Abstract
Many glycoside hydrolases possess carbohydrate-binding modules (CBMs) that help target these enzymes to appropriate substrates and increase their catalytic efficiency. The Vibrio cholerae sialidase contains two CBMs, one of which is designated as a family CBM40 module and has been shown through structural and calorimetry studies to recognize the α-anomer of sialic acid with a KD of ∼30 μm at 37 °C. The affinity of this V. cholerae CBM40 module for sialic acid is one of the highest reported for recognition of a monosaccharide by a CBM. As Nature often increases a weak substrate affinity through multivalency, we have explored the potential of developing reagents with an increased affinity for sialic acid receptors through linking CBM40 modules together. The V. cholerae CBM40 was subcloned and crystallized in the presence of sialyllactose confirming its ability to recognize sialic acid. Calorimetry revealed that this CBM40 demonstrated specificity to α(2,3)-, α(2,6)-, and α(2,8)-linked sialosides. Polypeptides containing up to four CBM40 modules in tandem were created to determine if an increase in affinity to sialic acid could be achieved through an avidity effect. Using SPR and a multivalent α(2,3)-sialyllactose ligand, we show that increasing the number of linked modules does increase the affinity for sialic acid. The four-CBM40 module protein has a 700- to 1500-fold increase in affinity compared with the single-CBM40 module. Varying the linker length of amino acids between each CBM40 module had little effect on the binding of these polypeptides. Finally, fluorescence-activated cell sorting analysis demonstrated that a green fluorescent protein fused to three CBM40 modules bound to subpopulations of human leukocytes. These studies lay the foundation for creating high affinity, multivalent CBMs that could have broad application in glycobiology.
Sialic acids are a family of carbohydrates with a nine-carbon backbone and are typically located at the distal end of glycan chains. There are over 50 natural derivatives within this family, and structural diversity is achieved through modifications such as acetylation, glycosylation, lactonization, and methylation at C4, C5, C7, C8, and C9, the most common molecule being N-acetylneuraminic acid (1). Sialic acids are found linked α(2,3) or α(2,6) to Gal and GalNAc, or α(2,8) or α(2,9) to another sialic acid. Such is their structural diversity that they are a fundamental component of many biological processes such as cell recognition, adhesion, defense, activation of cellular pathways, sialylation and desialylation events, and cell surface modification. Furthermore, the presence of sialic acids at the termini of glycoconjugates can be exploited by pathogens to allow entry into host cells (2).
Sialic acid recognition is mediated via lectins or lectin-like molecules and their corresponding receptors (3). In most cases, sialic acid binding lectins, which also include several viral glycoproteins and bacterial toxins (2, 4), as well as the mammalian lectin superfamilies such as the siglecs (5) and selectins (6), bind to their receptor with relatively high affinity due to the multivalent nature of these molecules, thus alleviating the low intrinsic affinity that most protein-carbohydrate interactions are associated with (7, 8). Generally, association constants (Ka) for the binding of monovalent and divalent sialosides by such lectins can reach 104 m-1. However, by virtue of their multivalency, some sialic acid binding lectins can interact with multivalent cell surface glycans to achieve affinities reaching 109 m-1 by an avidity effect. These enhanced affinities have been shown in part to be due to improved structural packing of proteins promoted by ligand binding, associated with favorable binding energetics (9-11). One of the best studied multivalent lectin-sialic acid interactions is the influenza virus trimeric hemagglutinin, which can achieve affinities up to 108 m-1 compared with around 4 × 102 m-1 when one or both of the entities are not in a multivalent state (12).
Sialidases, or neuraminidases, catalyze the hydrolysis of sialic acids from a variety of glycoconjugates and are often modular enzymes, containing accessory modules attached to the catalytic core of the protein. Some of these modules have been identified as carbohydrate binding modules (CBMs).3 CBMs are found widely in glycoside hydrolases and are discrete, non-catalytic modules that primarily exist to target the parent enzyme to its substrate for efficient hydrolysis by increasing the concentration of the enzyme at the substrate surface (13). The modules can be single, tandem, or in multiples within the glycosyl hydrolase architecture. Studies have shown that they can bind to their specific glycans independently when isolated from the parent molecule and can behave in a cooperative manner when isolated in tandem (14, 15). Currently, CBMs are grouped into 52 families based upon primary sequence similarity. Subtle differences in the structures of CBMs can lead to diverse ligand specificity, which make CBMs an attractive system for elucidating protein-carbohydrate mechanisms.
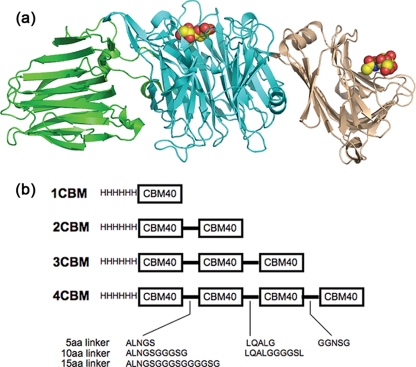
The sialidase from Vibrio cholerae possesses two CBMs that flank the central catalytic domain (15) (Fig. 1a). One CBM, whose ligand is unknown, is inserted into the catalytic β-pro-peller domain. We previously reported from a structural study on the complete V. cholerae sialidase that the N-terminal CBM (classified as a family 40 CBM) recognizes sialic acid with a relatively high affinity, Kd ∼ 30 μm, the highest reported affinity for a sialic acid-binding protein that recognizes a single sialic acid moiety (16). The N-terminal CBM from Clostridium perfringens NanJ sialidase has also been characterized structurally as a CBM40-recognizing sialic acid, but in contrast to the V. cholerae CBM40, displays high millimolar affinity toward its glycan (17).
FIGURE 1.
a, schematic drawing of the V. cholerae sialidase showing the central catalytic domain flanked by the lectin-like CBMs. The sialic acid recognizing CBM, CBM40, is on the right. b, schematic view of the constructs made in this study.
Inspired by Nature's multivalency approach to gaining increased binding affinity through avidity, we have engineered polypeptides containing tandem repeats of the V. cholerae CBM40. Here we report the subcloning of the V. cholerae CBM40 and the determination of its crystal structure in complex with 3′-sialyllactose (SL). Calorimetry was used to monitor the binding of a variety of sialic acid-containing substrates to the single isolated CBM40. Constructs containing two, three, or four CBM40 modules were then made with varying linker lengths between the modules (Fig. 1b). Calorimetry was used to measure the binding of 3′-sialyllactose to the engineered two, three, and four CBM40 polypeptides to show that the affinity was unaffected by the tandem repeat of the CBM40 unit. Also, using surface plasmon resonance, we demonstrate that a large gain in affinity is achieved through an avidity effect, with the four CBM40 polypeptide gaining up to three orders of increased binding compared with the single CBM40. We also show that, by adding a GFP molecule to one of the multidomain CBM40 constructs, the binding to human leukocytes can be assessed for cell surface sialylation, as well as employing it in a glycan microarray screen. These multivalent polymers could be further developed as tools to deliver other proteins to sialic acid-rich cell surfaces. Other CBMs could be repeated in a similar way to create other tools for use in glycobiology.
