Abstract
Plexins, comprising Plexin-A, -B, -C, and -D subfamilies, are receptors for semaphorins governing cell adhesion, migration, and axon guidance. Among plexin subfamilies, Plexin-A1 and Plexin-B1 have been shown to function as an R-Ras GAP, inducing repulsive responses, and the expression of R-Ras GAP activity requires the binding of Rnd1, a member of Rnd subfamily of Rho GTPases. However, signaling pathways of Plexin-D1 and Plexin-C1 still remain obscure. Here, we found that Plexin-D1 displayed R-Ras GAP activity and inhibited migration of COS-7 cells, and these actions required Rnd2, another Rnd subfamily GTPase. Rnd2 bound to Plexin-D1 in cortical neurons, and Sema3E/Plexin-D1-induced inhibition of axon outgrowth of cortical neurons required Rnd2 and down-regulation of R-Ras activity. On the other hand, Plexin-C1 displayed R-Ras GAP activity and inhibited cell migration of COS-7 cells without Rnd proteins. Therefore, R-Ras GAP activity is a common function of plexin subfamilies but the regulation of R-Ras GAP activity of plexins by Rnd proteins is different among plexin subfamilies.
Semaphorins (Sema)2 are a large family of secreted or membrane-bound molecules that play central roles in axon guidance in developing nervous system (1–3). In addition to the nervous system, they are widely expressed in embryonic and adult tissues and mediate diverse biological processes such as cardiac and skeletal development (4), tumor growth, and metastasis (5), and the immune response (6). The functions of semaphorins are mediated by plexins, which can be classified into four subfamilies: Plexin-A1–4, PlexinB1–3, Plexin-C1, and Plexin-D1 (2, 7). PlexinA1–4, together with ligand-binding neuropilins, transduce repulsive signaling for class3 semaphorins (8). Plexin-B1 is a receptor for Sema4D and mediates the Sema4D-induced growth cone collapse of hippocampal neurons and inhibition of various cell migration (9, 10). Plexin-C1, also named VESPR due to its viral origin, serves as a receptor for the virally encoded SemaVA, and Plexin-C1 stimulation induces inhibition of integrin-mediated adhesion and chemokine-induced migration of dendritic cells (2, 11, 12). Although it has been proposed that Sema7A is a ligand for Plexin-C1, the effect of Sema7A on neuronal cells is independent of Plexin-C1, and anti-Plexin-C1 monoclonal antibody did not affect Sema7A binding to Plexin-C1, suggesting that Sema7A is not likely to be a physiological binding partner of Plexin-C1 (13). Plexin-D1 was recently found to bind to Sema3E (14). Plexin-D1 is expressed in vascular endothelium, and Sema3E acts as a repulsive cue for the endothelial cells (14). Plexin-D1 is also highly expressed in ventrolateral cortical neurons, and Sema3E induces axonal repulsion of the neurons, guiding corticofugal and stritonigral projection (15).
The Rho family of small GTPases are signal transduction molecules that remodel the actin cytoskeleton and play fundamental roles in numerous cellular processes initiated by extracellular stimuli (16, 17). Activation of Rho family GTPases requires GDP-GTP exchange catalyzed by guanine nucleotide exchange factors, whereas the activation of GTPases is down-regulated by GTPase-activating proteins (GAPs), which stimulate the intrinsic GTPase activities. The Rnd proteins, Rnd1, Rnd2, and Rnd3/RhoE, comprise a new branch of Rho family in that they lack intrinsic GTPase activities (18). Rnd1 and Rnd3 are known to inhibit RhoA activity through p190-RhoGAP activation (19, 20), while Rnd2 stimulates RhoA activity through binding to its effector, Pragmin (21), showing that Rnd1 and Rnd2 oppositely regulate RhoA activity. In addition, Rnd1 has been shown to stimulate signaling of Plexin-A1 and Plexin-B1 through direct binding to the cytoplasmic domains of the plexins (22–24).
We recently reported that Plexin-B1 encodes a GAP for R-Ras in the cytoplasmic domain, and the Plexin-B1-Rnd1 complex mediates the Sema4D-induced repulsive response in hippocampal neurons and inhibition of cell migration by stimulating the intrinsic GTPase activity of R-Ras (25–27). Furthermore, Plexin-A1 has also been shown to exhibit R-Ras GAP activity, and Plexin-A1 and Rnd1 complex induces repulsive response through R-Ras GAP activity (28). The cytoplasmic domains of plexins are highly conserved among all plexin subfamilies. In contrast to Plexin-A1 and Plexin-B1, showing R-Ras GAP activity in the presence of Rnd1, signaling pathways of Plexin-C1 and Plexin-D1 remain to be elusive. We here demonstrate that Plexin-C1 exhibits R-Ras GAP activity without Rnd subfamily while Plexin-D1 requires Rnd2 for displaying R-Ras GAP activity.
EXPERIMENTAL PROCEDURES
Plasmid Constructs and Antibodies—cDNAs encoding full-length of mouse Plexin-D1, cytoplasmic domains of mouse Plexin-C1 (Plexin-C1-cyt; amino acids 972–1575), and Plexin-D1 (Plexin-D1-cyt; amino acids 1293–1926) were obtained by PCR from E16 mouse brain. Cytoplasmic domains of Plexin-B1 and Plexin-A1 were obtained as described (22). N-cyt and C-cyt fragments of Plexin-C1 (amino acids 972–1360 and 1361–1894, respectively) were amplified by PCR from the cytoplasmic domain of Plexin-C1. N-cyt and C-cyt fragments of Plexin-D1 (amino acids 1293–1707 and 1708–1926, respectively) were also amplified by PCR. RA mutants of both plexins were generated by PCR-mediated mutagenesis. Full-length of Plexin-D1 with Igκ signal peptide and Myc tag at the N terminus was subcloned into pcDNA3.1 (Invitrogen). Myc-tagged cytoplasmic domains of Plexin-C1 and D1 were subcloned into pcDNA3 (Invitrogen). To generate membrane-targeting forms of Myc-tagged Plexin-C1-cyt and Plexin-D1-cyt, they were fused to Src myristoylated (Myr) signal (MGSSKS). To generate glutathione S-transferase (GST)-fused proteins, these fragments were subcloned into pGEX-6P-1 or 6P-3 (GE Healthcare). Chimeric Plexin-B1/C1 encoding extracellular domain of Plexin-B1 (amino acids 1–1544) and cytoplasmic domain of Plexin-C1 (amino acids 1003–1575) was obtained by PCR. Hemagglutinin (HA)-tagged Rnd1, Rnd2, Rnd3, and green fluorescent protein (GFP)-tagged Rnd1, Rnd2, Rnd3, and R-Ras were obtained as described previously (19, 29). HA-tagged human R-Ras, GST-tagged Ras-binding domain of c-Raf-1 (RBD; amino acids 53–130), and Sema4D fused to human IgG1-Fc were obtained as described (25). M-Ras and TC21 were obtained by PCR from rat brain and mouse fibroblast, respectively, and they were subcloned into pEFBOS. Rat Plexin-D1 cytoplasmic domain (rPlexin-D1-cyt) was obtained by PCR from rat brain and subcloned into pcDNA3.
