhance recruitment of the basal transcription machinery and/or as factors that have the capacity to actively remodel repressive chromatin (7, 8).
The identification and characterization of AR cofactors have played a central role in advancing our understanding of molecular mechanism of transcriptional regulation by AR. An increasing number of AR coactivators has been reported, including the coactivators initially identified as coactivators for other NRs such as the SRC/p160 family members (9–14), CBP/p300 (15, 16), TRAP220 (17, 18), SWI/SNF (19, 20) as well as AR-interacting proteins, including ARA24 (21), ARA70 (22), FHL2 (23), and p44 (24). Most of the cofactors are initially identified as interacting proteins for one or more NRs through yeast two-hybrid screenings using either NR LBDs or N-terminal domains as baits (9–14). However, biochemical approaches also play an important role in identification of NR cofactors. For example, the mediator complex, also known as TRAP and DRIP, was isolated as liganded thyroid hormone receptor and vitamin D receptor-associated protein complex, respectively (17, 25). The mediator complex is believed to coordinate with other cofactors to regulate the recruitment of RNA polymerase II. In this regard, biochemical purification of AR-associated protein complex(es) so far has resulted in only limited success. Only one protein, p44, was identified in a previous effort of biochemical purification of AR-associated proteins (24). p44 was shown to enhance AR transcriptional activation both in vitro and in cells. This limited success may result from dissociation of AR-interacting proteins during cell lysis and/or purification process.
In this study, we have employed a reversible cross-linking-based approach to capture AR·protein complexes in cells treated with or without synthetic AR ligand R1881. We then isolated AR complex(es) by immunoaffinity purification and identified AR-associated proteins by mass spectrometry. In addition to known coactivators such as the human BAF complex (SWI/SNF), we identified DBC1, a protein initially identified as “deleted in breast cancer 1” (26, 27), as an AR-interacting protein. We demonstrated that DBC1 interacts with AR in a ligand-stimulated manner and facilitates AR transcriptional activity. The interaction between DBC1 and AR was mapped to the AR ligand binding domain and DBC1 N-terminal region. RNA interference experiments showed that DBC1 was required for optimal transcriptional activation of AR target genes in LNCaP cells and binding of AR to the PSA enhancer. In vitro, recombinant DBC1 facilitated AR DNA-binding activity in gel mobility shift assays. Taken together, our data have identified DBC1 as a novel AR coactivator.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Antibodies—The reporter plasmids MMTV-LTR-CAT and MMTV-Luc were previously described (28). To generate a doxycycline-inducible stable cell line expressing C-terminal FLAG-tagged AR, the full-length AR was cloned into a modified pcDNA5/FRT/TO vector (Invitrogen) containing two tandem FLAG tags. Full-length DBC1 in pcDNA3.1 was also cloned into the pSG5-FLAG vector for expression of FLAG-DBC1, MS2 vector for expression of proteins in Xenopus oocytes (29), and pFastBac for expression of recombinant DBC1 in SF9 cells. DBC1-(aa1–265) was generated by cloning the corresponding cDNA fragment into pSG5-FLAG, whereas DBC1-(aa266–923) and DBC1-(aa1–793) were cloned into pSG5-HA vector. For knocking down of DBC1 in HeLa cells, we also generated sh-DBC1–1 and sh-DBC1–2 constructs in pSUPER vector (OligoEngine) according to the company protocol using the following sequences: shDBC1–1F (GATCCCCCCAGCTTGCATGACTACTTTTCAAGAGAAAGTAGTCATGCAAGCTGGTTTTTA), shDBC1–1R (GGGGGTCGAACGTACTGATGAAAAGTTCTCTTTCATCAGTACGTTCGACCAAAAATTCGA), shDBC1–2F (GATCCCCGCAGACACTTCTAGACGGATTCAAGAGATCCGTCTAGAAGTGTCTGCTTTTTA), and shDBC1–2R (GGGCGTCTGTGAAGATCTGCCTAAGTTCTCTAGGCAGATCTTCACAGACGAAAAATTCGA). For in vitro pulldown assays, different truncated DBC1 and ARLBD were cloned into the pGEX4T-1 vector, and GST fusion proteins were purified using a kit from Amersham Biosciences. All plasmids were verified by DNA sequencing.
DBC1 antibody was generated by immunizing rabbit using recombinant GST-DBC1 (aa 266–375). The AR antibodies N-20 and C-19 were purchased from Santa Cruz Biotechnology. BRG1 (07–476) and HA tag antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). FLAG tag antibody was purchased from Sigma. Antibodies against SRC-1, SRC2, and SRC-3 were as previously described (30).
Generation of the AR-inducible Stable Cell Line—A stable FLAG-AR cell line was generated using the 293T Flp-In T-Rex cell line and Flp-In™-T-REx™ system from Invitrogen according to the manufacturer's instructions essentially as described (31).
Protein-Protein Cross-linking Using DSP—To capture AR·cofactor complexes in cells, we made use of short arm cross-linking reagent dithiobis[succinimidylpropionate] (DSP) from Pierce (no. 22585). DSP is a water-insoluble, membrane-permeable homobifunctional N-hydroxysuccimide ester, which can cross-link different proteins by primary amines in cells. The FLAG-AR 293T Flp-In T-Rex stable cell line was cultured in the Dulbecco's modified Eagle's medium supplemented with charcoal-stripped serum for 2 days to consume steroids in the medium. Doxycycline was added to the medium at a final concentration of 0.5 μg/ml, and the cells were incubated overnight to induce FLAG-AR expression. The cells were then treated without or with R1881 (50 nm) for 2 h to induce the formation of AR·cofactor complexes. The cells were collected by EDTA-trypsin and washed twice with cold 1× phosphate-buffered saline buffer. The cross-linking with DSP (20 mm) was carried out in 1× phosphate-buffered saline buffer at room temperature for 30 min. Tris-HCl (pH 7.5) was then added at the final concentration of 20 mm and incubated for 15 min to stop the cross-linking reaction. The cells were collected by centrifugation and lysed with high salt EBC buffer (20 mm Tris (pH 8.0), 500 mm NaCl, 0.2% SDS, 1% Nonidet P-40, 2 mm EDTA, and 20% glycerol plus a mixture of proteinase inhibitors (Roche Applied Science)). The lysates were sonicated three times for 15 s each time on ice at 40% output (Branson Sonifier 250), and clean extracts were obtained after centrifugation at 15,000 rpm for 30 min.