EXPERIMENTAL PROCEDURES
Recombinant DNA Techniques—The DNA fragment encoding the family 40 CBM of V. cholerae sialidase/neuraminidase, residues 25-216, was amplified by PCR from the construct pET30b+ containing the nanH gene (16) using oligonucleotide primer pair 1F and 1R based on the sequence outline in Table 1. The amplified DNA fragment of 573 bp was digested with NcoI and XhoI and cloned into the pEHISTEV vector (an engineered variant of pET30 with an N-terminal His6 tag that is cleaved by tobacco etch virus (TEV) protease) (18), digested with the same enzymes and used to transform Escherichia coli DH5a. The construct, p1CBM, was verified by DNA sequencing (University of Dundee Sequencing Service, UK).
TABLE 1.
Oligonucleotide primers used to amplify DNA fragments
| Primer | Oligonucleotide sequence |
|---|---|
| F | CGTCCCATGGCACTTTTTGACTATAACGC (NcoI) |
| 1R | CCGGCTCGAGCTAGTCGCCTTGAATTTCAAAC (XhoI) |
| 2F(5) | CGTCGGATCCATGGCACTTTTTGAC (BamHI) |
| 2R(5) | GCACGGATCCGTTCAGGGCGTCGCCTTGAATTT (BamHI) |
| 2F(10) | GCCTGGATCCGGTATGGCACTTTTTGACTATAAC (BamHI) |
| 2R(10) | GACCGGATCCACCTCCTGATCCGTTCAGGGCGTCGCC (BamHI) |
| 2R(15) | GACCGGATCCACCTCCACCTGATCCACCTCCTGATCC (BamHI) |
| 3F(5) | CTGCAAGCTTTGGGAATGGCACTTTTTGAC (HindIII) |
| 3R(5) | GCACTTCCAAAGCTTGCAGGTCGCCTTGAATTTC (HindIII) |
| 3F(10) | GGTGGAAGCTTGATGGCACTTTTTGACTATAAC (HindIII) |
| 3R(10) | GTCCAAGCTTCCACCTCCTCCCAATGCTTGCAGGTCGCC (HindIII) |
| 4F(5) | GGTAGGGAATTCGGGAATGGCACTTTTTGACTATAAC (EcoRI) |
| 4R(5) | GCACTCCCGAATTCCCTCCGTCGCCTTGAATTTC (EcoRI) |
The DNA fragment, encoding the V. cholerae CBM40, was modified at the 5′- and 3′-termini to incorporate different restriction endonuclease sites through the use of different primer pairs. This was performed to allow ligation of individual copies of the DNA fragment to generate 2, 3, and 4 copies in tandem (Fig. 1b). Moreover, the primer pairs allowed the insertion of 5, 10, or 15 codons, to represent the length of amino acids linking the individual modules. All these modifications were achieved by PCR amplification using different primer pairs outlined in Table 1 and p1CBM as template. The resulting fragments were cloned into an appropriately digested pEHISTEV vector until the desired number of modules was achieved. These were labeled p2CBM(5), p2CBM(10), p2CBM(15), p3CBM(5), and p3CBM(10), representing 2 and 3 repeating sialic acid-binding domains with the number of amino acids in the linker in parenthesis, respectively. For p4CBM(5), HindIII-EcoRI-modified and EcoRI-XhoI-modified DNA fragments were initially cloned into pET17b digested with the appropriate enzymes before assembling the final gene in pEHISTEV. For the construction of a GFP-fused 3CBM fragment (GFP-3CBM), the 3CBM fragment was digested with NcoI-XhoI from p3CBM(5) and inserted into a similarly digested pEHISTEV vector containing the gene encoding the enhanced GFP downstream of the N-terminal histidine tag (18). All constructs were propagated in E. coli DH5a. Positive clones were verified by DNA sequencing before transforming expression hosts E. coli BL21(DE3) (1CBM, 2CBM(5), 3CBM(5), 3CBM(10), and 4CBM(5)) and E. coli BL21(DE3) Gold (2CBM(10) and 2CBM(15)) for protein production.
Protein Production and Purification—All constructs were grown and expressed as described for V. cholerae sialidase/neuraminidase (16). Briefly, 1-liter aliquots of Luria-Bertani broth media containing 30 μg/ml kanamycin were inoculated with single colonies and grown at 37 °C until the optical density of the culture reached 0.4-0.6 at 600 nm. Cells were subjected to heat shock for 20 min in a 42 °C water bath before being cooled down to 25 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and left to shake overnight at the same temperature. Cells were harvested by centrifugation (8000 × g, 4 °C), and pellets were frozen at -20 °C, until required.
All polypeptides were purified by nickel affinity chromatography as previously described (16). Samples were analyzed using SDS-PAGE and partially purified polypeptides, with the exception of the GFP-3CBM, were dialyzed into TEV protease cleavage buffer (phosphate-buffered saline, 0.3 m NaCl, 1 mm dithiothreitol, 0.5 mm EDTA, 20 mm imidazole) and digested overnight with TEV protease. Each polypeptide was then further dialyzed with phosphate-buffered saline, 0.3 m NaCl, 10 mm imidazole buffer before a second round of purification on a nickel-charged column to remove undigested His-tagged polypeptides. All polypeptide samples were dialyzed extensively in 10 mm HEPES buffer, pH 7.4, 0.15 m NaCl and further purified using size-exclusion chromatography, which was performed in the same buffer on a HiPrep 16/60 Sephacryl S200HR column. Peak fractions containing purified CBM40 polypeptides were pooled and concentrated before use.
Crystallization and Structure Determination—Crystals of the single V. cholerae CBM40 were obtained in 0.1 m MOPS, pH 6.5, 1.1 m lithium sulfate, 0.6 m ammonium sulfate as precipitant, using the sitting drop method. Crystals were cryoprotected by transferring crystals first into 10% (v/v) glycerol-precipitant mix before leaving in 20% (v/v) glycerol precipitant. Diffraction data extending to 2.5 Å were collected on beamline BM-14 of the European Synchrotron Radiation Facility. The structure was solved using the molecular replacement program PHASER (19), which found three CBM40 monomers in the asymmetric unit. Refinement was carried out using REF-MAC (20) with COOT being used for model building (21). Data collection and refinement statistics are shown in Table 2. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession number 2w68.
TABLE 2.
X-ray data collection and refinement statistics
Numbers in parentheses refer to the highest resolution shell.
| Data collection | |
| Space group | C2221 |
| Unit cell edges (Å) | a = 138.6, b = 197.6, c = 83.0 |
| X-ray source | ESRF BM-14 |
| Resolution range (Å) | 30-2.5 (2.57-2.50) |
| No. of unique observations | 37,806 |
| Completeness (%) | 95.4 (88.6) |
| Redundancy | 4.9 (3.2) |
| Rmergea | 0.064 (0.414) |
| <I/σI> | 15.8 (2.7) |
| Refinement | |
| No. of reflections work/test | 35,893/1,911 |
| No. of protein atoms | 4,433 |
| No. of ligand atoms | 129 |
| No. of waters | 137 |
| Average B-factors (Å2) | |
| Protein monomers A/B/C | 42.4/43.9/50.1 |
| 3′SL ligands/waters | 66.7/43.5 |
| Rcrystb | 0.224 |
| Rfree | 0.263 |
| Root mean square deviation bond distance (Å) | 0.017 |
| Root mean square deviation bond angle (°) | 1.716 |
Rmerge = ∑hkl ∑i|Ihkl, i - 〈Ihkl〉|/∑hkl〈Ihkl〉.
