Abstract
Signaling initiated by the BCR-ABL1 kinase causes chronic myelogenous leukemia (CML). Recently, we reported that expression of Fyn, a Src kinase, is heightened in CML cells and patient specimens and confers in vitro and in vivo proliferative advantages. Fyn is regulated by redox, and because BCR-ABL1 raises intracellular oxidant levels, which have been implicated in CML progression, we explored the molecular regulation of Fyn. Here we identify the transcription factors that drive redox- and BCR-ABL1-dependent Fyn expression. Promoter deletion analysis in 293T, BaF3, BaF3-p210, and K562 cells identified the region essential for basal transcriptional activity. Mutation of Sp1 and Egr1 binding sites within the essential region diminished Fyn promoter activity and identified Egr1 as conferring redox sensitivity. Gel shift and chromatin immunoprecipitation assays confirmed the binding of Sp1 and Egr1 to the promoter fragments. Importantly, knockdown of Sp1 or Egr1 with small interference RNA or inhibition of Sp1 binding by mithramycin A repressed Fyn protein expression. Our work is the first to define transcription factors that are responsible for endogenous, oxidative stress-dependent and BCR-ABL1-dependent Fyn expression.
The molecular defect responsible for chronic myelogenous leukemia (CML)2 is the BCR-ABL1 oncogene, which arises from a t(9;22) (q34;q11) reciprocal translocation commonly referred to as the Philadelphia chromosome (1). The encoded BCR-ABL1 kinase is a potent activator of numerous proliferative signaling pathways (2). Improved understanding of this signaling network has enabled first and second generation kinase inhibitors to revolutionize the treatment of CML, particularly chronic phase disease (3). However, the terminal blast crisis phase of the disease remains difficult to treat, and mutant forms of BCR-ABL1, particularly the T315I mutant, are refractory to all three of the currently FDA approved BCR-ABL1 kinase inhibitors (4, 5).
A less studied but intriguing feature of the BCR-ABL1 kinase is its ability to raise levels of intracellular reactive oxygen species (ROS) (6-8). These increased ROS levels have been implicated in progression of CML to blast crisis (9). Further studies have shown that these ROS can lead to point mutations that confer resistance to imatinib mesylate (10), the first FDA-approved BCR-ABL1 kinase inhibitor, which is now frontline therapy for CML. Low levels of ROS, such as those stemming from BCR-ABL1, have been well documented to transducer mitogenic signals (11) and promote genomic instability (a feature seen in blast crisis CML) (12). Because elucidation of signaling pathways in CML has impacted the treatment of this disease so significantly, further understanding of oxidant-dependent signaling (13) will be useful in delineating new treatment strategies for refractory or progressed CML.
Our recent work has highlighted a role for the up-regulation of the Src family member, Fyn, in BCR-ABL1-expressing cells and CML patient material (14). Knockdown of Fyn impacted in vitro and in vivo proliferation and caused increased sensitivity to imatinib. Fyn is ubiquitously expressed across tissue types and is among three Src family members known to be responsive to oxidative stress (15-18). The Fyn kinase has been extensively studied as promoting cell growth and differentiation in a number of signal transduction pathways, such as T cell receptor signaling, integrin-mediated signaling, cell adhesion, and hydrogen peroxide-mediated Ras activation (19, 20). To date, only three studies (including ours) have examined the role of Fyn in BCR-ABL1-mediated signal transduction (14, 21, 22). In general, activation of Src family kinases has been targeted by therapeutics. Dasatinib, a dual Src/Abl inhibitor, has been approved for CML and demonstrates specificity for Fyn kinase activity (23-25). In Philadelphia positive acute lymphocytic leukemia, Fyn was identified as a hub for signaling using gene expression and systems biology approaches (21). Recently, an in vitro study using recombinant wild-type and mutant BCR-ABL1, demonstrated that Fyn can phosphorylate the SH3 or SH2 domain of BCR-ABL1 providing compelling evidence that Fyn may be upstream of BCR-ABL1, thereby initiating oncogenic kinase signaling (22). Our published results confirmed this finding but also placed Fyn downstream of BCR-ABL1, because Fyn mRNA and protein expression is increased in a BCR-ABL1-dependent manner (14). These data suggest a cyclical model of Fyn up-regulation (driven by both BCR-ABL1 and oxidative stress) that could further promote BCR-ABL1 leukemogenesis. In keeping with this idea, we have reported increased Fyn expression in blast crisis CML patients as compared with chronic phase patients (14). Taken together, these data highlight an important role for increased Fyn expression as a feature of CML progression (14); therefore, understanding the molecular basis for this up-regulation is necessary.
The present study provides the first molecular description of regulation of the Fyn promoter, which lacks a TATA box but is characterized by several GC-rich regions. Endogenous, BCR-ABL-1-dependent, and oxidant-dependent Fyn expression were examined, and the transcription factors Sp1 and Egr1 were found to regulate expression of Fyn. Using antioxidants in combination with promoter mutation and Egr1 DNA binding assays, we found that Egr1 was responsible for oxidant-dependent up-regulation of Fyn. Interestingly, Egr1 itself is also increased in response to oxidative stress; thereby further piquing interest in understanding redox-modulated signaling in CML.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals—Monoclonal Fyn antibody was purchased from Alexis (San Diego, CA); Egr1 and Sp1 polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); anti-mouse IgG peroxidase conjugate, N-acetylcysteine (NAC), and mithramycin A were purchased from Sigma-Aldrich. Anti-rabbit IgG horseradish peroxidase-linked whole antibody (from donkey) and ECL plus Western blotting detection system were obtained from Amersham Biosciences. Imatinib was kindly provided by Dr. Elisabeth Buchdunger at Novartis Pharmaceuticals (Basel, Switzerland). [γ-32P]ATP was obtained from Amersham Biosciences. Restriction enzymes were from Promega (Madison, WI) and New England Biolabs (Beverly, MA). The psiCHECK-2 plasmid vector was purchased from Promega.
Cell Lines—293T (an easily transfectable human kidney epithelial cell line expressing SV40 T-antigen) and K562 (human CML expressing p210 BCR-ABL1) cell lines were obtained from the American Type Culture Collection (Manassas, VA). Murine growth factor-dependent pro-B lymphoid BaF3 cell lines transformed with vector (BaF3 vector), wild-type BCR-ABL1 (BaF3 p210) were kindly provided by Dr. Charles Sawyers. 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. K562 and BaF3 cells were cultured in the log phase of cell growth by culture in RPMI 1640 medium containing 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. For the BaF3 vector cells, 1 ng/ml interleukin-3 (purchased from Peprotech, Rocky Hill, NJ) and 2 μg/ml puromycin (purchased from Sigma Chemicals) were added.
Measurement of Intracellular Peroxide Levels—Cells were incubated with dihydrodichlorofluorescein diacetate (Molecular Probes, Eugene, OR) at final concentrations of 10 μm, for 30 min at 37 °C (26). Fluorescence was measured on the FL-1 channel of a FACSCalibur (BD Biosciences, Palo Alto, CA). Data were analyzed using CellQuest Pro-analysis software (BD Biosciences).
Measurement of Hydrogen Peroxide Released from Cells—Per the manufacturer's directions, 60,000 cells were resuspended in 120 μl of reaction buffer containing 50 μm Amplex Red reagent, 0.1 unit/ml horseradish peroxidase, 145 mm NaCl, 5.7 mm sodium phosphate, 4.86 mm KCl, 0.54 mm CaCl2, 1.22 mm MgSO4, and 5.5 mm glucose, pH 7.35. Each sample was pipetted equally in quadruplet (120 μl/well) on a 96-well plate and incubated at 37 °C for 10 min. Absorbance at 560 nm was monitored. For a standard curve, 0, 1, 2, 4, 8, and 16 pmol of purified H2O2 were used, and an absorbance value of 0.003 was found to correspond with 1 pmol of H2O2.
Western Blotting—Cells were lysed in 20 mm Tris-HCl buffer (pH 7.6) containing 1 mm EDTA, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1% Nonidet P-40, and 0.1% Triton X-100 with 1% protease inhibitor mixture and rotated at 4 °C for 1 h. Supernatants were obtained by centrifuging the samples at 12,000 rpm for 15 min at 4 °C. Protein concentrations were determined using the Bio-Rad DC protein assay. After boiling for 10 min in SDS sample buffer (containing 100 mm dithiothreitol), the samples were subjected to a 12% SDS-PAGE gel, and the proteins were electrotransferred onto nitrocellulose membranes. Blots were incubated at room temperature for 1 h in TBS-T blocking buffer containing 5% nonfat milk. Membranes were then probed overnight at 4 °C with primary antibody. Bound antibody was detected using a 1:5000 dilution of horseradish peroxidase-conjugated secondary antibody and ECL-enhanced chemiluminescent reagents. The protein bands from at least three experiments per figure were analyzed by the Gel Documentation and Analysis System (Bio-Rad).