Purification of Human AR Complex from a Stable Cell Line—The clean extracts were diluted 2-fold with dilution buffer (20 mm Tris (pH 8.0), 100 mm KCl, 20% glycerol, 1 mm EDTA) before being used for complex purification (2 mg of proteins/ml). Approximately 50 μl of ANTI-FLAG® M2-agarose (Sigma) was added to the extract (5 ml), and the mixture was incubated at 4 °C with rotation for 2 h. The affinity resins were washed five times with Wash Buffer I (20 mm Tris-Cl, pH 8.0, 400 mm KCl, 0.5 mm EDTA, 10% glycerol, 0.25% Nonidet P-40, plus protease inhibitor mixture) and once with Wash Buffer II (20 mm Tris-Cl, pH 8.0, 100 mm KCl, 0.5 mm EDTA, 10% glycerol, 0.25% Nonidet P-40, and protease inhibitors). The cross-linking was reversed upon treatment at 95 °C for 5 min in SDS-loading buffer. The proteins were separated with a 4–20% SDS-PAGE, stained with Imperial Protein Stain (Pierce cat. no. 24615) and processed for identification by mass spectrometry as described previously (32).
Immunoprecipitation and Western Blotting—Immunoprecipitation and Western blotting were essentially as described previously (31).
Microinjection of Xenopus Oocytes and Primer Extension—Preparation and microinjection of mRNA and reporter DNA into stage VI Xenopus oocytes were performed as previously described (33). The preparation of single-stranded reporter DNA for Xenopus oocyte injection was essentially as described (34). All capped polyadenylated mRNAs used for injection were synthesized using a SP6 Message Machine kit (Ambion, Austin, TX). In vitro synthesized mRNA was injected at a concentration of 100–400 ng/μl (18.4 nl/oocyte), and reporter plasmid in single strand DNA form was injected at a concentration of 50 ng/μl (18.4 nl/oocyte) according to the experimental scheme described in each figure. Primer extension was used to analyze the quantity of RNA transcripts produced form reporter genes in Xenopus oocytes. The procedure used for primer extension has been previously described (33). The Xenopus oocyte storage histone H4 mRNA was used as an internal control in all primer extension assays.
Cell Culture and Short Hairpin RNA in a Stable Cell Line—HeLa cells were routinely maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% antibiotics at 37 °C under 5% CO2. HeLa cells were seeded the night before transfection at such a density that cells reach ∼30–40% confluence by the time of transfection. The transfection of pSUPER vector, sh-DBC1–1, and sh-DBC1–2 constructs against DBC1 were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Two days after transfection, the cells were re-plated and selected for stable clones in the presence of antibiotics puromycin. The pool of clones was selected, and the efficiency of the short hairpin RNA knockdown was determined by Western blotting analysis using DBC1 antibody.
For luciferase assay, the stable control pSUPER- and shDBC1-1-transfected cells were transfected with pSG5-AR and MMTV-luciferase reporter. 6 h after transfection, the cells were treated with R1881 (50 nm) for overnight, and the cells were harvested for measuring the luciferase activity.
Real-time PCR—Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the ABI Prism 7900 sequence detection system (Applied Biosystems). Quantitative PCR reactions were performed under conditions standardized for each primer. Standard curves were generated using 10-fold dilutions of standard plasmids. Primers for quantitative RT-PCR analysis of the PSA, NKX3.1 (NK3 homeobox 1), and TMPRSS2 (transmembrane protease, serine 2) mRNAs were as follow: PSA, 5-GGAAATGACCAGGCCAAGAC-3 and 5′-CAACCCTGGACCTCACACCTA-3′; NKX3.1, 5-TGAAGGCGCAGGCTTACTG-3 and 5′-TAGGCTGCCTTCTTTTCCATGT-3′; and TMPRSS2, 5-TGGCAGGGCGCCAA-3 and 5′-TCAATTTCCAGTGACTAGCAG-3′. The 18 S RNA was used as internal control, and all samples were first normalized to 18 S RNA. The SMARTpool siRNA against DBC1 was purchased from Dharmacon. To knock down DBC1 in LNCaP cells, LNCaP cells were transfected with 20 nm siDBC1 for 2 days. A scrambled siRNA (siCon) served as a control. The transfected cells were treated with or without 50 nm R1881 for 8 h before being collected for quantitative RT-PCR analysis or 2 h for ChIP assays.
Gel Mobility Shift Assay—DNA binding was determined by electrophoretic mobility shift assay for AR under the conditions as described previously (28). Briefly, The androgen response element oligonucleotide was labeled by using [α-32P]dATP. The binding reactions also contained 0.5 μg of competitor poly(dI-dC) (Sigma). AR and DBC1 were purified from Sf9 cells (amounts per assay are indicated in the figure legends) were added into 12 μl of DNA binding buffer (20 mm Tris-base (pH 7.5), 100 mm KCl, 5 mm dithiothreitol, 5 mm MgCl2, 1 mm EDTA, 0.1% Triton, 10% glycerol and 0.2 nm phenylmethylsulfonyl fluoride). All components of the binding reaction were preincubated for 10 min at 4 °C prior to addition of labeled probe. After 20 min, DNA binding reactions were electrophoresed on 4% polyacrylamide (40:1 acrylamide/bisacrylamide ratio) gels in 0.5× Tris acetate/EDTA buffer in cold room. Gels were dried and autoradiographed.
Chromatin Immunoprecipitation Assay—ChIP assays using injected Xenopus oocytes were performed exactly as previously described (33). The PCR primers for ChIP assay of MMTV-LTR-CAT reporter are 5′-CCAATCTTGGTTCCCAAGGTTT-3′ (forward) and 5′-GTTAGGACTGTTGCAAGTTTACT-3′ (reverse).
For ChIP assay in LNCaP cells, the cells were cultured in charcoal-stripped serum medium for 3 days before treatment with R1881 (50 nm) for 2 h. The ChIP assay was performed essentially as described (35).
Standard PCR was performed in 20-μl volumes with the inclusion of 1 μCi of [32P]dATP. The products were visualized by autoradiography. The PCR (94 °C for 45 s, 63 °C for 45 s, and 72 °C for 45 s) consisted of 20 cycles for DNA from oocytes and 25 cycles for DNA from mammalian cells.