Rcryst and Rfree = (∑||Fo| - |Fc||)/(∑|Fo|).
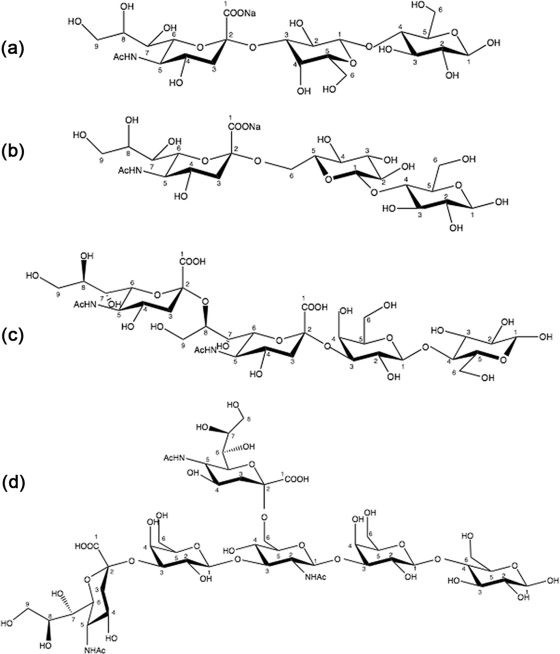
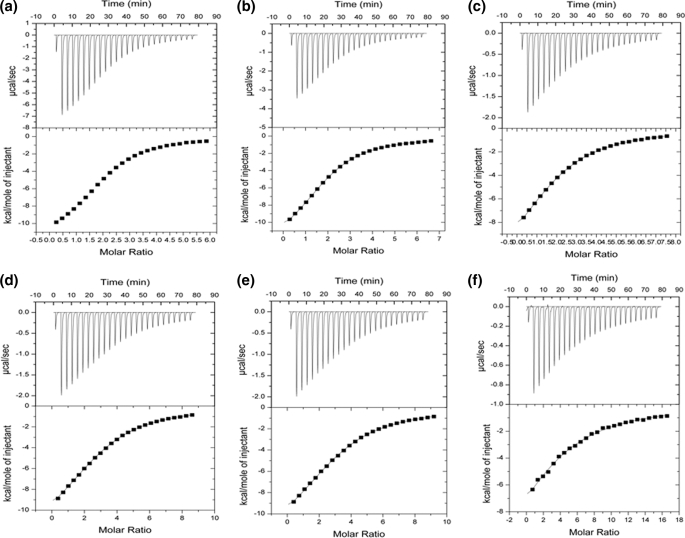
Isothermal Titration Calorimetry—ITC experiments were performed on a VP-ITC microcalorimeter from MicroCal Inc. (Northampton, MA) with a cell volume of 1.4 ml. Unless stated otherwise, all ITC titrations were performed at 25 °C in 10 mm HEPES buffer, pH 7.4, containing 0.15 m NaCl. For the characterization of the isolated CBM40 (1CBM), the following ligands were used: 3′-sialyllactose (3′SL), 6′sialyllactose, (6′SL), and disialyllacto-N-tetraose (DSLNT), purchased from Dextra Labs (Reading, UK) and disialyllactose (DSL) from Sigma-Aldrich (Fig. 2). The lyophilized sialoside ligands were resuspended in degassed, filtered buffer that was used for the dialysis of the peptide construct. For all other polypeptide constructs, 3′SL was used as the ligand throughout. Protein concentrations were determined at A280, using calculated molar extinction coefficients for 1CBM (38410 m-1 cm-1), 2CBM constructs (75860 m -1 cm-1), 3CBM constructs (113790 m-1 cm-1), and 4CBM(5) (151720 m-1 cm-1), respectively. The concentrations of CBM40 polypeptides were 0.007-0.084 mm, and the sialosides were 0.45-2.14 mm. Aliquots of sialosides (10 μl, unless stated otherwise) were titrated into each polypeptide solution. The heats of dilution were subtracted from binding isotherm data before data were fitted by means of a nonlinear regression analysis using a one-binding site model from MicroCal Origin software.
FIGURE 2.
Sialosides used in the ITC study. a, 3′SL; b, 6′SL; c, DSL; and d, DSLNT.
Surface Plasmon Resonance—Binding kinetics were determined by SPR using a BIACORE 3000 biosensor instrument (GE Biosystems). Prior to use, a streptavidin-coated biosensor chip was docked into the instrument and preconditioned with three consecutive 1-min injections of 1 m NaCl in 50 mm NaOH. Multivalent, biotinylated 3′SL-PAA (Glycotech) was diluted to 1 μg/ml in HBS-P (10 mm HEPES, pH 7.4, 0.15 m NaCl, and 0.005% surfactant P20) before being injected over three of the four flow cells in the chip. Typical immobilization levels of the ligand for each cell were ∼500 response units. A reference surface was also prepared for subtraction of bulk effects and nonspecific interactions with streptavidin. The running buffer consisted of 10 mm HEPES, pH 7.4, at a flow rate of 100 μl/min.
Interaction analysis for each peptide construct with immobilized 3′SL was performed in running buffer at 15, 25, and 35 °C. Purified peptide constructs were diluted into HBS-P to give a series of concentrations for 1CBM constructs, 0.625 μm, 1.25 μm, 2.5 μm, 5 μm, and 10 μm; 2CBM constructs, 20 nm, 125 nm 250 nm, 500 nm, and 1000 nm; 3CBM constructs, 1 nm, 5 nm, 20 nm, 62.5 nm, and 125 nm; and for 4CBM constructs, 0.18 nm, 0.5 nm, 1.6 nm, 5 nm, and 15 nm. All analytes were injected over the flow cell surface at 30 μl/min. The dissociation of analyte from the surface was achieved in running buffer at the same flow rate for 3-5 min. Surfaces were regenerated with two consecutive 30-s injections of 10 mm glycine-HCl, pH 2.5, at 30 μl/min. The affinity, as described by the equilibrium dissociation constant (KD) was determined globally by fitting to the kinetic simultaneous ka/kd model, assuming Langmuir (1:1) binding, using BIAevaluation software (BIAcore).
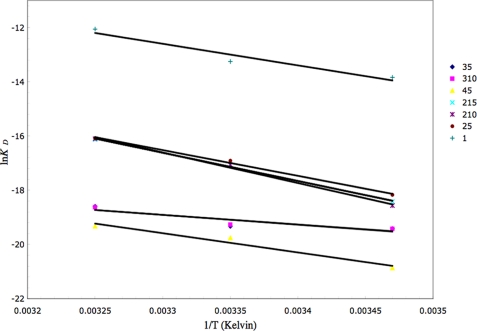
The free energy change of the interaction of the different CBM polypeptide constructs with biotinylated 3′SL-PAA was determined by using the equilibrium dissociation constants provided by the ratios of the kinetic rate constants for each temperature. van't Hoff plots of lnKD versus 1/T yielded values for ΔH°/R from the slope and -ΔS°/R from the y intercept.