Detection of Fyn mRNA—Total RNA of cells was extracted using the RNeasy mini kit (Qiagen). First-strand cDNA was synthesized using an Omniscript RT kit (Qiagen). Quantitative PCR was used to amplify Fyn. Fyn mouse primer pair A was purchased from Santa Cruz Biotechnology, cDNA generated from 0.2 μg of total RNA was added to 25 μl of iQ SYBR Green PCR master mix (Bio-Rad) with 200 nm primers and water to a total of 50 μl. PCR cycles were performed with the following temperature profile: 95 °C for 2 min, then 60 cycles consisting of 1 min at 94 °C, 30 s at 55 °C, 1 min elongation at 72 °C, and finally, elongation at 72 °C for 10 min. Actin was amplified under the same conditions using the following primers: forward, 5′-TGTGCCCATCTACGAGGGGTATGC, and reverse, 5′-GGTACATGGTGGTGCCGCCAGACA. PCR products were then resolved on a 1% agarose gel. Expression of Fyn was analyzed by calculating Ct values and normalized to BaF3 vector Fyn expression.
Cloning of Human Fyn Promoter Region DNA—The human Fyn promoter was cloned from K562 cells. The 3151-bp DNA (2268-bp upstream, 485-bp supposed first exon, and 398-bp intron) was cloned from genomic DNA by using primers GAGAGCATATGACACTTAGTC (forward) and GCATCAATTATTAAGTGATTACATC (reverse), which were designed based on the Human Chromosome 6 Project Genomic Sequences. The PCR conditions were: denaturing at 94 °C for 2 min, followed by 30 cycles of denaturing at 94 °C for 15 s, annealing at 55 °C for 30 s, and extension at 68 °C for 3 min, and a final extension at 68 °C for 15 min. The DNA polymerase used was KOD Hot Start DNA polymerase (Novagen, Gibbstown, NJ). The PCR product was isolated by electrophoresis and ligated into the plasmid pCR2 (Invitrogen). The nucleotide sequences of the DNA in the pCR2 plasmid were determined by the 16-capillary genetic analyzer ABI 3100.
Construction of Dual-luciferase Expression Plasmid pFynPromoterCheck Vector—Full-length and deletion constructs of the Fyn promoter (-2268/+883, -2000/-1, -2000/-501, -1500/-1, -1500/-501, -1000/-1, -1000/-501, -500/-1, -300/-1, -200/-1, -200/-51, -150/-1, -100/-1, and -50/-1 constructs) were generated by PCR with primers containing a BglII site and HindIII site (detailed in supplemental Table S1). The site-directed mutations in Sp1 A- and B-binding sites of the Fyn promoter were introduced into the -150/-1 and the -50/-1 constructs separately. A deletion mutant and a substitution mutant were generated in the -150/-1 construct for the Egr1 binding site; all mutants were made by PCR using the high fidelity KOD Hot Start DNA polymerase (Novagen) with primers listed in supplemental Table S2. As an internal control, we constructed the Renilla luciferase reporter plasmids containing the firefly luciferase gene in the same plasmid thereby allowing normalization of the Renilla luciferase data. Three DNA fragments: a 4.6-kb XhoI/BglII DNA fragment from psiCHECK-2 plasmid (Promega), a 1.2-kb XhoI/HindIII DNA fragment from psiCHECK-2 plasmid, and various BglII/HindIII Fyn promoter DNA fragments of different lengths from the plasmid pCR2-3.1-kb full length, were ligated together to construct the dualluciferase (firefly and Renilla luciferases) expression plasmids. This vector, renamed pFynPromoterCheck, allows the Fyn promoter region DNA to function as the Renilla luciferase gene's promoter. As a separate positive control, the 0.4-kb SV40 early promoter was used.
Transfection of Plasmid DNAs and siRNA and Dual-luciferase Assay—The dual-luciferase expression plasmid pFynPromoterCheck was transfected into 293T cells using the lipofection reagent, FuGENE 6 (Roche Applied Science), according to the protocol recommended by the manufacturer. Briefly, 97 μl of serum-free medium and 3 μl of the reagent were combined and vortexed. After 5 min at room temperature, 1 μg of plasmid DNA was added and vortexed for 1 s. The reagent-DNA complex was incubated for 20 min at room temperature and then added to 1 million cells. Medium was changed after 6 h. For the K562 and BaF3 cells, transfections were performed using the Nucleofector system (Amaxa Biosystems, Cologne, Germany) according to the manufacturer's instructions. Nucleofector kit V and transfection programs T-16 and X-01 were used, respectively. Renilla and firefly luciferase activities were determined 24 h post transfection using a dual-luciferase assay kit (Promega) and the Monolight 3010 luminometer (BD Pharmingen). The Renilla luciferase luminescence values were normalized to firefly luciferase luminescence values. The Sp1 siRNAs were purchased from Santa Cruz Biotechnology, targeting mRNA sequence (GGAACAGAGUCCCAACAGU); the Egr1 siRNA were ordered from Dharmacon, Inc. (Lafayette, CO), targeting 4 mRNA sequences (GGACAUGACAGCAACCUUU, GUUACUACCUCUUAUCCAU, GAUGAACGCAAGAGGCAUA, and CCACAGGGCUUUCGGACAU); 10 μl of 10 μm siRNA duplex or control siRNA was transfected into 1 million cells. After 24 h, a second transfection was performed; and target RNA and protein assays were processed 24 h after the second transfection.
Electrophoretic Mobility Shift Assay—Nuclear extracts were prepared from 10 million K562 cells by washing the cells in phosphate-buffered saline and resuspending the cell pellet in 0.4 ml of a low salt buffer (10 mm HEPES (pH 7.9), 10 mm KCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride, with 1% protease inhibitor mixture) (27). These isolated nuclei were incubated on ice for 15 min, 20 μl of Nonidet P-40 was added, and the lysates were centrifuged at 13,000 rpm for 20 s. Supernatants were discarded, and the nuclear pellets were re-suspended in 20 μl of a high salt buffer (20 mm HEPES (pH 7.9), 420 mm NaCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, and 0.2 mm phenylmethylsulfonyl fluoride, with 1% protease inhibitor mixture). Nuclear lysates were incubated at 4 °C for 20 min with vortexing every 5 min. Lysates were then centrifuged at 13,000 rpm for 5 min, and the protein concentration was measured in the supernatant by using the Bio-Rad DC protein assay. The probes shown in supplemental Table S3 were prepared by annealing complementary sense and antisense oligonucleotides, followed by end labeling with [γ-32P]ATP using T4 polynucleotide kinase (Promega) and purification through a Sephadex G-25 column (Bio-Rad). EMSAs were performed according to the instructions for the Gel Shift Assay System (Promega). Reactions containing 18 μg of K562 nuclear extract, 0.5 μg of poly(dI-dC), with or without unlabeled competitor probes in binding buffer (10 mm Tris·HCl (pH 7.5), 75 mm NaCl, 1 mm EDTA, 0.5 mm dithiothreitol, and 5% glycerol) were incubated at room temperature for 10 min. Next, 0.2 ng of the labeled probes was added to the reaction, and the mixture was incubated for 20 min. For supershift assays, 1 μg of antibody was added 10 min before the addition of the labeled probes. The DNA-protein complexes were separated on a 6% polyacrylamide gel in 0.5× Tris-Borate-EDTA buffer. Gels were dried and exposed to x-ray film for 1 h to overnight at -80 °C to obtain autoradiographs.
