Abstract
A protein known to regulate both lipid metabolism and vesicular transport is the phosphatidylcholine/phosphatidylinositol transfer protein Sec14 of Saccharomyces cerevisiae. Sec14 is thought to globally affect secretion from the trans-Golgi. The results from a synthetic genetic array screen for genes whose inactivation impaired growth of cells with a temperature-sensitive SEC14 allele implied Sec14 regulates transport into and out of the Golgi. This prompted us to examine the role of Sec14 in various vesicular transport pathways. We determined that Sec14 function was required for the route followed by Bgl2, whereas trafficking of other secreted proteins, including Hsp150, Cts1, Scw4, Scw10, Exg1, Cis3, and Ygp1, still occurred, indicating Sec14 regulates specific trans-Golgi export pathways. Upon diminution of Sec14 function, the v-SNARE Snc1 accumulated in endosomes and the trans-Golgi. Its accumulation in endosomes is consistent with Sec14 being required for transport from endosomes to the trans-Golgi. Sec14 was also required for trafficking of Ste3 and the lipophilic dye FM4-64 from the plasma membrane to the vacuole at the level of the endosome. The combined genetic and cell biology data are consistent with regulation of endosome trafficking being a major role for Sec14. We further determined that lipid ligand occupancy differentially regulates Sec14 functions.
The composition of lipids within a membrane affects vesicle fission, transport, and fusion. Information on proteins that integrate lipid metabolism with vesicular transport is sparse. One protein that does so is Sec14 from Saccharomyces cerevisiae. Sec14 is an essential protein that extracts phosphatidylcholine (PC)2 and phosphatidylinositol (PI) from membranes in vitro and regulates PC and phosphoinositide metabolism in cells (1). Decreasing Sec14 function results in a reduction in Golgi PI 4-phosphate levels, an increase in the rate of PC synthesis, increased turnover of PC via the PC-phospholipase D Spo14, and decreased PC turnover by the PC-phospholipase B Nte1 (Fig. 1A) (2-5). The mechanisms by which Sec14 regulates PI 4-phosphate and PC metabolism are not known; however, this appears to be a major function in its regulation of cell biology as inactivation of numerous genes that mediate PC and phosphoinositide metabolism enhances or impairs growth of cells with defective Sec14 function (2, 4-6, 8-11).
FIGURE 1.
Sec14 regulation of phospholipid metabolism. A, regulation of phospholipid metabolism by Sec14 is thought to be lipid ligand-dependent. Sec14 bound to PC regulates PC metabolism, whereas Sec14 bound to PI controls phosphoinositide metabolism. PC-bound Sec14 is thought to inhibit PC synthesis at the rate-limiting step of Pct1 while at the same time inhibiting PC turnover by the phospholipase D Spo14 (to decrease the production of choline to prevent supply of PC precursor). At the same time Sec14-PC shunts PC turnover toward the phospholipase B Nte1, away from supplying choline for PC synthesis. In this way, Sec14 acts as a homeostat for regulation of PC levels. Sec14 bound to PI maintains PI-4P levels, although whether this is by activating the PI 4-kinase Pik1 is not known. The solid arrows indicate metabolic pathways; the dotted arrows activation, and the dotted lines inhibition. GPC, glycerophosphocholine. B, pathways in and out of the trans-Golgi. A subset of vesicles destined to fuse with the trans-Golgi do so via the GARP complex (contains Vps51, Vps52, Vps53, and Vps54). The GARP complex binds Ypt6, a Rab regulated by the GEF pair Ric1/Rgp1. The GARP complex also directly interacts with the Arf-like GTPase Arl1 that is anchored to the trans-Golgi via Arl3 and Sys1. Fusion to the trans-Golgi is mediated in part by the t-SNARE Tlg2 and its accessory protein Vps45. A second Golgi-associated complex, the transport protein particle II (TRAPP II) complex (contains the essential subunits Bet3, Bet5, Trs20, Trs23, Trs31, Trs120, and Trs130 and the nonessential subunits Trs33, Trs65, and Trs85), acts as a GEF for the Rab pair Ypt31/32 and mediates transport both in and out of the trans-Golgi.
Sec14 is known to be required for the transport of vesicles from the trans-Golgi to the plasma membrane and vacuole (equivalent to the lysosome in mammalian cells) as reduced Sec14 function results in a rapid decrease in Golgi-derived transport of invertase and acid phosphatase out of the cell and carboxypeptidase Y to the vacuole (2-6, 8-10, 12-15). Regulation of PC and phosphoinositide metabolism by Sec14 is hypothesized to provide the appropriate membrane environment for fission of vesicles from the trans-Golgi, although the precise step(s) where Sec14 regulates vesicular transport has yet to be determined.
The human genome contains at least 29 Sec14 domain-containing genes that code for more than 45 proteins (16). Mutations in several human Sec14 domain-containing proteins result in the onset of human diseases, including neurodegeneration, blindness, and cancer (17-22). Sec14 domains from higher eukaryotes are often embedded as part of a larger protein, many of which are guanine exchange factors (GEFs) and GTPase-activating proteins (GAPs), indicating that regulation of small G protein activity is a unifying theme for several members of the Sec14 superfamily. Human Sec14 domains bind hydrophobic ligands, including phospholipids (16).
In this study, we used synthetic genetic array (SGA) analysis to determine more precisely how S. cerevisiae Sec14 regulates vesicular transport. Genetic interactions were observed between a SEC14 allele with reduced function and GTPases that regulate vesicular transport in and out of the Golgi via endosomes. This is the first instance of Sec14 regulating import into the Golgi.
A main route for fusion of endosome-derived vesicles with the trans-Golgi is facilitated by the t-SNARE Tlg2 (23). Tlg2, along with Vps45 (a protein of the Sec1/Munc18 family), tethers endosome-derived vesicles to the trans-Golgi to facilitate membrane fusion (23) (Fig. 1B). The tethering complex that directly interacts with Tlg2 is the Golgi-associated retrograde protein (GARP) complex. The GARP complex is composed of four subunits Vps51-54 (23). The GARP complex directly interacts with Ypt6 (a Rab GTPase) and Arl1 (an Arf-like GTPase) that together comprise the two main routes for endosome fusion with the trans-Golgi. Ypt6 is activated by a heterodimeric GEF comprised of Ric1 and Rgp1. The GEF for Arl1 is unclear; however, the localization of Arl1 to the Golgi is dependent on Arl3 and Sys1 (24, 25). Arl3 is also an Arf-like GTPase, whereas Sys1 is an integral membrane protein that targets Arl3 to the Golgi.
A second Golgi-associated tethering complex is the transport protein particle (TRAPP). TRAPP I regulates early Golgi events in part by acting as a GEF for the Rab Ypt1. TRAPP II is specific for transport in and out of the trans-Golgi and is a GEF for the Rab pair Ypt31/32 (Fig. 1B) (26). TRAPP I and II share seven subunits (Bet3, Bet5, Trs20, Trs23, Trs31, Trs33, and Trs35) with three subunits (Trs65, Trs120, and Trs130) added to form TRAPP II.
Sec14 has previously been implied to globally regulate vesicular transport out of the Golgi. Our genetic screen dramatically expanded the network of genes required for vesicular transport when Sec14 function is impaired, including those comprising GARP and TRAPP complexes. The results from the genetic screen prompted us to examine the role of Sec14 in multiple vesicular transport routes. We report that Sec14 regulates a subset of vesicular transport pathways out of the trans-Golgi. The genetic analysis, coupled with assessments of vesicular transport pathways, go on to indicate that a primary function of Sec14 is regulation of vesicular transport through endosomes. We also demonstrate that ligand binding by Sec14 differentially regulates function.
MATERIALS AND METHODS
Synthetic Genetic Array Screen—SGA analysis was performed essentially as described (27, 28) with the below modifications. CMY503 (contains a temperature-sensitive SEC14 allele, sec14ts) was mated with 4,800 S. cerevisiae single gene deletion strains at 25 °C; diploids were selected, and cells were sporulated for 5 days at 25 °C. To ensure that the haploid cells obtained were from mated diploids, cells were selected for histidine prototrophy followed by growth on medium containing G418 and nourseothricin. The resulting haploids were incubated at 25 or 35 °C. Three independent screens were performed. Mutants whose inactivation resulted in decreased growth when in combination with the sec14ts allele in at least two of three screens were subjected to random spore analysis to determine whether the genetic interactions observed were true.
