Abstract
Alternative translation is an underappreciated post-transcriptional regulation mechanism. Although only a small number of genes are found to be alternatively translated, most genes undergoing alternative translation play important roles in tumorigenesis and development. Protein phosphatase 2A (PP2A) is involved in many cellular events during tumorigenesis and development. The specificity, localization, and activity of PP2A are regulated by B regulatory subunits. B56ε, a member of the B56 regulatory subunit family, is involved in multiple signaling pathways and regulates a number of developmental processes. Here we report that B56ε is alternatively translated, leading to the production of a full-length form and a shorter isoform that lacks the N-terminal 76 amino acid residues of the full-length form. Alternative translation of B56ε occurs through a cap-dependent mechanism. We provide evidence that the shorter isoform is required for Wnt signaling and regulates the midbrain/hindbrain boundary formation during Xenopus embryonic development. This demonstrates that the shorter isoform of B56ε has important biological functions. Furthermore, we show that the N-terminal sequence of B56ε, which is not present in the shorter isoform, contains a nuclear localization signal, whereas the C terminus of B56ε contains a nuclear export signal. The shorter isoform, which lacks the N-terminal nuclear localization signal, is restricted to the cytoplasm. In contrast, the full-length form can be localized to the nucleus in a cell type-specific manner. The finding that B56ε is alternatively translated adds a new level of regulation to PP2A holoenzymes.
Recent progress in genome projects has led to the unexpected finding that the number of protein-coding genes does not account for the increasing complexity or diversity in eukaryotes. For example, the number of protein-coding genes in a sea urchin genome (1) is comparable with that in mammalian genomes (2). This highlights the importance of post-transcriptional regulation in generating the tremendous biological complexity in eukaryotes. Alternative translation is a post-transcriptional regulation mechanism that occurs at the level of translation. Alternative translation was first discovered in viruses. It is unique in that multiple protein isoforms are generated from a single mRNA through selective usage of translation initiation sites. Alternative translation isoforms may be targeted to different subcellular compartments or expressed in different tissues. In some cases, alternative translation isoforms have distinct functions. Even though alternative translation is an important post-transcriptional regulation mechanism, only a small number of vertebrate genes have been found to be alternatively translated (for review see Ref. 3).
Alternative translation has been studied for decades. Based on the “scanning model,” which was proposed by Kozak 30 years ago (4), the first step of translation initiation is the assembly of the 40 S ribosomal subunit, which carries tRNAMet, on the 5′ end of mRNA. Ribosomes then migrate on the mRNA, a process known as “scanning.” Once the first AUG codon in a context favorable for initiating translation is recognized via pairing with anti-codon in tRNAMet (5′-CAU-3′), ribosomes stop scanning and initiate peptide synthesis (5). In some cases, however, ribosomes skip the first AUG and initiate translation from an alternative site(s).
There are two mechanisms for alternative translation. The first mechanism is known as re-initiation or the internal ribosome entry site (IRES)3-dependent mechanism, meaning that ribosomes bind to IRES and initiate translation downstream of the IRES. Because initiation complex assembly on the 5′ mRNA cap is not required in this case, this mechanism is also known as a “cap-independent mechanism.” The second mechanism for escaping the first AUG is the context-dependent leaky scanning mechanism, meaning that because of the mRNA sequence flanking the first AUG, only a fraction of ribosomes initiate translation at the first AUG, whereas others continue scanning and initiate translation further downstream. As context-dependent leaky scanning requires assembly of ribosomes on the 5′ cap, it is often called “cap-dependent alternative translation.” So far, only a small number of genes have been found to be alternatively translated (3, 6). Surprisingly, many of them play important roles during tumorigenesis and development.
As one of the most abundantly expressed protein phosphatases, protein phosphatase 2A (PP2A) is involved in numerous biological processes, including tumorigenesis and development (7-9). The holoenzyme is a heterotrimeric complex consisting of a catalytic subunit (C), a structural subunit (A), and a regulatory subunit (B). The catalytic activity, the substrate specificity, and the subcellular localization of PP2A are controlled by the regulatory subunits. Three PP2A regulatory subunit families have been identified, including B55/B, B56/B′, and PR72/B″. Mammalian B56 family is composed of five members (B56α, -β,-δ,-ε, and -γ). B56 family members have a highly conserved central core region and variable N- and C-terminal sequences (10). B56 family members are involved in multiple signaling cascades, including the Wnt (11-15), Hedgehog (16-18), extracellular signal-regulated kinase (ERK) (19-22), Akt (17, 22, 23), and planar cell polarity pathways (24). Loss-of-function studies have revealed that B56 family members are critical for cell survival (19, 24, 25) and play essential roles in chromosome separation during mitosis (26, 27).
During vertebrate embryonic development, B56ε is required for cell movement during gastrulation (24), axis specification, midbrain/hindbrain boundary formation (15), eye induction, and eye field separation (17). Here, we report that B56ε is alternatively translated. This results in the production of a 56-kDa full-length form (B56ε-fl) and a 48-kDa isoform, lacking the N-terminal 76-amino acid residues (B56ε-s) through a cap-dependent alternative translation mechanism. We provide evidence that B56ε-s is required for midbrain/hindbrain boundary formation and is involved in the Wnt pathway. Furthermore, we have identified a nuclear localization signal (NLS) from the N-terminal 76 amino acid residues of B56ε-fl and a nuclear export signal (NES) from the C-terminal sequence of B56ε. We show that B56ε-s, which lacks the N-terminal NLS, is restricted to the cytoplasm, whereas B56ε-fl can be localized to the nucleus in a cell type-specific manner. Hence, our studies have identified an important mechanism for post-transcriptional regulation of B56ε.
EXPERIMENTAL PROCEDURES
Embryo Manipulations—Embryos were obtained and micro-injected as described (28). The dosage of RNA or morpholino for microinjection was indicated in the figure legends or text. For animal cap assay, animal caps were dissected at late blastula stage with a microsurgery device (Gastromaster, Nepagene, Japan). Caps were cultured in 1× modified Marc's Ringer and harvested when the sibling control reached stage 16.
Tissue Culture and Confocal Microscopy Analysis—The culture medium for 293 cells was Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. The culture medium for MB-MDA-231 cells was RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells were cultured at 37 °C in a humidified 5% CO2 atmosphere. Transfection was performed with Lipofectamine™ 2000 (Invitrogen). For confocal microscopy analysis, transfected cells were plated on glass coverslips. After coverslip attachment, cells were fixed in 4% paraformaldehyde and mounted in mounting medium with DAPI (Vector Shield). For leptomycin B treatment, cells were exposed to leptomycin B (2 ng/ml; Sigma) for 7 h before fixation. Images were collected by LSM 510 confocal microscopy (Zeiss) at the Morphology Core Facility at the Research Institute at Nationwide Children's Hospital.
Plasmids and Morpholinos—Myc-EGFP (17) and ε-FLAG (15) were described. The open reading frames of human B56ε, human B56α, mouse B56ε, Xenopus B56δ, and Drosophila wdb, were PCR-amplified and cloned between the BamHI and ClaI of pCS2-FLAG. Templates for PCRs were as follows: pCEP4-hB56ε and pCEP4-hB56α (gifts from Dr. D. Virshup) (10), pcDNA3-dwdb (a gift from Dr. A. Sehgal) (29), Xenopus B56δ (IMAGE, 6864669; ATCC), and mouse B56ε (IMAGE, 30941997; ATCC). HA-hB56β and HA-hB56γ1 were obtained from Addgene. M77A, M77V, M85G, and M77A/M85G were generated by site-directed mutagenesis. ΔN, Myc-ε-FLAG, GFP-ε-s, GFP-ε-fl, εN-Myc-EGFP, GFP-1/76, GFP-28/76, GFP-36/76, GFP-1/35, GFP-1/54, GFP-28/54, GFP-NLS, and GFP-NLS-ε-s were constructed through standard cloning approaches. Detailed information about constructs will be provided upon request.