Chromatin Immunoprecipitation—The assay was performed using an EZ ChIP chromatin immunoprecipitation kit (Upstate Biotechnology, Charlottesville, VA) with the protocol recommended by the manufacturer. Briefly, 10 million K562 cells were cross-linked at 37 °C for 10 min in 1% formaldehyde, and then sonicated in buffer containing 1% SDS, 10 mm EDTA, 50 mm Tris (pH 8.1), and 1% protease inhibitor mixture. After centrifugation at 10,000 rpm at 4 °C for 10 min, the supernatant was divided into three tubes for subsequent immunoprecipitation. Dilution buffer (0.01% SDS, 1.1% Triton, 1.2 mm EDTA, 16.7 mm Tris, pH 8.1, 167 mm NaCl, and 1% protease inhibitor mixture) was added to each tube, and protein A-Sepharose beads were added for preclearing. Lysates were rotated at 4 °C for 1 h. After centrifugation at 3,000 rpm for 1 min, 5 μg of anti-Sp1, anti-Egr1, or rabbit IgG was added to the supernatant, and the mixture was incubated on a rotator at 4 °C overnight. Protein A-Sepharose beads were added for 1 h at 4 °C. After centrifugation at 3,000 rpm for 1 min, the immune complex was sequentially washed with low salt buffer, high salt buffer, LiCl buffer, and TE buffer that were provided by the manufacturer. The immune complexes were eluted by rotation with 1% SDS and 0.1 m NaHCO3 for 15 min at room temperature. Cross-linked immunoprecipitate and input was reversed using 200 mm NaCl at 65 °C for 5 h. RNase A was added to the samples for 30 min at 37 °C, then incubated 2 h at 45 °C with 10 mm EDTA, 40 mm Tris, pH 6.5, and 50 μg/ml proteinase K. After purifying the DNA by spin column, DNA was amplified by PCR using the primers shown in supplemental Table S3 with the KOD Hot Start DNA polymerase (Novagen) at a temperature cycle of 15 s at 94 °C, 30 s at 59 °C, and 30 s at 68 °C for 30 cycles.
RESULTS
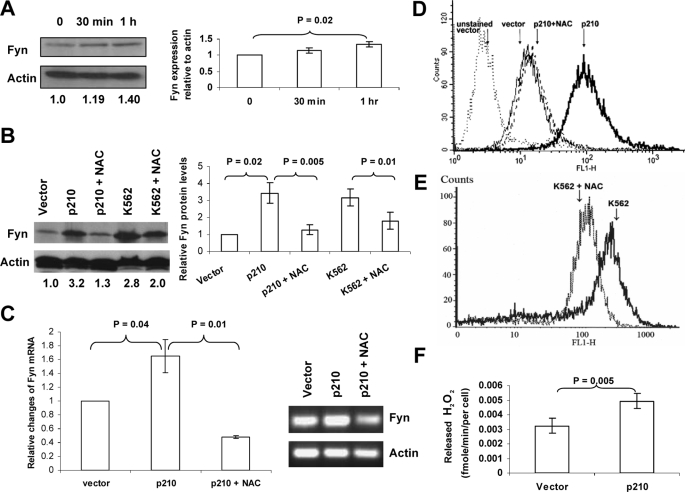
Up-regulation of Fyn Protein and mRNA in BCR-ABL1-expressing Cells is Dependent on Intracellular ROS Levels—Previous work from our laboratory demonstrated an increase in Fyn expression in BCR-ABL1-expressing cells (14). RNA interference-mediated knockdown revealed a role for Fyn in mediating proliferation in vitro and in vivo. Because the BCR-ABL1 oncogene is known to generate ROS (8), and because Fyn is known to be activated by ROS in fibroblasts and hepatocytes (15, 18), we examined the relationship between BCR-ABL1 expression, Fyn expression, and its regulation by ROS. In keeping with the oxidant dependence of Fyn described in other cell types (15), K562 cells expressing BCR-ABL1 pulsed with 1 mm hydrogen peroxide for 1 h showed increased Fyn protein expression by ∼1.4-fold as measured by Western blotting and densitometric analysis (Fig. 1A). This short exposure to hydrogen peroxide did not cause cell death as measured by trypan blue positivity (data not shown). To test whether suppressing oxidative stress would suppress Fyn expression, we used the antioxidant, NAC, which is thought to exert its effects by bolstering levels of glutathione (GSH), in K562 and in BaF3 cells transduced with BCR-ABL1 (BaF3 p210). Lysates from these cells were generated in the presence or absence of NAC, and Fyn protein was measured by Western blotting (Fig. 1B). Consistent with our previous findings, a 3-fold increase in Fyn protein expression was observed in BaF3 p210 cells as compared with BaF3 vector-transduced cells (lane 2 versus lane 1 of Fig. 1B). In two distinct BCR-ABL1-expressing cells (BaF3 p210 and K562), treatment with 24 mm NAC for 24 h lowered Fyn expression significantly (p = 0.005 and p = 0.01) without lowering expression of actin, which was used as a loading control (lanes 3 and 5, Fig. 1B). To determine if the repression of Fyn expression by NAC would be seen at the mRNA level, quantitative PCR was conducted using Fyn-specific primers (Fig. 1C). Mirroring the protein expression pattern, mRNA of Fyn was increased in BaF3 p210-expressing cells as compared with BaF3 vector-expressing cells and blunted in the presence of NAC (Fig. 1C). A possible explanation reconciling these Fyn mRNA alterations with the rapid changes in Fyn expression induced by hydrogen peroxide suggests that multiple mechanisms may contribute to oxidant-dependent Fyn expression, including transcriptional and post-translational regulation. The same dose of NAC that was utilized in these experiments lowered ROS levels in BaF3 cells transduced with p210 (Fig. 1D) and in K562 cells (Fig. 1E). To verify these results using an alternate measure of peroxide levels, we used an Amplex Red-based assay. Fig. 1F shows that the vector expressing BaF3 cells released ∼0.003 fm hydrogen peroxide/cell/minute, whereas BCR-ABL1-expressing cells released 0.005 fm. Thus, two different assays for cellular peroxide levels confirmed that BCR-ABL1 expression is linked to increased ROS. Taken together, these results suggest that the increased expression of Fyn in BCR-ABL1-expressing cells occurs in an ROS-dependent manner.
FIGURE 1.
The up-regulation of Fyn protein and mRNA in BCR-ABL1-expressing cells occurs in a ROS-dependent manner. A, hydrogen peroxide causes up-regulation of Fyn. K562 cells were treated with 1 mm H2O2 for 30 min or 1 h. Fyn protein levels were assessed by Western blot, and actin was used as a loading control. Trypan blue staining indicated that this short pulse of H2O2 was not cytotoxic. Densitometry, depicted in the bar graph, showing the ratio of Fyn to actin was normalized to control and was compiled from three independent experiments. B, NAC blocks up-regulation of Fyn in CML cells. K562 cells and BaF3 cells transduced with p210 BCR-ABL1 were treated with 24 mm NAC for 24 h. Fyn protein levels were assessed by Western blotting. The blot was then reprobed using an anti-actin antibody to ensure equal loading. The values below each lane represent the ratio of Fyn:actin protein expression. These values were calculated using densitometry, and each value was normalized to the non-BCR-ABL1 expressing BaF3 vector. The bar graph depicts densitometric values from three independent experiments. C, NAC decreases Fyn mRNA in BaF3 cells expressing BCR-ABL1. Fyn mRNA was measured in BaF3 p210 cells treated with diluent or 24 mm NAC for 24 h using RT-qPCR (p < 0.04). Relative change in expression was measured by calculating Ct values. The inset shows PCR products from that reaction resolved electrophoretically on an agarose gel. D and E, NAC lowers intracellular peroxide levels in BaF3 p210 and K562 cells. Cells were treated with diluent (solid line) or 24 mm NAC (dashed line) for 24 h followed by staining with dihydrodichlorofluorescein diacetate and analysis on the FL-1 channel of a flow cytometer to detect levels of intracellular peroxides. F, BCR-ABL1 p210-expressing cells release higher amounts of hydrogen peroxide than vector-expressing cells. BaF3 cells transduced with vector or BCR-ABL1 p210 were added to reaction buffer containing Amplex Red reagent and horseradish peroxidase, the absorbance at 560 nm was measured. Values depicted graphically were generated using a standard curve of absorbance using fixed increasing doses of hydrogen peroxide as described under “Experimental Procedures.”