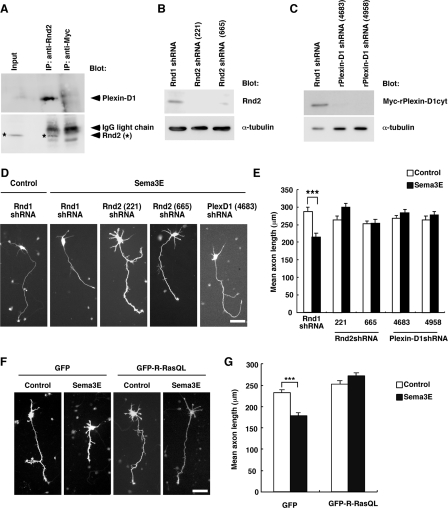
The short hairpin RNAs (shRNAs) for Rnd2 and Plexin-D1 were designed to target 19 nucleotides of rat transcripts and were expressed by using an shRNA expression vector, pSilencer (Ambion). The target sequences for Rnd2 and Plexin-D1 shRNA constructs are as follows: Rnd2 shRNA (221) (nucleotides 221–239, 5′-GGCCTACCCAGATTCTGAT-3′); Rnd2 shRNA (665) (nucleotides 665–683, 5′-CCAAGAGCTGTAACCTCAT-3′); Plexin-D1 shRNA (4683) (nucleotides 4683–4701, 5′-CTTCTGGGTGAACATCTTA-3′); Plexin-D1 shRNA (4958) (nucleotides 4958–4976, 5′-GGAAATACCAGAATGAGTT-3′). The target sequence for Rnd1 shRNA construct was obtained as described previously (25).
Antibodies used were as follows: a mouse monoclonal anti-Myc antibody 9E10, a rabbit polyclonal anti-HA antibody Y-11, a goat polyclonal anti-Plexin-D1 C-18, a mouse monoclonal anti-GFP antibody B-12 (Santa Cruz Biotechnology); a rat monoclonal anti-HA antibody 3F10 (Roche Applied Science); a mouse monoclonal anti-tau-1 antibody (Chemicon); horseradish peroxidase-conjugated secondary antibodies (DAKO); a rabbit polyclonal anti-GFP antibody, Alexa-conjugated secondary antibodies (Molecular Probes, Inc., Eugene, OR). A rabbit polyclonal antibody against Rnd2 has been described previously (30).
Cell Culture and Transfection—COS-7 cells and were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), 4 mm glutamine, 100 units/ml penicillin, and 0.2 mg/ml streptomycin under humidified conditions in 95% air and 5% CO2 at 37 °C. Transient transfections were performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Primary neurons from ventrolateral cortex were isolated from E19 rats and transfected with indicated plasmids using Rat Neuron Nucleofector kit (Amaxa Biosystems) following the manufacturer's instructions. Transfected neurons were plated onto poly-d-lysine- and laminin-coated coverslips (circular, 13 mm in diameter) or plastic dishes (60 mm in diameter) in DMEM containing 10% FBS, 4 mm glutamine, 100 units/ml penicillin, and 0.2 mg/ml streptomycin. After 8 h, the medium was replaced with Neurobasal medium supplemented with 0.5 mm glutamine and 1:25 B-27 supplement (Gibco). For quantification of axon growth, 1 nm mouse Sema3E/Fc chimera (R&D systems) or control human Fc (Jackson ImmnoResearch Laboratories) was added to Neurobasal medium.
Immunoblotting—Proteins were separated by 7.5% or 12.5% SDS-PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 3% low fat milk in Tris-buffered saline and then incubated with primary antibodies. The primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and a chemiluminescence detection kit (Chemi-lumi One; Nacalai Tesque or ECL Plus Western blotting Detection System; GE Healthcare). For detection of Rnd2 and Plexin-D1, primary and secondary antibodies were diluted in Can Get Signal (TOYOBO).
Measurement of Activity of Ras Family GTPases—Measurement of activity of Ras family GTPases in cells was performed as described previously (25). For measurement of GAP activity of full-length of Plexin-D1, COS-7 cells (5 × 105 cells) were plated on fibronectin (FN)-coated dishes (15 μg/ml) and maintained in DMEM containing 1% FBS after transfection. At 27–30 h after transfection, cells were stimulated with 0.5 nm mouse Sema3E/Fc chimera (R&D systems) or control human Fc at 37 °C for 7 min. For measurement of Ras family GAP activity of Myr-fused Plexin-C1- or Plexin-D1-cyt, COS-7 cells (R-Ras, 2.5 × 105; TC21 and M-Ras, 7.0 × 105) transfected with indicated plasmids were maintained in DMEM containing 5% FBS for 27 h. Cells were lysed with ice-cold cell lysis buffer (25 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 10% glycerol, 10 mm MgCl2, 25 mm NaF, 1 mm sodium vandate, 1 mm EDTA, 1 mm dithiothreitol, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) containing 75 μg of GST-RBD.
Pull-down Assays—COS-7 cells transfected with indicated plasmids were washed once with phosphate-buffered saline (PBS) and lysed with ice-cold cell lysis buffer (20 mm Tris-HCl, pH 7.4, 2 mm MgCl2, 1 mm dithiothreitol, 0.2% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Cell lysates were then centrifuged for 10 min at 4 °C. The supernatants were incubated for 10 min at 4 °C with 10 μg of GST fusion proteins and subsequently incubated with glutathione-Sepharose beads for 2 h at 4 °C. After the beads were washed with the ice-cold cell lysis buffer without Triton X-100, the bound proteins were eluted in Laemmli sample buffer and analyzed by SDS-PAGE and immunoblotting.
Immunoprecipitation—COS-7 cells transfected with indicated plasmids were lysed with ice-cold cell lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 5 mm MgCl2, 5 mm EGTA, 0.01% bovine serum albumin, 1 mm dithiothreitol, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Cell lysates were then centrifuged for 10 min at 16,000 × g at 4 °C. The supernatants were incubated with indicated antibodies for 2 h at 4 °C and then with protein G-Sepharose beads (GE Healthcare) for 1 h. The beads were washed with ice-cold lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting. For the endogenous protein binding assay, ventrolateral cortex from newborn rats at P2 was homogenized in ice-cold cell lysis buffer. After centrifugation, the supernatants were incubated with anti-Rnd2 polyclonal antibody for 3 h and then with protein A-Sepharose beads (GE healthcare) for 1 h at 4 °C. The beads were washed with the lysis buffer, and bound proteins were analyzed by SDS-PAGE and immunoblotting.