Flow Cytometry (FACS) of GFP-3CBM Binding to Human Leukocytes—Whole human blood leukocytes were collected from healthy adult volunteer donors and prepared as described from standard protocols (22). Leukocytes were then resuspended in 0.1 m sodium acetate buffer, pH 6.0, 10 mm CaCl2, where one-half of cells was incubated with 0.05 unit/ml of V. cholerae sialidase (Sigma), and the other half with buffer alone, for 1 h at 37 °C. Cells were then washed with RPMI buffer (10% fetal bovine serum, 250 units·ml-1 of penicillin and 250 μg·ml-1 streptomycin), before being aliquoted into a 96-well plate. Cells were washed extensively with phosphate-buffered saline/bovine serum albumin buffer and then incubated on ice with dilutions of GFP-3CBM in the same buffer for 30-45 min. Unbound probe was removed by extensive washing with phosphate-buffered saline/bovine serum albumin. Cells were then analyzed using a FACS LSR flow cytometer with CellQuest software to determine binding of probe to subset populations of leukocytes (BD Biosciences). A minimum of 1 × 104 cells was counted in all analyses. For the exclusion of non-viable cells during FACS analysis, 7-amino-actinomycin D was added to each sample.
Glycan Array—The sugar binding specificity of GFP-3CBM was analyzed by the Consortium for Functional Glycomics using Glycan array v3.1. The GFP-3CBM was assayed on the slide array at 200 μg/ml. The dilution was made in a standard binding buffer (TSM, 1% (w/v) bovine serum albumin and 0.05% (v/v) Tween 20). Protein (70 μl) was incubated on the printed array under a coverslip in a dark humidified chamber for 1 h, before images were read in a PerkinElmer Life Sciences Microscanarray XL4000 scanner.
RESULTS
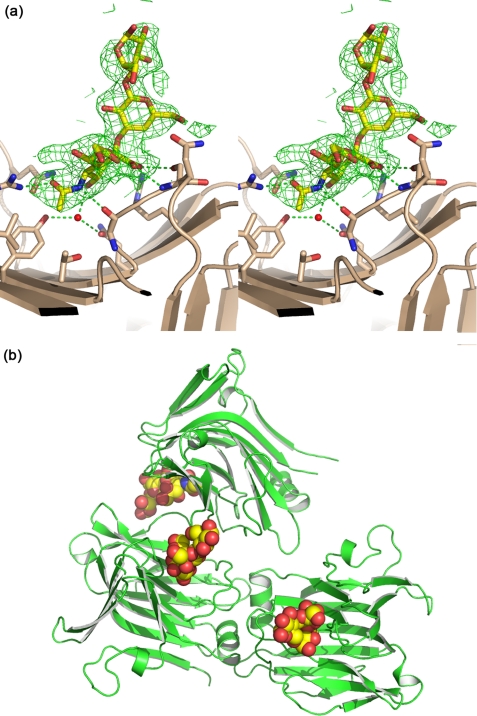
Structure of the Isolated CBM40 Module—The gene fragment encoding the sialic acid binding module CBM40 from the V. cholerae sialidase was subcloned into pEHISTEV and expressed in E. coli to generate 1CBM, which was subsequently purified for binding and structural studies. Initially, 1CBM was expressed insolubly albeit at very high levels, but a heat shock of cultures at 42 °C during log phase of growth improved the solubility of this CBM such that up to 50-70 milligram quantities per liter were produced consistently. Crystals of V. cholerae CBM40 were grown in the presence of 3′SL. The asymmetric unit contained three CBM40 monomers, and each had clear electron density for all three sugar moieties of the ligand (Fig. 3a). Only sialic acid makes interactions with the protein, and these are the same as those made by sialic acid alone in complex with the whole V. cholerae sialidase (16). There are three CBM40 modules in the asymmetric unit of the crystal, with monomers A and B burying an interface of ∼750Å2 (Fig. 3b).
FIGURE 3.
Crystal structure of V. cholerae CBM40 in complex with 3′SL. a, interactions of the ligand with the CBM with hydrogen-bonds drawn as dashed lines. The sigma-a-weighted Fo - Fc electron density map is drawn at 3σ. b, the relationship between the three CBM40 modules in the asymmetric unit of the crystal.
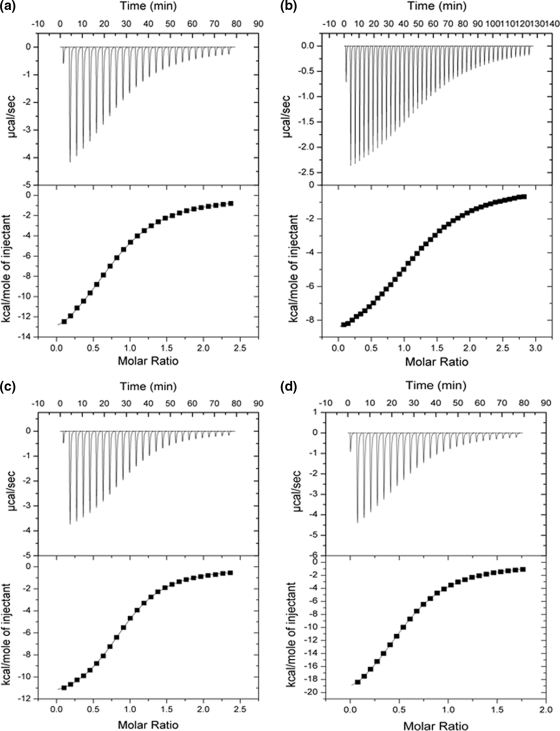
Determination of Sialoside and Linkage Specificity of V. cholerae CBM40—Studies were performed with 1CBM to determine its specificity toward different monovalent and divalent sialosides. Using ITC, the binding constant Ka and changes in enthalpy (ΔH) and entropy (ΔS) were measured directly for each interaction, and stoichiometries (n, number of binding sites) were determined from non-linear least squares fit of the data to the one-site binding model as described above. Data obtained from heat of dilution-corrected binding isotherms demonstrate the preference of 1CBM for α-sialic acids (Fig. 4), exhibiting broad binding specificity to α(2,3)-, α(2,6)-, and α(2,8)-linked sialosides. All sialosides tested displayed similar affinity with dissociation constants, Kd, ranging between 10 and 19 μm (at 25 °C) (Table 3 and Fig. 4). The disialosides, DSLNT and DSL, show 1.5-2 times greater affinity to sialic acid compared with monovalent ligands. DSLNT showed a large change in enthalpy, with a ΔH° value of -24 kcal/mol, compared with DSL (-12.5 kcal/mol). When comparing the stoichiometries for binding DSLNT and DSL, corresponding n values of 0.57 and 0.92 were observed, indicating that, for the DSLNT, two molecules of 1CBM are required to bind to one molecule of DSLNT, whereas for DSL there is a 1:1 interaction. The difference here is due, not just to the number of sialic acid moieties present, but to their position within the sialoside. Structurally, DSLNT is a branched divalent sialoside, unlike DSL, which has two sialic acid moieties linked together in a linear fashion, so that only the terminal sialic acid moiety is recognized (Fig. 2). This result indicates that the ΔH value of DSLNT is approximately the sum of the individual binding enthalpies of 3′SL, 6′SL, and DSL, which have a stoichiometry of one.
FIGURE 4.
ITC isotherms showing the binding of various ligands to the isolated CBM40. a, 3′SL; b, 6′SL; c, DSL; and d, DSLNT.
TABLE 3.