Yeast Strain Construction—The sec14ts allele linked to the nourseothricin drug resistance cassette was constructed as follows (mutated sites in primer sequences are underlined). Plasmid-borne SEC14 was subjected to site-directed mutagenesis to convert Gly-266 to Asp using primers 5′-CTTACCAGTCAAATTTGGCGATAAGTCTGAAGTTGATGATCA-3′ and 5′-GATTCATCAACTTCAGACTTATCGCCAAATTTGACTGGTAAG-3′. This mutation confers temperature sensitivity to Sec14. To insert the natMX4 cassette next to the sec14ts gene, an SphI site was generated 300 bp downstream of the SEC14 stop codon by site-directed mutagenesis with the primers 5′-GGCAGTATATACATTTATCTTATAGAATACGCATGCACGTATTAAACAATAAAAAGAACATAAG-3′ and 5′-CTTATGTTCTTTTTATTGTTTAATACGTGCATGCGTATTCTATAAGATAAATGTATATACTGCC-3′. The natMX4 cassette was amplified with SphI sites at each end using primers 5′-CTGAGAGCATGCACATGGAGGCCCAGAATACCC-3′ and 5′-CTGAGAGCATGCCAGTATAGCGACCAGCATTCAC-3′. An SphI site in the natMX4 DNA sequence was removed by site-directed mutagenesis (without changing the amino acid sequence) using primers 5′-CGCTCTACATGAGTATGCCCTGCCCCTA-3′ and 5′-TAGGGGCAGGGCATACTCATGTAGAGCG-3′, and this DNA fragment was subcloned into the SphI site downstream of the sec14ts open reading frame. The sec14ts:natMX4 disruption cassette was PCR-amplified from the resulting vector using primers 5′-CCTTTCTTGGATCCAGTCACTG-3′ and 5′-GGCATGGACTTGATATCCTAG-3′ to give a fragment containing both the sec14ts mutation and the natMX4 cassette along with 5′ and 3′ SEC14-flanking DNA. This DNA fragment was transformed into the Y2454 yeast strain to replace the SEC14 gene with the sec14ts allele linked to the natMX4 cassette to generate strain CMY503 (MATα mfa1Δ::MFA1pr-HIS3 can1Δ0 ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 sec14ts:natMX4). The sec14ts:natMX4 cassette was transformed into the indicated strains for the construction of isogenic sets of sec14ts containing yeast (Table 1). Strain genotypes were confirmed by genomic PCR and/or genomic Southern blot using the digoxygenin High Prime DNA Labeling and Detection Kit II (Roche Applied Science).
TABLE 1.
Yeast strains used in this study
UCSF is University of California, San Francisco.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | EUROSCARF |
| Y2454 | MATa mfa1Δ::MFA1pr-HIS3 can1Δ0 his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | 49 |
| CMY503 | Y2454 sec14ts:natMX4 | This study |
| CMY505 | BY4741 sec14ts:natMX4 | This study |
| CMY506 | BY4742 sec14ts:natMX4 | This study |
| Y01846 | BY4741 gyp1Δ::kanMX4 | EUROSCARF |
| CMY514 | BY4741 gyp1Δ::kanMX4 sec14ts:natMX4 | This study |
| Y06583 | BY4741 ypt31Δ::kanMX4 | EUROSCARF |
| CMY515 | BY4741 ypt31Δ::kanMX4 sec14ts:natMX4 | This study |
| Y02371 | BY4741 trs33Δ::kanMX4 | EUROSCARF |
| CMY516 | BY4741 trs33Δ::kanMX4 sec14ts:natMX4 | This study |
| Y01709 | BY4741 tlg2Δ::kanMX4 | EUROSCARF |
| CMY517 | BY4741 tlg2Δ::kanMX4 sec14ts:natMX4 | This study |
| Y05102 | BY4741 spo14Δ::kanMX4 | EUROSCARF |
| CMY518 | BY4741 spo14Δ::kanMX4 sec14ts:natMX4 | This study |
| Y06437 | BY4741 ric1Δ::kanMX4 | EUROSCARF |
| CMY526 | BY4741 ric1Δ::kanMX4 sec14ts:natMX4 | This study |
| CMY553 | BY4741 rgp1Δ::LEU2 | This study |
| CMY554 | BY4741 rgp1Δ::LEU2 sec14ts:natMX4 | This study |
| Y04462 | BY4741 vps45Δ::kanMX4 | EUROSCARF |
| CMY527 | BY4741 vps45Δ::kanMX4 sec14ts:natMX4 | This study |
| Y05171 | BY4741 ypt6Δ::kanMX4 | EUROSCARF |
| CMY528 | BY4741 ypt6Δ::kanMX4 sec14ts:natMX4 | This study |
| Y05091 | BY4741 vps51Δ::kanMX4 | EUROSCARF |
| CMY530 | BY4741 vps51Δ::kanMX4 sec14ts:natMX4 | This study |
| Y04318 | BY4741 vps52Δ::kanMX4 | EUROSCARF |
| CMY531 | BY4741 vps52Δ::kanMX4 sec14ts:natMX4 | This study |
| FRY107 | BY4742 vps53Δ::kanMX4 | 50 |
| CMY532 | BY4742 vps53Δ::kanMX4 sec14ts:natMX4 | This study |
| Y03966 | BY4742 vps54Δ::kanMX4 | EUROSCARF |
| CMY533 | BY4741 vps54Δ::kanMX4 sec14ts:natMX4 | This study |
| Y06772 | BY4741 sys1Δ::kanMX4 | EUROSCARF |
| CMY547 | BY4741 sys1Δ::kanMX4 sec14ts:natMX4 | This study |
| Y03304 | BY4741 arl1Δ::kanMX4 | EUROSCARF |
| CMY548 | BY4741 arl1Δ::kanMX4 sec14ts:natMX4 | This study |
| Y02777 | BY4741 arl3Δ::kanMX4 | EUROSCARF |
| CMY549 | BY4741 arl3Δ::kanMX4 sec14ts:natMX4 | This study |
| Y04576 | BY4741 ypt32Δ::kanMX4 | EUROSCARF |
| CMY550 | BY4741 ypt32Δ::kanMX4 sec14ts:natMX4 | This study |
| Y04796 | BY4741 trs65Δ::kanMX4 | EUROSCARF |
| CMY551 | BY4741 trs65Δ::kanMX4 sec14ts:natMX4 | This study |
| Y04738 | BY4741 trs85Δ::kanMX4 | EUROSCARF |
| CMY552 | BY4741 trs85Δ::kanMX4 sec14ts:natMX4 | This study |
| CMY566 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-SNF7 | UCSF |
| CMY557 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-CHC1 | UCSF |
| CMY559 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-ANP1 | UCSF |
| CMY563 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-SEC13 | UCSF |
| CMY562 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-COP1 | UCSF |
| CMY564 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-ERG6 | UCSF |
| CMY537 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-SNF7 sec14ts:natMX4 | This study |
| CMY535 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-CHC1 sec14ts:natMX4 | This study |
| CMY534 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-ANP1 sec14ts:natMX4 | This study |
| CMY536 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-SEC13 sec14ts:natMX4 | This study |
| CMY538 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-COP1 sec14ts:natMX4 | This study |
| CMY539 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 (S288C) RFP-ERG6 sec14ts:natMX4 | This study |
| X2180-1A | MATamal mel gal2 CUP1 SUC2 | Randy Sheckman |
| SF292-1A | MATasec14-3 (sec14ts) isogenic to X2180-1A | Randy Sheckman |
| SF282-1D | MATasec18-1 (sec18ts) isogenic to X2180-1A | Randy Sheckman |
| CBY926 | MATα ura3-52 his3Δ200 lys2-801 leu2-3,112 trp1Δ901 suc2Δ9 osh1Δ::kanMX4 osh2Δ::kanMX4 | 30 |
| osh3Δ::LYS2 osh4Δ::HIS3 osh5Δ::LEU2 osh6Δ::LEU2 osh7Δ::HIS3 [pCB255 (osh4-1 TRP1)] |
Monitoring Bulk Protein Secretion and Mass Spectrometry Identification of Secreted Proteins—Cells were grown to mid-log phase at 25 °C, and an equal cell number was shifted to 37 °C for 10 min and incubated with [35S]methionine/cysteine for 10 min and then with 10 mm unlabeled methionine/cysteine for 15 or 30 min. Proteins in the medium were precipitated with 10% trichloroacetic acid and separated by SDS-PAGE, and the gel was exposed to x-ray film. Similarly, equal numbers of log phase cells were shifted to 37 °C for 1 h in fresh YPD medium and proteins precipitated with 10% trichloroacetic acid, separated by SDS-PAGE, and stained with Gelcode (Thermo Scientific). Proteins contained in gel slices were reduced with dithiothreitol, carboxamidomethylated with iodoacetamide, and digested with trypsin. Peptides were extracted with 70% acetonitrile, 1% formic acid. The extraction solvent was removed under vacuum, and the tryptic peptides were resuspended in 5% methanol, 0.5% formic acid.