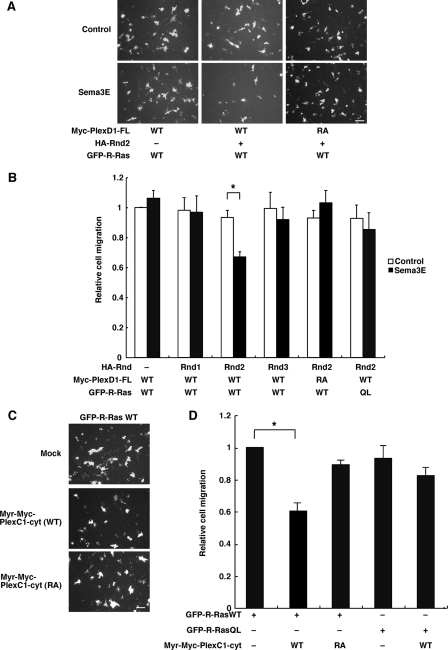
Transwell Cell Migration Assay—Cells were detached with 1 mm EDTA in PBS, and resuspended in serum-free DMEM containing 0.1% bovine serum albumin. The cells were replated at density of 2 × 104 cells onto the upper side of 8-μm pore filters of Transwell chambers (Costar) coated with 10 μg/ml of FN (Sigma-Aldrich). In the bottom chamber, mouse Sema3E/Fc chimera or control Fc was added at the concentration of 0.5 nm. After 4 h, non-migrated cells on the upper side of the filters were removed with cotton swab, and cells on the lower side were fixed with 4% paraformaldehyde. The numbers of migrated cells through the filter were counted by the fluorescence of GFP (A). At the same time, the cells were plated onto 24-well plastic plate to count the total number of transfected cells (B). Relative cell migration was then determined by the numbers of migrated cells normalized to the total numbers of transfected cells (A/B). Images were captured at room temperature in PBS using a microscope (Eclipse TE2000U, Nikon).
Quantification of Axon Growth—After 2–3 days in vitro, cultures were fixed with 4% paraformaldehyde and 0.4 m sucrose in PBS, and immunostained with anti-GFP and antitau-1 antibodies. Axon growth was analyzed by measuring the length of the longest neurite of each GFP and tau-1 double positive neuron. Digital images were taken with a Leica DC350F digital camera system equipped with a Nikon Eclipse E800 microscope, and axon length was determined using Image-Pro Plus image analysis software (Media Cybernetics).
RESULTS
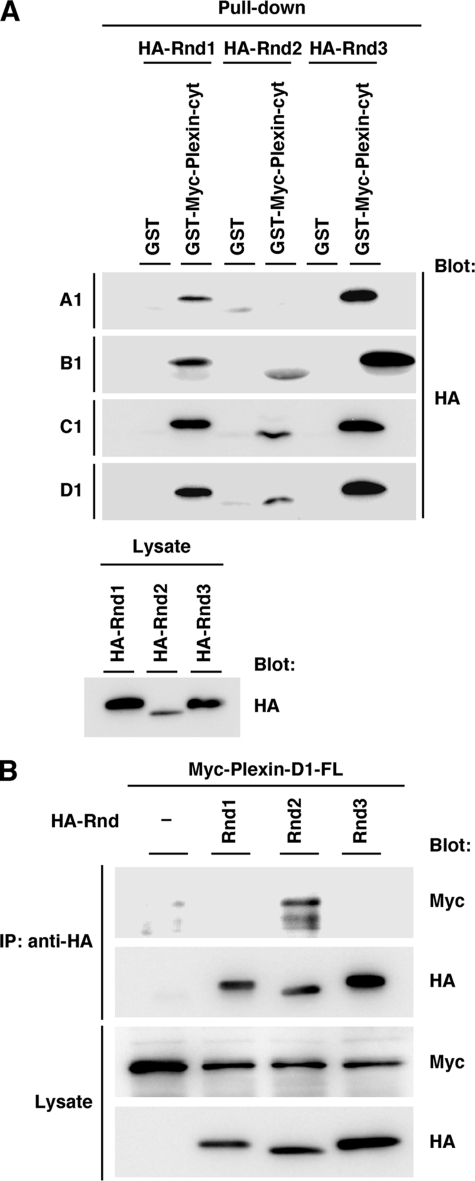
Interaction of Plexin-C1 and Plexin-D1 with Rnd Subfamilies—We and other groups (22, 24, 28) have previously shown that Plexin-A1 and Plexin-B1 interact with Rnd1 and that the interactions are required for the expression of R-Ras GAP activity of both plexins. To examine whether R-Ras GAP activity of plexins and requirement of Rnd1 are common signaling systems among plexin subfamilies, we first examined the interaction between plexin subfamilies and Rnd GTPases by a GST pull-down assay with GST-fused Myc-tagged cytoplasmic domains of plexin subfamilies. As previously reported (23, 31), Plexin-A1 bound to Rnd1 and Rnd3 but not to Rnd2. Plexin-B1 was confirmed to bind to all Rnd subfamily GTPases as we have already demonstrated (22). Plexin-C1 and Plexin-D1 interacted with three Rnd subfamily GTPases (Fig. 1A).
FIGURE 1.
Interaction between Rnd GTPases and plexin subfamilies. A, COS-7 cells were transfected with HA-tagged Rnd1, Rnd2, or Rnd3, and cell lysates were incubated with GST or GST-fused Myc-tagged cytoplasmic domain of Plexin-A1 (Plexin-A1-cyt, amino acids 1296–1894), Plexin-B1 (Plexin-B1-cyt, amino acids 1543–2136), Plexin-C1 (Plexin-C1-cyt, amino acids 972–1575), and Plexin-D1 (Plexin-D1-cyt, amino acids 1293–1926). Lysate inputs (lower panel) and bound proteins (upper panels) were analyzed by immunoblotting with anti-HA antibody. B, lysates from COS-7 cells transfected with the indicated plasmids were immunoprecipitated with anti-HA antibody. Lysate inputs and bound proteins were analyzed by immunoblotting with anti-HA or anti-Myc antibody.
To examine whether full-length of Plexin-D1 interacts with Rnd subfamily GTPases in mammalian cells, Myc-tagged full-length of Plexin-D1 and HA-tagged Rnd proteins were co-expressed in COS-7 cells. As shown in Fig. 1B, Plexin-D1 preferentially bound to Rnd2 among Rnd subfamily GTPases, suggesting that Plexin-D1 prefers Rnd2 among Rnd subfamilies as a binding partner.
