Abstract
tRNAs that read codons starting with U are usually modified at their A37 by isopentenyl-tRNA transferases to minimize peptidyl-tRNA slippage in translation. The consensus substrate requirements of the isopentenyl-tRNA transferase of Escherichia coli, MiaA, have been the focus of extensive study. However, the molecular basis of tRNA-MiaA recognition remains unknown. Here we describe the 2.5Å crystal structure of MiaA in complex with substrate tRNA(Phe). Comparative structural analysis reveals that the enzymatic reaction involves an RNA-protein mutually induced fit mechanism in which large domain movements in MiaA provoke the partial unfolding of the substrate tRNA anticodon loop. In addition, we show how substrate tRNAs are recognized by MiaA through a combination of direct and indirect sequence readouts.
In all kingdoms of life, mechanisms are in place to ensure the fidelity and efficiency of tRNA function. To this end, many nucleosides in tRNA undergo enzymatic base modification, thereby expanding the limited vocabulary of the canonical four bases. In fact, more than 70 different modifications have been described, and almost all tRNAs have multiple modifications (1). Several tRNA modifications affecting the anticodon region of tRNA are important for avoiding frameshift errors introduced by peptidyl-tRNA slippage (2). Specifically, there is extensive evidence that position 37 is important for the anticodon loop structure and to preposition the anticodon for interaction with the codon. Bulky hydrophobic modified nucleotides are often used at this position, presumably to facilitate stacking interactions (1–3). Almost all tRNAs that read codons starting with U have the modified nucleotide N6-isopentenyladenosine (or a further modified derivative) at position 37. The modification is formed by the transfer of an isopentenyl moiety from dimethylallyl pyrophosphate (DMAPP)3 to A37 of tRNAs. This transfer is catalyzed by isopentenyl-tRNA transferases (IPTs; E.C. 2.5.1.8). The Escherichia coli IPT (MiaA) has been the subject of extensive kinetic and mutagenesis studies (4–6). Specifically, several studies have attempted to pinpoint the tRNA requirements for MiaA catalysis (4, 5). Of particular interest is the observation that the canonical triple A36-A37-A38 is necessary but not sufficient for modification; for example, E. coli tRNA(Ser) (GGA) contains A36-A37-A38 but is not modified by MiaA (4, 7). The wobble G30*U40 base pair of tRNA(Ser)(GGA) has been pinpointed as a major reason for this tRNA not being a substrate for MiaA (4, 7). However, in addition, the two neighboring base pairs had to be mutated to the MiaA substrate consensus G29-C41 and A31-U39 base pairs in order for tRNA(Ser)(GGA) to be effectively modified by MiaA (4). Similar conclusions on the sequence requirements of the immediate stem can be drawn from steady-state kinetic studies of tRNA(Phe) anticodon stem loop minimal substrate mutants (5). Although A36-A37-A38 appears to be a conserved requirement for bacterial tRNAs analyzed to date, the yeast mitochondrial tRNA(Gly1), which has a C36-A37-A38 setup, has been shown to have the isopentenyl modification on A37 (8). The yeast IPT, MOD5, is active in the nucleus, cytosol, and mitochondria through the use of two translational starts (9). As MOD5 is presumably also responsible for the tRNA(Gly1) A37-isopentenyl modification, it should have a slightly different substrate recognition mode than its bacterial counterparts. A recently published structure of the main part of IPT from Pseudomonas aeruginosa featured a central channel where the A37 and the co-substrate (DMAPP) were speculated to bind from different sides of the molecule (10). A rather large part of the sequence was missing in the model because of the presumed disorder of a separate domain, referred to as the insertion domain. Since then, two structures of IPT from Staphylococcus epidermis (3D3Q) and Bacillus halodurans (3EXA) have been released to the PDB by structural genomics centers. These structures include an ordered insertion domain, but the insertion domains do not pack toward the major domain (core domain) in a functionally relevant mode, presumably due to electrostatic repulsion. In a recent structure of MOD5 in complex with yeast tRNA(Cys) determined to 2.95 Å resolution, MOD5 was shown to bind the tRNA between the core domain and the insertion domain (11). Furthermore, a number of DMAPP co-substrate soaks were studied structurally to probe the mechanism of isopentenyl transfer. To understand the molecular basis of MiaA substrate recognition, we set out to determine a co-crystal structure of MiaA with a tRNA substrate. Here, we present the three-dimensional structure of E. coli MiaA in complex with a substrate, tRNA(Phe) determined at 2.5 Å resolution. By comparing the structure of MiaA with the structures of other bacterial IPTs without bound tRNA, we show that the insertion domain moves in toward the core domain as a result of induced fit. In this process, the anticodon loop of tRNA unfolds. Importantly, the unfolded anticodon loop enables both direct and indirect substrate sequence recognition by MiaA. We also compare the MiaA-tRNA co-crystal structure with the MOD5-tRNA(Cys) structure (11) and discuss important regions where the bacterial IPT and eukaryotic IPT differ in light of their different substrate requirements.