Liquid chromatography MS/MS was performed using an Ultimate pump and Famos auto-sampler (LC Packings, Amsterdam, Netherlands) interfaced to the nanoflow electrospray ionization source of a hybrid triple quadrupole linear ion trap mass spectrometer (QTrap, Applied Biosystems). Samples were injected onto a capillary column (0.10 × 150 mm Chromolith C18, monolithic, Merck) at a flow rate of 1.2 μl per min. Solvent A consisted of 98% water, 2% acetonitrile, 0.1% formic acid, and Solvent B consisted of 2% water, 98% acetonitrile, 0.1% formic acid, and the linear gradient was as follows: 5% solvent B to 35% solvent B over 35 min then 90% B for 6 min before re-equilibration at 5% solvent B. The sample was sprayed through distal coated fused silica emitter tips, 75-μm inner diameter with 15-μm inner diameter tip (New Objectives PicoTip). The capillary voltage was 2.10 kV with a declustering potential of 60 V, and the curtain gas was set to 15 (arbitrary units). Spectra were acquired using the information-dependent acquisition mode. The two most intense ions from the survey scan (375-1200 m/z) were selected for tandem MS, and the collision energy was set based on the mass of the precursor as determined by the m/z and charge. The raw MS/MS data were searched against NCBI yeast entries and against all SwissProt entries using the MASCOT algorithm (Matrix Science). Search parameters were peptide mass and fragment mass tolerance 0.8 and 0.5 Da, respectively, with one missed cleavage allowed. Oxidized methionines and carboxamidomethylated cysteines were chosen as variable modifications.
Invertase and Bgl2 Secretion Assays—The invertase secretion assay was performed as described (29) with modifications (13). Bgl2 secretion was determined as described (30). Mid-log phase cells were incubated at 25 °C or shifted to 37 °C for 15-60 min. Cells were then harvested by centrifugation at 1000 × g for 5 min. Cell pellets were resuspended in 10 mm NaN3, 10 mm KF solution, incubated on ice for 10 min, and then transferred to microcentrifuge tubes. Cells were centrifuged at 10,000 × g for 1 min, and pellets were resuspended in fresh pre-spheroplasting buffer (100 mm Tris-H2SO4, pH 9.4; 50 mm β-mercaptoethanol; 10 mm NaN3; 10 mm KF), incubated on ice for 15 min, centrifuged as before, washed with 0.5 ml of spheroplast buffer (50 mm KH2PO4-KOH, pH 7; 1.4 m sorbitol; 10 mm NaN3), and pelleted. Cells were resuspended in spheroplast buffer containing 167 μg/ml Zymolyase 100T and incubated for 30 min at 25 °C. Spheroplasts were then pelleted at 5,000 × g for 10 min and resuspended in 2× SDS-PAGE sample buffer. Proteins were separated using a 10% SDS-polyacrylamide gel, and Bgl2 was detected by Western blot using a rabbit polyclonal antibody against Bgl2. Bgl2 antibodies were the kind gifts of Randy Schekman (University of California, Berkeley) and Wei Guo (University of Pennsylvania).
Ste3 Internalization—MATα cells were grown to mid-log phase at 25 °C and shifted to 37 °C for 2 h. Proteins were extracted, separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with antibodies for Ste3 and Pgk1 (Molecular Probes). Ste3 antibodies were the kind gift of George Sprague (University of Oregon).
Microscopy—Live cells were observed using a Zeiss Axiovert 200 M microscope fitted with a plan-neofluor 100× oil immersion lens. Images were captured using a Zeiss Axio Cam HR using Axiovision 4.5 software. Cells were visualized using differential interference contrast (DIC), green fluorescent protein, or rhodamine filters as required. GFP-Snc1 subcloned into the plasmid pRS416 (pMJL1) was the kind gift of Hugh Pelham (MRC Laboratory of Molecular Biology, Cambridge, UK). The plasmid expressing the Vps27-GFP fusion was from Christopher Stefan (Cornell University, Ithaca, NY). The yeast strains expressing red fluorescent protein-tagged organelle marker proteins were from Erin O'Shea (University of California, San Francisco).
RESULTS
SGA Analysis of sec14ts Cells Uncovers Novel Genetic Interactions—S. cerevisiae contains 6607 genes of which ∼5000 are not essential. A haploid query strain containing a temperature-sensitive sec14ts allele was mated to ∼4800 viable single gene deletion haploid yeast strains of the opposite mating type using SGA technology (28). The resulting diploids were isolated and forced to undergo meiosis, and haploid cells were selected that contained the sec14ts allele in combination with each single gene deletion. The resulting strains, obtained from three separate screens, were incubated at permissive (25 °C) and semipermissive (35 °C) temperatures for growth of cells containing the sec14ts allele alone. We considered only strains with the sec14ts gene in combination with each single gene deletion able to grow at 25 °C, but not 35 °C, to eliminate genes unable to grow because of defective mating, sporulation, or restoration of vegetative growth from spores.
Forty genes were isolated from the SGA screen whose inactivation aggravated growth of sec14ts-containing cells (Fig. 2A and Table 2). Inactivation of SPO14 (PC phospholipase D) had been previously determined to diminish growth of sec14ts cells (1, 5, 9), and it was isolated from our screen. Of the 40 genes isolated, the largest group contained genes known to regulate Golgi-dependent vesicular transport processes consistent with Sec14 acting as an essential regulator of this process (Fig. 2A). As genetic interactions can be representative of direct regulation of a protein/process, or may indicate processes whose functions are linked but by several degrees of separation, we focused on the genes isolated from the SGA screen with known roles in vesicular transport.
FIGURE 2.
Genes whose inactivation aggravates growth of sec14ts cells. A, pie chart of the functions of the 40 genes identified by the SGA screen whose inactivation decreased growth of sec14ts cells. A subset of these genes (SPO14, YPT31, TRS33, TLG2, GYP1, and VPS1) had been identified previously as genetic interactors in cells with decreased Sec14 function (2). B, isogenic mutant strains with the indicated genotypes were grown to mid-log phase in synthetic complete medium at 25 °C; identical numbers of cells were serial diluted, spotted onto agar plates, and grown for 3 days at the indicated temperatures. Invertase secretion is the ratio of external invertase enzyme activity to total (internal + external) invertase activity. Invertase data are the mean of three individual experiments performed in triplicate. Standard errors of the mean are indicated in the figure.
TABLE 2.