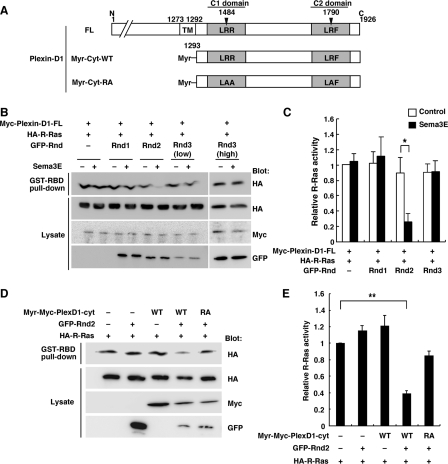
Requirement of Rnd Proteins for the Expression of R-Ras GAP Activity of Plexin-D1 and Plexin-C1—To examine whether Plexin-D1 and Plexin-C1 show R-Ras GAP activity, we measured GTP-bound R-Ras levels by pull-down assays with GST-RBD fusion protein in COS-7 cells. COS-7 cells, expressing Myc-tagged Plexin-D1, GFP-tagged Rnd proteins, and HA-tagged R-Ras-wild-type (WT), were plated onto FN-coated dishes and stimulated with recombinant Sema3E. Sema3E stimulation decreased R-Ras activity in COS-7 cells expressing Plexin-D1 and Rnd2, but the down-regulation of R-Ras activity was not observed in cells expressing Plexin-D1 alone or with Rnd1 or Rnd3 (Fig. 2, B and C). Ectodomains of Plexin-A1 and Plexin-B1 have been known to be an autoinhibitory domain (32), and ectodomain deletion mutant of Plexin-B1 has been shown to exhibit R-Ras GAP activity in the absence of Sema4D (33). We then generated membrane-targeting form of cytoplasmic domain of Plexin-D1, harboring myristoylation signal at N terminus, Myr-Plexin-D1-cyt (Fig. 2A). As shown in Fig. 2, D and E, Myr-Plexin-D1-cyt expressed with Rnd2 significantly decreased R-Ras activity in the absence of Sema3E, but Myr-Plexin-D1-cyt-RA, which has mutations at arginine motifs of C1 and C2 domains essential for GAP activity (Fig. 2A), had little effect on the R-Ras activity.
FIGURE 2.
Plexin-D1 inactivates R-Ras activity in the presence of Rnd2. A, schematic representation of Plexin-D1 constructs used in this study. Letters indicate specific amino acid residues within domains (A, Ala; F, Phe; L, Leu; R; Arg), and numbers indicate amino acid positions within the sequences. TM, transmembrane; Myr, myristoylated. B, COS-7 cells plated onto FN-coated dishes were transfected with Myc-tagged Plexin-D1, GFP-tagged Rnd proteins, and HA-tagged R-Ras. The cells were stimulated with 0.5 nm recombinant Sema3E (+) or control Fc (–) for 7 min. The cell lysates were incubated with GST-RBD, and bound R-Ras and total cell lysates were analyzed by immunoblotting. D, COS-7 cells were transfected with the indicated plasmids. The cell lysates were incubated with GST-RBD, and bound R-Ras and total cell lysates were analyzed by immunoblotting. C and E, relative R-Ras activity was determined by the amount of R-Ras bound to GST-RBD normalized to the amount of R-Ras in cell lysates analyzed by NIH Image software. The value from Myc-Plexin-D1 or Myr-Myc-Plexin-D1-cyt with HA-R-Ras-transfected cells was defined as 1. Results are the means ± S.E. of more than three independent experiments. The statistical significance was determined by two-sided one-sample t test or two-sided Student's t test. *, p < 0.05; **, p < 0.001.
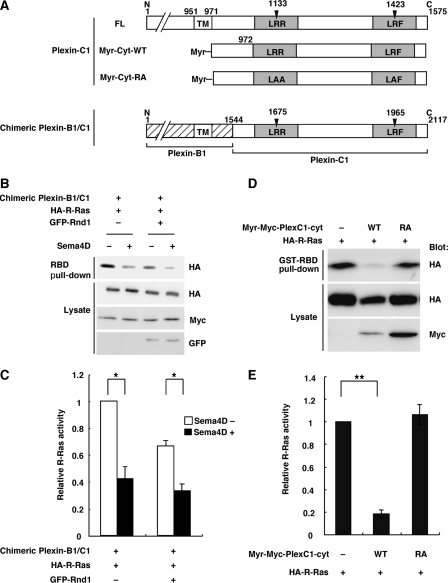
We next examined whether Plexin-C1 shows R-Ras GAP activity. Because physiological endogenous ligand for Plexin-C1 has not been yet identified, we constructed chimeric Plexin-B1/C1, having extracellular domain of Plexin-B1 and intracellular domain of Plexin-C1, responding to Sema4D as a ligand (Fig. 3A). The chimeric Plexin-B1/C1 induced R-Ras GAP activity in response to Sema4D regardless of the presence of Rnd GTPase (Fig. 3, B and C). The membrane-targeting form of cytoplasmic domain of Plexin-C1, harboring myristoylation signal at N terminus, Myr-Plexin-C1-cyt, alone decreased R-Ras activity, but Myr-Plexin-C1-cyt-RA mutant failed to decrease R-Ras activity, indicating that Plexin-C1 shows R-Ras GAP activity without Rnd proteins (Fig. 3, D and E). These results indicate that R-Ras GAP activity is a common function among plexin subfamilies, while the regulation of R-Ras GAP activity by Rnd proteins is different among plexin subfamilies.
FIGURE 3.
Plexin-C1 inactivates R-Ras activity regardless of the presence of Rnd GTPase. A, schematic representation of Plexin-C1 constructs used in this study. The R-Ras GAP domains are indicated by gray columns. B, COS-7 cells were transfected with Myc-tagged chimeric Plexin-B1/C1 and HA-tagged R-Ras in the absence or presence of GFP-tagged Rnd1. The cells were stimulated with (+) or without (–) 0.5 nm Sema4D (+) for 5 min. The cell lysates were incubated with GST-RBD, and bound R-Ras and total cell lysates were analyzed by immunoblotting. D, COS-7 cells were transfected with the indicated plasmids. The cell lysates were incubated with GST-RBD, and bound R-Ras and total cell lysates were analyzed by immunoblotting. C and E, relative R-Ras activity was determined as described in the legend to Fig. 2, C and E. The value from HA-R-Ras-transfected cells was defined as 1. The statistical significance was determined by two-sided one-sample t test or two-sided Student's t test. *, p < 0.05; **, p < 0.001.