EXPERIMENTAL PROCEDURES
Cloning and Protein Production—MiaA (gi:1790613) was cloned by ligation-independent cloning into a pET-28-based expression vector incorporating an N-terminal hexa-His tag fusion (pNIC28-Bsa4;gi:EF198106). After transformation and liquid culture growth using standard methods, recombinant expression of MiaA in E. coli strain KRX (Promega, Nacka, Sweden) was induced at 291 K by the addition of 0.5 mm isopropyl-1-thio-β-d-galactopyranoside and 0.1% rhamnose to Terrific Broth. Induction was maintained for 18 h before harvesting. MiaA was purified using IMAC (12) on a nickel-charged IMAC-Sepharose 6 resin. The N-terminal tag with the His tag fusion and residual cloning elements were excised through incubation with tobacco etch virus protease (1:20 protease:protein ratio) for 12 h at 277 K. After protease digestion, the IMAC purification was repeated to remove non-cleaved protein and the His-tagged tobacco etch virus protease. A synthetic E. coli tRNA(Phe) gene was cloned into pRav23 for native (MS2 stem loop and glmS ribozyme-based) purification (13). After large scale T7 RNA polymerase-based in vitro transcription and native purification, tRNA(Phe) and MiaA were mixed in a 1:4 molecular ratio (in TMT buffer: 30 mm Tris-HCl, 10 mm MgCl2, 2 mm tris(2-carboxyethyl) phosphine, pH 8.0), incubated at 310 K for 30 min, and then purified by gel filtration on a 120-ml Sephacryl S-300 column in TMT buffer. All columns used were from GE Healthcare, Uppsala, Sweden.
Crystallization and Structure Solution—Crystals of MiaA-tRNA(Phe) complex were grown by hanging drop vapor diffusion at 293 K by mixing the purified complex (at 4 mg/ml in TMT buffer) with an equal amount (0.6 μl) of reservoir solution (15% polyethylene glycol 4000, 0.2 m calcium chloride). After 1 week, 10-μm rhombohedral crystals appeared; crystal size could be improved up to 75 μm after several rounds of seeding. Crystals were swept through a reservoir solution complemented with 20% (2R,3R)-(-)-2,3-butanediol and flash-frozen in liquid-nitrogen. Data were collected using a wavelength of 0.97 Å at beam line I911-3 MAX-lab (Lund, Sweden). The data were processed to 2.5 Å resolution with MOSFLM and scaled with SCALA in space group P212121 with a = 89.24 Å, b = 91.26 Å, c = 152.36 Å (Table 1). The structure was solved by molecular replacement with PHASER (14) using a model based on the PDB entry 3D3Q (from S. epidermis). In this process, it was essential to search for the two domains in the structure separately as their relative orientation is totally different in our MiaA-tRNA(Phe) complex structure (see Fig. 1D). The tRNA(Phe) was located using the fully modified tRNA(Phe) from Thermus aquaticus in 1TTT (15) as a search probe (excluding nucleotides 27–43). Two protein molecules and two tRNA molecules were located in the asymmetric unit. The tRNA part was built anew by hand in the resulting electron density. The model was completed and improved through extensively repeated sessions of model building in COOT (16) interspersed by refinement in REFMAC5 (17). The resulting model and map is of good quality to 2.5 Å resolution (see Table 1 and Fig. 1C).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Resolution (Å) | 30-2.5 (2.6-2.5)a |
| Rsym | 9.7 (49.3) |
| <I / σI> | 6.8 (2.1) |
| Completeness (%) | 99.8 (100) |
| Unique/observed reflections | 43749/285072 (4650/20528) |
| Wilson B-factor (Å2) | 51 |
| Refinement | |
| Resolution (Å) | 30-2.5 (2.6-2.5) |
| Rwork/Rfree | 24.3/26.8 (38.2/41.4) |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.28 |
| Average B-factors (Å2) | 35.8 |
| Ramachandran plotb: favored, allowed (%) | 98.2, 1.8 |
Values in parentheses are for highest-resolution shell.