Yeast genes isolated from the synthetic genetic array screen whose inactivation aggravated growth of sec14ts cells
| Gene | Open reading frame | Description | Cellular process |
|---|---|---|---|
| SPO14 | YKR031C | Phospholipase D, catalyzes hydrolysis of phosphatidylcholine to phosphatidic acid and choline | Vesicular transport/phospholipid metabolism |
| YPT31 | YER031C | GTPase of Rab family involved in the endocytic and exocytic pathways | Vesicular transport |
| TRS33 | YOR115C | Protein of the TRAPPII complex involved in endocytic and exocytic pathways; GEF activity toward Ypt31 | Vesicular transport |
| GYP1 | YOR070C | Golgi GTPase-activating protein for Rab family members | Vesicular transport |
| TLG2 | YOL018C | t-SNARE involved in endocytosis and maintenance of resident proteins in the trans-Golgi | Vesicular transport |
| UBP3 | YER151C | Ubiquitin-specific protease, interacts with Bre5 | Vesicular transport |
| BRE5 | YNR051C | Required by Ubp3 to form an active de-ubiquitination complex which protects Sec23, a COPII subunit, from degradation | Vesicular transport |
| KEX2 | YNL238W | Calcium-dependent serine protease involved in the activation of proproteins of the secretory pathway | Vesicular transport/Golgi function |
| VPS1 | YKR001C | Dynamin-like GTPase for vacuolar protein sorting and high density vesicle production for secretion of invertase | Vesicular transport |
| TCB2 | YNL087W | Synaptotagmin like protein; contains 3 C2 domains | Golgi localized |
| INP51 | YIL002C | PI-4,5-bisphosphate 5-phosphatase | Phospholipid metabolism |
| YBR030W | YBR030W | Protein of unknown function. Likely involved in phospholipid metabolism as it has Ino2/4-binding sites in promoter | Phospholipid metabolism |
| CLN2 | YPL256C | G1/S-specific cyclin, interacts with Cdc28 protein kinase to control events at START | Cell cycle progression |
| CDH1 | YGL003C | Cell cycle-regulated activator of the anaphase-promoting complex required for exit from mitosis | Cell cycle progression |
| WHI2 | YOR043W | Protein involved in stress response and growth regulation; negative regulator of G1 cyclin expression | Cell cycle progression/stress response |
| RIM9 | YMR063W | Protein of unkonwn function; involved in proteolytic activation of Rim101 in response to pH | Stress response |
| TUS1 | YLR425W | GDP-GTP exchange factor that functions to modulate Rho1 activity as part of the cell integrity pathway | Stress response/cell wall organization |
| SLG1 | YOR008C | Plasma membrane protein required for maintenance of cell wall integrity and for the stress response | Cell wall organization |
| ECM33 | YBR078W | Glycosylphosphatidylinositol anchor protein of unknown function; possible role cell wall organization | Cell wall organization |
| NCS2 | YNL119W | Role in urmylation | Cell polarity |
| NCS6 | YGL211W | Protein of unknown function; role in urmylation and invasive/pseudohyphal growth | Cell polarity |
| PEA2 | YER149C | Coiled-coil polarisome protein | Cell polarity |
| CRN1 | YLR429W | Coronin, actin and microtubules associated protein | Actin patch assembly |
| ICE2 | YIL090W | Protein of unknown function; integral endoplasmic reticulum membrane protein | Endoplasmic reticulum organization |
| PER1 | YCR044C | Vacuolar membrane protein; mutant is dependent on activation of the unfolded protein response | Protein Processing |
| DSD1 | YGL196W | d-Serine dehydratase | Amino acid metabolism |
| HXT8 | YJL214W | PM protein with strong similarity to hexose transporters; expression induced by low glucose | Hexose transport |
| LSM1 | YJL124C | Protein involved in mRNA degradation in cytoplasm | mRNA catabolism |
| CBF1 | YJR060W | Helix-loop-helix protein that binds to promoters in CDE1 and MET genes; required for chromosome stability | DNA/chromosome organization |
| RAD27 | YKL113C | Single-stranded DNA endonuclease and 5′-3′-exonuclease, 5′-flap endonuclease | DNA/chromosome organization |
| EAF7 | YNL136W | Subunit of the NuA4 histone acetyltransferase complex | DNA/chromosome organization |
| RAD34 | YDR314C | Nucleotide excision repair | DNA repair |
| YSA1 | YBR111C | Nucleosidediphosphate sugar hydrolase | Unknown |
| CUE3 | YGL110C | Protein of unknown function; has CUE domain that may facilitate intramolecular monoubiquitination | Unknown |
| YFR045W | YFR045W | Protein with similarity to mitochondrial transporter family | Unknown |
| YMR003W | YMR003W | Protein of unknown function | Unknown |
| YGR064W | YGR064W | Protein of unknown function | Unknown |
| YPL261C | YPL261C | Protein of unknown function | Unknown |
| YJL007C | YJL007C | Protein of unknown function | Unknown |
| YMR052C-a | YMR052C-a | Protein of unknown function | Unknown |
To independently confirm the results of the SGA screen, a set of isogenic strains was constructed whereby the wild type SEC14 gene was replaced with the sec14ts allele in strains with inactivated genes for SPO14 (PC phospholipase D) (1, 5, 9), YPT31 (Rab that regulates Golgi vesicle export and import) (31), TRS33 (a component of TRAPP complex that acts as a GEF for Ypt31 and its homologue Ypt32) (32, 33), GYP1 (GAP for the essential Rab Ypt1) (34), and TLG2 (syntaxin-like t-SNARE that mediates fusion of endosome-derived vesicles with the trans-Golgi) (34). Growth of cells containing the sec14ts allele alone was slightly slower than wild type at the semi-permissive temperature for this allele (35 °C) (Fig. 2B). This growth defect was exacerbated when a cell containing the sec14ts allele had in combination an inactivated gene for SPO14, YPT31, TRS33, GYP1, or TLG2. Inactivation of any of these genes alone did not affect cell growth. The SGA analysis successfully revealed previously unknown genetic interactions with SEC14.
Secretion Pathways Are Differentially Affected in Cells Defective in Sec14 Function—Sec14 regulates fission of vesicles from the trans-Golgi. A hallmark of Sec14 deficiency is reduced invertase secretion from the cell. Invertase follows a pathway from the trans-Golgi via endosomes on its route to the plasma membrane (35, 36). We monitored invertase secretion in sec14ts cells and compared it with sec14ts cells containing inactivated genes isolated from our SGA screen. In sec14ts cells invertase secretion was reduced to 66% at 35 °C (Fig. 2B) (compared with ∼30% at 37 °C, which is the non-permissive temperature for the sec14ts allele; data not shown). In sec14ts cells that also contained inactivated SPO14, YPT31, TRS33, GYP1, or TLG2 invertase secretion was reduced to 22-48% of wild type at 35 °C (Fig. 2B). All strains secreted invertase similar to wild type at 25 °C and in addition all single gene deletions secreted invertase at wild type levels at 35 and 37 °C (data not shown). Therefore, inactivation of SPO14, YPT31, TRS33, GYP1, or TLG2 decreases Sec14-dependent vesicular transport of invertase from the trans-Golgi.
Two types of secretory vesicles have been isolated from cells based on differences in density (35, 36). The endosome-dependent route fractionates with higher density vesicles that contain invertase and acid phosphatase. These are the only two proteins that have been shown to be inefficiently secreted in sec14ts cells.
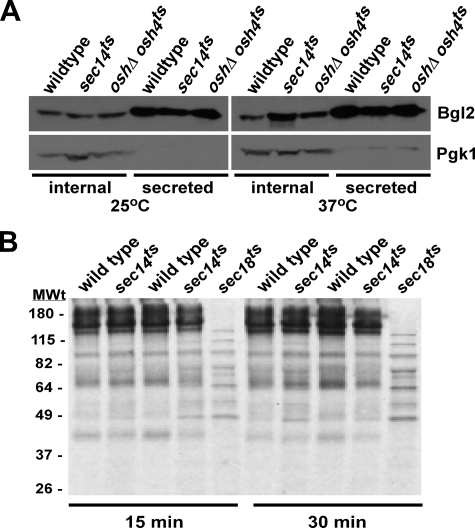
A second route to the plasma membrane is found in lower density vesicles containing Bgl2 and Pma1 and represent an endosome-independent route from the trans-Golgi to the plasma membrane. To assess if a deficiency in Sec14 function also affects secretion through the endosome-independent route, we monitored Bgl2 secretion. Bgl2 accumulated in sec14ts cells compared with wild type after incubation at the non-permissive temperature for the sec14ts allele. We observed a build up of Bgl2 in cells with defective Sec14 function as early as 15 min after temperature shift, and this was obvious within 1 h (Fig. 3A). The oshΔ osh4ts strain is known to accumulate Bgl2 and served as a positive control (30). Bgl2 observed outside the cell in all strains is standard for this assay as Bgl2 is present in the periplasmic space prior to onset of the secretory defect.
FIGURE 3.
Cells with reduced Sec14 function do not have a major defect in bulk protein secretion. A, mid-log phase cells were incubated at 25 °C or shifted to 37 °C for 1 h. Cells were harvested and resuspended in 10 mm NaN3, 10 mm KF and incubated on ice. Cells were washed and resuspended in buffer containing 1.4 m sorbitol and zymolyase. Resulting spheroplasts were pelleted and proteins separated by SDS-PAGE. Bgl2 was detected by Western blot. B, wild type, sec14ts, and sec18ts cells (strains used were BY4741 (wild type), CMY505 (sec14ts), X2180 (wild type), SF292-1A (sec14ts), and SF282-1D (sec18ts)) were grown to mid-log phase at 25 °C, shifted to 37 °C for 10 min, incubated with [35S]methionine/cysteine for 10 min, and then with 10 mm unlabeled methionine/cysteine for 15 and 30 min at 37 °C. Proteins in the medium were separated by SDS-PAGE, and the gel was exposed to x-ray film.