Morpholinos were purchased from Gene Tools (Philomath, OR). The sequences of the morpholino oligonucleotides are as follows: MO1, 5′-GAG GAG TGG TTG GTG CTG AGG ACA T-3′ (15); MO2, 5′-GAG CTT TGT GAC CTC TTC TGC CTG G-3′; MO3, 5′-GTA ATC CAC CAG TTC ATT CAG A GT-3′; and MO4, 5′-TAT ATT GCA GGA TAC CAT TCT AAC T-3′.
RT-PCR and Whole Mount in Situ Hybridization—RNA extraction and RT-PCR methods were described (15). Primers for ODC, en2, and slug were described (15, 30). Whole mount in situ hybridization was performed as described (31).
Western Blots—For Western blot analysis, embryos or oocytes were homogenized (40 μl per embryo or oocyte) in cold lysis buffer (125 mm NaCl, 20 mm Tris, pH 7.5, 1% Nonidet P-40, 1 mm EDTA, proteinase inhibitor mixture (Sigma)). Lysates were centrifuged at 500 × g for 5 min and at 14,000 × g for 5 min in a tabletop centrifuge at 4 °C. Cleared lysates were collected. After SDS sample buffer was included, each sample was boiled at 100 °C for 5 min and separated by SDS-PAGE. Western blots were performed according to the standard protocol. Antibodies are as follows: anti-FLAG (1:1000; M2, Sigma), anti-Myc (1:1000; 9E10, Sigma), anti-B56γ (1:200; 39-3600, Invitrogen), and anti-PP2Ac (1:5000; Millipore). A keyhole limpet hemocyanin-coupled peptide (Ac-CELKRGLRRDGIIPT-OH) was synthesized at New England Peptide and was used to immunize rabbits for generating anti-B56ε (human and mouse) antiserum. A B56ε peptide affinity column was prepared using the SulfoLink immobilization kit (Pierce). The B56ε antiserum was loaded to the affinity column, washed, and eluted with 100 mm glycine, pH 2.5. The eluate, which contains anti-B56ε antibody, was neutralized and dialyzed against phosphate-buffered saline. For Western blot, the purified anti-B56ε antibody was used at a concentration of 2.5 μg/ml.
RESULTS
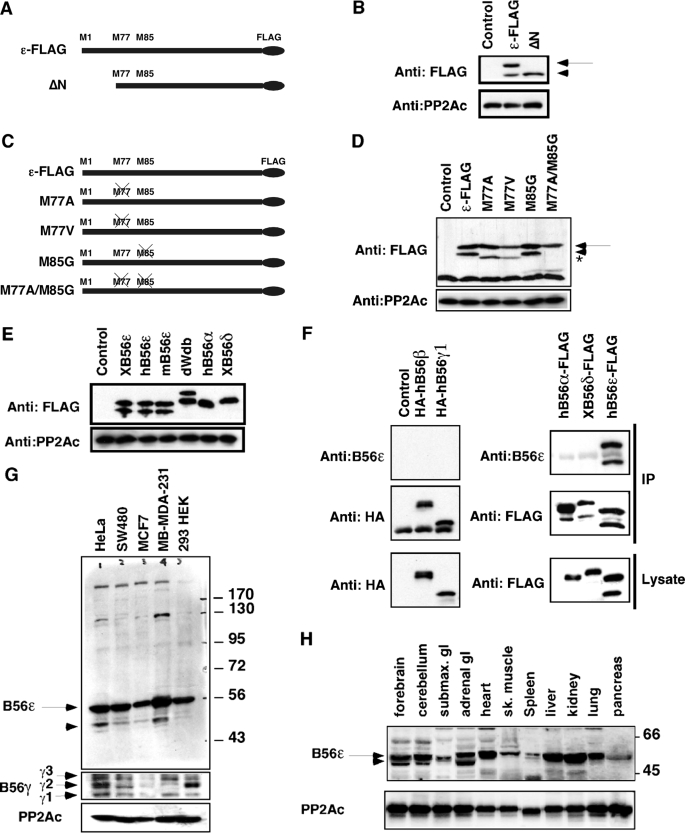
Alternative Translation of B56ε—B56ε regulatory subunit of PP2A is involved in several signaling pathways and functions in multiple developmental processes (11, 13-15, 17-19, 23-27, 29). We noticed that overexpression of a C-terminal FLAG-tagged Xenopus B56ε (ε-FLAG) in Xenopus embryos resulted in the appearance of two polypeptides on the FLAG Western blot (Fig. 1B) (17). According to their migration rates, the estimated molecular mass of the slower migrating polypeptide is ∼56-kDa, which is the calculated molecular mass of ε-FLAG. The faster migrating polypeptide is ∼48-kDa. A similar observation was made when ε-FLAG RNA was translated in vitro using rabbit reticulocyte lysate (data not shown). The 5′-coding sequence of vertebrate B56ε mRNA contains three in-frame AUGs, which encode Met1, Met77, and Met85 (Fig. 1A). The internal methionines are conserved in all vertebrate orthologs of B56ε but not in other B56 family members. To test whether the 48-kDa polypeptide could be an alternative translation product initiated from the second AUG, we generated ΔN, a truncation mutant lacking the initial AUG and the following 108-bp sequence. As a consequence, translation of ΔN is started at the second AUG of B56ε, which corresponds to Met77. When analyzed on SDS-PAGE, the migration rate of the ΔN translation product is identical to that of the 48-kDa polypeptide derived from ε-FLAG (Fig. 1B). This raises the possibility that the 48-kDa polypeptide is an alternative translation isoform of B56ε.
FIGURE 1.
Alternative translation of B56ε. A, schematic representation of constructs used in B. B, Western blot showing the expression of ε-FLAG (100 pg) and ΔN (100 pg) in Xenopus embryos. C, schematic representation of mutation constructs used in D. D, Western blot showing that the Met77 is the downstream translation initiation site. Note that overexpression of ε-FLAG resulted in the production of a 56-kDa polypeptide (arrow) and a 48-kDa polypeptide (arrowhead). Overexpression of M77A and M77V resulted in the production of the 56-kDa polypeptide and a polypeptide migrating slightly faster than the 48-kDa polypeptide (indicated by asterisk). Neither the 48-kDa polypeptide nor the polypeptide migrating slightly faster than the 48-kDa polypeptide was produced when M77A/M85G is expressed. E, Western blot results showing that when expressed in Xenopus embryos, B56ε and Drosophila Wdb, but not other B56 family members, were alternatively translated. Note that Drosophila Wdb contains 524 amino acid residues, whereas vertebrate B56ε has only 467 amino acid residues. All constructs were C-terminal FLAG-tagged. Overexpressed proteins were probed with the anti-FLAG antibody. F, specificity of the anti-B56ε antibody. hB56ε-FLAG, hB56α-FLAG, xB56δ-FLAG, HA-hB56β, and HA-hB56γ1 were overexpressed. Expression levels of overexpressed B56 proteins were determined by FLAG and HA Western blots (bottom panels). Overexpressed B56 proteins were immunoprecipitated with anti-FLAG and anti-HA antibodies, respectively. Immunoprecipitation of overexpressed B56 proteins was first verified by HA and FLAG Western blots (middle panels). Immunoprecipitates were then probed with the affinity-purified anti-B56ε antibody. Note that B56ε antibody recognizes B56ε, without cross-reacting with other B56 family members. G, Western blot results showing the expression of endogenous B56ε and B56γ in tissue culture cell lines. H, expression of endogenous B56ε in adult mouse tissues. Catalytic subunit of PP2A, which is abundantly expressed in most cells, was used as the loading control. Arrows and arrowheads in B, D, G, and H indicate B56ε-fl and B56ε-s, respectively.