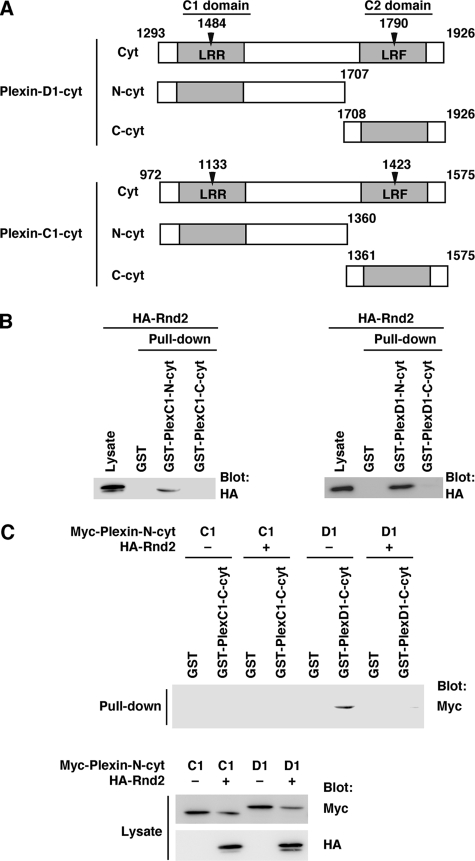
Interaction between the N- and C-terminal Regions within the Cytoplasmic Domains of Plexin-C1 and Plexin-D1—Rnd1 binds to the region between C1 and C2 domains of Plexin-B1 and Plexin-A1, and this binding is required for the expression of R-Ras GAP activity of both plexins. In addition, we have shown that the N-terminal region of the cytoplasmic region of Plexin-B1 containing C1 domain interacts with the C-terminal region containing C2 domain, and Rnd1 disrupts this interaction (33). We examined the Rnd2-binding region within the cytoplasmic domain of Plexin-D1. COS-7 cells were transfected with Rnd2 and used in a pull-down assay with GST-fused N-terminal region (N-cyt; amino acids 1293–1707) and C-terminal region (C-cyt; amino acids 1708–1926) of Plexin-D1 cytoplasmic domain, which contain C1 and C2 domains, respectively (Fig. 4A). As shown in Fig. 4B, Rnd2 interacted with N-cyt but not with C-cyt. We next examined the interaction between the N-terminal region and C-terminal region. Myc-tagged N-cyt of Plexin-D1 was expressed in COS-7 cells with or without Rnd2, and a pull-down assay was performed with GST-C-cyt of Plexin-D1. N-cyt interacted with C-Cyt in the absence of Rnd2, but this interaction was blocked by Rnd2 (Fig. 4C). We also examined whether Rnd2 regulates the interaction between N-cyt and C-cyt of Plexin-C1. Although Rnd2 bound to N-cyt of Plexin-C1, N-cyt of Plexin-C1 did not interact with C-cyt of Plexin-C1 regardless of Rnd2 binding (Fig. 4, B and C).
FIGURE 4.
Interactions of N-terminal and C-terminal regions within the cytoplasmic domains of Plexin-D1 and Plexin-C1. A, schematic representation of the Plexin-D1 and Plexin-C1 constructs used in this study. The R-Ras GAP domains are indicated by gray columns. B, lysate from COS-7 cells transfected with HA-tagged Rnd2 was used in a pull-down assay with GST-fused C1 domain-containing N-terminal region (N-cyt) and C2 domain-containing C-terminal region (C-cyt) of Plexin-C1 and Plexin-D1. Bound and total HA-Rnd2 were analyzed by immunoblotting with anti-HA antibody. C, COS-7 cells were transfected with Myc-tagged Plexin-C1-N-cyt or Plexin-D1-N-cyt alone or in combination with HA-tagged Rnd2. The cell lysates were used in a pull-down assay with GST, GST-Plexin-C1-C-cyt, or GST-Plexin-D1-C-cyt. Bound proteins and total cell lysates were analyzed by immunoblotting with anti-HA and anti-Myc antibodies.
Plexin-D1 and Plexin-C1 Inhibit Extracellular Matrix (ECM)-mediated Cell Migration through R-Ras GAP Activity—Sema/plexin signaling controls migration of a variety of cells, and we have previously reported that Sema4D inhibits the integrin-mediated cell migration through R-Ras GAP activity of Plexin-B1 (26). We then examined whether Plexin-D1 and Plexin-C1 suppress the ECM-mediated cell migration through R-Ras GAP activity. COS-7 cells expressing Plexin-D1 and each Rnd protein with R-Ras were plated on the upper side of transwell chambers, which were coated on the lower side with FN. Coexpression of Plexin-D1-WT and Rnd2 blocked the ECM-mediated cell migration in the presence of Sema3E at the lower well, whereas Plexin-D1-WT alone or coexpressed with Rnd1 or Rnd3 did not. Plexin-D1-RA, which lacks R-Ras GAP activity, failed to show the Sema3E-dependent inhibition of cell migration. In addition, Sema3E did not suppress the cell migration in the presence of constitutively activated R-Ras, R-Ras-QL (Fig. 5, A and B). We also examined the effect of Plexin-C1 on the ECM-mediated cell migration (Fig. 5, C and D). Myr-Plexin-C1-cyt-WT suppressed the ECM-mediated cell migration without Rnd proteins, while Myr-Plexin-C1-cyt-RA did not. Myr-Plexin-C1-cyt-WT did not suppress the cell migration in the presence of R-Ras-QL. These results suggest that Plexin-D1 and Plexin-C1 inhibit the cell migration through R-Ras GAP activity.
FIGURE 5.
Plexin-D1 and Plexin-C1 inhibit the ECM-mediated cell migration through R-Ras GAP activity. A, COS-7 cells expressing the indicated plasmids were tested in a transwell assay using the chambers coated with FN (10 μg/ml) in the presence of 0.5 nm Sema3E or control Fc. Migrated cells were visualized by the fluorescence of GFP. Bars, 100 μm. B, relative cell migration was determined by the number of the migrated cells normalized to the total number of the transfected cells. Expression levels of the constructs were verified by immunoblot analysis. Results are the means ± S.E. of three independent experiments. The value from Myc-Plexin-D1-transfected cells that were stimulated with control Fc was defined as 1. The statistical difference was determined by two-sided Mann-Whitney's U test. *, p < 0.05. C, COS-7 cells transfected with the indicated plasmids were tested in transwell assay using the chambers coated with FN (10μg/ml). Bars, 100μm. D, relative cell migration was determined. Results are the means ± S.E. of five independent experiments. The value from GFP-R-Ras-WT-transfected cells was defined as 1. The statistical difference was determined by two-sided onesample t test or two-sided Student's t test. *, p < 0.01.