The high and low density vesicles that contain invertase/acid phosphatase and Bgl2/Pma1, respectively, were isolated from cells with defects in fusion of Golgi-derived vesicles with the plasma membrane (36). These experiments did not rule out other routes to the plasma membrane from the trans-Golgi. To determine the extent and specificity of the protein secretion defect in sec14ts cells, wild type and sec14ts cells (from two different genetic backgrounds) were grown at 37 °C for 10 min, incubated with [35S]methionine/cysteine for 10 min to radiolabel cellular protein, and chased with 10 mm unlabeled methionine/cysteine for various times, and radiolabeled proteins secreted into the medium were separated by SDS-PAGE. Wild type and sec14ts cells secreted a similar amount of protein. The pattern of proteins secreted was similar but not completely identical (Fig. 3B). As a control we monitored the amount of radiolabeled protein secreted from sec18ts cells that possess a well characterized defect in secretion (Sec18 encodes the ATPase-N-ethylmaleimide-sensitive fusion protein that primarily regulates endoplasmic reticulum to Golgi transport). The sec18ts cells secreted significantly less protein.
To determine whether the radiolabeled bands could be due to protein breakdown, or whether they represented a series of proteins, proteins secreted into the medium were acid-precipitated and separated by SDS-PAGE, and gels were stained with Coomassie Blue and silver nitrate. Like the secretion of [35S]methionine/cysteine-labeled proteins, the banding pattern and amount were similar between wild type and sec14ts cells (data not shown). Identity was determined by mass spectrometry and included Hsp150, Cts1, Scw4, Scw10, Exg1, Cis3, and Ygp1, proteins known to be secreted by S. cerevisiae.
Decreasing Sec14 function does not appear to diminish all vesicular transport routes from the trans-Golgi, but it affects specific vesicular transport pathways emanating from this organelle.
GFP-Snc1 Trafficking Is Defective in Cells with Diminished Sec14 Function—Invertase transits through early endosomes on its way to the cell surface (35, 36). A possible explanation for the defect in delivery of invertase to the cell surface is that Sec14 is required for trafficking via endosomes. Consistent with this notion, genes isolated from the SGA screen (YPT31, TRS33, TLG2, and VPS1) regulate vesicular transport pathways through endosomes (Table 2). The endosome is a central organelle required for transit of a subset of proteins from the trans-Golgi to the plasma membrane, as well as trafficking of proteins from the plasma membrane back to the Golgi as well as the vacuole. To further assess if Sec14 regulated endosome requiring pathways we first monitored trafficking of the v-SNARE Snc1.
Snc1, the S. cerevisiae homologue of synaptobrevin, continually cycles between the plasma membrane and trans-Golgi. Snc1 is found in both low and high density Golgi-derived vesicles during transit to the plasma membrane (37), and thus the SNARE machinery is thought to be shared among vesicular transport pathways emanating from the trans-Golgi. Subsequently, Snc1 is transported from the plasma membrane back to the trans-Golgi via endosomes (36, 38). At steady state Snc1 is primarily found at the plasma membrane (34, 38), and consistent with prior studies GFP-Snc1 was primarily localized at the plasma membrane as well as a few punctate structures in the cytoplasm (endosomes and trans-Golgi) in wild type cells (Fig. 4A). A similar localization was observed in sec14ts cells grown at the permissive temperature for function of this allele (25 °C); however, when sec14ts cells were shifted to the non-permissive temperature (37 °C), GFP-Snc1 rapidly localized to large intracellular punctate spots. Localization of GFP-Snc1 with organellar marker proteins revealed that in the sec14ts cells shifted to 37 °C for 30 min GFP-Snc1 largely colocalized with the trans-Golgi with a smaller fraction colocalizing with endosomes (Fig. 4B). GFP-Snc1 accumulation in endosomes indicates reduced endosome trafficking to the trans-Golgi.
FIGURE 4.
Sec14 regulates Snc1 cycling. A, wild type and sec14ts cells expressing GFP-Snc1 were grown to early log phase at 25 °C; a portion was shifted to 37 °C, and live cells were imaged using DIC and fluorescence microscopy at the indicated time points. B, wild type SEC14 gene was replaced with the sec14ts allele in yeast strains where RFP was fused to the organellar markers Chc1 (trans-Golgi) and Snf7 (endosomes). These strains were transformed with the plasmid expressing GFP-Snc1, and colocalization of GFP-Snc1 was determined in sec14ts cells after cells were shifted to 37 °C for 30 min. C, wild type and sec14ts cells (strains BY4741 and CMY505) were shifted to 37 °C for 30 min resulting in GFP-Snc1 localization to the trans-Golgi in sec14ts cells. Lat-A was added for 10 min to prevent GFP-Snc1 cycling back into the cell from the plasma membrane, and cells were grown for a further 15-30 min at 37 °C. Live cells were visualized using DIC and fluorescence microscopy. The number of cells from random fields (four fields from two separate experiments totaling at least 100 cells) was visually assessed for GFP-Snc1 at the plasma membrane using a double-blinded protocol.
Accumulation of GFP-Snc1 at the trans-Golgi could be due to either reduced fusion to or reduced transit from this organelle. To assess whether the GFP-Snc1 accumulating in the trans-Golgi upon reduction of function of Sec14 can exit this organelle, we treated cells with latrunculin-A (Lat-A). Lat-A blocks actin polymerization and blocks endocytosis from the plasma membrane to endosomes in S. cerevisiae. These conditions allowed us to monitor trans-Golgi to plasma membrane transport in the absence of recycling of GFP-Snc1 out of the plasma membrane. Cells were grown at 37 °C for 30 min to localize GFP-Snc1 to the trans-Golgi in sec14ts cells, and Lat-A was added to inhibit endocytosis, and cells were grown for a further 15-30 min at 37 °C to determine whether GFP-Snc1 could exit the trans-Golgi and traffic to the plasma membrane. As expected, wild type cells exhibit only plasma membrane GFP-Snc1 after treatment with Lat-A. A significant number of the Lat-A-treated sec14ts cells (58-67%) were able to transport GFP-Snc1 from the trans-Golgi to the plasma membrane (Fig. 4C). This is consistent with a Sec14-independent vesicular transport route from the trans-Golgi to the plasma membrane.
Endosome-dependent Routes from the Plasma Membrane Are Defective in Cells with Reduced Sec14 Function—Endosomes are also required for transport of vesicles from the plasma membrane to the vacuole. Ste3 is a mating factor receptor that, in the absence of mating factor, is transported from the plasma membrane via endosomes to the vacuole for degradation (39). Ste3 was observed in wild type cells at 25 °C with reduced levels at 37 °C (Fig. 5A). At increased temperature the rate of endocytosis is increased (40) resulting in the lower steady-state levels of Ste3 observed in wild type cells at 37 °C. In cells with defective Sec14, Ste3 was stabilized at 37 °C. Sec14 functions in the transport of Ste3 from the plasma membrane to the vacuole.
FIGURE 5.
Sec14 is required for trafficking of endosomes to the vacuole. A, wild type and sec14ts MAT α cells (BY4742 and CMY506) were grown to mid-log phase at 25 °C and shifted to 37 °C for 2 h. Proteins were extracted, separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with antibodies for Ste3 and Pgk1. B, wild type (BY4741) and sec14ts (CMY505) cells were grown to mid-logarithmic phase at 25 °C, shifted to 37 °C for 15 min, labeled for 2 min with 40 μm FM4-64, and incubated post-label in fresh pre-warmed 37 °C medium for the indicated times. Live cells were visualized using DIC and fluorescence microscopy. C, FM4-64 was colocalized with the endosome marker Vps27-GFP at various time points after the shift to 37 °C.
We also monitored internalization of the lipophilic dye FM4-64 as a separate assay to determine whether there were defects in endosome to vacuole trafficking upon reduction of function of Sec14. FM4-64 incorporates into the plasma membrane and traffics to the vacuole via early and late endosomes (40). In wild type cells FM4-64 was transported to the vacuole in a matter of minutes at 37 °C, whereas in sec14ts cells shifted to 37 °C, FM4-64 accumulated in punctate spots in the cytoplasm and was unable to transit out of these punctate regions (Fig. 5B). These punctate structures colocalized with the endosomal marker Vps27-GFP (Fig. 5C). The inhibition of FM4-64 trafficking and stabilization of Ste3 upon inactivation of Sec14 is consistent with Sec14 regulating plasma membrane-derived endosome trafficking pathways.