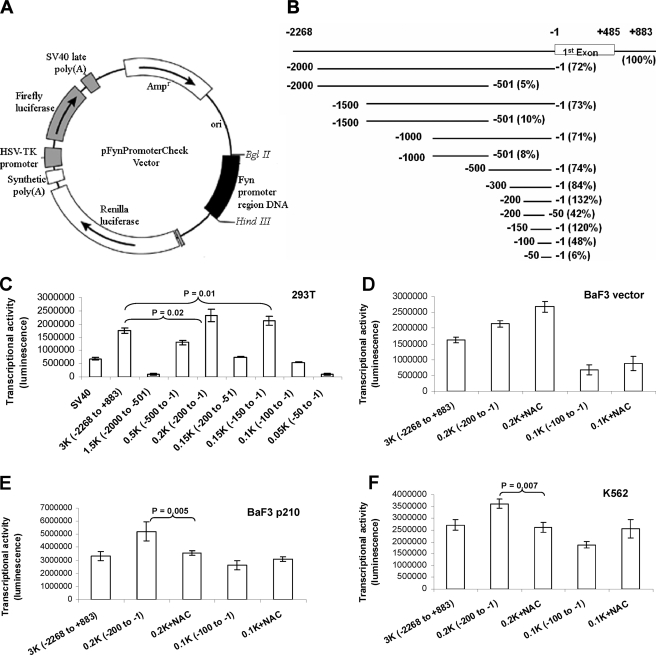
Characterization of the Human Fyn Promoter Reveals a 200-bp Region Lacking a TATA Box as the Basal Promoter—Despite abundant knowledge of the role of Fyn in signal transduction, no analysis of the Fyn promoter has been conducted to date. Therefore, the factors responsible for Fyn transcription and the molecular mechanism of its mRNA up-regulation in response to ROS are unknown. To elucidate this topic, we cloned the human Fyn promoter region from -2268 to +883 bp and constructed a dual-luciferase expression plasmid containing Renilla luciferase linked to Fyn promoter fragments and firefly luciferase as an internal control (Fig. 2A), thus enabling normalization with values from the same vector. To identify the minimal promoter region required for basal transcriptional activity, a series of deletion constructs (Fig. 2B) were transfected into 293T cells, and dual-luciferase activity was quantitated (Fig. 2, B-F). This initial characterization of the Fyn promoter was carried out in 293T cells, which are easily transfected, because Fyn is ubiquitously expressed across tissue types. Transfection with the longest reporter constructs (-2268/+883) resulted in a 1.5-fold increase in Renilla luciferase activity as compared with SV40 early promoter, demonstrating the robust nature of the Fyn promoter and suggesting that luciferase values from this construct represent a viable control for comparison of other promoter fragments. Constructs lacking the -500 to -1 region had very low luciferase activities, suggesting that the minimal promoter is in this region (Fig. 2B and C). The -200 to -1 construct had the strongest promoter activity, even higher than that seen with the full-length construct, suggesting the presence of repressive elements in the upstream regions. The rapid drop (>70%) of luciferase activities of the constructs -200 to -51, -100 to -1, and -50 to -1, suggests that the -200 to -1 region is the essential region for basal promoter transcriptional activity (Fig. 2, B and C). Because that construct retained almost the same level of activity as the -150 to -1 construct and the full promoter construct, we concluded that the region between -150 and -1 contains key sites for Fyn expression in 293T cells. Sequences in this region indicated the absence of a TATA-box. However, three GC-rich sites were identified that represented putative sites for transcription factor binding (Fig. 3A).
FIGURE 2.
Identification of transcriptional activities of various human Fyn promoter regions and a ROS-responsive fragment in the Fyn promoter. A, the dual-luciferase (firefly and Renilla luciferase) expression plasmid pFynPromoterCheck vector map. The 4.6-kb XhoI/BglII and 1.2-kb XhoI/Hind fragments from the psiCHECK-2 plasmid (Promega) were ligated with the BglII/HindIII promoter fragment to construct the pFynPromoterCheck dual-luciferase expression plasmid. In these vectors, the Fyn promoter region DNAs act as the promoters for the Renilla luciferase gene. B, a schematic of Fyn promoter fragment positions, and relative transcriptional activities. The transcriptional activities of the promoter fragments listed were determined using luciferase assays of transfected 293T cells and normalized to the full-length Fyn promoter. C, identification of transcription activities using deletion analysis of the human Fyn promoter in 293T cells. The full length and a series of deleted Fyn promoter constructs were transfected into 293T cells for 24 h followed by dual-luciferase assays in which transcriptional activity of Fyn (Renilla luciferase activity) was normalized to firefly luciferase activity. The 0.4-kb SV40 early promoter was used as a positive control (all p values < 0.04). D, NAC increases Fyn promoter transcription activity in BaF3 cells. Diluent or 24 mm NAC was added to cell culture medium 2 h before the transfection of pFynPromoterCheck 0.2- and 0.1-kb vectors (all p values < 0.05). Luciferase assays were used to measure Fyn promoter activity. E and F, identification of a ROS-responsive fragment in the Fyn promoter. BaF3 p210 cells (F) or K562 cells (G) transfected with either -200 to -1 or -100 to -1 Fyn promoter fragments were treated with either diluent or 24 mm NAC followed by measurement of luciferase activity (all p values < 0.01 in E; all p values < 0.04 in G).
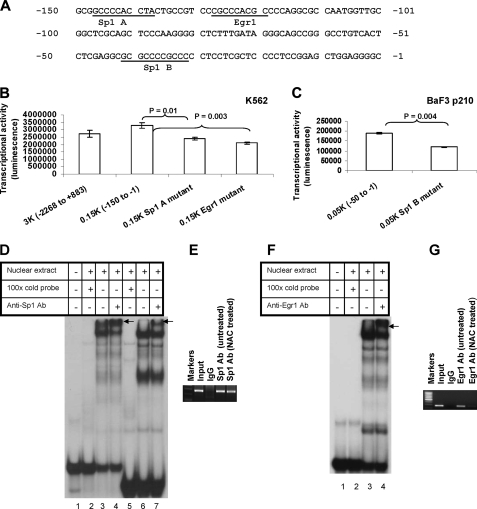
FIGURE 3.
Analysis of Sp1 and Egr1 binding sites in the Fyn promoter and their ROS dependence. A, the nucleotide sequence of the human Fyn promoter region (-150 to -1) that has high basal transcription activity. Numbering is relative to the transcription start site (+1). Putative binding sites for transcription factors Sp1 and Egr1 are indicated on the sequences. B, mutational analysis of the putative Sp1 A-binding site and Egr1-binding site of the Fyn promoter. Individual mutagenesis that destroyed Sp1 A and Egr1 putative binding sites were introduced to the pFynPromoterCheck plasmid by method described under “Experimental Procedures.” The Egr1 binding site was deleted, whereas the Sp1 binding site was mutated as described under “Experimental Procedures.” The mutant constructs were transfected into K562 cells. Renilla luciferase activity was normalized to firefly luciferase activity. C, mutational analysis of the putative Sp1 B-binding site of Fyn promoter in BaF p210 cells. Site-directed mutagenesis destroyed the Sp1 B putative binding site in the -50 to -1 region of the Fyn promoter construct. Following transfection of the mutant sequence into BaF3 p210 cells, dual-luciferase assays were used to measure promoter activity. D, gel shift assay reveals K562 cell nuclear proteins bind to the probe containing a putative Sp1 binding site. The 32P-labeled oligonucleotide probe (-150 to -121 bp) was incubated alone, with nuclear extract, with nuclear extract in the presence of excess unlabeled oligonucleotide, or with nuclear extract and a Sp1 antibody. Arrows indicate the supershifted complexes. Lanes 1-4 contain a 30-bp probe of the Fyn promoter, while lanes 5-7 contain a 22-bp Sp1 consensus probe (from Promega). E, chromatin immunoprecipitation of Sp1 with the Fyn promoter in K562 cells. Bands indicate PCR products targeting the -250 to -1 region of the promoter. 5 μg of anti-Sp1 or rabbit IgG antibody or 1/100 of input was separately added in the assay. F, gel shift assay of K562 cell nuclear proteins bind to a probe containing a putative Egr1 binding site. The 32P-labeled oligonucleotide probe (-140 to -111 bp) was incubated alone, with nuclear extract, with nuclear extract in the presence of excess unlabeled oligonucleotide, or with nuclear extraction and anti-Egr1 antibody. Arrows indicate the supershifted complexes. G, chromatin immunoprecipitation of Egr1 with the human Fyn promoter in K562 cells. Bands indicate PCR products targeting the -250 to -1 region of the Fyn promoter. K562 cells were untreated or treated with 24 mm NAC for 24 h. 5 μg of anti-Egr1 or rabbit IgG antibody or 1/100 of input was separately added in the assay. Every result shown is representative of three independent experiments.