Rnd2 Is Required for Sema3E/Plexin-D1-mediated Inhibition of Axon Growth—To address the biological role of interaction between Plexin-D1 and Rnd2, we first examined the endogenous interaction of Plexin-D1 with Rnd2 in neurons. Endogenous Plexin-D1 was coimmunoprecipitated with Rnd2 by anti-Rnd2 antibody but not by anti-Myc antibody used as a negative control in ventrolateral cortex of P2 rats (Fig. 6A). It has been reported that Sema3E/Plexin-D1 signaling inhibits axon elongation of neurons in the ventrolateral region of the cortex (15). To determine whether R-Ras GAP activity of Plexin-D1/Rnd2 is involved in Sema3E-induced suppression of axon elongation, we reduced the expression of Rnd2 in neurons by RNA interference. We designed two Rnd2-specific shRNA vectors to target different coding region of Rnd2, Rnd2 shRNA (221) and Rnd2 shRNA (665), and confirmed that both Rnd2 shRNA (221) and Rnd2 shRNA (665) significantly reduced expression of Rnd2 in dissociated neurons (Fig. 6B). The two Rnd2 shRNAs were coelectroporated with GFP reporter vector, and the cells were cultured in the presence or absence of recombinant Sema3E for 3 days. Both Rnd2 shRNA (221) and Rnd2 shRNA (665) but not Rnd1 shRNA abrogated the inhibition of axon elongation by Sema3E (Fig. 6, D and E). To confirm the involvement of the endogenous Plexin-D1 in Sema3E-induced inhibition of axon outgrowth, we constructed two Plexin-D1-specific shRNAs, Plexin-D1 shRNA (4683) and Plexin-D1 shRNA (4958), and examined the effects of these shRNAs on Sema3E-induced inhibition of axon outgrowth. Two shRNAs blocked the inhibition of axon elongation by Sema3E, indicating that the Sema3E-induced inhibition is mediated by Plexin-D1 (Fig. 6, C–E). To examine whether inactivation of R-Ras activity is required for the Sema3E/Plexin-D1-induced inhibition of axon outgrowth, cortical neurons were nucleofected with GFP or GFP-tagged R-RasQL and cultured in the presence or absence of Sema3E for 2 days. Expression of R-RasQL blocked the Sema3E-induced suppression of axon outgrowth (Fig. 6, E and F). These results indicate that Sema3E/Plexin-D1 induces the inhibition of axon outgrowth through R-Ras GAP activity, and this inhibition requires Rnd2.
FIGURE 6.
Rnd2 is required for Sema3E-mediated inhibition of axon outgrowth. A, lysate from ventrolateral cortex of P2 rats was immunoprecipitated with anti-Rnd2 antibody or with anti-Myc antibody as a control. The lysate input and immunoprecipitates were analyzed by immunoblotting with anti-Plexin-D1 or anti-Rnd2 antibody. For detection of immunoprecipitates of Plexin-D1, 3/4 volume of the total immunoprecipitates was applied. B, neurons from ventrolateral cortex of E19 rats were nucleofected with GFP together with Rnd1 shRNA or Rnd2 shRNA (shRNA-221 or shRNA-665) and cultured for 3 days. The cell lysates were immunoblotted with anti-Rnd2 or anti-α-tubulin antibody. C, COS-7 cells were transfected with Myc-rPlexin-D1-cyt together with Rnd1 shRNA or Plexin-D1 shRNA (shRNA-4683 or shRNA-4958) and cultured for 3 days. The cell lysates were immunoblotted with anti-Myc or anti-α-tubulin antibody. D, neurons of ventrolateral cortex were nucleofected with GFP together with Rnd1 shRNA, Rnd2 shRNA (shRNA-221 or shRNA-665), or Plexin-D1 shRNA (shRNA-4683 or shRNA-4958) and cultured for 3 days in the presence or absence of 1 nm Sema3E. Scale bar, 50 μm. F, neurons of ventrolateral cortex were nucleofected with GFP or GFP-R-RasQL and cultured for 2 days in the presence or absence of 1 nm Sema3E. Scale bar, 50 μm. E and G, quantification of axon length. Results are the means ± S.E. of 100 neurons from three independent experiments. The statistical difference was determined by two-sided Mann-Whitney's U test. ***, p < 0.0005.
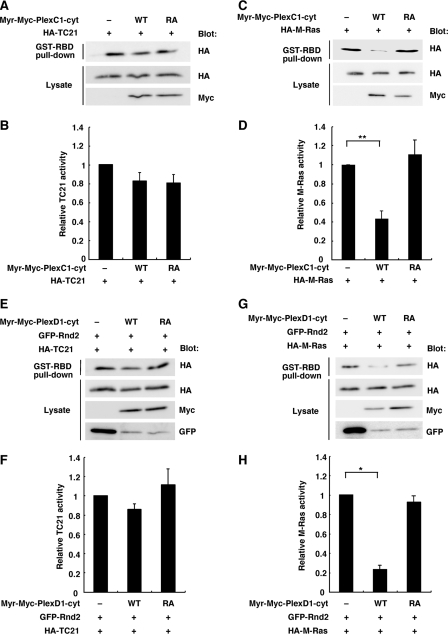
Plexin-C1 and Plexin-D1 Display M-Ras GAP Activity—R-Ras subfamily comprises R-Ras, TC21, and M-Ras. We then examined GAP activity of Plexin-C1 and Plexin-D1 for TC21 and M-Ras, by using a pull-down assay with GST-RBD in COS-7 cells expressing Myr-Plexin-C1-cyt and Myr-Plexin-D1-cyt. Myr-Plexin-C1-cyt showed a GAP activity for M-Ras but not for TC21, while this M-Ras GAP activity was not observed with its RA mutant (Fig. 7, A–D). Myr-Plexin-D1-cyt also showed a GAP activity for M-Ras in the presence of Rnd2 but not for TC21, while this M-Ras GAP activity was not observed with its RA mutant (Fig. 7, E–H). Thus, Plexin-C1 and Plexin-D1 display GAP activity for two R-Ras subfamily GTPases, R-Ras, and M-Ras.
FIGURE 7.
Plexin-C1 and Plexin-D1 display M-Ras GAP activity. A and C, COS-7 cells were transfected with Myr-Plexin-C1-cyt (wild-type or RA mutant) and either HA-TC21 (A) or HA-M-Ras (C). The cell lysates were incubated with GST-RBD, and bound Ras GTPases and total lysates were analyzed by immunoblotting. E and G, COS-7 cells were transfected with Myr-Plexin-D1-cyt (wild-type or RA mutant), GFP-Rnd2, and either HA-TC21 (E) or HA-M-Ras (G). The cell lysates were incubated with GST-RBD, and bound Ras GTPases and total lysates were analyzed by immunoblotting. B, D, F, and H, relative Ras activity was determined as described in the legend to Fig. 2, C and E. The value from Ras-transfected or Ras and Rnd2-transfected cells was defined as 1. The statistical significance was determined by two-sided one-sample t test or two-sided Student's t test. *, p < 0.01; **, p < 0.001.