Genetic Interactions of sec14ts with Genes That Regulate Endosome Trafficking—Our data are consistent with Sec14 regulating endosome function, and genes isolated from our SGA screen participate in this process. A main strength of the SGA strategy is comprehensiveness; however, SGA screens can miss genetic interactions depending on several factors, including the allele used in the screen, the nature of the screen, the fitness of each single gene deletion strain, and the effect of the gene(s) on mating, sporulation, and the transition from spore to growing cell (41). Therefore, we tested all nonessential genes involved in regulation of tethering and fusion of endosome-derived vesicles at the trans-Golgi for aggravating effects on growth of sec14ts cells (see Fig. 1B).
The wild type SEC14 gene was replaced with the sec14ts allele in an isogenic set of strains containing inactivated genes that regulate GARP-mediated fusion from endosome to the trans-Golgi, including VPS51, VPS52, VPS53, VPS54, VPS45, YPT6, RIC1, RGP1, ARL1, ARL3, or SYS1. Inactivation of any of these genes in sec14ts cells decreased growth at 35 and 33 °C compared with cells containing the sec14ts allele alone, except for cells containing an inactivated RGP1 gene that was inviable at any temperature (Fig. 6).
FIGURE 6.
Tethering complex defects aggravate Sec14 function. Isogenic mutant strains with the indicated genotypes were grown to mid-log phase in synthetic complete medium at 25 °C, and identical numbers of cells were serial diluted, spotted onto agar plates, and grown for 3 days at the indicated temperatures.
Inactivation of YPT31 was found by our SGA screen to aggravate growth of sec14ts cells. Ypt31/Ypt32 and the TRAPP II complex regulate both export from the trans-Golgi to the plasma membrane and endosome trafficking to the trans-Golgi (26, 31, 32). Inactivation of any of the nonessential subunits of the TRAPP complexes, including TRS65 which is specific to the TRAPP II complex, aggravated growth of sec14ts cells (Fig. 6).
Inactivation of YPT32 did not aggravate growth of sec14ts cells. Ypt32 is found in cells at lower levels than Ypt31 (42), and thus inactivation of the YPT32 gene may have its phenotype masked by the Ypt31 still present in the cell. The converse would not be true as the low level of Ypt32 could not buffer the effect of inactivation of the YPT31 gene.
For all of the genes tested, growth of isogenic strains containing inactivating mutations for each gene in the presence of the wild type SEC14 gene was determined at 35, 33, and 25 °C. All grew at rates similar to wild type except strains with inactivated TRS85, YPT6, or RIC1 genes that displayed moderately reduced growth at 35 °C. However, the combination of the sec14ts allele along with inactivation of TRS85, YPT6, or RIC1 resulted in much more severe growth defects than inactivation of any one gene alone (data not shown). Combined, the cell biology and genetic evidence clearly shows that Sec14 regulates endosome trafficking from the plasma membrane to both the trans-Golgi and the vacuole.
Specific Lipid Ligands Regulate Discrete Vesicular Transport Processes—Sec14 extracts PC and PI from membranes, and this activity is essential for its function in cells (4, 10, 43). A mutant version of Sec14 (Sec14K66A,K239A known as Sec14-PC) that transfers PC with much higher affinity than PI in vitro was found to restore growth and secretion to cells lacking Sec14 function (7, 43). Yeast contain five other Sec14 homologues (Sfh1-5) capable of PI (but not PC) transfer, and overexpression of two of these, Sfh2 and Sfh4, restored growth and secretion to cells lacking Sec14 function.
To facilitate analysis of roles for PC versus PI binding by Sec14, we transformed plasmids expressing wild type Sec14, the Sec14-PC allele, or Sfh4 into the strains containing the sec14ts allele in combination with every gene deletion determined in this study to diminish Sec14-dependent growth. If there are separate functions for PC and PI binding by Sec14, then a subset may be rescued by only one or more of wild type Sec14, Sec14-PC and Sfh4.
Growth of sec14ts cells lacking VPS51-53, three of the four nonessential components of the endosome to Golgi tethering complex GARP, required Sec14 or Sfh4, whereas Sec14-PC was insufficient (Fig. 7). Growth of cells inactivated for the fourth member of the GARP complex, VPS54, was only restored by wild type Sec14. Inactivation of the GARP complex as a whole requires Sec14 that can exchange PC and PI in cis within the same Sec14 molecule.
FIGURE 7.
Regulation of specific vesicular transport processes based on Sec14 lipid ligand occupancy. A, isogenic mutant strains with the indicated genotypes transformed with low copy plasmids expressing Sec14, Sec14-PC, or a high copy plasmid expressing Sfh4 were grown to mid-log phase in synthetic complete medium at 25 °C, and identical numbers of cells were serial diluted, spotted onto agar plates, and grown for 3 days at the indicated temperatures. B, tabulation of the ability of Sec14, Sec14-PC, and Sfh4 to rescue cell growth of sec14ts cells with the indicated inactivated genes.
Endosome to Golgi fusion by the GARP complex is facilitated by the Rab Ypt6, and sec14ts cells lacking YPT6 were similar to those lacking VPS54 in that growth was supported only by wild type Sec14 but not by Sec14-PC or Sfh4 (Fig. 7). Ypt6 is activated by the GEF Ric1, and growth of sec14ts cells with inactivated RIC1 was supported by Sfh4 but not by wild type Sec14 or Sec14-PC. A similar sec14ts rescue phenotype associated with loss of RIC1 was observed for loss of function of VPS45. Vps45 is required for the t-SNARE Tlg2 to tether the GARP complex to the Golgi. The sec14ts cells with inactivated TLG2 could grow normally when expressing wild type Sec14 or Sfh4, with some growth observed if Sec14-PC was expressed.
The second complex capable of activating GARP for fusion of endosomes with the Golgi is mediated by Arl1, Arl3, and Sys1. Growth was restored upon inactivation of ARL3 or SYS1 in sec14ts cells by Sec14, Sec14-PC, and Sfh4 (Fig. 7). ARL1 was unique in that its inactivation in sec14ts cells was not restored by wild type Sec14, but it was restored by either Sec14-PC or Sfh4. The two pathways that mediate GARP complex fusion of endosomes with the Golgi have specific lipid ligand occupancy requirements by Sec14. Growth of sec14ts cells lacking YPT31 or TRS33 was restored by Sec14, Sec14-PC, or Sfh4, whereas growth of sec14ts cells with inactivated TRS85 required wild type Sec14 or Sec14-PC (although growth was restored to a lesser extent).
Growth of sec14ts cells lacking the GAP Gyp1 (has specificity for several small G proteins in vitro, and the essential Rab Ypt1 in vivo; with Ypt1 activity activated by the TRAPP complex) was restored by wild type Sec14, Sfh4, and to a lesser extent Sec14-PC. Processes influenced by Sec14 function are differentially affected depending on Sec14 lipid ligand occupancy.
DISCUSSION
Sec14 Regulates Specific trans-Golgi Export Pathways—Sec14 was known to be required for secretion through the endosome-dependent route followed by invertase and acid phosphatase. In this study we demonstrate that Sec14 also regulates the endosome-independent route from the trans-Golgi to the plasma membrane used by Bgl2. Moreover, not all secretion was inhibited upon diminution of Sec14 function as many proteins were still secreted at a similar level to wild type, including Hsp150, Cts1, Scw4, Scw10, Exg1, Cis3, and Ygp1. This implies that there is at least one more route from the trans-Golgi to the plasma membrane, and it is independent of Sec14 (Fig. 8).
FIGURE 8.