According to Molprobity (20).
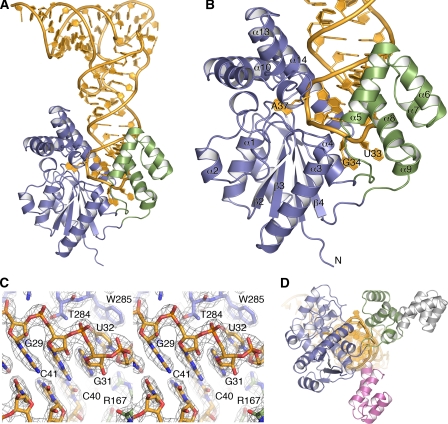
FIGURE 1.
Overall structure of the RNA-protein complex and display of mutual induced fit. A, schematic diagram with the core domain in blue, the insertion domain in green, and tRNA(Phe) in gold. B, close-up of the partly unfolded anticodon loop and of MiaA with numbered secondary structure elements. C, stereo representation of a simulated annealing composite omit map that shows parts of the anticodon stem region. The map is a SIGMAA-weighted 2Fo – Fc synthesis at 1.5 σ. D, superimpositioning of E. coli MiaA (same coloring as in A) and the apo-structures of IPTs from B. halodurans (gray; 3EXA) and S. epidermis (pink; 3D3Q). The core domains superimpose closely, whereas in the absence of tRNA, the insertion domains are scattered about, displaying a large variety of biologically irrelevant packing interactions. For clarity, only the insertion domains are shown here. The view is rotated 90° when compared with A and B, looking along up the axis of the anticodon loop.
Protein Structure Accession Numbers—The atomic coordinates and structure factors for E. coli MiaA in complex with tRNA(Phe) have been deposited with the Research Collaboratory for Structural Bioinformatics (RCSB) under PDB code 3FOZ.
RESULTS AND DISCUSSION
Overall Structure of the MiaA-tRNA(Phe) Complex and Comparison with Bacterial Counterparts—MiaA features two domains: a large core domain, reminiscent of small kinase domains (residues 7–118; 191–311), and a small helix bundle insertion domain (residues 119–190) (Fig. 1, A and B). The core domain is highly similar (1 Å r.m.s. deviation on Cα:s) to the previously determined IPT from P. aeruginosa (10). A deep, highly basic rift is formed between the two domains. The tRNA substrate is bound in the rift with the anticodon stem loop wedged between the two domains. The tRNA anticodon loop rests upon the floor of the rift formed by two extensive connections between the domains (Fig. 1, A and B). Comparing the present tRNA-complex structure with the two available structures of bacterial full-length IPTs in the absence of tRNA (Fig. 1D), it becomes clear that the rift is formed as a result of the interaction between the insertion and core domains only upon tRNA binding. This conformation of the insertion domain results in partial unfolding of the anticodon loop. Hence, the enzymatic reaction involves an RNA-protein mutually induced-fit mechanism.