Next, we generated several point mutations, including M77A (Met77 to Ala), M77V (Met77 to Val), M85G (Met85 to Gly), and M77A/M85G (Fig. 1C). We reasoned that mutation of Met77 would prevent the production of the 48-kDa polypeptide, if this 48-kDa polypeptide is indeed an alternative translation product of B56ε. RNAs encoding these mutants and ε-FLAG were synthesized in vitro and injected into fertilized eggs. Control and injected embryos were harvested at stage 9 and analyzed by FLAG Western blot. Consistent with previous results, ε-FLAG produced the 48- and the 56-kDa polypeptides. M77A and M77V produced the 56-kDa polypeptide but not the 48-kDa polypeptide. Strikingly, a polypeptide migrating slightly faster than the 48-kDa polypeptide was detected. This is likely a translation product initiated at Met85. When both Met77 and Met85 were mutated (M77A/M85G), neither the 48-kDa polypeptide nor the faster migrating polypeptides were detected (Fig. 1D). These results indicate that the 48-kDa polypeptide is a product of alternative translation initiated from the second AUG that corresponds to Met77 of the full-length B56ε. Hereafter, we designate the full-length and the shorter B56ε isoforms as B56ε-fl and B56ε-s, respectively.
B56 family members are highly similar to each other. We asked whether B56ε from other species and other B56 family members are alternatively translated as well. C-terminal FLAG-tagged human B56ε (hB56ε), mouse B56ε (mB56ε), Drosophila wdb, human B56α ((hB56α), and Xenopus B56δ (xB56δ) were constructed. When expressed in Xenopus embryos, only xB56ε, hB56ε, mB56ε, and wdb were alternatively translated, as judged by the appearance of two polypeptides on the FLAG Western blot (Fig. 1E). Thus, B56ε and Drosophila wdb, but not other B56 family members, were alternative translated.
To determine whether endogenous B56ε is alternatively translated, a polyclonal antibody against a C-terminal epitope of mammalian B56ε was generated. This antibody recognized B56ε, without cross-reacting with overexpressed hB56α, hB56β, xB56δ, and hB56γ1 (Fig. 1F). Using this antibody, we examined the expression of B56ε in several cell lines and adult mouse tissues. As shown in Fig. 1G, both B56ε-s and B56ε-fl were detectable in HeLa, SW480, MCF7, and MB-MDA-231 cells. In HEK 293 cells, however, the expression level of B56ε-s was below the limit of detection. B56γ is alternatively spliced (10, 32, 33). When the expression of B56γ was analyzed, all three splicing isoforms were detected in HeLa, SW480, and HEK 293 cells. The MB-MDA-231 cell only expressed B56γ1 and B56γ3. MCF7 cells expressed very low level of B56γ (Fig. 1G). Additionally, we analyzed the expression of B56ε in adult mouse tissues. Both isoforms were detected in the forebrain, cerebellum, and adrenal gland, whereas heart, muscle, liver, kidney, and lung expressed predominantly B56ε-fl (Fig. 1H). These results demonstrate that endogenous B56ε undergoes alternative translation, and the relative abundance of two isoforms is cell type-dependent.
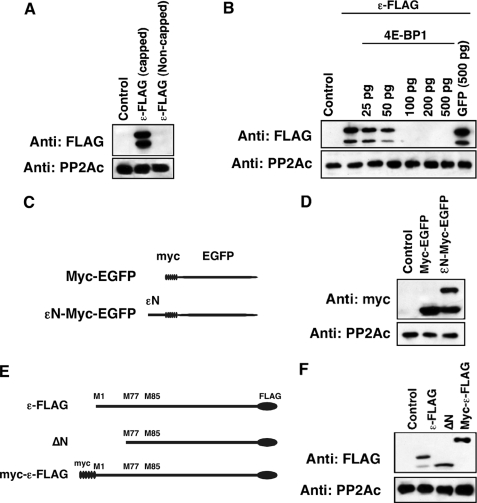
Alternative Translation of B56ε Occurs through a Cap-dependent Mechanism—As described above, alternative translation may be through the IRES-dependent mechanism or the cap-dependent mechanism. The cap-dependent alternative translation requires assembly of ribosomes on the 5′ cap of mRNA, whereas ribosomes bind directly to IRES in the case of IRES-dependent alternative translation. To distinguish which mechanism applies to alternative translation of B56ε, we asked whether assembly of ribosomes on the 5′ cap of B56ε mRNA is required for translation of B56ε-s. Capped and non-capped mRNAs of ε-FLAG were synthesized in vitro and injected into Xenopus oocytes. Thirty minutes after injection, oocytes were harvested and subjected to Western blot analysis. Injection of capped, but not non-capped, ε-FLAG RNA resulted in production of B56ε-fl and B56ε-s (Fig. 2A). In addition, we found that translation of both B56ε-fl and B56ε-s was blocked by overexpression of the elF4E-binding protein (4E-BP) but not by overexpression of GFP (Fig. 2B). 4E-BP inhibits assembly of elF4F complex, a multisubunit complex interacting directly with the 5′ cap structure of mRNA (34). These results indicate that assembly of ribosomes on the 5′ end of B56ε mRNA is required for translation of B56ε isoforms and suggest that alternative translation of B56ε is cap-dependent.
FIGURE 2.
Alternative translation of B56ε occurs through a cap-dependent mechanism. A, Western blot result to show that the 5′ cap is required for translation of both B56ε isoforms. Samples from left to right are uninjected oocytes, oocytes injected with a capped ε-FLAG RNA (100 pg), and oocytes injected with a non-capped ε-FLAG RNA (100 pg). B, Western blot result to show that overexpression of 4E-BP1 blocked the translation of ε-FLAG RNA (100 pg) in oocytes in a dose-dependent manner. Injection of GFP RNA (500 pg) had no effect on ε-FLAG translation. C, schematic representation of constructs used in D. D, Western blot result to show that εN-Myc-EGFP is alternatively translated. E, schematic representation of constructs used in F. F, Western blot result to show that improving the context of upstream AUG by engineering the Myc tag to the 5′ of B56ε prevented alternative translation of B56ε. Catalytic subunit of PP2A was used as the loading control.
In the case of cap-dependent alternative translation, the sequence around the first AUG of an mRNA determines whether ribosomes initiate translation or continue scanning. If B56ε-s is generated through a cap-dependent mechanism, one might predict that the first AUG of B56ε mRNA is in a context unfavorable for translation initiation. This AUG and its context, when transferred to the 5′ of a nonrelated sequence, would cause alternative translation. To test this, we generated an εN-Myc-EGFP, which contains the 5′-coding sequence of B56ε (Met1-Phe76) followed in-frame with a Myc-EGFP (Fig. 2C). Indeed, when expressed in Xenopus embryo, εN-Myc-EGFP produced two polypeptides. One migrated with the same rate as Myc-EGFP did on SDS-PAGE. The other migrated about 8 kDa larger than Myc-EGFP, which corresponds to the calculated molecular weight of the N-terminal 76 amino acid residues of B56ε-fl (Fig. 2D). This result indicates that the first AUG of B56ε is in a context unfavorable for translation initiation.
Cap-dependent alternative translation can be prevented by improving the context of the upstream translation initiation site (3, 6). To further test the hypothesis that alternative translation of B56ε is through a cap-dependent mechanism, we constructed a Myc-ε-FLAG. Myc-ε-FLAG contains six copies of the Myc tag fused at the N terminus of B56ε (Fig. 2E). We reasoned that engineering an AUG in a context favorable for translation initiation upstream of the B56ε open reading frame would prevent the production of B56ε-s. Indeed, overexpression of Myc-ε-FLAG in Xenopus embryo only produced one polypeptide. The translation of B56ε-s was completely blocked. The estimated molecular mass of the polypeptide is about 75 kDa, which corresponds to the calculated molecular mass of Myc-ε-FLAG (Fig. 2F). Thus, improving the context of upstream translation initiation site blocks the translation of B56ε-s. These results demonstrate that alternative translation of B56ε occurs through a cap-dependent mechanism.