RESULTS
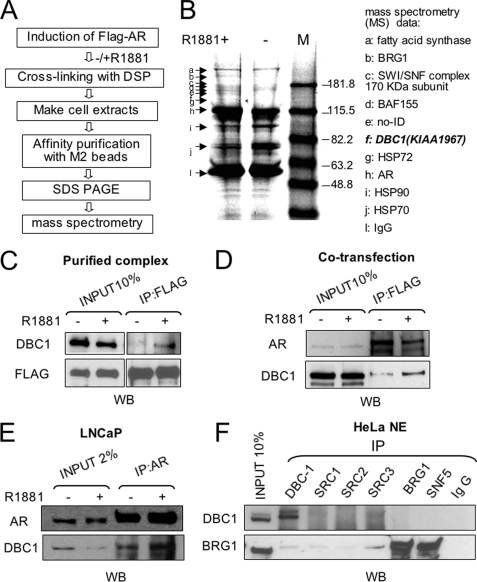
Identification of DBC1 as an AR-interacting Protein—To isolate AR cofactors through a biochemical approach, we first established a stable inducible FLAG-AR expression cell line using the 293T Flp-In T-Rex cell line and Flp-In™-T-REx™ system from Invitrogen (data not shown) and a scheme outlined in Fig. 1A. The expression of FLAG-AR (see “Experimental Procedures”) was induced by 0.5 μg/ml doxycycline overnight and followed by treatment with or without 50 nm AR synthetic ligand R1881 for 2 h. The cells were then treated with 20 mm DSP, a reversible cross-linking reagent, and processed for affinity purification of FLAG-AR and associated proteins using anti-FLAG M2-agarose beads. After reversing cross-linking, the resulting FLAG-AR complexes were resolved by a 4–20% SDS-PAGE and visualized by Coomassie Blue staining (Fig. 1B). A number of bands were found to differentially associate with liganded and unliganded AR. These bands were excised, in gel digested with trypsin, and their identities were determined by mass spectrometry. The results (Fig. 1B, right panel) show that the proteins that were preferentially associated with liganded AR are mainly BRG1 and BAF proteins, the components of human BAF or SWI/SNF complexes (36, 37). The ligand-dependent interaction between AR and SWI/SNF has been reported previously (20, 38). Consistent with the general concept that unliganded AR forms complexes with heat shock proteins and ligand treatment induces the dissociation of heat shock proteins, the bands (i and j) that became weaker after ligand treatment were identified as Hsp90, Hsp72, and Hsp70. Together these data validated our experimental approach.
FIGURE 1.
Purification and identification of DBC1 as a liganded AR-interacting protein. A, experimental scheme for isolation of AR-interacting proteins. B, immunopurified FLAG-AR and associated were resolved by 4–20% SDS-PAGE and stained with Coomassie Blue. The bands a–k were excised and subjected to mass spectrometry analysis. The identity of each band was shown in the right panel. C, Western blot analysis showing the presence of DBC1 in the immunopurified liganded AR preparation. Input, 10% equivalent whole cell extract. D, DBC1 and AR were co-transfected into HeLa cells, and the interaction was examined by IP with anti-FLAG antibody followed by Western analysis with DBC1 and AR antibodies. E, IP-Western analysis using whole cell extracts from LNCaP cells treated without or with 50 nm R1881 for 2 h. F, HeLa nuclear extracts were used for IP using various antibodies as indicated and followed by Western analysis using DBC1 or Brg1 antibody.
In addition to aforementioned proteins, we identified DBC1 (band f) as a protein co-purified only with liganded FLAG-AR (band h). The DBC1 gene, also called KIAA1967, was initially isolated in an effort to identify candidate tumor suppressor gene(s) located in the human chromosome 8p21 region that is frequently deleted in breast cancer. Subsequent study revealed that deletion of DBC1 is not linked to breast cancer but rather implicated another gene deleted in breast cancer 2 (DBC2) as a breast tumor suppressor (26). DBC1 encodes a 923-amino acid nuclear protein. Interestingly, DBC1 was shown to be processed during apoptosis, and this caspase-dependent processing of DBC1 may act as a feed-forward mechanism to promote apoptosis (27).
We first verified the mass data of DBC1 by Western analysis using a DBC1-specific antibody. Consistent with the mass data, Fig. 1C shows that DBC1 was detected in the purified FLAG-AR complexes derived from R1881-treated cells. To further validate this association, we co-transfected FLAG-AR and DBC1 into HeLa cells and analyzed their interaction by IP-Western analysis in the absence of DSP treatment. The result in Fig. 1D shows that more DBC1 was co-precipitated with FLAG-AR in IP with FLAG antibody from the cells treated with R1881.
To further test the interaction between the endogenous DBC1 and AR, we also performed IP-Western analysis using cell extracts derived from prostate cancer LNCaP cells treated with or without R1881. The result in Fig. 1E shows again an R1881-stimulated association between DBC1 and AR.
DBC1 Interacts Directly with AR—Next we analyzed whether DBC1 interacts directly or indirectly with AR. As abundant SWI/SNF components were co-purified with liganded AR (Fig. 1B), DBC1 might indirectly interact with AR through its association with SWI/SNF or an unidentified protein. Although mass analysis did not detect the presence of SRC/p160 family proteins in the liganded AR complexes, we detected their presence by Western analysis (data not shown). We thus immunoprecipitated HeLa nuclear extracts with antibodies against members of SRC/p160 family coactivators and SWI/SNF and analyzed the presence of DBC1. Fig. 1F shows that DBC1 did not co-precipitate with SRC-1, SRC-2, or SRC-3, or with BRG1 or SNF5, two subunits of human SWI/SNF. As a positive control, BRG1 was abundantly detected in the precipitates with antibodies against BRG1 and SNF5. Together these results indicate that DBC1 is neither associated with the SRC/p160 members nor SWI/SNF, suggesting that DBC1 is likely to interact directly with liganded AR.
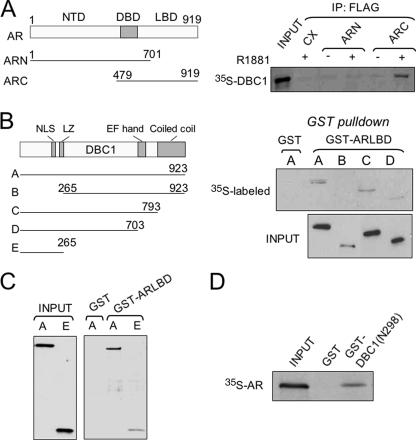
To test whether DBC1 interacts directly with liganded AR, we first expressed FLAG-tagged AR mutants with either deletion of the AR N-terminal domain or the C-terminal ligand binding domain (Fig. 2A) in HeLa cells. The resulting cell extracts were mixed with 35S-labeled, in vitro synthesized DBC1 in the presence of absence of 100 nm R1881 and immunoprecipitation was performed using anti-FLAG antibody. We found that 35S-DBC1 was co-precipitated with the C-terminal ligand binding domain but not with AR N-terminal domain (Fig. 2A). In addition, we found that DBC1 bound to GST-ARLBD but not to control GST in in vitro pulldown assays (data not shown, see Fig. 2, B and C). These data support a direct interaction between AR and DBC1.
FIGURE 2.