Structural Basis for Substrate tRNA Recognition and Partial Anticodon Loop Unfolding—In a canonical anticodon loop, the bases 34–37 form a continuous stack and U33 interacts with the phosphate of A36 (U-turn motif). However, here, U33, G34, A35, and the nucleotide to be modified, A37, flip out (Fig. 2A). As a result, the base of A37 moves into a channel that runs through the core domain as predicted previously (10). The interaction surface between the unfolded RNA and the protein is quite extensive (4700 Å2 with 1.4 Å probe; Fig. 2B). By comparing the unfolded anticodon loop found in the present structure with the canonical anticodon loop, we observe that U33 has moved the farthest distance from its canonical position (Fig. 2A). Nevertheless, it only interacts with the backbone nitrogen and side chain hydroxyl group of Ser120 (Fig. 2B).
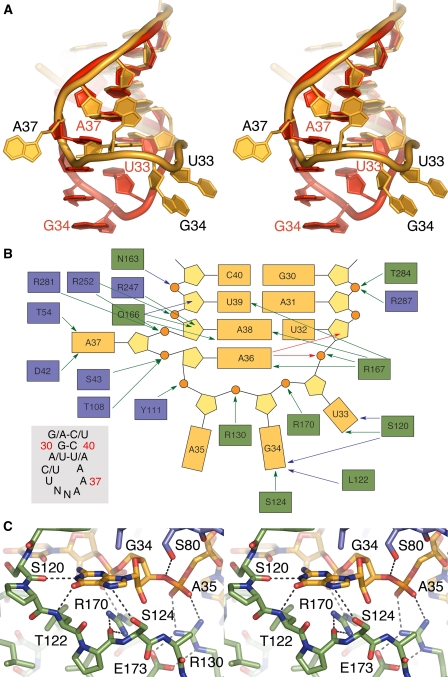
FIGURE 2.
tRNA conformation and tRNA-protein interactions. A, stereo overlay of the unfolded anticodon loop in the present complex (gold) with a canonical anticodon-loop structure in red exemplified by yeast tRNA(Phe) (from 1EHZ; Ref. 18). For clarity, the bases in yeast tRNA(Phe) are shown unmodified. B, summary of RNA-protein interactions. Protein and nucleic acid are colored as in Fig. 1. Green arrows for side chain-RNA interactions, blue for backbone-RNA interactions, and red for intra-RNA interactions (excluding standard pairing). S80 is excluded for clarity. Gray insert, consensus sequence of the eight tRNAs in E. coli that are modified by MiaA. Nucleotide numbering is in red. N is any nucleotide. C, close-up in stereo of the tight interaction between G34 and the “bottom” loop.
At the position 33, G or A would be sterically hindered, and a C could be accommodated; however, it is not compatible with a functional tRNA as it requires the U-turn motif for proper function at the ribosome. Moreover, G34 interacts closely with the backbone oxygen of Ser120 and the backbone nitrogen of Leu122, whose positions are fixed with an intervening Pro121 (Fig. 2, B and C). Furthermore, Arg170 interacts strongly with the G34 phosphate and, at the same time, fixes the position of the segment Ser120 to Ser124 by forming a strong bifurcated hydrogen bond with the backbone of Pro123 (Fig. 2C). Ser124, in turn, interacts with the N-7 of G34 (Fig. 2C). This stretch of amino acids, whose conformation is strongly imposed by Pro121 and Pro123, is of considerable importance for tRNA recognition. We propose that the very strong and specific interactions formed by this segment with nucleotides at positions 33 and 34 contribute, to a large extent, to the interaction energy needed to unfold the substrate tRNA anticodon loop.