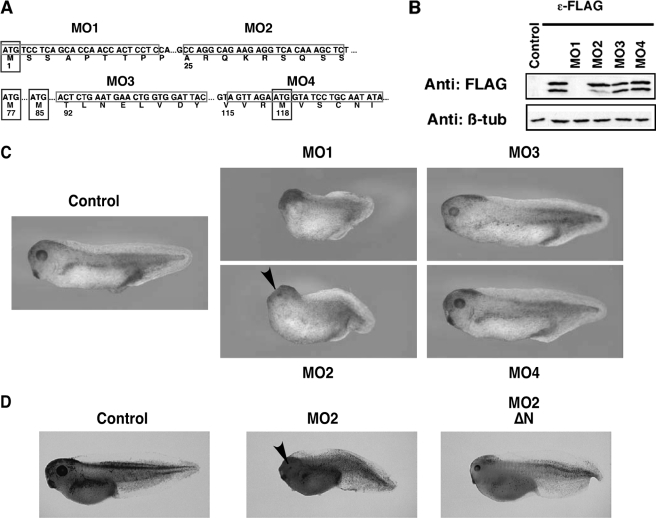
B56ε-s Is Essential for the Midbrain/Hindbrain Boundary Formation—Knowing that B56ε is alternatively translated, we wanted to know whether B56ε-s, which lacks the N-terminal 76 amino acid residues of B56ε-fl, is biologically functional. We took advantage of morpholino antisense oligonucleotides, which, upon binding to their mRNA target, inhibit translation initiation by acting as a “roadblock” for the scanning ribosome. As alternative translation of B56ε occurs through a cap-dependent mechanism, we hypothesized that a morpholino, which targets the sequence between the first and the second AUG of Xenopus B56ε mRNA, should block the translation of B56ε-s, without affecting B56ε-fl. Four morpholinos that target to different B56ε sequences were synthesized (Fig. 3A). As expected, MO1 (15), which binds to the first AUG region, efficiently blocked translation of both B56ε isoforms. B56ε-s translation, but not B56ε-fl translation, was significantly reduced by MO2, which targets the sequence of B56ε mRNA between the first AUG and the second AUG. MO3 and MO4, which target sequences downstream of the second AUG, had no effect on the translation of B56ε-fl and B56ε-s (Fig. 3B).
FIGURE 3.
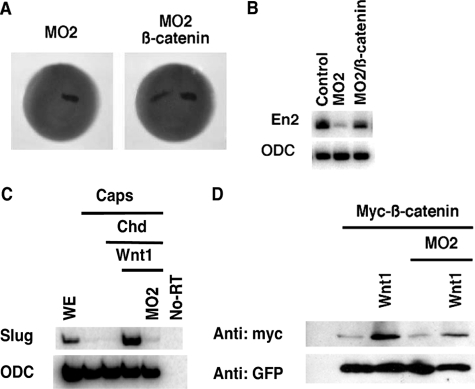
Depletion of B56ε-s interferes with Xenopus embryonic development. A, schematic drawing to show the design of morpholinos that target B56ε translation isoforms. Four methionine residues are highlighted by boxes. Morpholino targeting sequences are highlighted by boxes. MO1, which binds to the upstream AUG region, has been reported (15). MO2 targets the sequence between the first and the second AUG. MO3 and MO4 target the sequences downstream of the second AUG. B, Western blot result to show the effects of morpholinos on translation of B56ε isoforms.ε-FLAG RNA was injected into control oocytes or oocytes previously injected with 20 ng of MO1, MO2, MO3, or MO4. Translation of B56ε isoforms were evaluated by FLAG Western. β-Tubulin (β-tub) Western blot serves as a control for loading. MO1 blocked translation of both B56ε isoforms. MO2 reduced the amount of B56ε-s, without affecting B56ε-fl. MO3 and MO4 had no effect on translation of B56ε isoforms. C, effects of morpholino injection on Xenopus embryonic development. MO1 (5 ng), MO2 (10 ng), MO3 (10 ng), or MO4 (10 ng) were bilaterally injected into dorsal animal blastomeres. Injection of MO1 and MO2 caused defective embryonic development. D, developmental defects caused by MO2 injection could be partially rescued by ΔN, which lacks MO2-binding sequence. From left to right are a control tadpole stage embryo, an embryo injected with MO2 (10 ng), and an embryo injected with MO2 (10 ng) and ΔN (100 pg). Arrowheads in C and D indicate eyes in MO2-injected embryos.
Taking advantage of MO2, which specifically interferes with the translation of B56ε-s, we investigated the function of B56ε-s during Xenopus development. Thus, MO2 was injected into both dorsal animal blastomeres at the 8-cell stage. Based on fate map studies, these injected blastomeres will give rise to neural ectoderm and some axial mesoderm tissues (35). MO1, MO3, and MO4 were injected as controls.
Consistent with our previous reports (15, 17), the anterior/posterior axes of MO1 (5 ng)-injected embryos were very short. These embryos lacked eye formation and showed severely reduced heads (Fig. 3C). In contrast, MO2 (10 ng)-injected embryos showed a less severe phenotype. Their heads were relatively small. Although the size of their eyes was reduced, most MO2-injected embryos had two eyes (Fig. 3C). Injection of a higher dose of MO2 (15-20 ng) blocked blastopore closure permanently (data not shown). Little if any defects were observed in embryos injected with MO3 (10 ng) or MO4 (10 ng) (Fig. 3C), suggesting that the defects in MO2-injected embryos were caused by depletion of B56ε-s. To further prove the specificity of MO2, we determined whether developmental defects induced by MO2 injection could be rescued by coinjection of ΔN, which lacks MO2 binding sequence. When overexpressed, ΔN can restore the expression of B56ε-s in MO2-injected embryos. Indeed, coinjection of MO2 with ΔN (100 pg) partially rescued head development and the length of anterior/posterior axis (Fig. 3D). Collectively, the above results demonstrate that B56ε-s plays important roles during embryonic development.
Depletion of both B56ε-fl and B56ε-s by injection of MO1 causes defects in multiple developmental processes. These include failures in eye induction at the gastrula stage, eye field separation at the tailbud stage (17), and defective midbrain/hindbrain boundary formation (15). We thus determined whether these developmental processes were affected when B56ε-s was selectively depleted.
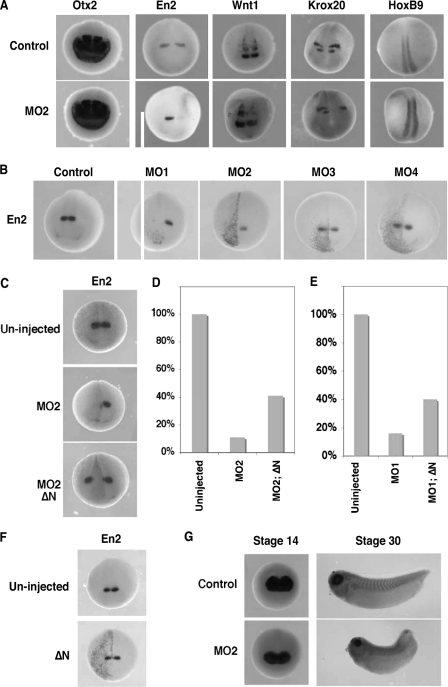
Embryos were unilaterally injected with MO2 (10 ng) at the 8-cell stage and analyzed for the expression of regional neural ectodermal markers at stage 20. These markers are as follows: otx2, a forebrain marker (36); en2 (37) and wnt1 (38), midbrain/hindbrain boundary markers; krox20 (39), a hindbrain marker; and HoxB9 (40), a spinal cord marker. As shown in Fig. 4A, expression of otx2 was not affected by MO2 injection (n = 86), indicating that depletion of B56ε-s did not affect forebrain specification. In contrast, en2 expression was severely reduced by MO2 injection (89%, n = 101). The expression of wnt1 was decreased in 68% (n = 72) of MO2-injected embryos. The expression of krox20 was reduced in 69% (n = 48) of embryos. Notably, the r3 expression domain of krox20 was affected more severely than the r5 expression domain, which was only slightly reduced. The expression of HoxB9 remained unchanged in 94% (n = 47) of embryos. Thus, injection of MO2 impairs the formation of the midbrain/hindbrain boundary.