Next, we applied these findings to CML model systems to determine if the minimum promoter region identified in 293T cells was conserved in hematopoietic cell lines expressing BCR-ABL1. In both BaF3 vector and BaF3 p210 cells, the same 0.2-kb region elicited a strong luciferase signal as compared with transfection of the largest promoter piece (Fig. 2, D and E). Similar experiments in the human BCRABL1-expressing K562 cells highlighted the same region as the essential Fyn promoter (Fig. 2F). Interestingly, comparison of the transcriptional activity of the full-length construct in BaF3 vector cells (first bar, Fig. 2D) and BaF3 p210 cells (first bar, Fig. 2E) indicates that BCR-ABL1-expressing cells had higher activity.
ROS-dependent Transcription of Fyn Is within the Promoter Region Spanning -200 to -100 bp—Our previous studies, as well as data in Fig. 1 (B and C), show that the expression of BCR-ABL1 promotes Fyn mRNA and protein expression. We also found that an antioxidant can blunt this up-regulation. We therefore investigated whether there was a region of the Fyn promoter responsible for Fyn expression in an oxidant-dependent manner. To identify that sequence, a series of deletion constructs were transfected into BaF3 vector or p210-expressing cells, and dual-luciferase activity was measured in the presence or absence of NAC. Interestingly, our results showed that the luciferase activities of both -200 to -1 and -100 to -1 constructs in the BaF3-vector cells were slightly increased upon NAC treatment (Fig. 2D). However, in the BaF3-p210 or K562 cells, luciferase activity in the -200 to -1 construct was decreased upon NAC treatment, but in the -100 to -1 construct activity was increased with NAC exposure (Fig. 2, E and F). These data suggest that the region spanning -200 to -100 bp in the promoter is responsible for ROS sensitivity only in BCR-ABL1-expressing cells. In both the BaF3-p210 and K562 cells, NAC caused a significant reduction (p = 0.005 and 0.007) in Fyn promoter activity.
DNA Binding of Sp1 and Egr1 Regulate Fyn Transcription—To identify specific transcription factors that could be driving Fyn expression, we analyzed the promoter region spanning -150 to -1 kb using the Transcription Element Search System, which can predict transcription factor binding sites based on DNA sequence. The analysis revealed that the three GC-rich sites present on the minimal promoter (Fig. 3A) are potential binding sites for the transcription factors Sp1 and Egr1. Two putative Sp1 binding sites appear in this region, Sp1 site A (-147 to -138) and Sp1 site B (-41 to -32), along with one binding site for Egr1 (-128 to -120) (Fig. 3A). To determine whether these sites were important for Fyn promoter activity, the Sp1-A (-147 to -138), Sp1-B (-41 to -32), and Egr1 binding sites were mutated using site-directed mutagenesis (Fig. 3A) and transfected into K562 and BaF3 p210 cells. For the Egr1 site, both a deletion mutant and a substitution mutant were generated, whereas for the Sp1 sites, only substitution mutants were made. Base pair sequences were altered as described under “Experimental Procedures.” As shown in Fig. 3 (B and C), mutations in Sp1-A or Sp1-B binding sites caused a 20-30% reduction in luciferase activity as compared with unmutated promoter sequences. Luciferase activity was also reduced by ∼20% when the Egr1 binding site was replaced by a substitution mutant (data not shown) and was reduced by 36% when an Egr1 deletion mutant was introduced into the promoter sequence (Fig. 3B). Collectively, these results indicate that the Sp1-A, -B, and Egr1 binding sites regulate Fyn promoter activity.
DNA Binding of Egr1 to the Fyn Promoter Is Redox-sensitive whereas Sp1 DNA Binding Is Not—Because the Fyn reporter gene assay data indicated that mutation of Sp1 and Egr1 binding sites reduced promoter activity, we sought to determine if Sp1 and Egr1 actually bind to the Fyn promoter. Two approaches were adopted: EMSA (Fig. 3, D and F) and chromatin immunoprecipitation assays (Fig. 3, E and G). EMSA using 32P-labeled oligonucleotide probes with the nuclear extract from K562 cells was conducted using three sequences from the Fyn promoter: -150 to -121, -140 to -111, and -50 to -21. The 30-bp -150 to -121 probe, whose sequence contains a putative Sp1 binding site, formed a DNA-protein complex with the nuclear extract, whereas no complex was found in the absence of nuclear extract (Fig. 3D, lanes 1 and 3). The formation of a complex was completely inhibited with the addition of an excess amount of unlabeled oligonucleotide (Fig. 3D, lane 2), suggesting that the complex is binding to the Sp1-A site. The presence of Sp1 in the complex was confirmed by the formation of a supershifted band (as indicated by the arrow) upon incubation with an anti-Sp1 antibody (Fig. 3D, lane 4). The EMSA assay was also conducted with a Sp1 consensus probe (Fig. 3D, lanes 5-7), and this migrated at a slightly smaller size than the 30-bp probe representative of the Fyn promoter due to its shorter length of 22 bp. Similar experiments investigating the Sp1-B binding site, represented by the -50 to -21 probe (data not shown), yielded similar results. Egr1 was also found to bind to the Fyn promoter by EMSA (Fig. 3F). The probe used was the DNA sequence of the Fyn promoter from -140 to -111 bp, which encompasses a putative Egr1 binding site. This probe was capable of binding protein from the nuclear extract of K562 cells, and the band was supershifted when incubated with anti-Egr1 antibody, demonstrating a functional effect of Egr1 on the Fyn promoter (Fig. 3F).
To further demonstrate that Sp1 and Egr1 can bind to the Fyn promoter in an environment more physiologically representative of chromatin interactions, we performed chromatin immunoprecipitation assays using K562 cells. For both Sp1 and Egr1, antibodies for these transcription factors bound to PCR amplified promoter regions (Fig. 3E, lane 4 and 3G, lane 4), confirming our EMSA results. To test the redox dependence of this binding, cells were treated with 24 mm NAC for 24 h followed by immunoprecipitation of the cross-linked cell lysates with anti-Sp1 and anti-Egr1 antibodies (lane 5, Fig. 3, E and G). Decreased binding of Egr1 was observed in the NAC-treated K562 cells (Fig. 3G), whereas binding of Sp1 was not changed in NAC-treated cells (Fig. 3E). Therefore, Egr1 is likely responsible for the decrease in Fyn mRNA and protein expression seen in BCR-ABL1-expressing cells treated with NAC (Fig. 1, B and C). Taken together, our results indicate that Sp1 and Egr1 both promote Fyn expression, although Egr1 does so in a redox-dependent manner.
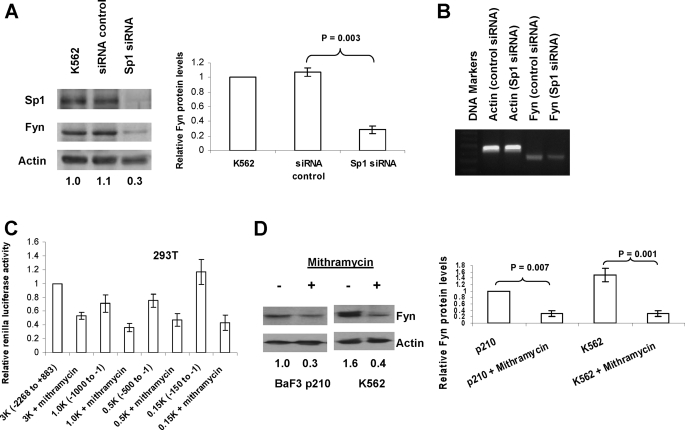
Knockdown of Sp1 and Inhibition of Sp1 Binding Causes a Decrease in Fyn Expression—Sp1 is a well characterized transcription factor that binds GC-rich regions in promoters, thereby recruiting TATA-binding proteins and prepping transcription start sites (28). Our data (Fig. 3, D and E) implicates Sp1 in binding to key portions of the Fyn promoter. To connect Sp1 more directly with Fyn expression we investigated the effect of Sp1 knockdown and inhibition of Sp1 binding on the protein expression of Fyn and on Fyn promoter activity (Fig. 4). Knockdown of Sp1 by siRNA in K562 cells significantly decreased (70%) Fyn mRNA levels (Fig. 4B) and protein levels after two sequential electroporations (Fig. 4A), whereas nonspecific siRNA did not affect levels of Fyn or Sp1 protein. Mithramycin A is a compound known to bind to GC boxes and inhibit Sp1 binding (29). The effect of mithramycin A on Fyn promoter activity was investigated using the different promoter constructs transfected into 293T cells (Fig. 4C). Luciferase activity was markedly reduced (40% to 70%) in all the constructs containing the -150 to -1 essential promoter region when cells were incubated with mithramycin A. Treatment of BaF3 cells expressing BCR-ABL1 p210 and K562 cells with mithramycin A also led to a decrease of more than 60% in Fyn protein levels (Fig. 4D). These data provide further evidence that Sp1 is a critical factor for Fyn expression.