Focusing on A35 and A36, A35 is flipped outwards, and its phosphate backbone is strongly bound by Arg130, which in turn is held in place by Glu173 (Fig. 2C). In contrast to A35, A36 folds inward to make non-sequential stacking interactions with A38 and form hydrogen bonds with its base N-1 to the ribose O2 of U32 and to the NH2 of Arg167. Furthermore, the A36 N6 atom interacts with the phosphate moiety of U33. Here, a U36 or C36 would not be able to form these intra-RNA interactions and would not enable the observed shape of the anticodon loop necessary to bind the enzyme. A G36, on the other hand, would be able to form the intra-RNA interactions, but would, instead, most likely clash with U32 and Trp285 with its N2. Also, in this case, the shape of the unfolded part of the tRNA would be different. Hence, the consensus A36 (Fig. 2B, inset) is specified indirectly by the enzyme through the MiaA-induced structure of the anticodon loop. Furthermore, the base of A37 is firmly bound via its N6 and N-7 atoms to Asp42 and to the backbone of Thr54 through its N6. Mutagenesis of Asp42 and Thr54 highlighted possible roles in catalysis rather than in substrate binding (6). This is in agreement with the proposed role of Asp42 (by extension also the backbone carbonyl of Thr54) to facilitate a nucleophilic attack of N6 on the co-substrate DMAPP (10). The phosphate of A37 is firmly bound from opposite sides by Ser43 and Thr108. Similarly, the phosphate of A38 is sequestered from opposite directions, in this case by Arg281 and Arg252. Hence, the A37 is rotated ∼180° about the phosphate-phosphate axis, with the phosphates as ball bearings that are, in turn, fixed by intimately interacting amino acid side chains. Moreover, A38 interacts through its N6 with the O2 of U32 and through its N-1 with NH2 of Arg167. A purine at nucleotide position 32 would clash into A38; hence, MiaA achieves indirect sequence readout to select for the consensus pyrimidine at position 32 (Fig. 2B, inset). Furthermore, Gln166 forms close interactions with the A38 N-3 and ribose O2, whereas Arg247 interacts intimately with ribose O2 from the opposite side. In this position, U38 or C38 would not form the necessary interactions, whereas a G38 would fit with only minor rearrangement. It should be noted that, at least in E. coli, there are no tRNAs with G38 in this position; however, a tRNA stem loop mutant with G38 (5) was shown to be a substrate, albeit poor.
Arg167 and Arg170 play a particularly prominent role in tRNA binding by MiaA. Specifically, Arg167 coordinates the positions of U33, A36, A38, and U39, whereas Arg170 fixates the position of both the phosphate backbone of G34 and the important Ser120–Ser124 segment. Inevitably, mutating either of these residues renders MiaA virtually incapable of substrate binding (6).
Implications for Substrate Selectivity—What about tRNA(Ser) with CGA codon that has the canonical triple-A motif but is not modified by MiaA? One of the differences of tRNA(Ser) to the tRNAs that are actually modified by MiaA is the presence of a C31-G39 Watson-Crick bond instead of the A/U-U/A Watson-Crick bond found in the tRNAs being modified. Modeling this change in the present structure reveals a possible clash between Arg167 and a G39, and hence, the crucial Arg167 would need to undergo rearrangement. Another difference between tRNA(Ser) and the tRNAs modified by MiaA lies in that the anti-codon proximal stem of tRNA(Ser) has a wobble G30-U40 base pair that would distort the local geometry. In fact, this part of the anti-codon proximal stem appears to be very important because, from the 29–41 base pair and downwards to the 31–39 base pair, the rift formed between the two protein domains is very narrow, allowing only tight base pairing in the antiodon proximal stem. This is especially pronounced at the rift entrance at the position/level of the G30-C40 base pair. This agrees with the observation that tRNA(Ser), having the wobble G30*U40 base pair, is not a substrate. Furthermore, the N-7 of G29 is firmly bound by Lys280. Because (i) the replacement Lys280 → Ala renders MiaA inactive (6) and (ii) the G29 N-7-Lys280 interaction observed here is only possible with purines, we propose that this interaction explains the strong selection for purines at position 29 (4).