FIGURE 4.
Depletion of B56ε-s impairs midbrain/hindbrain boundary formation, without disrupting eye induction and eye field separation. A, whole mount in situ hybridization to show the expression of otx2, en2, wnt1, krox-20, and hoxB9 in stage 20 control embryos and embryos unilaterally injected with 10 ng of MO2. One of the dorsal animal blastomeres was injected at the 8-cell stage. Upper panels are control embryos. Lower panels are MO2-injected embryos. Right side was injected. B, whole mount in situ hybridization to show en2 expression in a stage 20 control embryo and embryos unilaterally injected with MO1 (5 ng), MO2 (10 ng), MO3 (10 ng), or MO4 (10 ng). Left side was injected. C, inhibitory effect of MO2 on the expression of en2 was rescued by ΔN. Upper panel shows en2 expression in a control embryo. Middle panel shows en2 expression in an embryo injected with MO2 (10 ng). Bottom panel shows en2 expression in an embryo injected with MO2 (10 ng) and ΔN (100 pg). Left side was injected. D, summary of results shown in C. Values represent percent of embryos with normal en2 expression. E, coinjection of ΔN (100 pg) rescued en2 expression in MO1 (5 ng)-injected embryos. Values represent percent of embryos with normal en2 expression. F, whole mount in situ hybridization showing en2 expression in a stage 20 control embryo and an embryo unilaterally injected with ΔN (100 pg). Left side was injected. G, whole mount in situ hybridization showing the expression of rx in control (top panels) and MO2-injected (bottom panels) embryos. Embryos were bilaterally injected at the 8-cell stage and harvested at the early neurula stage (left panels) and the early tadpole stage (right panels). In some of these experiments, nuclear β-galactosidase was coinjected as the lineage tracer.
To determine whether the abnormal midbrain/hindbrain boundary formation in MO2-injected embryos was because of loss of B56ε-s, we first compared the expression of en2 in embryos injected with MO1 (5 ng), MO2 (10 ng), MO3 (10 ng), and MO4 (10 ng). As shown in Fig. 4B, 66% of MO1-injected embryos lacked en2 expression completely (n = 44). Injection of MO2 reduced en2 expression in 59% of embryos and completely blocked the en2 expression in 24% of embryos (n = 63). The expression of en2 was normal in MO3- (93%, n = 82) and MO4 (97%, n = 73)-injected embryos. Thus, only MO1 and MO2, which are capable of blocking B56ε-s translation, reduced the expression of en2.
To confirm further that B56ε-s is indeed critically important for the formation of the midbrain/hindbrain boundary, we performed two sets of rescue experiments. In the first experiment, we asked whether restoring the expression of B56ε-s in MO2-injected embryos could rescue en2 expression. In this experiment, injection of MO2 (10 ng) completely blocked en2 expression in 26% (n = 102) of embryos and decreased en2 expression in 63% of embryos. Only 11% of injected embryos exhibited normal en2 expression. Coinjection of ΔN (100 pg) with MO2 (10 ng) rescued en2 expression, with 41% (n = 96) of injected embryos exhibiting normal en2 expression. Only 2% of embryos lacked en2 expression on the injected side completely (Fig. 4, C and D). In the second experiment, we determined whether en2 expression in MO1-injected embryos could be rescued by restoring the expression of B56ε-s. MO1 blocks the translation of both B56ε-fl and B56ε-s (Fig. 3B). Injection of MO1 (5 ng) severely affected the expression of en2. Despite a few MO1-injected embryos that showed normal en2 expression (16%, n = 70), the majority of embryos either lacked en2 expression completely (38%) or exhibited reduced en2 expression (46%). Coinjection of ΔN with MO1 partially rescued en2 expression. In this group, 40% of embryos showed normal en2 expression. Only 13% of embryos lacked en2 expression completely (n = 85) (Fig. 4E). We also examined the effect of B56ε-s overexpression and found that the expression of en2 remained normal in ΔN (100 pg)-injected embryos (97%, n = 38) (Fig. 4F). Based on these results, we conclude that B56ε-s is critically important for the midbrain/hindbrain boundary formation.
To evaluate the effects of B56ε-s depletion on eye development, we analyzed expression of rx, an eye-specific transcription factor (41). MO2 (10 ng) was bilaterally injected into both dorsal animal blastomeres at the 8-cell stage. At stage 14, both control (n = 47) and MO2-injected embryos (n = 72) exhibited a single rx expression domain in the anterior neural plate. No difference was noticed between control and injected groups (Fig. 4G), suggesting that depletion of B56ε-s does not affect the process of eye induction. At stage 30, all MO2-injected embryos showed two eyes (n = 52), as judged by two separated rx expression domains. This indicates that eye field separation occurred in these embryos. Interestingly, although rx was clearly expressed at this stage, the size of rx expression domains was reduced in 78% of injected embryos (Fig. 4G). Later on, the eye formation became more severely affected (Fig. 3, C and D). Thus, depletion of B56ε-s interferes with later stage of eye development, without affecting eye induction and eye field separation.
B56ε Is Required for Wnt Signaling during Xenopus Development—Depletion of both B56ε isoforms impairs canonical Wnt signaling (15). The above results demonstrate that B56ε-s regulates the expression of en2, a well characterized Wnt target (42, 43), suggesting that B56ε-s is required for canonical Wnt signaling. Thus, we tested whether overexpression of β-catenin could rescue en2 expression in B56ε-s-depleted embryos. Injection of MO2 reduced en2 expression in 75% of embryos (n = 56). When 100 pg of β-catenin was coinjected with MO2, only 25% of embryos (n = 48) exhibited decreased en2 expression (Fig. 5A), suggesting that en2 expression was rescued by β-catenin. This conclusion was further confirmed by RT-PCR. As shown in Fig. 5B, bilateral injection of MO2 severely reduced en2 expression. The expression of en2 was clearly rescued by coinjection of β-catenin RNA (100 pg). These observations support the hypothesis that B56ε-s is required for the canonical Wnt pathway.
FIGURE 5.
B56ε-s is required for canonical Wnt signaling. A, whole mount in situ hybridization showing the expression of en2 in stage 20 embryos. The left panel is an embryo injected with 10 ng of MO2. The right panel is an embryo injected with MO2 (10 ng) and β-catenin RNA (100 pg). One of the dorsal animal blastomeres was injected at the 8-cell stage. Nuclear β-galactosidase was coinjected as a lineage tracer. B, RT-PCR result showing that β-catenin rescued en2 expression. Embryos were bilaterally injected with MO2 (10 ng) or a mixture of MO2 (10 ng) and β-catenin (100 pg). Injected embryos were harvested at stage 20. Ornithine decarboxylase (ODC) serves as a control for loading. C, RT-PCR results show that depletion of B56ε-s by MO2 injection blocked slug expression induced by Wnt1 (10 pg) in neutralized animal caps. Animal caps were neutralized by injection of 50 pg of Chordin (Chd) RNA. Ornithine decarboxylase is the loading control. WE, whole embryo. D, Western blot showing that Wnt1 (10 pg) stabilized a Myc-tagged β-catenin in control animal caps but not in B56ε-s depleted animal caps. GFP was used as a control for both injection and loading.