Thermodynamic parameters of interaction of isolated CBM40 from V. cholerae sialidase with different linkage sialosides
| Sialoside | Linkage specificity | n | ΔH | TΔS | ΔG | Ka | Kd |
|---|---|---|---|---|---|---|---|
| kcal/mol | kcal/mol | kcal/mol | 10−4m | μm | |||
| 3′SL | α2,3 | 0.96 ± 0.002 | −16.3 ± 0.05 | −9.8 | −6.5 | 5.48 ± 0.05 | 18 |
| 6′SL | α2,6 | 1.02 ± 0.001 | −9.9 ± 0.002 | −3.5 | −6.4 | 5.16 ± 0.03 | 19 |
| DSL | α2,8 α2,3 | 0.92 ± 0.001 | −12.5 ± 0.003 | −5.7 | −6.8 | 10.2 ± 0.01 | 9.8 |
| DSLNT | α2,6, α2,3 | 0.57 ± 0.001 | −24.0 ± 0.007 | −17.3 | −6.7 | 7.8 ± 0.06 | 13 |
Engineering of Multivalent, Sialic Acid-specific CBM40 Polypeptides—To investigate whether sialic acid binding would occur if identical copies of 1CBM were linked together, copies of the gene encoding CBM40 were tethered together with a DNA linker (representing up to 15 amino acids) to create polypeptides of two, three, and four modules in tandem, designated as 2CBM(5), 2CBM(10), 2CBM(15), 3CBM(5), 3CBM(10), and 4CBM(5), respectively (the number in parenthesis indicates the number of linker amino acids) (Fig. 1b). Expression of these gene constructs was performed in E. coli, and all demonstrated insolubility until cultures were subjected to heat shock as for the isolated 1CBM. After purification with nickel affinity chromatography, SDS-PAGE analysis of all peptide constructs demonstrated monomeric molecular masses of ∼21 kDa, ∼42 kDa, ∼63 kDa, and ∼85 kDa for 1CBM, 2CBM, 3CBM, and 4CBM constructs, respectively (data not shown).
The binding isotherms of the various engineered CBM40 polypeptides with monovalent 3′SL are shown in Fig. 5. Using ITC, it was revealed that the binding of this sialoside to the designed CBM40 polypeptides is enthalpically driven, with ΔH° values being very similar ranging from -12.3 to -16.3 kcal/mol at 25 °C (Table 4). There is also very little difference in all of the other thermodynamic parameters measured for each polypeptide interaction. In fact, the binding affinity of the multivalent CBM40s to sialic acid appeared to be similar to that of the 1CBM. Furthermore, the length of the linker between modules was not shown, thermodynamically, to contribute significantly to this interaction (Table 4). Based on the one-site binding model, the n values demonstrate the appropriate number of sites for each CBM40 polypeptide, interacting with 3′SL. The fact that no significant increase in affinity is seen as we increase the number of linked modules, suggests that the sialic acid-multivalent polypeptide interaction is similar to that of a monovalent-monomeric one, indicating a simple bimolecular association.
FIGURE 5.
ITC isotherms showing the binding of 3′SL to various CBM40 constructs. a, 2CBM(5); b, 2CBM(10); c, 2CBM(15); d, 3CBM(5); e, 3CBM(10); and f, 4CBM(5).
TABLE 4.
ITC results of binding of CBM40 peptides to 3′SL (duplicate measurements)
| Peptide | [P] | [3′SL] | n | ΔH | TΔS | ΔG | Ka | Kd |
|---|---|---|---|---|---|---|---|---|
| mm | mm | kcal/mol | kcal/mol | kcal/mol | 10−4m | μm | ||
| 1CBM | 0.084 | 1.04 | 0.96 ± 0.002 | −16.3 ± 0.05 | −9.8 | −6.5 | 5.48 ± 0.05 | 18 |
| 2CBM(5) | 0.071 | 2.19 | 2.00 ± 0.003 | −12.3 ± 0.03 | −6.2 | −6.1 | 3.18 ± 0.02 | 31 |
| 2CBM(10) | 0.033 | 1.14 | 1.93 ± 0.007 | −13.7 ± 0.07 | −7.3 | −6.4 | 4.36 ± 0.05 | 22 |
| 2CBM(15) | 0.026 | 0.79 | 1.99 ± 0.014 | −13.8 ± 0.01 | −7.5 | −6.3 | 3.49 ± 0.04 | 28 |
| 3CBM(5) | 0.018 | 0.75 | 2.96 ± 0.003 | −13.5 ± 0.03 | −7.2 | −6.3 | 3.63 ± 0.04 | 27 |
| 3CBM(10) | 0.016 | 0.72 | 3.09 ± 0.024 | −13.5 ± 0.01 | −7.1 | −6.4 | 4.54 ± 0.09 | 22 |
| 4CBM(5) | 0.007 | 0.45 | 3.96 ± 0.322 | −15.8 ± 0.02 | −9.6 | −6.2 | 3.56 ± 0.03 | 28 |
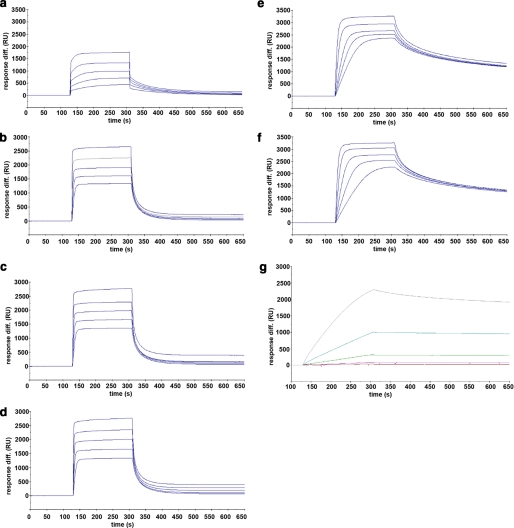
Enhanced Binding Affinity of Multivalent CBM40 Polypeptides for Multivalent 3′SL—To test whether an avidity effect for sialic acid can be achieved with multivalent CBM40 polypeptides, SPR was performed using a commercially available multivalent, biotinylated 3′SL immobilized on a streptavidin chip. Sensorgrams for all the CBM40 polypeptides injected over immobilized 3′SL are shown in Fig. 6. The affinity for each CBM40 -3′SL interaction, described here as the equilibrium dissociation constant Kd, was determined by a global fit model derived from the ratio of association/dissociation rate constants (ka/kd), assuming Langmuir 1:1 binding (Table 5). An increase in affinity toward sialic acid is observed as the number of modules is increased. For the 1CBM-3′SL interaction at 25 °C, there is a 10-fold increase in binding (Kd ∼ 1.8 μm) compared with that of the corresponding monomeric-monovalent interaction (Kd ∼ 18 μm) measured by ITC. This disparity is due to the different physical properties of the two assays employed to measure binding. Despite this, enhanced affinity is observed, using SPR, when the number of modules increases from one to two, where there is an approximate 400- to 500-fold increase in binding, resulting in affinities between 38 and 45 nm at 25 °C (Table 5). The linker length appears to have a marginal influence on the avidity in this case, because only a 1.2-fold increase in affinity is seen when increasing the number of amino acids from 5 to 15. With three and four CBM40 modules there is a further 10- to 20-fold enhancement in affinity to sialic acid, reaching affinities of around 4 nm for the 5-amino acid- and 10-amino acid-linked 3CBM modules, and 2.6 nm for the 4CBM module at 25 °C. The highest affinity was 4CBM(5) with a Kd of ∼861 pm when binding multivalent 3′SL at 15 °C. Thus, using SPR, it appears that a 700- to 1500-fold increase in affinity can be achieved by going from a monovalent interaction to a multivalent one, depending on the temperature of the interaction. Data derived from van't Hoff plots for each CBM40 polypeptide-sialic acid interaction measured at 15, 25, and 35 °C (Fig. 7 and Table 6), demonstrate ΔG values of -7.8 kcal/mol for 1CBM, around -10 kcal/mol for 2CBM, -11.3 kcal/mol for 3CBM, and -12 kcal/mol for 4CBM. The large difference in ΔG values between 1CBM and 2CBM is also reflected in the changes in enthalpy and entropy of the interaction. Out of all the CBM40 polypeptides, the 2CBM polypeptides gave the largest enthalpic change with a ΔH° value of around -20 kcal/mol, which compensated a large unfavorable entropic contribution (Table 6). This large difference in the energetics between 1CBM and 2CBM interactions is also shown in the enhanced affinity of 2CBM to sialic acid, compared with 1CBM, 3CBM, and 4CBM polypeptides, suggesting cooperativity of the ligand-receptor interaction.