DBC1 binds to AR LBD in a ligand-dependent manner through its N-terminal region. A, DBC1 was synthesized and labeled with 35S-met by in vitro transcription/translation-coupled reaction and mixed with AR deletion mutants expressed in HeLa cells. DBC1 was detected by autoradiography after IP AR mutant proteins with anti-FLAG antibody. B, the left panel illustrates the structure of full-length DBC1 and deletion mutants. GST-ARLBD was purified from E. coli cultured with addition of 10 nm R1881. In vitro synthesized, 35S-met labeled DBC1 and deletion mutants were analyzed for binding of GST-ARLBD by pulldown assay. No binding of DBC1-(265–923) was detected even after prolonged exposure. C, the N-terminal region 1–265 of DBC1 interacts with AR LBD. D, AR bound to GST-DBC1(N298) but not control GST in an in vitro pulldown assay.
We next defined the AR interaction domain in DBC1. A series of DBC1 deletion mutants (27) was synthesized and labeled with [35S]methionine, and their interaction with AR was tested using GST-ARLBD in in vitro pulldown assays. The results show that deletion of the N-terminal region, including the nuclear localization signal and the leucine zipper abrogated the interaction, whereas deletion of the C-terminal coiled-coil and EF-hand domains had no effect (Fig. 2B). The N-terminal region (aa 1–265) was not only necessary (Fig. 2C) but also sufficient for binding of GST-ARLBD (Fig. 3B). In a reciprocal experiment, in vitro translated, [35S]methionine-labeled AR was able to bind GST-DBC1 fusion containing amino acids 1–298 but not to control GST (Fig. 2D). Taken together, we conclude that DBC1 interacts directly with AR, and this interaction involves AR LBD and the N-terminal region (aa 1–265) of DBC1.
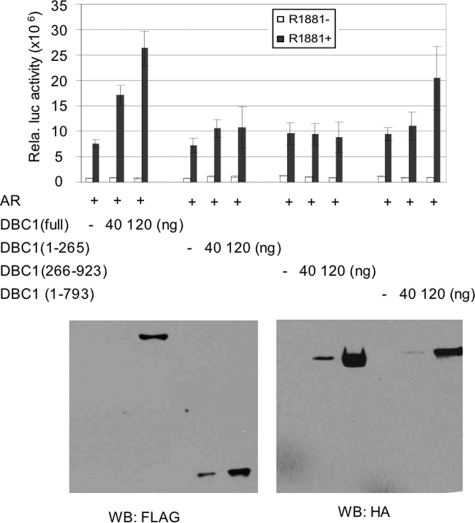
FIGURE 3.
DBC1 functions as an AR coactivator. HeLa cells were transfected with 5 ng pSG5-AR, 50 ng MMTV-luc reporter and indicated amount of pSG5-DBC1 or deletion mutants in 12 well plates. The luciferase activities were means + S.E. from three independent experiments. The group of samples with R1881 treatment was also analyzed by Western blot to detect the expression levels of DBC1 and deletion mutants.
DBC1 Functions as an AR Coactivator—Having established that DBC1 interacts with AR in a ligand-dependent manner, we next analyzed whether DBC-1 modulated AR transcriptional activity. We co-transfected HeLa cells with an MMTV-LTR-driven luciferase reporter and AR expression vector in the absence or presence of DBC1. Two days after transfection, the cells were treated with or without 50 nm R1881 overnight and assayed for luciferase activity. A representative result in Fig. 3 shows that addition of DBC1 enhanced AR transcriptional activity in a dose-dependent manner. Under the same conditions, we also analyzed the ability for various DBC1 deletion mutants to enhance AR activation. We found that, although DBC1-(1–265) is sufficient to interact with AR (Fig. 2), it was not able to facilitate AR activation (Fig. 3). Western analysis using FLAG antibody showed that DBC1 and DBC1-(1–265) were expressed to a similar level. On the other hand, deletion of the N-terminal AR interaction domain also impaired the DBC1 coactivator activity, whereas deletion of the C-terminal coiled-coil domain slightly reduced the coactivator activity. Again, Western analysis showed that both deletion mutants were expressed to comparable levels (Fig. 3, lower panel). Together these results indicate that interaction with AR is required for DBC1 to facilitate AR transcriptional activation.
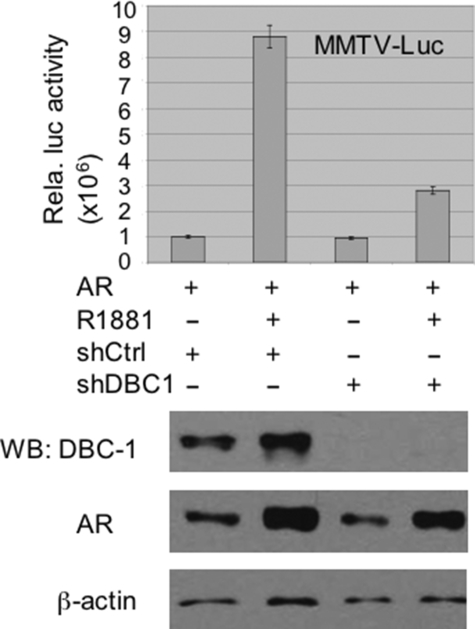
To further assess the role of DBC1 in AR transcriptional activation, we attempted to generate stable DBC1 knockdown HeLa cells using two short hairpin RNAs specific for DBC1 (shDBC1-1 and shDBC1-2) (see “Experimental Procedures”). We found that the pool of stable shDBC1-1 but not shDBC1-2 HeLa cells showed a significant reduction of DBC1 (data not shown, see Fig. 4, lower panel). We thus transfected the stable shDBC1-1 cells with AR and MMTV-LTR-luc and assayed for AR transcriptional activity. We found that knockdown of DBC1 substantially reduced the AR transcriptional activity (Fig. 4, upper panel). Western analysis revealed that the expression of AR was not significantly affected by the absence of DBC1 (Fig. 4, lower panel). An increase in the levels of AR protein upon 24-h R1881 treatment was likely a result of AR stabilization by R1881. These data provide first evidence that DBC1 is required for optimal transcriptional activation by AR.
FIGURE 4.
Knocking down DBC1 in HeLa cells impaired AR transcriptional activation. HeLa cells stably transfected with the control short hairpin RNA vector or shDBC1 were transfected with pSG5-AR and MMTV-Luc in 6-well plates. After overnight treatment with or without 50 nm R1881, the cells were collected for luciferase assay. The luciferase activities were means ± S.E. from three independent experiments. The knockdown of DBC1 was confirmed by Western analysis, and the levels of AR were revealed by Western blot. Western blot for β-actin served as a loading control.