It is well established that overexpression of Wnt1 induces the expression of slug, a direct target of Wnt signaling, in neutralized animal caps (44). We took advantage of this assay to determine whether B56ε-s is required for Wnt1-induced slug expression. Animal caps were neutralized by overexpression of Chordin. As expected, overexpression of Wnt1 induced the expression of slug in animal caps. In contrast, Wnt1 failed to induce slug expression in MO2-injected animal caps (Fig. 5C). This indicates that B56ε-s is required for Wnt-induced target gene expression. Wnt-induced β-catenin stabilization is key to Wnt signaling (45). We further tested whether B56ε-s is required for Wnt1 induced β-catenin stabilization in an animal cap assay. As shown in Fig. 5D, overexpression of Wnt1 increased the amount of myc-β-catenin in wild type animal caps. In contrast, overexpression of Wnt1 failed to stabilize myc-β-catenin in MO2-injected animal caps. The amount of GFP, which was coexpressed with myc-β-catenin, was not altered by Wnt1 overexpression or B56ε-s depletion (Fig. 5D). Taken together, we conclude that B56ε-s is required for the Wnt pathway.
Identification of NLS in the N Terminus of B56ε-fl—The above observations demonstrate that B56ε-s plays important roles during development. To explore whether B56ε translational isoforms may have distinct activities, we investigated whether the N-terminal 76 amino acid residues of B56ε-fl, which is truncated in B56ε-s, plays a role in targeting B56ε-fl to a specific subcellular compartment.
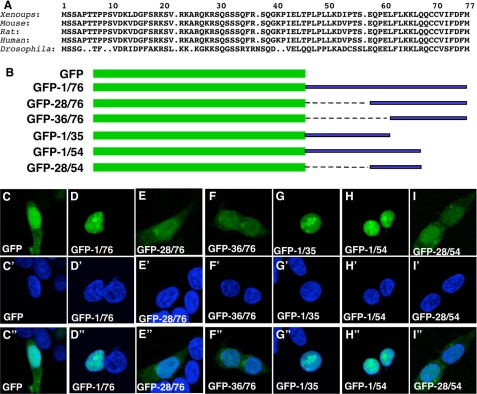
As shown in Fig. 6A, B56ε N-terminal sequences are highly conserved. We fused the N-terminal 76 amino acid residues of Xenopus B56ε-fl and various deletions to the C terminus of GFP (Fig. 6B). When expressed in HEK 293 cells, GFP-1/76 (Fig. 6D), GFP-1/35 (Fig. 6G), and GFP-1/54 (Fig. 6H) were restricted to the nuclei, whereas GFP (Fig. 6C), GFP-28/76 (Fig. 6E), GFP-36/76 (Fig. 6F), and GFP-28/54 (Fig. 6I) were detected in both the cytoplasm and nucleus. Identical results were obtained when the above constructs were transfected into MB-MDA-231 cells (data not shown). These results indicate that the N terminus of B56ε-fl contains an NLS.
FIGURE 6.
Identification of an NLS from the N terminus of B56ε-fl. A, alignment of N-terminal sequences of vertebrate B56ε-fl and Drosophila Wdb. Only the sequence between the first and the second Met was selected for alignment. B, schematic drawing of constructs used in C. From C-I″ are subcellular localizations of GFP (C, C′, and C″), GFP-1/76 (D, D′, and D″), GFP-28/76 (E, E′, and E″), GFP-36/76 (F, F′, and F″), GFP-1/35 (G, G′, and G″), GFP-1/54 (H, H′, and H″), and GFP-28/54 (I, I′, and I″). HEK 293 cells were transfected with the above constructs, stained with DAPI, and subjected to confocal microscopy analysis. From C-I are images for GFP-tagged proteins. From C′-I′ are DAPI stainings showing the nuclei. From C″-I″ are merged images. Identical subcellular distribution patterns were obtained when the above constructs were transfected into MB-MDA-231 cells (data not shown).
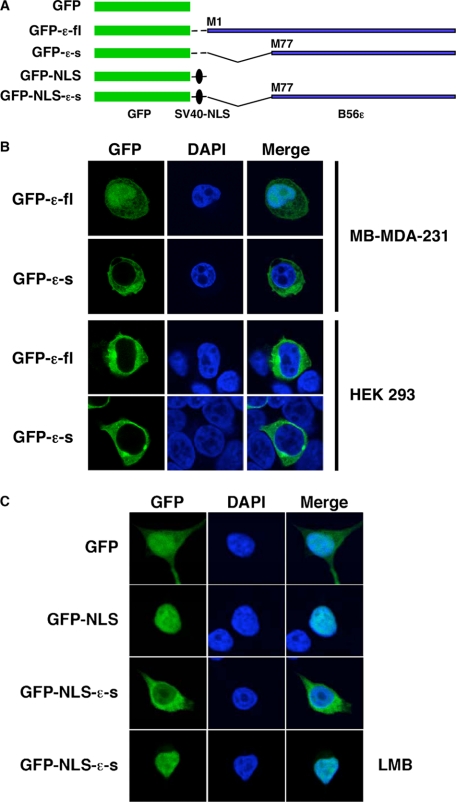
Subcellular Localization of B56ε-fl and B56ε-s—It has been reported that B56ε was located in the cytoplasm of CV-1 cells (46). Promoted by the observation that the N terminus of B56ε-fl contains an NLS, we examined the subcellular localization of B56ε translation isoforms. N-terminal GFP-tagged B56ε-fl (GFP-B56ε-fl) and B56ε-s (GFP-B56ε-s) were transfected into MB-MDA-231 breast cancer cells and HEK 293 cells. The subcellular localization of GFP-B56ε-fl and GFP-B56ε-s was analyzed by confocal microscopy. We found that both GFP-B56ε-fl and GFP-B56ε-s were localized in the cytoplasm in HEK 293 cells. In MB-MDA-231 cells, however, GFP-B56ε-fl was detected in both the nucleus and cytoplasm, whereas GFP-B56ε-s was mainly restricted to the cytoplasm (Fig. 7B). Thus, the N-terminal NLS causes nuclear localization of B56ε-fl in a cell type-specific manner.
FIGURE 7.
Subcellular localization of B56ε isoforms. A, schematic drawing of constructs used in B and C. B, N-terminal GFP-tagged B56ε-fl and N-terminal GFP tagged B56ε-s were transfected into MB-MDA-231 breast cancer and HEK 293 cells. Transfected cells were stained with DAPI and subjected to confocal microscopy analysis. Columns from left to right are GFP-tagged proteins, DAPI staining, and merged images. GFP-B56ε-s is restricted to the cytoplasm in both cell lines. In MB-MDA-231, GFP-B56ε-fl was detected in both cytoplasm and nucleus. In HEK 293 cells, it is mainly localized in the cytoplasm. C, subcellular localization of GFP, GFP-NLS, and GFP-NLS-ε-s in HEK 293 cells. The bottom panels are GFP-NLS-ε-s in cells treated with leptomycin B (LMB). Columns from left to right are the distribution of GFP or GFP-tagged proteins, DAPI staining, and merged images. Note that GFP-NLS was nucleus-localized, whereas GFP-NLS-ε-s was detected in both nucleus and cytoplasm. When treated with leptomycin B, GFP-NLS-ε-s became nucleus-localized.