FIGURE 6.
SPR sensorgrams showing the binding of the CBM40 modules to immobilized 3′SL receptors. a, 1CBM; b, 2CBM(5); c, 2CBM(10); d, 2CBM(15); e, 3CBM(5); f, 3CBM(10); and g, 4CBM(5).
TABLE 5.
Biacore kinetic parameters for the different CBM polypeptides interacting with multivalent 3′SL (n = 3 determinations)
| Peptide | T | ka | kd | KD |
|---|---|---|---|---|
| m−1 s−1 | s−1 | m | ||
| 1CBM | 35 | (3.1 ± 0.05) × 103 | (1.8 ± 0.3) × 10−2 | 5.8 × 10−6 |
| 25 | (4.3 ± 0.7) × 103 | (7.6 ± 0.4) × 10−3 | 1.8 × 10−6 | |
| 15 | (3.7 ± 0.4) × 103 | (3.6 ± 0.9) × 10−3 | 9.7 × 10−7 | |
| 2CBM(5) | 35 | (3.5 ± 0.2) × 105 | (3.6 ± 0.04) × 10−2 | 1.0 × 10−7 |
| 25 | (5.6 ± 0.7) × 105 | (2.5 ± 0.1) × 10−2 | 4.5 × 10−8 | |
| 15 | (7.4 ± 0.13) × 105 | (9.5 ± 0.07) × 10−3 | 1.3 × 10−8 | |
| 2CBM(10) | 35 | (2.0 ± 0.1) × 104 | (2.6 ± 0.2) × 10−3 | 1.3 × 10−7 |
| 25 | (5.9 ± 0.3) × 105 | (2.3 ± 0.23) × 10−2 | 3.9 × 10−8 | |
| 15 | (8.1 ± 0.17) × 105 | (6.9 ± 0.05) × 10−3 | 8.5 × 10−9 | |
| 2CBM(15) | 35 | (5.3 ± 0.6) × 105 | (5.2 ± 0.3) × 10−2 | 9.8 × 10−8 |
| 25 | (8.8 ± 0.5) × 105 | (3.4 ± 0.08) × 10−2 | 3.8 × 10−8 | |
| 15 | (8.7 ± 0.5) × 105 | (8.7 ± 0.3) × 10−3 | 1 × 10−8 | |
| 3CBM(5) | 35 | (5.2 ± 0.35) × 105 | (4.4 ± 0.2) × 10−3 | 8.44 × 10−9 |
| 25 | (5.6 ± 0.15) × 105 | (2.3 ± 0.08) × 10−3 | 4.0 × 10−9 | |
| 15 | (3.0 ± 0.3) × 105 | (1.1 ± 0.04) × 10−3 | 3.7 × 10−9 | |
| 3CBM(10) | 35 | (5.1 ± 0.01) × 105 | (4.1 ± 0.48) × 10−3 | 8.07 × 10−9 |
| 25 | (4.5 ± 0.4) × 105 | (1.95 ± 0.03) × 10−3 | 4.26 × 10−9 | |
| 15 | (3.3 ± 0.2) × 105 | (1.21 ± 0.02) × 10−3 | 3.63 × 10−9 | |
| 4CBM(5) | 35 | (2.2 ± 0.8) × 105 | (9.2 ± 0.07) × 10−4 | 4.01 × 10−9 |
| 25 | (2.8 ± 0.5) × 105 | (7.4 ± 0.9) × 10−4 | 2.62 × 10−9 | |
| 15 | (2.9 ± 0.4) × 105 | (2.5 ± 0.01) × 10−4 | 8.61 × 10−10 |
FIGURE 7.
van't Hoff plot derived from the SPR experiments at different temperatures.
TABLE 6.
Thermodynamic parameters for the interaction of designed sialic acid binding peptide modules with multivalent 3′SL-PAA-biotin derived from van't Hoff plots
| ΔG | ΔH | ΔS | TΔS | ||
|---|---|---|---|---|---|
| kcal/mol | |||||
| 1CBM | −7.8 | −15.9 | −27.3 | −8.1 | |
| 2CBM(5) | −10.0 | −18.8 | −29.4 | −8.8 | |
| 2CBM(10) | −10.3 | −22.4 | −40.8 | −12.1 | |
| 2CBM(15) | −10.1 | −20.7 | −35.4 | −10.6 | |
| 3CBM(5) | −11.3 | −7.2 | 13.8 | 4.1 | |
| 3CBM(10) | −11.3 | −7.1 | 14.2 | 4.2 | |
| 4CBM(5) | −11.9 | −14.1 | −7.5 | −2.2 | |
In contrast, the 3CBM polypeptides showed small favorable entropic gains but the change in free energy of the interaction was still strongly negative due to a better favorable enthalpic penalty (Table 6). Interestingly, both 3CBM and 4CBM polypeptides demonstrated less negative contributions to both ΔH° and TΔS° on ligand binding compared with 1CBM and 2CBM polypeptides, despite the fact that free energy of the interactions increased with increasing number of modules, which probably contributed to the gain in affinities. This observation could be based on a number of factors such as the interaction being sensitive to linker flexibility due to the conformational arrangement of modules, the accessibility of ligand binding sites, the modes of binding such as intra-, and/or intermolecular binding, and internal structural packing of the modules. The influence of linker length between the 3CBM polypeptides, however, was negligible in terms of binding energy and affinity, similar to the different 2CBM polypeptides. Because no relevant gain in affinity was achieved with 4CBM(5) at 25 °C, it was decided that no further design of polypeptides would be undertaken.