FIGURE 4.
Sp1 is a basal transcriptional regulator of human Fyn promoter. A and B, siRNA-mediated inhibition of Sp1 leads to decreased Fyn expression. K562 cells were transfected with Sp1 siRNA or control once followed by a second transfection after 24 h. Protein levels of Sp1 and Fyn were assessed by Western blotting, Fyn mRNA levels were measured by PCR, and actin was used as a loading control. Densitometry (Fyn:actin ratio normalized to control) was conducted on three independent experiments and is depicted in the bar graph. C, the Sp1 inhibitor mithramycin A inhibits Fyn promoter transcriptional activity. 293T cells were transfected with the -2268/+883, -1000/-1, -500/-1, and -150/-1 constructs of the Fyn promoter; 50 nm mithramycin A was added to the cells just after transfection. Renilla luciferase activity was normalized to firefly luciferase activity (all p values < 0.007). D, mithramycin A decreases Fyn protein. K562 and BaF3 p210 cells were treated with/without 100 nm mithramycin A for 24 h, and Fyn protein levels were assessed by Western blotting. Densitometric values are shown below the bands.
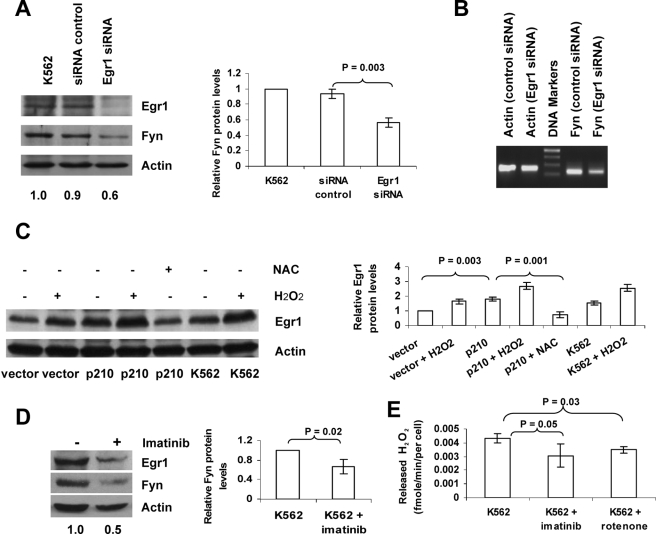
ROS-induced Egr1 Mediates Up-regulation of Fyn in BCR-ABL1 Expressing Cells—Datta et al. (30) previously observed that ROS generated by ionizing radiation induced Egr1 activation in human myeloid leukemia cells. In the present study, the contribution of Egr1 to Fyn expression was assessed using siRNA directed toward Egr1. Two sequential transfections of Egr1 siRNA (containing four different sequences) in K562 cells resulted in effective knockdown of Egr1 protein (Fig. 5A). This RNA interference mediated reduction in Egr1 expression led to a 40% decrease in Fyn protein expression and a noticeable decrease in Fyn mRNA (Fig. 5, A and B), thereby linking Egr1 expression to Fyn expression. Because Egr1 has been shown to be redox-dependent by others in c-abl-expressing cells (31), we examined whether BCR-ABL1-mediated ROS might contribute to Egr1 expression. In BaF3 p210 cells, Egr1 protein expression was doubled as compared with BaF3 vector cells (Fig. 5C). Exposure to 50 μm hydrogen peroxide increased Egr1 protein expression in all cell lines tested (BaF3 vector, BaF3 p210, and K562) and NAC treatment caused a reduction in Egr1 in BaF3 p210 cells as measured by Western blot (Fig. 5C), highlighting the oxidant dependence of Egr1 expression. To place Egr1 and Fyn expression within the context of BCR-ABL1, K562 cells were treated with the BCR-ABL1 kinase inhibitor, imatinib, and Egr1 and Fyn protein levels were assessed. Imatinib treatment caused a 40% reduction in Egr1 and Fyn protein expression (Fig. 5D). To establish a link between this result and ROS generation by BCR-ABL1, K562 cells were treated with 0.25 μm imatinib to inhibit BCR-ABL1 kinase or with 0.1 μm rotenone to suppress mitochondrial ROS production arising from electron transport. Both imatinib and rotenone caused reduced release of hydrogen peroxide from K562 cells as measured by Amplex Red (Fig. 5E). This is consistent with a published report of imatinib and rotenone reducing intracellular hydrogen peroxide as measured by dichlorofluoroscein fluorescence (8). These results indicate that the kinase activity of BCR-ABL1 and mitochondrial electron transport contribute to the increased ROS observed in BCR-ABL1-expressing cells, which in turn up-regulates Egr1 and Fyn.
FIGURE 5.
Increased Fyn expression in Bcr-Abl1-expressing cells results from ROS-dependent activation of Egr1. A and B, siRNA-mediated inhibition of Egr1 leads to decreased Fyn expression. K562 cells were transfected once with Egr1 siRNA or control, followed by a second transfection after 24 h. Protein levels of Egr1 and Fyn were assessed by Western blotting, Densitometry was conducted (Fyn:actin ratio normalized to control) on three independent experiments and is depicted in the bar graph. Fyn mRNA levels were measured by PCR. C, hydrogen peroxide induces Egr1, whereas NAC blocks up-regulation of Egr1 in BaF3 and K562 cells. After treatment with 50 μm H2O2 or with 24 mm NAC for 24 h, Egr1 protein levels were assessed by Western blot. Densitometric values from three independent experiments are depicted in the bar graph to the right of the Western blot. D, inhibition of BCR-ABL1 decreases Egr1 and Fyn protein expression in K562 cells. Cells were treated with diluent or 0.25 μm imatinib mesylate for 24 h. Levels of Egr1 and Fyn protein were measured by Western blotting. All blots above were reprobed with an actin antibody to ensure equal loading. The numerical values listed below the bands represent a ratio of Fyn protein expression to actin protein expression. E, inhibition of BCR-ABL1 or electron transfer decreases hydrogen peroxide released from K562 cells. Cells were treated with 0.25 μm imatinib mesylate for 24 h or treated with 0.10 μm rotenone for 1 h, released hydrogen peroxide levels measured by the Amplex Red and horseradish peroxidase method described under “Experimental Procedures.”
DISCUSSION
The current study builds upon our previous work showing that Fyn up-regulation in BCR-ABL1-expressing cells is an important mediator of in vitro and in vivo proliferation (14) and sought to identify the mechanism of heightened levels of Fyn. BCR-ABL1 is one of several oncogenes such as Ras (32), Flt3 (33), and Myc (34) that raises levels of ROS in addition to potently promoting signaling via kinase cascades (6-8). Here, we report that ROS are responsible for up-regulation of Fyn mRNA and protein levels by inducing expression of Egr1, a transcription factor that interacts with the Fyn promoter in an oxidant-dependent fashion. These data provide insight into the transcriptional regulation of Fyn, which is applicable to systems including but not limited to CML cells.
Interestingly, Egr1 and Sp family members have been linked to the promoter of another BCR-ABL1 signaling regulator, protein-tyrosine phosphatase 1B (PTP1B) (35). However, unlike Fyn, PTP1B opposes BCR-ABL1 transformation. Within the PTP1B promoter, a stress-responsive element was found where Sp1, Sp3, and Egr1 bind, however Sp3 binding and PTP1B promoter activation are antagonized by Egr1. In contrast, our findings suggest a positive feed-forward relationship between BCR-ABL1, Egr1, and Fyn. We show for the first time that Egr1 expression is decreased when BCR-ABL1 kinase activity is diminished by the potent and specific kinase inhibitor, imatinib (Fig. 5D). Therefore, signaling downstream of BCR-ABL1 is important for Egr1 expression. Of note is the fact that the conclusion by Fukada et al. (35) that p210 BCR-ABL1 inhibited Egr1 expression only addressed fibroblasts overexpressing BCR-ABL1 whereas our study utilized hematopoietic cells overexpressing BCR-ABL1 and a human CML cell line. Therefore our seemingly disparate observations could be due to cell type-specific differences. From these seemingly conflicting results, it is apparent that the contribution of Sp1 and Egr1 to BCR-ABL1-mediated leukemogenesis requires more detailed study, in particular in mouse models for CML.