This observation is striking and raises the possibility that the C-terminal sequence of B56ε may contain an NES or a cytoplasmic retention signal that is capable of antagonizing the function of the N-terminal NLS. To address this, we constructed a GFP-NLS-B56ε-s by fusing B56ε-s to the C terminus of GFP-NLS. GFP-NLS contains an NLS derived from the SV40 large T antigen (Fig. 7A). As expected, GFP-NLS was restricted to the nuclei in HEK 293 cells. When GFP-NLS-B56ε-s was analyzed, we observed a large amount of GFP-NLS-B56ε-s in the cytoplasm. When GFP-NLS-B56ε-s-overexpressing cells were treated with the nuclear export inhibitor leptomycin B, GFP-NLS-B56ε-s became nucleus-localized (Fig. 7C). Identical results were obtained when GFP-NLS-B56ε-s was tested in MB-MDA-231 cells (data not shown). This indicates that the C-terminal sequence of B56ε contains an NES. It appears that the subcellular localization of B56ε-fl is controlled by multiple factors, i.e. the N-terminal NLS and the C-terminal NES. The N-terminal NLS is required, but is not sufficient, for nuclear localization of B56ε-fl.
DISCUSSION
PP2A is involved in numerous biological processes. The regulatory subunits play important roles in controlling the activity, localization, and substrate specificity of PP2A. Not surprisingly, many PP2A regulatory subunits are regulated at post-transcriptional levels. Within the B56 superfamily, B56γ (10, 32, 33) is alternatively spliced. In this study, we report that B56ε undergoes alternative translation. To the best of our knowledge, this is the first time that a regulatory subunit of PP2A has been shown to be alternatively translated. B56ε is involved in multiple signaling transduction pathways and cellular events (13, 15-19, 24, 26, 27). These data provide an important clue to fully understand the mechanisms through which B56ε functions in many signaling cascades and cellular events.
Results presented here indicate that B56ε is alternatively translated. Using a site-specific mutagenesis approach, we were able to show that alternative translation of B56ε is initiated at the second AUG, which corresponds to Met77. Like vertebrate B56ε, Drosophila wdb is alternatively translated. Existing evidence suggests that the second AUG of Drosophila wdb may be used as the alternative translation initiation site as well. Hannus et al. (24) identified four alleles of wdb, which carry nonsense mutations. They found that Wdb protein remained detectable in alleles bearing mutations upstream of the second AUG. In contrast, mutations downstream of the second AUG prevented production of Wdb protein completely. It therefore appears that alternative translation of B56ε is evolutionarily conserved.
Interestingly, the expression level of B56ε-s varies in different tissues and cell lines. For example, B56ε-s expression falls below the limit of detection in kidney and HEK 293 cells, which is derived from the kidney. In contrast, the expression level B56ε-s is relatively high in neural tissues. Although this may be caused by regulated usage of translation initiation sites of B56ε, it may also reflect differences in the stabilities of B56ε isoforms in different tissues. Further experiments are needed to distinguish between these possibilities.
We have also addressed the mechanism by which alternative translation of B56ε occurs. First, we show that translation of B56ε-s requires a cap on the 5′ end of mRNA and can be blocked by 4E-BP, which inhibits assembly of ribosomes on the 5′ cap. In addition, we show that the N terminus of B56ε is sufficient for alternative translation when fused to enhanced GFP. We also show that improving the context of the upstream translation initiation site prevents translation of B56ε-s. Finally, MO2, a morpholino that binds to the sequence between the first and the second AUG of B56ε, blocks translation of B56ε-s without affecting B56ε-fl. These observations indicate that alternative translation of B56ε is cap-dependent.
The observation that B56ε is alternatively translated immediately raises two important questions. 1) Does the internal translation initiation variant, B56ε-s, have any biological functions? 2) What is the functional difference between two B56ε isoforms? To address whether B56ε-s is biologically active, we performed loss-of-function studies. Taking advantage of MO2, which selectively interferes with the translation of B56ε-s, we were able to show that B56ε-s is required for embryonic development. Depletion of B56ε-s impairs the midbrain/hindbrain boundary formation, causes shorter anterior/posterior axis, and reduces the size of the head. Our data argue that the defective midbrain/hindbrain boundary formation is caused by impaired canonical Wnt signaling in B56ε-s-depleted embryos. In agreement with this view, we observed that depletion of B56ε-s blocked Wnt1-induced β-catenin stabilization and slug expression in animal cap assays. Importantly, overexpression of β-catenin efficiently rescued en2 expression in B56ε-s-depleted embryos. In the case of eye development, we found that eye induction and eye field separation processes occurred in B56ε-s-depleted embryos. However, eye development was clearly affected during the later stage of development. At the early tadpole stage, B56ε-s-depleted embryos exhibited decreased rx expression. Later on, the size of the eye was dramatically reduced. This is reminiscent of the remarkable phenotypes in frizzled5-depleted embryos (47). It will be of interest to test whether B56ε-s regulates eye development through its effect on the canonical Wnt pathway. Nevertheless, our results demonstrate that B56ε-s plays essential roles during embryonic development.
Currently, it remains largely unclear whether B56ε isoforms have distinct functions. GFP fusion experiments show that the first 35 amino acid residues of B56ε-fl contain an NLS, whereas the C-terminal sequence of B56ε contains an NES. Likely due to a lack of the N-terminal NLS, B56ε-s is restricted to the cytoplasm. In contrast, the subcellular localization of B56ε-fl, which contains both NLS and NES, is cell type-specific. In MB-MDA-231 cells, B56ε-fl can be detected in both cytoplasm and nucleus, whereas in HEK 293 cells, it is located in the cytoplasm. Thus, the subcellular localization of B56ε-fl can be influenced by multiple factors. The N-terminal NLS appears to be required, but not sufficient, for the nuclear localization of B56ε-fl. Interestingly, the N terminus of B56ε-fl, which contains the NLS, is highly rich in Ser/Thr. These Ser/Thr residues are conserved from Drosophila to human. This raises the possibility that the activity of the N-terminal NLS may be differentially regulated by some protein kinases/phosphatases in different cells. Further experiments are needed to determine whether the activity of the N-terminal NLS is indeed regulated and how this NLS contributes to the function of B56ε-fl.
Although it has been clear that B56ε-s regulates Wnt signaling during midbrain/hindbrain boundary formation, we cannot rule out the possibility that B56ε-fl has an involvement in these processes as well. On the other hand, although depletion of B56ε-s has no effect on eye induction and subsequent eye field separation, this may be due to the functional redundancy between B56ε-fl and B56ε-s. Clearly, future work will be needed to clarify the functional difference between B56ε alternative translation isoforms. Nevertheless, results presented here uncover a novel mechanism for post-transcriptional regulation of B56ε.
Acknowledgments
We thank Drs. Nahum Sonenberg, David Virshup, Amita Sehgal, John Barnard, Jiayuh Li, Christopher Phiel, and Kai Ge for reagents; Dr. Christopher Phiel for sharing tissue culture equipments; Drs. Peter Klein and Christopher Phiel for helpful discussions; and Drs. John Barnard, Heithem El-Hodiri, and Scott Harper for reading the manuscript.