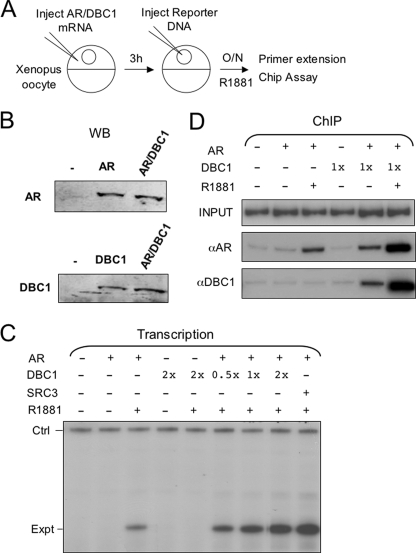
DBC1 Facilitates AR Transcriptional Activation in the Context of Chromatin—We and others have previously shown that Xenopus oocytes can be used as a convenient model system for studying transcriptional regulation in the context of chromatin (33, 38, 39). Reporter DNA microinjected into the nucleus of Xenopus oocytes in a single-stranded form is known to undergo a complementary strand synthesis-coupled chromatin assembly process and become chromatinized (34, 40). We thus tested whether DBC1 was able to stimulate AR transcriptional activation in the context of chromatin. We first attempted to express AR and DBC1 individually or in combination in Xenopus oocytes through microinjection of their in vitro synthesized mRNAs. Western analysis in Fig. 5B confirmed the expression of AR and DBC1 and revealed that expression of DBC1 did not affect the level of AR expression. We then tested whether expression of DBC1 affected AR transcriptional activity using an MMTV-LTR-driven CAT reporter (single-stranded MMTV-CAT was injected into the nucleus of Xenopus oocytes, which underwent complementary DNA strand synthesis and assembled into chromatin (data not shown, see Ref. 38). Transcriptional analysis by primer extension showed that expression of AR enhanced the level of transcription from MMTV-CAT reporter in the presence of R1881 (Fig. 5C, compare lane 3 with 2). Co-expression of DBC1 further enhanced transcriptional activation by AR in a dose-dependent manner (Fig. 5C, compare lanes 6–8 with 3). Note that expression of DBC1 alone did not have any effect on transcription. As a positive control, expression of SRC-3, a known AR coactivator, also enhanced AR transcriptional activation.
FIGURE 5.
DBC1 enhances AR transcriptional and DNA-binding activity in Xenopus oocytes. A, the experimental scheme for analyzing DBC1 coactivator for AR in the context of chromatin using Xenopus oocytes as a model. B, groups of oocytes were injected with mRNA encoding AR (300 ng/μl or DBC1 (200 ng/μl) or both and subjected to Western analysis after overnight incubation. C, groups of Xenopus oocytes were injected with mRNA encoding AR (300 ng/μl), DBC1 (1×= 200 ng/μl), or SRC3 (100 ng/μl) as indicated and then single-stranded reporter (MMTV-CAT). The oocytes were then treated with or without 50 nm R1881 overnight. Total RNAs were prepared from the oocytes, and the transcriptional activities from MMTV-CAT reporter were determined by primer extension (Expt). The control (Ctrl) is the primer extension product derived from oocyte storage histone H4 mRNA and serves as an internal loading control. D, the injection and R1881 treatment of Xenopus oocytes were as described above except the oocytes were processed for ChIP assay for MMTV-LTR region.
As a first step to understanding the mechanism by which DBC1 enhances AR transcriptional activation, we examined whether DBC1 affected binding of AR to chromatinized MMTV-CAT reporter by ChIP assays. Consistent with our previous results, R1881 treatment stimulated binding of AR to chromatin (Fig. 5D, compare lane 3 with 2). Significantly, expression of DBC1 substantially increased the binding of AR to chromatin. Note that expression of DBC1 also enhanced binding of AR to chromatin even in the absence of R1881. It remains to be determined if this R1881-independent effect is truly ligand-independent or dependent on the low level endogenous androgens. Consistent with DBC1 being an AR-interacting protein, ChIP assays using DBC1-specific antibody revealed an AR-dependent, R1881-stimulated association of DBC1 with chromatin. Together the results from Xenopus oocytes not only substantiate DBC1 as an AR coactivator but also reveal a role for DBC1 in promoting AR binding to chromatin.
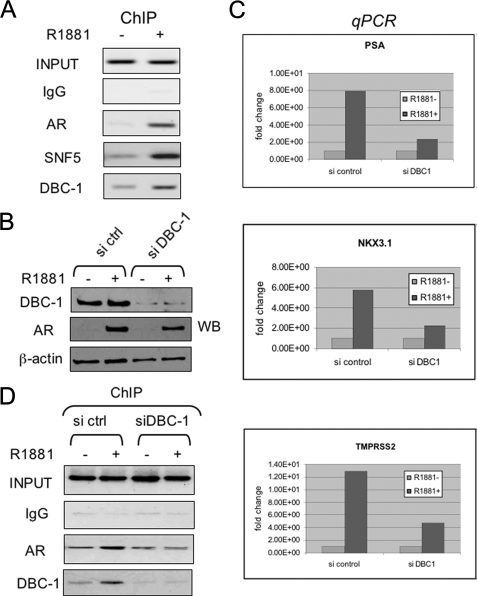
DBC1 Is Required for Transcriptional Activation by AR in LNCaP Cells—We and others have shown previously that R1881 treatment enhanced binding of AR to the PSA enhancer in LNCaP cells (35, 42, 43). To further test the role of DBC1 as an AR activator, we examined whether DBC1 was recruited to the AR target gene PSA in the LNCaP cells upon R1881 treatment. The results in Fig. 6A show that R1881 treatment indeed led to increased DBC1 association with the PSA enhancer. In addition, ChIP analysis also revealed an R1881-dependent recruitment of SNF5, a component of human BAF complex. This ChIP result nicely supports the co-purification of BAF components with liganded AR in Fig. 1.
FIGURE 6.
DBC1 is involved in the transcriptional activation of AR target genes in LNCaP cells. A, ChIP analysis revealed the recruitment of DBC1 to the PSA enhancer by liganded AR. Note that SNF5 was also recruited by liganded AR. B, Western analysis showing knockdown of DBC1 in LNCaP cells transfected with SMARTpool siDBC1 and control siRNA from Dharmacon. Note that knockdown of DBC1 did not significantly affect the R1881-induced stabilization of AR. C, knockdown of DBC1 in LNCaP by siRNA impaired R1881-induced activation of AR target genes. LNCaP cells were transfected with control or siDBC1 for 2 days, followed by treatment with or without 50 nm R1881 for 8 h. The levels of transcripts encoded by three AR target genes were determined by quantitative real-time PCR. The data were average of two independent experiments. D, ChIP analysis revealed that knockdown of DBC1 impaired R1881-induced binding of AR to the PSA enhancer. LNCaP cells were transfected with control siRNA and siDBC1 as above. After treatment with 50 nm R1881 for 2 h, the cells were processed for ChIP analysis using the antibodies indicated for the PSA enhancer.