Regulation of vesicular transport by Sec14. A, Sec14 facilitates nonendosomal and endosomal transport pathways from the trans-Golgi to the plasma membrane (pink and blue dotted lines). At least one other route from the trans-Golgi to the plasma membrane is not facilitated by Sec14. Transport from the plasma membrane to both the trans-Golgi and vacuole via endosomes is also regulated by Sec14. B, genetic evidence suggest Sec14 regulates endosome trafficking to the trans-Golgi via all major routes into this organelle by regulating Golgi PI-4P levels. Many of the gene deletions that aggravate growth of sec14ts cells also aggravate growth of cells containing a temperature-sensitive allele of the Golgi PI 4-kinase, PIK1; Golgi PI-4P levels are decreased in sec14ts and pik1ts cells (2). This implies that regulation of Golgi PI-4P levels by Sec14 is necessary for regulation of import and export of vesicles from the trans-Golgi. The processes affected could include GARP complex (Vps51-54) binding to the t-SNARE Tlg2 that, together with Vps45, aid in targeting and fusion of endosome-derived vesicles with the trans-Golgi. Fusion is promoted by interaction of the GARP complex with the Rab GTPase Ypt6 that is in turn activated by the heterodimeric GEF Ric1/Rgp1. GARP also binds the Arf-like GTPase Arl that is localized to the Golgi by a second Arf-like GTPase Arl3 that interacts with the Golgi resident protein Sys1. The function of the Rab GTPase Ypt31 appears to be to regulate trafficking both in and out of the Golgi. Ypt31 is activated by the 10-subunit TRAPP II complex of which Trs33. Trs65, and Trs85 are nonessential. Reduction of Sec14 function results in aberrant Golgi PI-4P levels directly confounding vesicle import and export. The defects in endosome trafficking from the plasma membrane to the vacuole could be due to improper endosome trafficking because of miscommunication between Rab cascades into and out of the trans-Golgi.
The v-SNARE Snc1 traffics from the trans-Golgi to the plasma membrane via both the endosome-dependent and -independent routes (35, 36). Decreasing Sec14 function resulted in ∼50% of Snc1 still being able to traffic from the trans-Golgi to the plasma membrane, implying Snc1 may also transit to the plasma membrane by the route used by Hsp150, Cts1, Scw4, Scw10, Exg1, Cis3, and Ygp1. This is consistent with the idea that the SNARE machinery required for secretion is shared by the trans-Golgi to plasma membrane routes.
Regulation of Endosome Trafficking by Sec14—We also determined in this study that Sec14 was required for trafficking from the plasma membrane to the vacuole via endosomes. FM4-64 internalization was inhibited at the endosome stage of transport, and consistent with this Ste3 was stabilized in cells with diminished Sec14 function. Indeed, trafficking of FM4-64 was dramatically reduced within 5 min of reduction of Sec14 function. These findings imply a role for Sec14 is regulation of endosome trafficking from the plasma membrane to the vacuole.
We observed that inactivation of Sec14 resulted in the accumulation of a portion of GFP-Snc1 in endosomes implying a defect in endosome to trans-Golgi transport. We also present substantive genetic evidence pointing to an important role for Sec14 in this pathway. Cells with defective Sec14 function were further impaired for growth by inactivation of nonessential components of the GARP or TRAPP II complexes and their regulators, including Rabs, Arf-like proteins, and their GEFs. In total, this study has revealed that endosome trafficking is likely a major role for Sec14 (Fig. 8).
Sec14 Affects Phospholipid-mediated Regulation of Vesicular Trafficking—Sec14 binds PC and PI and affects the metabolism of both these ligands, although precisely how is not known (1-6, 8, 9, 11, 44, 45). It is believed that occupancy of Sec14 by PC regulates PC synthesis and turnover, whereas occupancy by PI regulates PI-4P levels (2, 4, 16, 45, 46). Two separate SGA screens using different temperature-sensitive alleles of the Golgi PI 4-kinase PIK1 determined that inactivation of many of the genes in the current study (as aggravating growth of sec14ts cells) exacerbated Pik1-dependent vesicular trafficking from the Golgi, including YPT31 (but not YPT32), TRS33, TRS65, ARL1, SYS1, VPS51, VPS54, YPT6, RIC1, and VPS1 (47, 48). As inactivation of a similar set of genes was found to inhibit growth of pik1ts and sec14ts cells (this study), and the fact that Golgi PI-4P levels are reduced in cells with diminished Pik1 and Sec14 function (2, 11, 44), it is likely that inactivation of these genes inhibits growth when Golgi PI-4P levels are reduced. Indeed, the majority of the genes we identified in the sec14ts SGA screen could be rescued by wild type Sec14 or Sfh4, suggesting that they are dependent on the function of PI-bound Sec14.
Specifically, in the absence of Vps51-53, wild type Sec14 and Sfh4 restored life but Sec14-PC did not. This implies that normal phosphoinositide metabolism is required in the absence of these components of the GARP complex. Strains lacking Vps54, Ypt6, or Trs85 only grew when transformed with a plasmid expressing wild type Sec14 indicating that both PC and phosphoinositide metabolism have to be intact and/or Sec14 has to exchange PI and PC in cis within the same protein when these functions are absent. Remarkably, in the absence of some proteins (Arl1, Vps45, and Ric1) wild type Sec14 did not restore growth, with growth of cells lacking Arl1 restored by Sec14-PC or Sfh4, and growth of cells lacking Vps45 or Ric1 restored only by Sfh4. Although Sec14 and its analogues were expressed from their own promoters, they were plasmid-borne, with Sec14 and Sec14-PC on low copy plasmids (2-3 copies per cell) and Sfh4 on a high copy 2-μm plasmid (∼20 copies per cell). The inability of plasmid-borne Sec14 to restore growth to cells lacking Arl1, Vps45, or Ric1 could be due to inhibition of growth upon increased dose of a Sec14 that exchanges PI and PC in cis and regulates both PC and phosphoinositide metabolism, whereas either one of these functions alone is not growth-inhibitory. Regulation of components of these pathways by specific lipids or membrane properties needs to be investigated to allow for a more precise determination of how lipid metabolism interfaces with and regulates vesicular trafficking.
Sec14 Domain and Regulation of GTPases—The Sec14 domain of human p50RhoGAP is required for its association with endosomes and for its interaction with the small GTPase Rab11 (20). Rab11 associates with endosomes and the trans-Golgi and regulates trafficking through these organelles. Recruitment of p50RhoGAP to activated Rab11 does not result in GAP activity toward Rab11 but instead is thought to temporally coordinate Rab and Rho signaling events. Human Rab11 is the homologue of S. cerevisiae Ypt31, and interestingly Rab11 directly interacts with mammalian PI 4-kinase β (the homologue of S. cerevisiae Pik1). Abolishing the Rab11-PI kinase β interaction resulted in mislocalization of Rab11 implying a conserved role for PI-4P in regulation of Rab signaling as inactivation of YPT31 affected growth and vesicular transport in S. cerevisiae cells with reduced Pik1 or Sec14 function (7).
Indeed, a similar coordination in Rab signaling appears to operate in S. cerevisiae. Inactivation of the S. cerevisiae Rab YPT6 was also aggravating for growth of sec14ts cells. GTP-bound Ypt6 physically interacts with the GAP for Ypt31, Gyp2, while exhibiting no GAP activity toward Ypt6 (47). Regulation of timing of the Ypt31 and Ypt6 Rab cascades has been proposed to facilitate transport between endosomes and the trans-Golgi, similar to coordination of mammalian Rab and Rho pathways. There may be a conserved role for the Sec14 domain in regulation of Rab/Rho signaling cascades.
This work was supported by the Canadian Institutes for Health Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PC, phosphatidylcholine; PI, phosphatidylinositol; GEF, guanine exchange factor; GAP, GTPase-activating protein; TRAPP, transport protein particle; GARP, Golgi-associated retrograde protein; DIC, differential interference contrast; MS, mass spectrometry; Lat-A, latrunculin-A; SNARE, soluble NSF attachment protein receptors; PI-4P, phosphatidylinositol 4-phosphate; SGA, synthetic genetic array.