Comparison with the Yeast IPT—The recent structure of MOD5 in complex with tRNA(Cys) at 2.95 Å (11) shows a high overall similarity to the present structure with respect to the core domain (r.m.s. deviation 1.7Å on Cα:s); however, the position and structure of the insertion domain is different (Fig. 3A). Although the five-helix bundle of the insertion domain is similar in the yeast IPT, the length and orientation of the helices differ when compared with the present structure (Fig. 3A). The eukaryotic IPTs carry a conserved C-terminal zinc-finger domain (19) that forms interactions that span the entire length of one side of the anticodon stem, all the way up to the D-loop (11). In the bacterial structure described here, there is no equivalent interaction, and the function of this extension in the eukaryotic IPTs is unclear at this point. The opening of the rift at the position of the 30–40 pair is much more narrow in MiaA when compared with MOD5, thus lending further support to our hypothesis that this feature constitutes a criterion that selects against tRNA(Ser) in E. coli because in yeast, there is no tRNA with an A36-A37-A38 triple that has a wobble 30–40 base pair. In fact, only a minority of the RNA-protein interactions are shared between the MOD5-tRNA(Cys) and MiaA-tRNA(Phe) structures. This depends, at least partly, on the large structural differences in the insertion domain, although the overall docking arrangement of the tRNA to the protein is largely conserved. One direct protein-RNA interaction conserved is the coordination of the positions of U33, A38, and U39 in the yeast tRNA(Cys) by Arg171. The corresponding Arg167 in MiaA coordinates not only U33, A38, and U39 but also A36 in E. coli tRNA(Phe). However, this difference may relate to the lower resolution of the yeast structure because only a slight coordinate shift of the MOD5 Arg171 would bring it into hydrogen bonding distance to the N6 of A36.
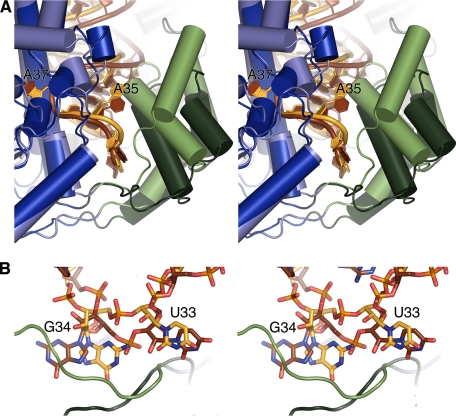
FIGURE 3.
Comparison with the yeast IPT (MOD5). A, schematic in stereo of superimposed MOD5 and MiaA. Note the large differences in the insertion domain. MiaA is colored as in Fig. 1. The yeast core domain is colored dark blue, the yeast insertion domain is dark green, the yeast tRNA is dark brown, and the yeast zinc finger is domain gray. B, close-up in stereo of differences between MiaA and MOD5 close to the bottom loop.
One particularly interesting difference in the overall RNA-protein interaction is found at the floor of the rift between the domains, where the loops that delineate this region take very different paths in MiaA and MOD5. This difference affects the conformation of G34 and U33 (Fig. 3B). Although MiaA forms several tight interactions with G34 (Fig. 2, B and C), MOD5 is devoid of interactions with the base in this position. In contrast, U33 forms a very firm, and in eukaryotes conserved (11), interaction with Gln193, whereas MiaA shows relatively weak interaction with the U33 base (Fig. 2B). Furthermore, the phosphate of nucleotide 35 (A35 in E. coli tRNA(Phe)) is firmly coordinated in MiaA, whereas the phosphate of C35 in yeast tRNA-(Cys) is not bound in the MOD5 co-crystal structure. We propose that these differences are needed in the eukaryotic IPTs to enable modification of mitochondrial tRNAs that harbor a cytosine at position 36, like yeast mitochondrial tRNA(Gly1). This can be rationalized such that in order for C36 to form the strong intra-RNA interactions with the phosphate and the U32 ribose O2 atom (Fig. 2B), the anticodon stem loop must follow a different path in the eukaryotic IPTs. Modeling a cytosine at position 36 in the yeast structure indicates that because the position of U33 is constrained/fixed, nucleotides 34 and 35 need to be accommodating and move away/out from the cleft. The reshaping of the anticodon loop required by yeast mitochondrial tRNA(Gly1) to bind is not possible if nucleotide 34 is too tightly bound and frozen conformationally, as shown here for MiaA.