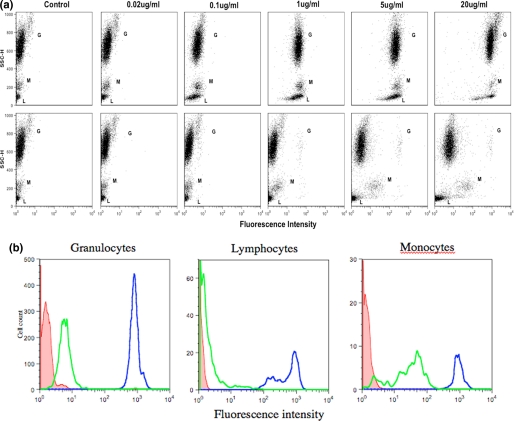
Binding of GFP-3CBM to Human Leukocytes—To test whether the engineered multivalent CBM40 polypeptides can bind to cell surface sialic acids, a GFP was fused to the N terminus of 3CBM and incubated with isolated human leukocytes. Flow cytometry analysis of subset populations of leukocytes demonstrated that the probe bound to sialic acids on the non-sialidase-treated surfaces of granulocytes, monocytes, and lymphocytes. Furthermore, it appears that sialic acid binding by the multivalent probe could be detected at concentrations much lower than 20 μg/ml, suggesting a high affinity for its receptor (Fig. 8a). When leukocyte cells were pretreated with sialidase, it is evident that the probe remained bound to monocytes, albeit to a lesser extent, and also slightly bound to granulocytes compared with non-sialidase treated cells (Fig. 8, a and b). This suggests that either the sialidase was not as efficient in removing all linked sialic acids, because it is has been shown that α(2,8)-linked substrates are cleaved much more slowly than α(2,3)- and α(2,6)-linked substrates with V. cholerae sialidase (23), or that the probe recognized and bound cryptic sialic acids, which are unmasked by treatment with V. cholerae sialidase. In contrast, lymphocytes treated with sialidase showed negligible binding by the GFP-3CBM probe.
FIGURE 8.
FACS analysis of GFP-3CBM polypeptide binding to human leukocytes. a, dot plots of leukocytes treated without (top panel) and with (bottom panel) sialidase prior to incubation with varying dilutions of GFP-3CMB. Letters G, M, and L represent granulocytes, lymphocytes, and monocytes populations, respectively. b, histogram representation of binding of probe (20 μg/ml) observed in all three subset populations of leukocytes. Dark and pale line histograms represent sialidase non-treated and treated cells, respectively. Shaded histograms represent control cells.
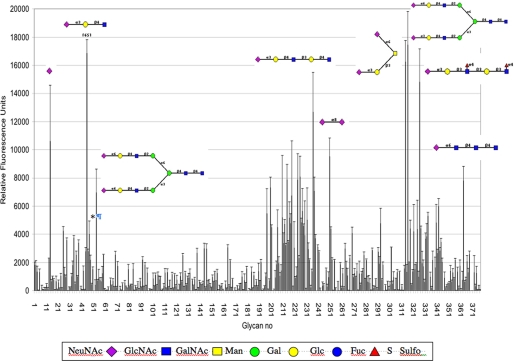
Characterization of GFP-3CBM—To characterize the extent of sialic acid specificity of the 3CBM, the GFP-fused probe was submitted to the Consortium of Functional Glycomics to be tested against a glycan array screen of 377 glycans, which contains a large number of sialosides. As expected, the GFP-3CBM bound exclusively to sialic acids, with broad linkage specificity, the main hits being those that fall within a cluster of sialosides between glycan numbers 197 and 262, and between 313 and 327. The high affinity ligands shown in Fig. 9 were identified to be mainly α(2,3)- and α(2,6)-linked and to a lesser extent, α(2,8)-linked sialosides. Moreover, strong signals were observed with some sialic acids that were branched irrespective of linkage. The probe also recognized 9NAcN-acetylneuraminic acid (glycan no. 47, see asterisk, Fig. 9), a 9-O-acetylated sialic acid, and a component of human colon mucus glycoproteins (24) as well as in human tonsil B lymphocytes (25).
FIGURE 9.
Glycan array profile of GFP-3CBM. The GFP-3CBM probe was tested against 377 glycans, listed on the Consortium for Functional Glycomics website. Some examples of α(2,3)-, α(2,6)-, and α(2,8)-linked sialosides are shown. *, binding to 9-O-acetyl-N-acetyl-neuraminic acid.
DISCUSSION
It has been shown that the sialic acid-specific CBM from V. cholerae sialidase can be successfully isolated from its parent enzyme and exploited to generate multivalent polypeptides that bind sialic acid with high affinity. Using a combination of ITC and SPR to analyze the binding of both monovalent and multivalent forms of V. cholerae CBM40 to sialic acid, the binding of the monovalent and divalent forms is shown to be enthalpically driven, as are many CBM-carbohydrate interactions (13), but when the number of linked modules is greater than two the binding is entropically driven. With respect to its ligand and linkage specificity, the isolated CBM40 binds α(2,3)-, α(2,6)-, and α(2,8)-linked sialic acids with micromolar affinity. This binding affinity is similar to that reported for V. cholerae sialidase interaction with a non-hydrolysable thiosialoside (16). The fact that there is little discrimination between different linked sialosides can be seen in the crystal structure of the CBM40 complexed with 3′SL. The structure reveals an extensive network of intermolecular interactions with sialic acid. The galactose and glucose moieties do not interact with the protein domain, and this is reflected in the similar Kd values and free energy of binding observed with different linked sialosides.
By exploiting the relatively high monovalent affinity of V. cholerae CBM40 for sialic acid, tandem-linked repeat polypeptides have been engineered to achieve sub-nanomolar avidity when interacting with a multivalent surface. This is possibly the first report of a CBM that has been manipulated by fusing identical copies using different length linker peptides. There have been previous reports of the use of isolated CBMs as molecular probes for analysis of plant cell wall polymers (26) as well as the study of CBMs naturally found in multimodular glycoside hydrolases, which have been isolated either as separate modules or as combinations with their neighboring modules to determine their function (14, 17, 27).
It is well known that linker length between protein subunits can influence the thermodynamics of an interaction, and in many cases, covalently linking subunits can enhance oligomeric stability of an interaction by reducing the entropic driving force for dissociation (28). Flexible peptide linkers were engineered between CBM modules ranging from 5 amino acids to 15 amino acids to determine whether linker length would significantly enhance the affinity of the engineered CBM polypeptides for sialic acid. In the case of 2-tandem-linked CBMs, increasing the peptide linker to 10 amino acids gave a significant reduction in conformational entropy despite observing a 1.5- to 2-fold increase in affinity to sialic acid between the different 2CBM constructs. This suggests that some degree of flexibility of the modules has occurred when two CBMs are linked together. The binding interaction of 2CBMs to sialic acid also showed an increase in binding enthalpy, which compensated the large unfavorable entropic contribution, resulting in an increase of free energy of binding as well as enhanced affinity for sialic acid, when compared with 1CBM-multivalent 3′SL interaction. This suggests that binding of 2CBM to sialic acid is a very stable, intramolecular interaction and, as for 1CBM, is enthalpically driven. However, for 3CBMs interacting with immobilized multivalent 3′SL, there was no significant difference between different length peptides. In fact, van't Hoff plots of the different 3CBM-sialic acid interactions all show an increase in entropy. In addition, a significant decrease in binding enthalpy and a 10-fold enhancement of affinity were observed when going from two to three CBMs. It is likely that the enhancement of affinity observed in the case of 3CBM binding to sialic acid could result from the probable formation of stable aggregates occurring at the surface of the SPR chip and that the large change in entropy could be due to structurally ordered water molecules being released from the hydration shells, influenced by tight protein-protein packing. This can be seen from the crystal structure of the 1CBM·3′SL complex, where three 1CBM monomers are closely packed together (Fig. 3b). Entropy-driven interactions have been seen in other multivalent protein-carbohydrate systems particularly where intermolecular binding occurs, leading to the formation of aggregates (29-31). Interestingly, increasing the number of linked modules to four demonstrated a less favorable entropic contribution to that of 3CBMs and favorable binding enthalpy, although this is slightly less than that of the 1CBM interaction with immobilized 3′SL. The free energy of binding was slightly greater than all the other CBMs corresponding to a slight increase in affinity from 3- to 4CBM. It is likely that the valency of the engineered tandem-linked polypeptides may play an important part in the stabilization of oligomers and their interaction. Poon (28) describes that increasing the valency of a tandem-linked single polypeptide chain can lead to a corresponding reduction in the molecularity of a tethered oligomer, which, thermodynamically speaking, becomes more stable. In practice, Poon (28) has stated that a tandem of valency higher than, or not divisible by the molecularity of the oligomer, will result in cross-linked oligomers. Thus for a tetrameric oligomer, such as in the case of 4CBM, a tandem dimer or tetramer would be desirable, whereas a trimeric or pentameric oligomer requires a tandem of the same respective valency.