References
- 1.Patton-Vogt, J. L., Griac, P., Sreenivas, A., Bruno, V., Dowd, S., Swede, M. J., and Henry, S. A. (1997) J. Biol. Chem. 272 20873-20883 [DOI] [PubMed] [Google Scholar]
- 2.Fairn, G. D., Curwin, A. J., Stefan, C. J., and McMaster, C. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 15352-15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henneberry, A. L., Lagace, T. A., Ridgway, N. D., and McMaster, C. R. (2001) Mol. Biol. Cell 12 511-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez-Murray, J. P., and McMaster, C. R. (2005) J. Biol. Chem. 280 8544-8552 [DOI] [PubMed] [Google Scholar]
- 5.Sreenivas, A., Patton-Vogt, J. L., Bruno, V., Griac, P., and Henry, S. A. (1998) J. Biol. Chem. 273 16635-16638 [DOI] [PubMed] [Google Scholar]
- 6.Cleves, A. E., McGee, T. P., Whitters, E. A., Champion, K. M., Aitken, J. R., Dowhan, W., Goebl, M., and Bankaitis, V. A. (1991) Cell 64 789-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaf, P., Zwart, W. T., van Dijken, R. A., Deneka, M., Schulz, T. K., Geijsen, N., Coffer, P. J., Gadella, B. M., Verkleij, A. J., van der Sluijs, P., and van Bergen en Henegouwen, P. M. (2004) Mol. Biol. Cell 15 2038-2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie, Z., Fang, M., and Bankaitis, V. A. (2001) Mol. Biol. Cell 12 1117-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie, Z., Fang, M., Rivas, M. P., Faulkner, A. J., Sternweis, P. C., Engebrecht, J. A., and Bankaitis, V. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12346-12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaaf, G., Ortlund, E. A., Tyeryar, K. R., Mousley, C. J., Ile, K. E., Garrett, T. A., Ren, J., Woolls, M. J., Raetz, C. R., Redinbo, M. R., and Bankaitis, V. A. (2008) Mol. Cell 29 191-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, X., Rivas, M. P., Fang, M., Marchena, J., Mehrotra, B., Chaudhary, A., Feng, L., Prestwich, G. D., and Bankaitis, V. A. (2002) J. Cell Biol. 157 63-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bankaitis, V. A., Aitken, J. R., Cleves, A. E., and Dowhan, W. (1990) Nature 347 561-562 [DOI] [PubMed] [Google Scholar]
- 13.Bankaitis, V. A., Malehorn, D. E., Emr, S. D., and Greene, R. (1989) J. Cell Biol. 108 1271-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha, B., Phillips, S. E., Bankaitis, V. A., and Luo, M. (1998) Nature 391 506-510 [DOI] [PubMed] [Google Scholar]
- 15.Chang, H. J., Jones, E. W., and Henry, S. A. (2002) Genetics 162 29-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curwin, A. J., and McMaster, C. R. (2008) Future Lipidol. 3 399-410 [Google Scholar]
- 17.D'Angelo, I., Welti, S., Bonneau, F., and Scheffzek, K. (2006) EMBO Rep. 7 174-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostenko, E. V., Mahon, G. M., Cheng, L., and Whitehead, I. P. (2005) J. Biol. Chem. 280 2807-2817 [DOI] [PubMed] [Google Scholar]
- 19.Phillips, S. E., Vincent, P., Rizzieri, K. E., Schaaf, G., Bankaitis, V. A., and Gaucher, E. A. (2006) Crit. Rev. Biochem. Mol. Biol. 41 21-49 [DOI] [PubMed] [Google Scholar]
- 20.Sirokmany, G., Szidonya, L., Kaldi, K., Gaborik, Z., Ligeti, E., and Geiszt, M. (2006) J. Biol. Chem. 281 6096-6105 [DOI] [PubMed] [Google Scholar]
- 21.Ueda, S., Kataoka, T., and Satoh, T. (2004) Cell. Signal. 16 899-906 [DOI] [PubMed] [Google Scholar]
- 22.Welti, S., Fraterman, S., D'Angelo, I., Wilm, M., and Scheffzek, K. (2007) J. Mol. Biol. 366 551-562 [DOI] [PubMed] [Google Scholar]
- 23.Panic, B., Whyte, J. R., and Munro, S. (2003) Curr. Biol. 13 405-410 [DOI] [PubMed] [Google Scholar]
- 24.Behnia, R., Panic, B., Whyte, J. R., and Munro, S. (2004) Nat. Cell Biol. 6 405-413 [DOI] [PubMed] [Google Scholar]
- 25.Setty, S. R., Strochlic, T. I., Tong, A. H., Boone, C., and Burd, C. G. (2004) Nat. Cell Biol. 6 414-419 [DOI] [PubMed] [Google Scholar]
- 26.Cai, H., Zhang, Y., Pypaert, M., Walker, L., and Ferro-Novick, S. (2005) J. Cell Biol. 171 823-833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairn, G. D., and McMaster, C. R. (2005) Methods (Orlando) 36 102-108 [DOI] [PubMed] [Google Scholar]
- 28.Tong, A. H., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., Chen, Y., Cheng, X., Chua, G., Friesen, H., Goldberg, D. S., Haynes, J., Humphries, C., He, G., Hussein, S., Ke, L., Krogan, N., Li, Z., Levinson, J. N., Lu, H., Menard, P., Munyana, C., Parsons, A. B., Ryan, O., Tonikian, R., Roberts, T., Sdicu, A. M., Shapiro, J., Sheikh, B., Suter, B., Wong, S. L., Zhang, L. V., Zhu, H., Burd, C. G., Munro, S., Sander, C., Rine, J., Greenblatt, J., Peter, M., Bretscher, A., Bell, G., Roth, F. P., Brown, G. W., Andrews, B., Bussey, H., and Boone, C. (2004) Science 303 808-813 [DOI] [PubMed] [Google Scholar]
- 29.Goldstein, A., and Lampen, J. O. (1975) Methods Enzymol. 42 504-511 [DOI] [PubMed] [Google Scholar]
- 30.Kozminski, K. G., Alfaro, G., Dighe, S., and Beh, C. T. (2006) Traffic 7 1224-1242 [DOI] [PubMed] [Google Scholar]
- 31.Chen, S. H., Chen, S., Tokarev, A. A., Liu, F., Jedd, G., and Segev, N. (2005) Mol. Biol. Cell 16 178-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morozova, N., Liang, Y., Tokarev, A. A., Chen, S. H., Cox, R., Andrejic, J., Lipatova, Z., Sciorra, V. A., Emr, S. D., and Segev, N. (2006) Nat. Cell Biol. 8 1263-1269 [DOI] [PubMed] [Google Scholar]
- 33.Liang, Y., Morozova, N., Tokarev, A. A., Mulholland, J. W., and Segev, N. (2007) Mol. Biol. Cell 18 2533-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lafourcade, C., Galan, J. M., Gloor, Y., Haguenauer-Tsapis, R., and Peter, M. (2004) Mol. Cell. Biol. 24 3815-3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harsay, E., and Schekman, R. (2002) J. Cell Biol. 156 271-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurunathan, S., David, D., and Gerst, J. E. (2002) EMBO J. 21 602-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lustgarten, V., and Gerst, J. E. (1999) Mol. Cell. Biol. 19 4480-4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson, M., Poon, P. P., Schindler, C., Murray, L. E., Kama, R., Gabriely, G., Singer, R. A., Spang, A., Johnston, G. C., and Gerst, J. E. (2006) Mol. Biol. Cell 17 1845-1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, L., and Davis, N. G. (2000) J. Cell Biol. 151 731-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vida, T. A., and Emr, S. D. (1995) J. Cell Biol. 128 779-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong, A. H., and Boone, C. (2006) Methods Mol. Biol. 313 171-192 [DOI] [PubMed] [Google Scholar]
- 42.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003) Nature 425 686-691 [DOI] [PubMed] [Google Scholar]
- 43.Phillips, S. E., Sha, B., Topalof, L., Xie, Z., Alb, J. G., Klenchin, V. A., Swigart, P., Cockcroft, S., Martin, T. F., Luo, M., and Bankaitis, V. A. (1999) Mol. Cell 4 187-197 [DOI] [PubMed] [Google Scholar]
- 44.Hama, H., Schnieders, E. A., Thorner, J., Takemoto, J. Y., and DeWald, D. B. (1999) J. Biol. Chem. 274 34294-34300 [DOI] [PubMed] [Google Scholar]
- 45.Skinner, H. B., McGee, T. P., McMaster, C. R., Fry, M. R., Bell, R. M., and Bankaitis, V. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 112-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mousley, C. J., Tyeryar, K. R., Vincent-Pope, P., and Bankaitis, V. A. (2007) Biochim. Biophys. Acta 1771 727-736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sciorra, V. A., Audhya, A., Parsons, A. B., Segev, N., Boone, C., and Emr, S. D. (2005) Mol. Biol. Cell 16 776-793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demmel, L., Gravert, M., Ercan, E., Habermann, B., Muller-Reichert, T., Kukhtina, V., Haucke, V., Baust, T., Sohrmann, M., Kalaidzidis, Y., Klose, C., Beck, M., Peter, M., and Walch-Solimena, C. (2008) Mol. Biol. Cell 19 1991-2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G., Pagé, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., Andrews, B., Tyers, M., and Boone, C. (2001) Science 294 2364-2368 [DOI] [PubMed] [Google Scholar]
- 50.Reggiori, F., Wang, C. W., Stromhaug, P. E., Shintani, T., and Klionsky, D. J. (2003) J. Biol. Chem. 278 5009-5020 [DOI] [PMC free article] [PubMed] [Google Scholar]