Like Fyn, Egr1 expression is up-regulated in BCR-ABL1-expressing cells and blunted by the antioxidant, NAC (Fig. 5C). The induction of Egr1 by ionizing radiation has been linked to ROS (30) and to c-abl (31), however, ours is the first work to link BCR-ABL1 kinase activity and oxidative stress to Egr1. Because the BCR-ABL1 kinase inhibitor, imatinib, blocks heightened ROS, Egr1, and Fyn expression, this places BCR-ABL1 at the apex of a network that activates Egr1 and Fyn, ultimately transducing a mitogenic signal that controls CML cell growth (14). The complexity of the regulation of this pathway by redox highlights the importance of understanding the source of oxidative stress seen in BCR-ABL1-expressing cells. Although the increased level of oxidative stress in several cell lines ectopically expressing BCR-ABL1 has been verified by two other independent groups (8, 36), only one study has addressed potential sources for these ROS (6). Inhibition of mitochondrial respiration and inhibitors of the phosphatidylinositol 3-kinase pathway partially block BCR-ABL1-mediated oxidative stress, suggesting that multiple endogenous sources of ROS may be hyperactive. In Fig. 5E, we use a different method to confirm that mitochondrial sources contribute to the increased ROS in BCR-ABL1-expressing cells. Because these ROS have been linked to genomic instability and mutations in CML, it is a potentially therapeutically relevant goal to elucidate the source of oxidative stress that is driving expression of Egr-1 and Fyn.
Our analysis of the Fyn promoter designated two Sp1 sites in controlling Fyn expression (Fig. 3, A-C). Like Egr1, Sp1 also recognizes GC-rich regions and also contains zinc finger domains (28). Unlike Egr1, Sp1-mediated Fyn expression and DNA binding were not ROS-dependent (Fig. 3E). Reports from many other systems indicate that Sp1 is a redox-sensitive transcription factor under specific circumstances. For example, COX-2 induction in cortical neurons by oxidative stress requires Sp1 and Sp3 binding to the COX-2 promoter (37). We find that Sp1 is required for Fyn protein expression, because RNA interference directed toward Sp1 caused a 70% reduction in Fyn levels (Fig. 4A), and mithramycin A (an agonist of Sp1 DNA binding) caused a 75% reduction in Fyn levels (Fig. 4D). From promoter reporter gene analysis in BaF3 cells expressing vector alone or BCR-ABL1, the region spanning -100 to -1 contains elements important for BCR-ABL1-dependent Fyn expression, implicating the second Sp1 binding site designated as Sp1 B, in this response. Interestingly, Sp1 along with Sp3 are reported to be downstream targets for imatinib in regulating vascular epidermal growth factor gene transcription (38), therefore reinforcing the BCR-ABL1-dependent role for Sp1 we are also describing. An increase in Fyn promoter activity was observed in the -200 to -1 fragment as compared with the full-length fragment (Fig. 2, B-F), indicating the potential presence of repressive elements. We cannot rule out the possibility that these negative regulators of Fyn expression may be redox-sensitive and/or BCR-ABL1-sensitive. Studies to address that possibility are underway. The present study focuses on Egr1 and Sp1 as positive regulators of Fyn expression, which are both GC box binding factors. Typically, transcription factors that bind GC-rich sequences are components of large transcriptional complexes containing co-activators and co-repressors. Little is known regarding which co-activators assist Egr1 to increase transcriptional activity, however it is interesting that there did not appear to be repression of Fyn reporter activity occurring within the identified (-128 to -120) Egr1 binding site. This lack of co-repression may explain the opposite role of Fyn in CML (where high expression is seen in progressed blast crisis patients) versus neuroblastoma (39) and prostate cancer (40) where down-regulation or silencing of Fyn is seen in advanced tumor stages.
This study provides the first functional characterization of the Fyn promoter, which is notable because Fyn is one of the Src family members that is expressed ubiquitously across tissue types (16). During the course of our functional deletion analysis, we did discover classic promoter elements such as CCAAT (-769 to -765) and TATA (-624 to -621) boxes within the region upstream of the first exon of Fyn spanning the -500- to -1000-bp regions. However, insertion of this promoter piece into our reporter gene assay showed extremely low luciferase activity (∼8%); therefore, this region does not represent a true promoter (Fig. 2, B and C). Instead, the -200 to -1 region more than compensated for full-length promoter activity, although it lacked a TATA box. The three GC-rich regions, which we proved to be binding sites for Sp1 and Egr1, constitute the promoter for Fyn in non-BCR-ABL1-expressing cells as well as those expressing BCR-ABL1. Analysis of the promoters of other Src family kinases reveals functional homology. Characterization of the promoter for Src also showed a lack of TATA or CAAT boxes but did contain high GC content with several consensus Sp1 and AP2 binding sites (41, 42). Promoter analysis of Lyn, another Src family member, revealed four GC box-like sequences but no TATA or CCAAT box (43). GC-rich regions are also prevalent and functional in the Hck (44) and Yes (45) promoters. Therefore, the promoter regions of several Src family members, including Fyn, share conserved features of having no functional TATA or CCAAT box, and of basal transcription being driven by Sp1. Our previously published work highlighted up-regulation of Fyn rather than activation of Fyn as targetable entities for advanced or refractory CML. The knowledge that Sp1 and Egr1 drive Fyn expression delineates an additional set of potential therapeutic targets. Given the pleiotropic effects of the BCR-ABL1 kinase, it is not surprising that multiple transcriptional and translational molecular alterations that could potentially be targeted are being uncovered. For example, the expression and activity of RNA-binding proteins with varied roles in regulation of mRNA metabolism (46), and targets of these RNA-binding proteins such as mdm2 (47), c/ebpα (48, 49), and E2F3 (50), are modulated by BCR-ABL1 and may play a role in CML development and progression. Because the treatment of blast crisis CML and management of kinase inhibitor-resistant disease remain problematic for CML patients, we anticipate that uncovering these novel downstream biological signals sent by BCR-ABL1 will aid in overcoming these persistent challenges.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA 115811 (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S3.
Footnotes
The abbreviations used are: CML, chronic myelogenous leukemia; ROS, reactive oxygen species; NAC, N-acetylcysteine; siRNA, small interference RNA; EMSA, electrophoretic mobility shift assay; PTP1B, protein-tyrosine phosphatase 1B.