Finally, to assess the biological relevance of these multivalent polypeptides, FACS studies demonstrated that GFP-3CBM can bind to different subpopulations of human leukocytes, which present a much higher valency of sialic acid at the cell surface compared with ligands of lower valency used in our ITC and SPR studies. Although these results only present the binding interaction of one construct, the fact remains that an engineered, high affinity multivalent polypeptide can bind to different groups of cells and that binding can be almost abolished, depending on cell type, by the action of sialidases.
These studies have shown that a sialic acid-recognizing CBM40 can be isolated and manipulated by fusing identical copies of it to enhance its affinity through avidity. It is therefore possible that other CBMs could be isolated and engineered for use as high affinity tools for glycan screening and profiling.
Acknowledgments
We thank Diana Gil for help with the FACS analysis, Dr. Uli Schwarz-Linek for advice on the ITC, Drs. Lester Carter and Ken Johnson for X-ray data collection, and the Consortium for Functional Glycomics for the glycan array data.
The atomic coordinates and structure factors (code 2w68) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Biotechnology and Biological Sciences Research Council. The staff at the European Synchrotron Radiation Facility and the European Union provided funds for access to the facility. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CBM, carbohydrate-binding module; GFP, green fluorescent protein; TEV, tobacco etch virus; MOPS, 4-morpholinepropanesulfonic acid; ITC, isothermal titration calorimetry; SL, sialyllactose; DSLNT, disialyllacto-N-tetraose; DSL, disialyllactose; SPR, surface plasmon resonance; FACS, fluorescence-activated cell sorting.
References
- 1.Angata, T., and Varki, A. (2002) Chem. Rev. 102 439-469 [DOI] [PubMed] [Google Scholar]
- 2.Lehmann, F., Tiralongo, E., and Tiralongo, J. (2006) Cell Mol. Life Sci. 63 1331-1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lis, H., and Sharon, N. (1998) Chem. Rev. 98 637-674 [DOI] [PubMed] [Google Scholar]
- 4.Mandal, C., and Mandal, C. (1990) Experientia 46 433-441 [DOI] [PubMed] [Google Scholar]
- 5.Crocker, P. R. (2002) Curr. Opin. Struct. Biol. 12 609-615 [DOI] [PubMed] [Google Scholar]
- 6.Ehrhardt, C., Kneuer, C., and Bakowsky, U. (2004) Adv. Drug Deliv. Rev. 56 527-549 [DOI] [PubMed] [Google Scholar]
- 7.Lee, R. T., and Lee, Y. C. (2000) Glycoconj. J. 17 543-551 [DOI] [PubMed] [Google Scholar]
- 8.Sacchettini, J. C., Baum, L. G., and Brewer, C. F. (2001) Biochemistry 40 3009-3015 [DOI] [PubMed] [Google Scholar]
- 9.Williams, D. H., O'Brien, D. P., Sandercock, A. M., and Stephens, E. (2004) J. Mol. Biol. 340 373-383 [DOI] [PubMed] [Google Scholar]
- 10.Williams, D. H., Stephens, E., O'Brien, D. P., and Zhou, M. (2004) Angew Chem. Int. Ed. Engl. 43 6596-6616 [DOI] [PubMed] [Google Scholar]
- 11.Williams, D. H., Stephens, E., and Zhou, M. (2003) J. Mol. Biol. 329 389-399 [DOI] [PubMed] [Google Scholar]
- 12.Mammen, M., Choi, S.-K., and Whitesides, G. M. (1998) Angew Chem. Int. Ed. Engl. 37 2754-2794 [DOI] [PubMed] [Google Scholar]
- 13.Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004) Biochem. J. 382 769-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boraston, A. B., McLean, B. W., Chen, G., Li, A., Warren, R. A., and Kilburn, D. G. (2002) Mol. Microbiol. 43 187-194 [DOI] [PubMed] [Google Scholar]
- 15.Crennell, S., Garman, E., Laver, G., Vimr, E., and Taylor, G. (1994) Structure 2 535-544 [DOI] [PubMed] [Google Scholar]
- 16.Moustafa, I., Connaris, H., Taylor, M., Zaitsev, V., Wilson, J. C., Kiefel, M. J., von Itzstein, M., and Taylor, G. (2004) J. Biol. Chem. 279 40819-40826 [DOI] [PubMed] [Google Scholar]
- 17.Boraston, A. B., Ficko-Blean, E., and Healey, M. (2007) Biochemistry 46 11352-11360 [DOI] [PubMed] [Google Scholar]
- 18.Liu, H., and Naismith, J. H. (2009) Protein Expression Purif. 63 102-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCoy, A., Grosse-Kunstleve, R. W., Adams, P., Winn, M., Storoni, L., and Read, R. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murshudov, G., Vagin, A., and Dodson, E. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 21.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 22.Biddison, W. E. (2001) Curr. Protoc. Cell Biol., Chap. 2, Unit 2.4 [DOI] [PubMed]
- 23.Corfield, T. (1992) Glycobiology 2 509-521 [DOI] [PubMed] [Google Scholar]
- 24.Rogers, C. M., Cooke, K. B., and Filipe, M. I. (1978) Gut 19 587-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamerling, J. P., Makovitzky, J., Schauer, R., Vliegenthart, J. F., and Wember, M. (1982) Biochim. Biophys. Acta 714 351-355 [DOI] [PubMed] [Google Scholar]
- 26.McCartney, L., Gilbert, H. J., Bolam, D. N., Boraston, A. B., and Knox, J. P. (2004) Anal. Biochem. 326 49-54 [DOI] [PubMed] [Google Scholar]
- 27.Guillen, D., Santiago, M., Linares, L., Perez, R., Morlon, J., Ruiz, B., Sanchez, S., and Rodriguez-Sanoja, R. (2007) Appl. Environ. Microbiol. 73 3833-3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon, G. M. (2007) Biochem. Soc. Trans. 35 1574-1578 [DOI] [PubMed] [Google Scholar]
- 29.Lundquist, J. J., and Toone, E. J. (2002) Chem. Rev. 102 555-578 [DOI] [PubMed] [Google Scholar]
- 30.Ambrosi, M., Cameron, N. R., Davis, B. G., and Stolnik, S. (2005) Org. Biomol. Chem. 3 1476-1480 [DOI] [PubMed] [Google Scholar]
- 31.Vyas, N. K., Vyas, M. N., Chervenak, M. C., Johnson, M. A., Pinto, B. M., Bundle, D. R., and Quiocho, F. A. (2002) Biochemistry 41 13575-13586 [DOI] [PubMed] [Google Scholar]