Having established that DBC1 was recruited to the PSA enhancer by liganded AR, we next tested how knocking down DBC1 affected AR target gene expression. We failed to generate stable shDBC1-1 LNCaP cells and thus turned to the use of siRNA for knockdown. The LNCaP cells were treated with a control siRNA or SMARTpool siDBC1 (Dharmacon) for 48 h, followed by overnight induction with 50 nm R1881 and subsequent quantitative RT-PCR analysis of three endogenous AR target genes, PSA, NKX3.1, and TMPRSS2 (35, 41). Western analysis in Fig. 6B confirmed a marked reduction of DBC1 in siDBC-1 but not the control siRNA-treated cells. Data in Fig. 6C showed that knockdown of DBC1 led to 60–80% reduction of all three AR target genes tested.
Given our observation that expression of DBC1 in Xenopus oocytes enhanced binding of AR to chromatin, we next tested if knockdown of DBC1 affected binding of AR to the PSA enhancer. Significantly, we found that knockdown of DBC1 impaired the R1881-enhanced AR binding to the PSA enhancer (Fig. 6D). The R1881-stimulated DBC1 association was not observed, confirming the efficacy of siDBC1 as well as the specificity of DBC1 antibody. Thus, we conclude that DBC1 is required for efficient binding of AR to the PSA enhancer in LNCaP cells.
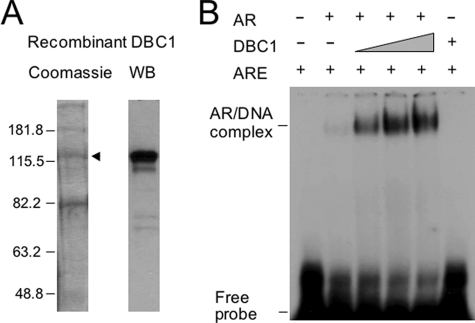
DBC1 Enhances AR DNA-binding Activity in Vitro—The finding that DBC1 promotes AR binding to chromatin in Xenopus oocytes and in LNCaP cells prompted us to investigate the potential underlying mechanism. To this end, we expressed and purified DBC1 from SF9 insect cells (Fig. 7A). We analyzed whether DBC1 promoted binding of AR to a 32P-labeled consensus androgen response element using a gel mobility shift assay. Fig. 7B shows a relatively weak DNA binding by AR alone (compare lane 2 with 1). Significantly, addition of recombinant DBC1 drastically enhanced AR DNA-binding activity in a dose-dependent manner (lanes 3–5). Note that DBC1 alone did not bind to DNA (lane 6), indicating that DBC1 enhanced the formation of DNA-protein complex by promoting AR DNA-binding activity. This effect was specific to DBC1, because addition of bovine serum albumin or GST proteins had no effect (data not shown).
FIGURE 7.
Recombinant DBC1 enhances AR DNA-binding activity in vitro. A, His6-tagged DBC1 was expressed in SF9 cells and purified using nickel-nitrilotriacetic acid resin. The resulting proteins were resolved by 8% SDS-PAGE and visualized by Coomassie Blue staining (left panel). The band marked by the arrowhead was confirmed as DBC1 by Western analysis (right panel). B, DBC1 substantially enhances AR DNA-binding activity in vitro. Gel mobility shift was performed with purified full-length AR, recombinant DBC1, and a 32P-labeled consensus androgen response element probe. Note that DBC1 itself did not show DNA-binding activity.
DISCUSSION
In this study we made use of a reversible cross-linking reagent DSP to preserve AR·cofactor complexes in cells. We then isolated AR-associated proteins under relatively stringent condition (500 mm NaCl, 0.2% SDS, and 1% Nonidet P-40) by immuno-affinity purification of AR. Under this condition, most non-cross-linked proteins were expected to dissociate from FLAG-AR. Thus, our approach is likely to yield one or more proteins that bind directly to AR, given that DSP is a short arm cross-linking reagent. Consistent with this idea, we identified Hsp90 and Hsp70 as major AR-associated proteins in the absence of ligand (Fig. 1A). It is well established that these heat shock proteins associate with unliganded AR to maintain AR in a ligand-competent state and that binding of hormone triggers dissociation of heat shock proteins and translocation of AR from cytoplasm into nucleus (3, 44).
Among the proteins that became associated with AR upon treatment with R1881, we identified DBC1 (Fig. 1A). Although DBC1 was named based on its first identification as a candidate tumor suppressor gene in the 8p21 region frequently deleted in breast cancers, later studies indicate it is not a breast cancer tumor suppressor (26). In contrast, DBC1 was found to be overexpressed in breast tumors (45, 46). Through multiple approaches, we confirmed a ligand-dependent interaction between DBC1 and AR. This interaction is direct and involves the ligand binding domain of AR and the N-terminal region of DBC1. Previous studies have identified two types of AR-LBD-interacting sequence motifs, LXXLL and FXXLF, within AR-interacting proteins (47, 48). Although LXXLL motif is commonly involved in ligand-dependent cofactors-NR interaction, FXXLF motif specifically mediates the interaction of a group of AR-specific cofactors with AR. Crystal structural analysis of the AR·LBD in complex with both motifs reveals that side chains unique to the AR·LBD rearrange to bind either the bulky FXXLF motifs or the more compact LXXLL motifs (49). Although the N-terminal region of DBC1-(aa 1–265) was sufficient for interaction with AR, we could identify neither an LXXLL nor a FXXLF motif in this region. Thus, the molecular mechanism that dictates the binding of DBC1 to liganded AR remains to be determined.
We also present three lines of evidence that DBC1 functions as an AR coactivator. First, DBC1 overexpression facilitated AR transcriptional activation and DBC1 knockdown impaired AR activation (Figs. 3 and 4). Second, in Xenopus oocytes, expression of DBC1 enhanced AR transcriptional activation as well as binding of AR to a chromatinized template (Fig. 5). Third, in LNCaP cells, knockdown of DBC1 by siRNA impaired R1881-induced transcriptional activation of three AR target genes, PSA, NKX3.1, and TMPRSS2 (Fig. 6). Furthermore, knockdown of DBC1 also impaired the binding of AR to the endogenous PSA enhancer. Together these data firmly established DBC1 as an AR coactivator that may be critically important for optimal transcriptional activity of AR. However, it is not yet clear if DBC1 enhanced AR transcriptional activity solely through its ability to facilitate AR DNA-binding activity.