Our results show that a mutually induced-fit mechanism ensures the formation of an intimate intermolecular interface that is, in turn, necessary for proper MiaA substrate recognition. In addition, our results explain elegantly existing biochemical data on MiaA tRNA substrate requirements (4, 5).
Acknowledgments
We are grateful to Susanne Bergstedt for excellent technical assistance and to Christer Svensson and Krister Larsson at MAX-lab (Lund, Sweden) for support during data collection.
The work was supported by grants from the Swedish Research Council, the Royal Swedish Academy of Sciences, Jeansson's Foundation, Längmanska kulturfonden, Helge Ax:son Johnson's Foundation, Magnus Bergvall's Foundation, O.E. and Edla Johansson's Foundation, Otto Swärd's and Ulrika Eklund's Foundation, and Karolinska Institutet (to B. M. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (code 3FOZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: DMAPP, dimethylallyl pyrophosphate; IPT, isopentenyl-tRNA transferase; PDB, Protein Data Bank; IMAC, immobilized metal ion affinity chromatography; r.m.s., root mean square.
References
- 1.Agris, P. F. (2008) EMBO Rep. 9 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Urbonavicius, J., Qian, Q., Durand, J. M., Hagervall, T. G., and Björk, G. R. (2001) EMBO J. 20 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agris, P. F., Vendeix, F. A., and Graham, W. D. (2007) J. Mol. Biol. 366 1–13 [DOI] [PubMed] [Google Scholar]
- 4.Motorin, Y., Bec, G., Tewari, R., and Grosjean, H. (1997) RNA (Cold Spring Harbor) 3 721–733 [PMC free article] [PubMed] [Google Scholar]
- 5.Soderberg, T., and Poulter, C. D. (2000) Biochemistry 39 6546–6553 [DOI] [PubMed] [Google Scholar]
- 6.Soderberg, T., and Poulter, C. D. (2001) Biochemistry 40 1734–1740 [DOI] [PubMed] [Google Scholar]
- 7.Grosjean, H., Nicoghosian, K., Haumont, E., Söll, D., and Cedergren, R. (1985) Nucleic Acids Res. 13 5697–5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sibler, A. P., Dirheimer, G., and Martin, R. P. (1986) FEBS Lett. 194 131–138 [DOI] [PubMed] [Google Scholar]
- 9.Gillman, E. C., Slusher, L. B., Martin, N. C., and Hopper, A. K. (1991) Mol. Cell Biol. 11 2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie, W., Zhou, C., and Huang, R. H. (2007) J. Mol. Biol. 367 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou, C., and Huang, R. H. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 16142–16147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porath, J., Carlsson, J., Olsson, I., and Belfrage, G. (1975) Nature 258 598–599 [DOI] [PubMed] [Google Scholar]
- 13.Batey, R. T., and Kieft, J. S. (2007) RNA (Cold Spring Harbor) 13 1384–1389 [Google Scholar]
- 14.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B. F., and Nyborg, J. (1995) Science 270 1464–1472 [DOI] [PubMed] [Google Scholar]
- 16.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 18.Shi, H., and Moore, P. B. (2000) RNA (Cold Spring Harbor) 6 1091–1105 [Google Scholar]
- 19.Golovko, A., Hjälm, G., Sitbon, F., and Nicander, B. (2000) Gene (Amst.) 258 85–93 [DOI] [PubMed] [Google Scholar]
- 20.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins 50 437–450 [DOI] [PubMed] [Google Scholar]