This work was supported by a Nationwide Children's Hospital internal startup fund (to J. Y.) and by The Madison Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IRES, internal ribosome entry site; PP2A, protein phosphatase 2A; NLS, nuclear localization signal; NES, nuclear export signal; RT-PCR, reverse transcriptase-PCR; GFP, green fluorescent protein; HA, hemagglutinin; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Cameron, R. A., Mahairas, G., Rast, J. P., Martinez, P., Biondi, T. R., Swartzell, S., Wallace, J. C., Poustka, A. J., Livingston, B. T., Wray, G. A., Ettensohn, C. A., Lehrach, H., Britten, R. J., Davidson, E. H., and Hood, L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9514-9518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennisi, E. (2003) Science 300 1484. [DOI] [PubMed] [Google Scholar]
- 3.Kozak, M. (2002) Gene (Amst.) 299 1-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak, M. (1978) Cell 15 1109-1123 [DOI] [PubMed] [Google Scholar]
- 5.Mayer, C., Kohrer, C., Kenny, E., Prusko, C., and RajBhandary, U. L. (2003) Biochemistry 42 4787-4799 [DOI] [PubMed] [Google Scholar]
- 6.Kozak, M. (2005) Gene (Amst.) 361 13-37 [DOI] [PubMed] [Google Scholar]
- 7.Janssens, V., and Goris, J. (2001) Biochem. J. 353 417-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mumby, M. (2007) Cell 130 21-24 [DOI] [PubMed] [Google Scholar]
- 9.Eichhorn, P. J., Creyghton, M. P., and Bernards, R. (2008) Biochim. Biophys. Acta 1795 1-15 [DOI] [PubMed] [Google Scholar]
- 10.McCright, B., and Virshup, D. M. (1995) J. Biol. Chem. 270 26123-26128 [DOI] [PubMed] [Google Scholar]
- 11.Seeling, J. M., Miller, J. R., Gil, R., Moon, R. T., White, R., and Virshup, D. M. (1999) Science 283 2089-2091 [DOI] [PubMed] [Google Scholar]
- 12.Gao, Z. H., Seeling, J. M., Hill, V., Yochum, A., and Virshup, D. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1182-1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, X., Yost, H. J., Virshup, D. M., and Seeling, J. M. (2001) EMBO J. 20 4122-4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratcliffe, M. J., Itoh, K., and Sokol, S. Y. (2000) J. Biol. Chem. 275 35680-35683 [DOI] [PubMed] [Google Scholar]
- 15.Yang, J., Wu, J., Tan, C., and Klein, P. S. (2003) Development (Camb.) 130 5569-5578 [DOI] [PubMed] [Google Scholar]
- 16.Nybakken, K., Vokes, S. A., Lin, T. Y., McMahon, A. P., and Perrimon, N. (2005) Nat. Genet. 37 1323-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rorick, A. M., Mei, W., Liette, N. L., Phiel, C., El-Hodiri, H. M., and Yang, J. (2007) Dev. Biol. 302 477-493 [DOI] [PubMed] [Google Scholar]
- 18.Jia, H., Liu, Y., Yan, W., and Jia, J. (2009) Development (Camb.) 136 307-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverstein, A. M., Barrow, C. A., Davis, A. J., and Mumby, M. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4221-4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letourneux, C., Rocher, G., and Porteu, F. (2006) EMBO J. 25 727-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, W., Silverstein, A. M., Shu, H., Martinez, B., and Mumby, M. C. (2007) Mol. Cell. Proteomics 6 319-332 [DOI] [PubMed] [Google Scholar]
- 22.Rocher, G., Letourneux, C., Lenormand, P., and Porteu, F. (2007) J. Biol. Chem. 282 5468-5477 [DOI] [PubMed] [Google Scholar]
- 23.Van Kanegan, M. J., Adams, D. G., Wadzinski, B. E., and Strack, S. (2005) J. Biol. Chem. 280 36029-36036 [DOI] [PubMed] [Google Scholar]
- 24.Hannus, M., Feiguin, F., Heisenberg, C. P., and Eaton, S. (2002) Development (Camb.) 129 3493-3503 [DOI] [PubMed] [Google Scholar]
- 25.Li, X., Scuderi, A., Letsou, A., and Virshup, D. M. (2002) Mol. Cell. Biol. 22 3674-3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, F., Archambault, V., Kar, A., Lio, P., D'Avino, P. P., Sinka, R., Lilley, K., Laue, E. D., Deak, P., Capalbo, L., and Glover, D. M. (2007) Curr. Biol. 17 293-303 [DOI] [PubMed] [Google Scholar]
- 27.Holland, A. J., Bottger, F., Stemmann, O., and Taylor, S. S. (2007) J. Biol. Chem. 282 24623-24632 [DOI] [PubMed] [Google Scholar]
- 28.Sive, H., Grainger, R., and Harland, R. (2000) Early Development of Xenopus laevis: A Laboratory Manual, 1st Ed., pp. 91-141, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 29.Sathyanarayanan, S., Zheng, X., Xiao, R., and Sehgal, A. (2004) Cell 116 603-615 [DOI] [PubMed] [Google Scholar]
- 30.Deardorff, M. A., Tan, C., Saint-Jeannet, J. P., and Klein, P. S. (2001) Development (Camb.) 128 3655-3663 [DOI] [PubMed] [Google Scholar]
- 31.Deardorff, M. A., Tan, C., Conrad, L. J., and Klein, P. S. (1998) Development (Camb.) 125 2687-2700 [DOI] [PubMed] [Google Scholar]
- 32.Csortos, C., Zolnierowicz, S., Bako, E., Durbin, S. D., and DePaoli-Roach, A. A. (1996) J. Biol. Chem. 271 2578-2588 [DOI] [PubMed] [Google Scholar]
- 33.Muneer, S., Ramalingam, V., Wyatt, R., Schultz, R. A., Minna, J. D., and Kamibayashi, C. (2002) Genomics 79 344-348 [DOI] [PubMed] [Google Scholar]
- 34.Gingras, A. C., Raught, B., Gygi, S. P., Niedzwiecka, A., Miron, M., Burley, S. K., Polakiewicz, R. D., Wyslouch-Cieszynska, A., Aebersold, R., and Sonenberg, N. (2001) Genes Dev. 15 2852-2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moody, S. A. (1987) Dev. Biol. 119 560-578 [DOI] [PubMed] [Google Scholar]
- 36.Lamb, T. M., Knecht, A. K., Smith, W. C., Stachel, S. E., Economides, A. N., Stahl, N., Yancopolous, G. D., and Harland, R. M. (1993) Science 262 713-718 [DOI] [PubMed] [Google Scholar]
- 37.Hemmati-Brivanlou, A., de la Torre, J. R., Holt, C., and Harland, R. M. (1991) Development (Camb.) 111 715-724 [DOI] [PubMed] [Google Scholar]
- 38.Christian, J. L., Gavin, B. J., McMahon, A. P., and Moon, R. T. (1991) Dev. Biol. 143 230-234 [DOI] [PubMed] [Google Scholar]
- 39.Bradley, L. C., Snape, A., Bhatt, S., and Wilkinson, D. G. (1993) Mech. Dev. 40 73-84 [DOI] [PubMed] [Google Scholar]
- 40.Sharpe, C. R., Fritz, A., De Robertis, E. M., and Gurdon, J. B. (1987) Cell 50 749-758 [DOI] [PubMed] [Google Scholar]
- 41.Mathers, P. H., Grinberg, A., Mahon, K. A., and Jamrich, M. (1997) Nature 387 603-607 [DOI] [PubMed] [Google Scholar]
- 42.Joyner, A. L. (1996) Trends Genet. 12 15-20 [DOI] [PubMed] [Google Scholar]
- 43.McGrew, L. L., Takemaru, K., Bates, R., and Moon, R. T. (1999) Mech. Dev. 87 21-32 [DOI] [PubMed] [Google Scholar]
- 44.Saint-Jeannet, J. P., He, X., Varmus, H. E., and Dawid, I. B. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13713-13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He, X., Semenov, M., Tamai, K., and Zeng, X. (2004) Development (Camb.) 131 1663-1677 [DOI] [PubMed] [Google Scholar]
- 46.McCright, B., Rivers, A. M., Audlin, S., and Virshup, D. M. (1996) J. Biol. Chem. 271 22081-22089 [DOI] [PubMed] [Google Scholar]
- 47.Van Raay, T. J., Moore, K. B., Iordanova, I., Steele, M., Jamrich, M., Harris, W. A., and Vetter, M. L. (2005) Neuron 46 23-36 [DOI] [PubMed] [Google Scholar]