Although a large number of proteins have been reported as AR coactivators (50), few have been shown to enhance AR DNA-binding activity. Among the short list of this type AR cofactors are HMG1/2 and nucleophosmin. HMG1/2 enhances DNA-binding activity of steroid hormone receptors such as progesterone receptor, glucocorticoid receptor, and AR (51, 52). Nucleophosmin was shown to enhance AR DNA-binding and transcriptional activity (53). Thus, an important finding in this study was that DBC1 not only enhanced AR DNA-binding activity in vitro, but also that it was required for efficient binding of AR to the PSA enhancer in LNCaP cells. Because recombinant DBC1 was able to enhance AR DNA-binding activity in gel shift assays, the direct protein-protein interaction between DBC1 and AR was sufficient to promote AR DNA-binding activity. In our hands, DBC1 alone did not appear to bind DNA directly in in vitro gel shift assays. Like HMG1/2 and nucleophosmin, how DBC1 enhanced AR DNA-binding activity is not yet clear. Based on direct interaction between AR and DBC1, two general models can be envisioned. First, DBC1 may interact with AR and help convert AR into a DNA binding-favorable conformation. Second, DBC1 may also make contact with DNA once associated with AR and thus stabilize the AR·DNA complex. Future study will aim to elucidate the molecular mechanism by which DBC1 facilitates AR DNA-binding activity.
During the preparation of this report, Trauernicht et al. (54) reported the identification of DBC1 as an ERα-interacting protein. DBC1 interacts with the ligand-binding domain of ERα, and this interaction is ligand-independent. Significantly, DBC1 was shown to play a critical role in determining the steady-state level of unliganded ERα and promotes survival of human breast cancer cells. However, there are clear differences in terms of DBC1 interaction and function between ERα and AR. First, DBC1 interacts with AR in a ligand-dependent manner, whereas it interacts with ERα in a ligand-independent manner. Second, DBC1 is clearly a critical AR coactivator, whereas it does not appear to affect ERα transcriptional activity. Third, DBC1 does not appear to significantly affect the stability of AR. Fourth, although several studies have reported overexpression of DBC1 in breast carcinoma (45, 46), we did not observe significant overexpression of DBC1 in more than 100 prostate cancer specimens we have analyzed (data not shown). Given its critical role in AR transcriptional activation, it will be interested to determine the role of DBC1 in prostate cancer. In addition, two very recent reports show that DBC1 interacts with and functions as a negative regulator of SIRT1 (55, 56). It is of interest to analyze whether SIRT1 is also involved in the regulation of AR transcriptional activity by DBC1.
Acknowledgments
We sincerely thank Drs. Lei Huo, Charles Guo, Qiantian Li, and Jianming Xu for analysis of DBC1 expression in prostate cancer specimens.
This work was supported by the Research Platform of Cell Signaling Networks from the Science and Technology Commission of Shanghai Municipality (Grant 06DZ22923) and the Educational Department of China (to J. W.) and by Grant 30371493 from Natural Science Foundation of China (to J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AR, androgen receptor; NR, nuclear receptor; LBD, ligand-binding domain; DBC1, deleted in breast cancer 1; aa, amino acid(s); GST, glutathione S-transferase; HA, hemagglutinin; DSP, dithiobis(succinimidyl propionate); MMTV, murine mammary tumor virus; siRNA, small interference RNA; RT, reverse transcription; ChIP, chromatin immunoprecipitation; IP, immunoprecipitation; LTR, long terminal repeat; ERα, estrogen receptor α.
References
- 1.Lee, H. J., and Chang, C. (2003) Cell Mol. Life Sci. 60 1613-1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Culig, Z. (2003) Urology 62 21-26 [DOI] [PubMed] [Google Scholar]
- 3.Gelmann, E. P. (2002) J. Clin. Oncol. 20 3001-3015 [DOI] [PubMed] [Google Scholar]
- 4.McKenna, N. J., Lanz, R. B., and O'Malley, B. W. (1999) Endocr. Rev. 20 321-344 [DOI] [PubMed] [Google Scholar]
- 5.Heinlein, C. A., and Chang, C. (2002) Endocr. Rev. 23 175-200 [DOI] [PubMed] [Google Scholar]
- 6.Wang, L., Hsu, C. L., and Chang, C. (2005) Prostate 63 117-130 [DOI] [PubMed] [Google Scholar]
- 7.Roeder, R. G. (2005) FEBS Lett. 579 909-915 [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld, M. G., Lunyak, V. V., and Glass, C. K. (2006) Genes Dev. 20 1405-1428 [DOI] [PubMed] [Google Scholar]
- 9.Onate, S. A., Tsai, S. Y., Tsai, M. J., and O'Malley, B. W. (1995) Science 270 1354-1357 [DOI] [PubMed] [Google Scholar]
- 10.Voegel, J. J., Heine, M. J., Zechel, C., Chambon, P., and Gronemeyer, H. (1996) EMBO J. 15 3667-3675 [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, H., Kohli, K., Trivedi, A., Johnson, D. L., and Stallcup, M. R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4948-4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, H., Gomes, P. J., and Chen, J. D. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 8479-8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., Lin, R. J., Schiltz, R. L., Chakravarti, D., Nash, A., Nagy, L., Privalsky, M. L., Nakatani, Y., and Evans, R. M. (1997) Cell 90 569-580 [DOI] [PubMed] [Google Scholar]
- 14.Torchia, J., Rose, D. W., Inostroza, J., Kamei, Y., Westin, S., Glass, C. K., and Rosenfeld, M. G. (1997) Nature 387 677-684 [DOI] [PubMed] [Google Scholar]
- 15.Chakravarti, D., LaMorte, V. J., Nelson, M. C., Nakajima, T., Schulman, I. G., Juguilon, H., Montminy, M., and Evans, R. M. (1996) Nature 383 99-103 [DOI] [PubMed] [Google Scholar]
- 16.Kamei, Y., Xu, L., Heinzel, T., Torchia, J., Kurokawa, R., Gloss, B., Lin, S. C., Heyman, R. A., Rose, D. W., Glass, C. K., and Rosenfeld, M. G. (1996) Cell 85 403-414 [DOI] [PubMed] [Google Scholar]