DISCUSSION
Plexin-A1 and Plexin-B1 function as an R-Ras GAP, inducing repulsive responses, and R-Ras GAP activity of these plexins requires Rnd1 binding to the cytoplasmic domains of the plexins (25, 28). In the present study, we have demonstrated that Plexin-C1 and Plexin-D1 display R-Ras GAP activity, and that the expression of R-Ras GAP activity of Plexin-D1 requires Rnd2, another member of Rnd subfamily, while that of Plexin-C1 does not require any Rnd proteins.
The cytoplasmic domains of plexins have highly conserved Ras GAP-homologous domains, C1 and C2 domains, which contain primary and secondary arginine motifs, respectively (25). These motifs are essential for GAP activity, and actually mutations of the arginine residues to alanine in the motifs of Plexin-A1 and Plexin-B1 made the receptors inactive in R-Ras GAP activities (25).3 Two arginine motifs are also conserved in the cytoplasmic domains of Plexin-C1 and Plexin-D1. We here demonstrated that RA mutants, having mutations at two arginine motifs, of Plexin-C1 and Plexin-D1 have no ability to inhibit not only R-Ras activity but also ECM-mediated cell migration. Thus, Plexin-C1 and Plexin-D1 display R-Ras GAP activity and inhibit cell migration. Considering these results and previous reports of Plexin-A1 and Plexin-B1, the highly conserved cytoplasmic domains of plexins encode R-Ras GAP and down-regulation of R-Ras activity through R-Ras GAP domains is likely to be a common signaling pathway for Sema/plexin-mediated repulsive responses.
R-Ras GAP activity of Plexin-A1 and Plexin-B1 requires Rnd1 binding to their cytoplasmic domains and the binding domains are located between C1 and C2 domains. We previously reported that the N-terminal region of the cytoplasmic domain of Plexin-B1 containing C1 domain interacts with the C-terminal region containing C2 domain, and Rnd1 disrupts this interaction by binding to the region between C1 and C2 domains. In addition, this Rnd1-induced open conformation of R-Ras GAP domains, C1 and C2 domains, is required for the expression of R-Ras GAP activity (33). Thus, the Rnd1-binding region is a regulatory domain for the expression of R-Ras GAP activity. We here demonstrated that among Rnd subfamily GTPases Plexin-D1 specifically requires Rnd2 for R-Ras GAP activity and subsequent inhibition of ECM-mediated cell migration. In addition, we showed that Rnd2 binds to the cytoplasmic domain of Plexin-D1 and disrupts the interaction of the N-terminal region containing C1 domain and C-terminal region containing C2 domain. From these results, it is inferred that Rnd2 binds to the region between the C1 and C2 domains of Plexin-D1 and induces open conformation of R-Ras GAP domains, inducing R-Ras GAP activity and inhibition of cell migration. On the other hand, we showed that Plexin-C1 does not require Rnd subfamily for R-Ras GAP activity and inhibition of cell migration, and that the N-terminal region containing C1 domain does not interact with the C-terminal region containing C2 domain, indicating that R-Ras GAP domains of Plexin-C1 show open conformation in the absence of Rnd proteins. Thus, this open conformation is ascribed to Rnd-independent R-Ras GAP activity of Plexin-C1. Therefore, the regulation of R-Ras GAP activity of plexin subfamilies is different in dependence on Rnd subfamily GTPases: Plexin-A1 and Plexin-B1 require Rnd1, Plexin-D1 requires Rnd2, and Plexin-C1 does not require Rnd proteins. In contrast to highly conserved C1 and C2 domains, the regions between C1 and C2 domains are not conserved among plexin subfamilies. This variety of the Rnd-binding regions may provide diverse regulation of R-Ras GAP activity of plexins.
Plexin-D1 is expressed in the developing central nervous system (34, 35), and Rnd2 is also primarily expressed in brain and plays an important role in developing central nervous system (36–38). Sema3E/Plexin-D1 was recently reported to regulate descending axon tracts in forebrain in vivo (15). Sema3E reduces cortical axon length through Plexin-D1 (15). We here demonstrated that Rnd2 shRNAs and constitutively activated R-Ras abrogate the inhibition of axon growth of cortical neurons by Sema3E. Association of Rnd2 with Plexin-D1 is essential for the expressing R-Ras GAP activity of Plexin-D1 in COS-7 cells. These results suggest that inactivation of R-Ras by the Plexin-D1-Rnd2 complex is necessary for the Sema3E-induced inhibition of axon outgrowth in cortical neurons.
In conclusion, we have demonstrated that Plexin-D1 displays R-Ras GAP activity in concert with Rnd2 while Plexin-C1 displays the activity without Rnd proteins. In addition, thinking over the requirement of Rnd1 for R-Ras GAP activity of Plexin-A1 and B1, selective coupling of Rnd subfamily GTPases may be critical for the expression of R-Ras GAP activity of plexin subfamilies.
Acknowledgments
We thank L. Tamagnone, H. Fujisawa, and H. Kikutani for providing Plexin-B1, Plexin-A1, and the soluble form of Sema4D expression plasmids, respectively. We also thank Y. Saito and G. Tasaka (Laboratory of Molecular Neurobiology, Graduate School of Biostudies, Kyoto University) for experimental help.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Sema, semaphorin; DMEM, Dulbecco's modified Eagle's medium; ECM, extracellular matrix; En, embryonic day n; FBS, fetal bovine serum; FN, fibronectin; GAP, GTPase-activating protein; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, hemagglutinin; Myr, myristoylated; PBS, phosphate-buffered saline; Pn, n days of postnatal age; RBD, Ras-binding domain of c-Raf-1; shRNA, short hairpin RNA; WT, wild-type.