References
- 1.Nowell, P. C. (2007) J. Clin. Investig. 117 2033-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlinghaus, R., and Sun, T. (2004) Cancer Treat. Res. 119 239-270 [DOI] [PubMed] [Google Scholar]
- 3.Deininger, M. W. (2007) Exp. Hematol. 35 144-154 [DOI] [PubMed] [Google Scholar]
- 4.Hochhaus, A., Erben, P., Ernst, T., and Mueller, M. C. (2007) Semin. Hematol. 44 S15-S24 [DOI] [PubMed] [Google Scholar]
- 5.Shah, N. P., and Sawyers, C. L. (2003) Oncogene 22 7389-7395 [DOI] [PubMed] [Google Scholar]
- 6.Kim, J. H., Chu, S. C., Gramlich, J. L., Pride, Y. B., Babendreier, E., Chauhan, D., Salgia, R., Podar, K., Griffin, J. D., and Sattler, M. (2005) Blood 105 1717-1723 [DOI] [PubMed] [Google Scholar]
- 7.Sattler, M., and Griffin, J. D. (2003) Semin. Hematol, 40 4-10 [DOI] [PubMed] [Google Scholar]
- 8.Sattler, M., Verma, S., Shrikhande, G., Byrne, C. H., Pride, Y. B., Winkler, T., Greenfield, E. A., Salgia, R., and Griffin, J. D. (2000) J. Biol. Chem. 275 24273-24278 [DOI] [PubMed] [Google Scholar]
- 9.Nowicki, M. O., Falinski, R., Koptyra, M., Slupianek, A., Stoklosa, T., Gloc, E., Nieborowska-Skorska, M., Blasiak, J., and Skorski, T. (2004) Blood 104 3746-3753 [DOI] [PubMed] [Google Scholar]
- 10.Koptyra, M., Falinski, R., Nowicki, M. O., Stoklosa, T., Majsterek, I., Nieborowska-Skorska, M., Blasiak, J., and Skorski, T. (2006) Blood 108 319-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkel, T. (2003) Curr. Opin. Cell Biol. 15 247-254 [DOI] [PubMed] [Google Scholar]
- 12.Penserga, E. T., and Skorski, T. (2007) Oncogene 26 11-20 [DOI] [PubMed] [Google Scholar]
- 13.Pendergast, A. M. (2002) Adv. Cancer Res. 85 51-100 [DOI] [PubMed] [Google Scholar]
- 14.Ban, K., Gao, Y., Amin, H. M., Howard, A., Miller, C., Lin, Q., Leng, X., Munsell, M., Bar-Eli, M., Arlinghaus, R. B., and Chandra, J. (2008) Blood 111 2904-2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe, J., and Berk, B. C. (1999) J. Biol. Chem. 274 21003-21010 [DOI] [PubMed] [Google Scholar]
- 16.Courtneidge, S. A., Fumagalli, S., Koegl, M., Superti-Furga, G., and Twamley-Stein, G. M. (1993) Dev. Suppl. 57-64 [PubMed]
- 17.Furumoto, Y., Gonzalez-Espinosa, C., Gomez, G., Kovarova, M., Odom, S., Parravicini, V., Ryana, J. J., and Rivera, J. (2004) Immunol. Res. 30 241-253 [DOI] [PubMed] [Google Scholar]
- 18.Sanguinetti, A. R., Cao, H., and Corley Mastick, C. (2003) Biochem. J. 376 159-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appleby, M. W., Gross, J. A., Cooke, M. P., Levin, S. D., Qian, X., and Perlmutter, R. M. (1992) Cell 70 751-763 [DOI] [PubMed] [Google Scholar]
- 20.Palacios, E. H., and Weiss, A. (2004) Oncogene 23 7990-8000 [DOI] [PubMed] [Google Scholar]
- 21.Juric, D., Lacayo, N. J., Ramsey, M. C., Racevskis, J., Wiernik, P. H., Rowe, J. M., Goldstone, A. H., O'Dwyer, P. J., Paietta, E., and Sikic, B. I. (2007) J. Clin. Oncol. 25 1341-1349 [DOI] [PubMed] [Google Scholar]
- 22.Meyn, M. A., 3rd, Wilson, M. B., Abdi, F. A., Fahey, N., Schiavone, A. P., Wu, J., Hochrein, J. M., Engen, J. R., and Smithgall, T. E. (2006) J. Biol. Chem. 281 30907-30916 [DOI] [PubMed] [Google Scholar]
- 23.Doggrell, S. A. (2005) Expert Opin. Investig. Drugs 14 89-91 [DOI] [PubMed] [Google Scholar]
- 24.Kamath, A. V., Wang, J., Lee, F. Y., and Marathe, P. H. (2008) Cancer Chemother. Pharmacol. 61 365-376 [DOI] [PubMed] [Google Scholar]
- 25.Shah, N. P., Tran, C., Lee, F. Y., Chen, P., Norris, D., and Sawyers, C. L. (2004) Science 305 399-401 [DOI] [PubMed] [Google Scholar]
- 26.Long, J., Manchandia, T., Ban, K., Gao, S., Miller, C., and Chandra, J. (2007) Cancer Chemother. Pharmacol. 59 527-535 [DOI] [PubMed] [Google Scholar]
- 27.Chandra, J., Niemer, I., Gilbreath, J., Kliche, K. O., Andreeff, M., Freireich, E. J., Keating, M., and McConkey, D. J. (1998) Blood 92 4220-4229 [PubMed] [Google Scholar]
- 28.Safe, S., and Abdelrahim, M. (2005) Eur. J. Cancer 41 2438-2448 [DOI] [PubMed] [Google Scholar]
- 29.Albertini, V., Jain, A., Vignati, S., Napoli, S., Rinaldi, A., Kwee, I., Nur-e-Alam, M., Bergant, J., Bertoni, F., Carbone, G. M., Rohr, J., and Catapano, C. V. (2006) Nucleic Acids Res. 34 1721-1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta, R., Taneja, N., Sukhatme, V. P., Qureshi, S. A., Weichselbaum, R., and Kufe, D. W. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2419-2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart, J. R., Kawai, H., Tsai, K. K., Chuang, E. Y., and Yuan, Z. M. (2005) Oncogene 24 8085-8092 [DOI] [PubMed] [Google Scholar]
- 32.Masutani, H. (2000) Int. J. Hematol. 71 25-32 [PubMed] [Google Scholar]
- 33.Sallmyr, A., Fan, J., Datta, K., Kim, K. T., Grosu, D., Shapiro, P., Small, D., and Rassool, F. (2008) Blood 111 3173-3182 [DOI] [PubMed] [Google Scholar]
- 34.Dang, C. V., Li, F., and Lee, L. A. (2005) Cell Cycle 4 1465-1466 [DOI] [PubMed] [Google Scholar]
- 35.Fukada, T., and Tonks, N. K. (2001) J. Biol. Chem. 276 25512-25519 [DOI] [PubMed] [Google Scholar]
- 36.Trachootham, D., Zhou, Y., Zhang, H., Demizu, Y., Chen, Z., Pelicano, H., Chiao, P. J., Achanta, G., Arlinghaus, R. B., Liu, J., and Huang, P. (2006) Cancer Cell 10 241-252 [DOI] [PubMed] [Google Scholar]
- 37.Lee, J., Kosaras, B., Aleyasin, H., Han, J.A., Park, D. S., Ratan, R. R., Kowall, N. W., Ferrante, R. J., Lee, S. W., and Ryu, H. (2006) FASEB J. 20 2375-2377 [DOI] [PubMed] [Google Scholar]
- 38.Legros, L., Bourcier, C., Jacquel, A., Mahon, F. X., Cassuto, J. P., Auberger, P., and Pages, G. (2004) Blood 104 495-501 [DOI] [PubMed] [Google Scholar]
- 39.Berwanger, B., Hartmann, O., Bergmann, E., Bernard, S., Nielsen, D., Krause, M., Kartal, A., Flynn, D., Wiedemeyer, R., Schwab, M., Schafer, H., Christiansen, H., and Eilers, M. (2002) Cancer Cell 2 377-386 [DOI] [PubMed] [Google Scholar]
- 40.Sorensen, K. D., Borre, M., Orntoft, T. F., Dyrskjot, L., and Torring, N. (2008) Int. J. Cancer 122 509-519 [DOI] [PubMed] [Google Scholar]
- 41.Bonham, K., and Fujita, D. J. (1993) Oncogene 8 1973-1981 [PubMed] [Google Scholar]
- 42.Ritchie, S., Boyd, F. M., Wong, J., and Bonham, K. (2000) J. Biol. Chem. 275 847-854 [DOI] [PubMed] [Google Scholar]
- 43.Uchiumi, F., Semba, K., Yamanashi, Y., Fujisawa, J., Yoshida, M., Inoue, K., Toyoshima, K., and Yamamoto, T. (1992) Mol. Cell Biol. 12 3784-3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hauses, M., Tonjes, R. R., and Grez, M. (1998) J. Biol. Chem. 273 31844-31852 [DOI] [PubMed] [Google Scholar]
- 45.Matsuzawa, Y., Semba, K., Kawamura-Tsuzuku, J., Sudo, T., Ishii, S., Toyoshima, K., and Yamamoto, T. (1991) Oncogene 6 1561-1567 [PubMed] [Google Scholar]
- 46.Iervolino, A., Santilli, G., Trotta, R., Guerzoni, C., Cesi, V., Bergamaschi, A., Gambacorti-Passerini, C., Calabretta, B., and Perrotti, D. (2002) Mol. Cell Biol. 22 2255-2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trotta, R., Vignudelli, T., Candini, O., Intine, R. V., Pecorari, L., Guerzoni, C., Santilli, G., Byrom, M. W., Goldoni, S., Ford, L. P., Caligiuri, M. A., Maraia, R. J., Perrotti, D., and Calabretta, B. (2003) Cancer Cell 3 145-160 [DOI] [PubMed] [Google Scholar]
- 48.Chang, J. S., Santhanam, R., Trotta, R., Neviani, P., Eiring, A. M., Briercheck, E., Ronchetti, M., Roy, D. C., Calabretta, B., Caligiuri, M. A., and Perrotti, D. (2007) Blood 110 994-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerzoni, C., Bardini, M., Mariani, S. A., Ferrari-Amorotti, G., Neviani, P., Panno, M. L., Zhang, Y., Martinez, R., Perrotti, D., and Calabretta, B. (2006) Blood 107 4080-4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eiring, A. M., Neviani, P., Santhanam, R., Oaks, J. J., Chang, J. S., Notari, M., Willis, W., Gambacorti-Passerini, C., Volinia, S., Marcucci, G., Caligiuri, M. A., Leone, G. W., and Perrotti, D. (2008) Blood 111 816-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.