- 17.Fondell, J. D., Ge, H., and Roeder, R. G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8329-8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, Q., Sharma, D., Ren, Y., and Fondell, J. D. (2002) J. Biol. Chem. 5 5. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao, P. W., Fryer, C. J., Trotter, K. W., Wang, W., and Archer, T. K. (2003) Mol. Cell Biol. 23 6210-6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link, K. A., Burd, C. J., Williams, E., Marshall, T., Rosson, G., Henry, E., Weissman, B., and Knudsen, K. E. (2005) Mol. Cell Biol. 25 2200-2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao, P. W., Lin, D. L., Nakao, R., and Chang, C. (1999) J. Biol. Chem. 274 20229-20234 [DOI] [PubMed] [Google Scholar]
- 22.Yeh, S., and Chang, C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5517-5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller, J. M., Isele, U., Metzger, E., Rempel, A., Moser, M., Pscherer, A., Breyer, T., Holubarsch, C., Buettner, R., and Schule, R. (2000) EMBO J. 19 359-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosohata, K., Li, P., Hosohata, Y., Qin, J., Roeder, R. G., and Wang, Z. (2003) Mol. Cell Biol. 23 7019-7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rachez, C., Suldan, Z., Ward, J., Chang, C. P., Burakov, D., Erdjument-Bromage, H., Tempst, P., and Freedman, L. P. (1998) Genes Dev. 12 1787-1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamaguchi, M., Meth, J. L., von Klitzing, C., Wei, W., Esposito, D., Rodgers, L., Walsh, T., Welcsh, P., King, M. C., and Wigler, M. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13647-13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundararajan, R., Chen, G., Mukherjee, C., and White, E. (2005) Oncogene 24 4908-4920 [DOI] [PubMed] [Google Scholar]
- 28.Huang, Z., Li, J., and Wong, J. (2002) Mol. Endocrinol. 16 924-937 [DOI] [PubMed] [Google Scholar]
- 29.Wong, J., Shi, Y. B., and Wolffe, A. P. (1997) EMBO J. 16 3158-3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, J., O'Malley, B. W., and Wong, J. (2000) Mol. Cell Biol. 20 2031-2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu, J., Yoon, H. G., Qin, J., and Wong, J. (2007) Mol. Cell Biol. 27 4641-4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin, J., and Chait, B. T. (1997) Anal. Chem. 69 4002-4009 [DOI] [PubMed] [Google Scholar]
- 33.Li, J., Fu, J., Toumazou, C., Yoon, H. G., and Wong, J. (2006) Mol. Endocrinol. 20 776-785 [DOI] [PubMed] [Google Scholar]
- 34.Wong, J., Shi, Y. B., and Wolffe, A. P. (1995) Genes Dev. 9 2696-2711 [DOI] [PubMed] [Google Scholar]
- 35.Yoon, H. G., and Wong, J. (2006) Mol. Endocrinol. 20 1048-1060 [DOI] [PubMed] [Google Scholar]
- 36.Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P. A., Biggar, S. R., Muchardt, C., Kalpana, G. V., Goff, S. P., Yaniv, M., Workman, J. L., and Crabtree, G. R. (1996) EMBO J. 15 5370-5382 [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, W., Xue, Y., Zhou, S., Kuo, A., Cairns, B. R., and Crabtree, G. R. (1996) Genes Dev. 10 2117-2130 [DOI] [PubMed] [Google Scholar]
- 38.Huang, Z. Q., Li, J., Sachs, L. M., Cole, P. A., and Wong, J. (2003) EMBO J. 22 2146-2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolffe, A. P., Collingwood, T. N., Li, Q., Yee, J., Urnov, F., and Shi, Y. B. (2000) Vitam. Horm. 58 449-492 [DOI] [PubMed] [Google Scholar]
- 40.Almouzni, G., and Wolffe, A. P. (1993) Genes Dev. 7 2033-2047 [DOI] [PubMed] [Google Scholar]
- 41.Nelson, P. S., Clegg, N., Arnold, H., Ferguson, C., Bonham, M., White, J., Hood, L., and Lin, B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11890-11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang, Y., Myers, M., and Brown, M. (2002) Mol. Cell 9 601-610 [DOI] [PubMed] [Google Scholar]
- 43.Kang, Z., Janne, O. A., and Palvimo, J. J. (2004) Mol. Endocrinol. 18 2633-2648 [DOI] [PubMed] [Google Scholar]
- 44.Pratt, W. B., Galigniana, M. D., Morishima, Y., and Murphy, P. J. (2004) Essays Biochem. 40 41-58 [DOI] [PubMed] [Google Scholar]
- 45.Radvanyi, L., Singh-Sandhu, D., Gallichan, S., Lovitt, C., Pedyczak, A., Mallo, G., Gish, K., Kwok, K., Hanna, W., Zubovits, J., Armes, J., Venter, D., Hakimi, J., Shortreed, J., Donovan, M., Parrington, M., Dunn, P., Oomen, R., Tartaglia, J., and Berinstein, N. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11005-11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson, A. L., Wang, Z. C., De Nicolo, A., Lu, X., Brown, M., Miron, A., Liao, X., Iglehart, J. D., Livingston, D. M., and Ganesan, S. (2006) Cancer Cell 9 121-132 [DOI] [PubMed] [Google Scholar]
- 47.Heery, D. M., Kalkhoven, E., Hoare, S., and Parker, M. G. (1997) Nature 387 733-736 [DOI] [PubMed] [Google Scholar]
- 48.He, B., Minges, J. T., Lee, L. W., and Wilson, E. M. (2002) J. Biol. Chem. 277 10226-10235 [DOI] [PubMed] [Google Scholar]
- 49.Estebanez-Perpina, E., Moore, J. M., Mar, E., Delgado-Rodrigues, E., Nguyen, P., Baxter, J. D., Buehrer, B. M., Webb, P., Fletterick, R. J., and Guy, R. K. (2005) J. Biol. Chem. 280 8060-8068 [DOI] [PubMed] [Google Scholar]
- 50.Chmelar, R., Buchanan, G., Need, E. F., Tilley, W., and Greenberg, N. M. (2007) Int. J. Cancer 120 719-733 [DOI] [PubMed] [Google Scholar]
- 51.Boonyaratanakornkit, V., Melvin, V., Prendergast, P., Altmann, M., Ronfani, L., Bianchi, M. E., Taraseviciene, L., Nordeen, S. K., Allegretto, E. A., and Edwards, D. P. (1998) Mol. Cell Biol. 18 4471-4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verrijdt, G., Haelens, A., Schoenmakers, E., Rombauts, W., and Claessens, F. (2002) Biochem. J. 361 97-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leotoing, L., Meunier, L., Manin, M., Mauduit, C., Decaussin, M., Verrijdt, G., Claessens, F., Benahmed, M., Veyssiere, G., Morel, L., and Beaudoin, C. (2008) Oncogene 27 2858-2867 [DOI] [PubMed] [Google Scholar]
- 54.Trauernicht, A. M., Kim, S. J., Kim, N. H., and Boyer, T. G. (2007) Mol. Endocrinol. 21 1526-1536 [DOI] [PubMed] [Google Scholar]
- 55.Kim, J. E., Chen, J., and Lou, Z. (2008) Nature 451 583-586 [DOI] [PubMed] [Google Scholar]
- 56.Zhao, W., Kruse, J. P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008) Nature 451 587-590 [DOI] [PMC free article] [PubMed] [Google Scholar]