K. Uesugi, I. Oinuma, H. Katoh, and M. Negishi, unpublished observation.
References
- 1.Kolodkin, A. L., Matthes, D. J., and Goodman, C. S. (1993) Cell 75 1389–1399 [DOI] [PubMed] [Google Scholar]
- 2.Tamagnone, L., Artigiani, S., Chen, H., He, Z., Ming, G. I., Song, H., Chedotal, A., Winberg, M. L., Goodman, C. S., Poo, M., Tessier-Lavigne, M., and Comoglio, P. M. (1999) Cell 99 71–80 [DOI] [PubMed] [Google Scholar]
- 3.Luo, Y., Raible, D., and Raper, J. A. (1993) Cell 75 217–227 [DOI] [PubMed] [Google Scholar]
- 4.Behar, O., Golden, J. A., Mashimo, H., Schoen, F. J., and Fishman, M. C. (1996) Nature 383 525–528 [DOI] [PubMed] [Google Scholar]
- 5.Christensen, C. R., Klingelhofer, J., Tarabykina, S., Hulgaard, E. F., Kramerov, D., and Lukanidin, E. (1998) Cancer Res. 58 1238–1244 [PubMed] [Google Scholar]
- 6.Kumanogoh, A., and Kikutani, H. (2003) J. Cell Sci. 116 3463–3470 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura, F., Kalb, R. G., and Strittmatter, S. M. (2000) J. Neurobiol. 44 219–229 [DOI] [PubMed] [Google Scholar]
- 8.Takahashi, T., Fournier, A., Nakamura, F., Wang, L. H., Murakami, Y., Kalb, R. G., Fujisawa, H., and Strittmatter, S. M. (1999) Cell 99 59–69 [DOI] [PubMed] [Google Scholar]
- 9.Swiercz, J. M., Kuner, R., Behrens, J., and Offermanns, S. (2002) Neuron 35 51–63 [DOI] [PubMed] [Google Scholar]
- 10.Barberis, D., Artigiani, S., Casazza, A., Corso, S., Giordano, S., Love, C. A., Jones, E. Y., Comoglio, P. M., and Tamagnone, L. (2004) Faseb. J. 18 592–594 [DOI] [PubMed] [Google Scholar]
- 11.Comeau, M. R., Johnson, R., DuBose, R. F., Petersen, M., Gearing, P., VandenBos, T., Park, L., Farrah, T., Buller, R. M., Cohen, J. I., Strockbine, L. D., Rauch, C., and Spriggs, M. K. (1998) Immunity 8 473–482 [DOI] [PubMed] [Google Scholar]
- 12.Walzer, T., Galibert, L., Comeau, M. R., and Smedt, T. D. (2005) J. Immunol. 174 51–59 [DOI] [PubMed] [Google Scholar]
- 13.Holmes, S., Downs, A., Fosberry, A., Hayes, P., Michalovich, D., Murdoch, P., Moores, K., Fox, J., Deen, K., Pettman, G., Wattam, T., and Lewis, C. (2002) Scand. J. Immunol. 56 270–275 [DOI] [PubMed] [Google Scholar]
- 14.Gu, C., Yoshida, Y., Livet, J., Reimert, D. V., Mann, F., Merte, J., Henderson, C. E., Jessell, T. M., Kolodkin, A. L., and Ginty, D. D. (2005) Science 307 265–268 [DOI] [PubMed] [Google Scholar]
- 15.Chauvet, S., Cohen, S., Yoshida, Y., Fekrane, L., Livet, J., Gayet, O., Segu, L., Buhot, M. C., Jessell, T. M., Henderson, C. E., and Mann, F. (2007) Neuron 56 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negishi, M., and Katoh, H. (2002) J. Biochem. 132 157–166 [DOI] [PubMed] [Google Scholar]
- 17.Etienne-Manneville, S., and Hall, A. (2002) Nature 420 629–635 [DOI] [PubMed] [Google Scholar]
- 18.Nobes, C. D., Lauritzen, I., Mattei, M. G., Paris, S., Hall, A., and Chardin, P. (1998) J. Cell Biol. 141 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh, H., Harada, A., Mori, K., and Negishi, M. (2002) Mol. Cell. Biol. 22 2952–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wennerberg, K., Forget, M. A., Ellerbroek, S. M., Arthur, W. T., Burridge, K., Settleman, J., Der, C. J., and Hansen, S. H. (2003) Curr. Biol. 13 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka, H., Katoh, H., and Negishi, M. (2006) J. Biol. Chem. 281 10355–10364 [DOI] [PubMed] [Google Scholar]
- 22.Oinuma, I., Katoh, H., Harada, A., and Negishi, M. (2003) J. Biol. Chem. 278 25671–25677 [DOI] [PubMed] [Google Scholar]
- 23.Zanata, S. M., Hovatta, I., Rohm, B., and Puschel, A. W. (2002) J. Neurosci. 22 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong, Y., Chugha, P., Hota, P. K., Alviani, R. S., Li, M., Tempel, W., Shen, L., Park, H. W., and Buck, M. (2007) J. Biol. Chem. 282 37215–37224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oinuma, I., Ishikawa, Y., Katoh, H., and Negishi, M. (2004) Science 305 862–865 [DOI] [PubMed] [Google Scholar]
- 26.Oinuma, I., Katoh, H., and Negishi, M. (2006) J. Cell Biol. 173 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, Y., Oinuma, I., Katoh, H., Kaibuchi, K., and Negishi, M. (2006) EMBO Rep. 7 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toyofuku, T., Yoshida, J., Sugimoto, T., Zhang, H., Kumanogoh, A., Hori, M., and Kikutani, H. (2005) Nat. Neurosci. 8 1712–1719 [DOI] [PubMed] [Google Scholar]
- 29.Aoki, J., Katoh, H., Mori, K., and Negishi, M. (2000) Biochem. Biophys. Res. Commun. 278 604–608 [DOI] [PubMed] [Google Scholar]
- 30.Fujita, H., Katoh, H., Ishikawa, Y., Mori, K., and Negishi, M. (2002) J. Biol. Chem. 277 45428–45434 [DOI] [PubMed] [Google Scholar]
- 31.Rohm, B., Rahim, B., Kleiber, B., Hovatta, I., and Puschel, A. W. (2000) FEBS Lett. 486 68–72 [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, T., and Strittmatter, S. M. (2001) Neuron 29 429–439 [DOI] [PubMed] [Google Scholar]
- 33.Oinuma, I., Katoh, H., and Negishi, M. (2004) J. Neurosci. 24 11473–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Zwaag, B., Hellemons, A. J., Leenders, W. P., Burbach, J. P., Brunner, H. G., Padberg, G. W., and Van Bokhoven, H. (2002) Dev. Dyn. 225 336–343 [DOI] [PubMed] [Google Scholar]
- 35.Molyneaux, B. J., Arlotta, P., Hirata, T., Hibi, M., and Macklis, J. D. (2005) Neuron 47 817–831 [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, K., Yamashita, Y., Tamamaki, N., Katoh, H., Kaneko, T., and Negishi, M. (2006) Neurosci. Res. 54 149–153 [DOI] [PubMed] [Google Scholar]
- 37.Nishi, M., Takeshima, H., Houtani, T., Nakagawara, K., Noda, T., and Sugimoto, T. (1999) Mol. Brain Res. 67 74–81 [DOI] [PubMed] [Google Scholar]
- 38.Decourt, B., Bouleau, Y., Dulon, D., and Hafidi, A. (2005) Dev. Brain Res. 159 36–54 [DOI] [PubMed] [Google Scholar]