Abstract
Vinculin is a highly conserved and abundant cytoskeletal protein involved in linking the actin cytoskeleton to the cell membrane at sites of cellular adhesion. At these sites of adhesion, vinculin plays a role in physiological processes such as cell motility, migration, development, and wound healing. Loss of normal vinculin function has been associated with cancer phenotypes, cardiovascular disease, and lethal errors in embryogenesis. The tail domain of vinculin (Vt) binds to acidic phospholipids and has been proposed to play a role in vinculin activation and focal adhesion turnover. To better characterize Vt-lipid specificity, we conducted a series of lipid co-sedimentation experiments and find that Vt shows specific association with phosphatidylinositol 4,5-bisphosphate (PIP2), compared with phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylserine (PS), or phosphatidylinositol (PI) in the context of mixed lipid vesicles. The C terminus of Vt has been proposed to be important for PIP2 association, as various mutations and deletions within the C-terminal reduce PIP2 association. Lipid co-sedimentation and NMR analyses indicate that removal of the hydrophobic hairpin does not alter Vt structure or PIP2 association. However, more extensive deletions within the C-terminal introduce Vt structural perturbations and reduce PIP2 binding. Intriguingly, a significant increase in PIP2 binding was observed for multiple Vt variants that perturb interactions between the N-terminal strap and helix bundle, suggesting that a rearrangement of this N-terminal strap may be required for PIP2 binding.
Vinculin is a highly conserved cytoskeletal protein which localizes to points of cell adhesion and is involved in linking the actin cytoskeleton to the cell membrane (1). Sites of adhesion in which vinculin is enriched include: focal adhesions (cell-extracellular matrix), adherens junctions (cell-cell), costameres in muscle cells, and intercalated discs in cardiac cells (1-3). At these sites, vinculin is believed to play an important role in cell adhesion processes involving regulation of the actin cytoskeleton. Moreover, vinculin has been linked to pathways that control cell growth, differentiation, motility, and survival (4-7). Vinculin is also critical for proper development in model organisms (6, 8-10), and its loss in cells leads to increased motility, invasiveness, and resistance to apoptosis (5, 11, 12). Decreased vinculin expression and mutations have also been associated with human cardiomyopathies (10, 13-15).
Vinculin is a 116-kDa cytoskeletal protein, and early studies by electron microscopy and proteolytic cleavage identified a globular head domain (Vh), a flexible neck, and a tail domain (Vt)2 (16-18). The full-length structure of vinculin has been solved by x-ray crystallography, and has been described as a “bundle of bundles” (19, 20). Vh is composed of 7 helical bundles organized into 3 tandem pairs of bundles (D1-D3) and one unpaired bundle (D4) while Vt consists of a single helical bundle. The head and tail domain are connected by a flexible proline-rich region and interact to form a closed, autoinhibited conformation with Vt held in a “pincer-like” state by Vh (Fig. 1) (19). Vinculin binds a number of cytoskeletal and adhesion proteins including: actin, talin, α-actinin, α-catenin, β-catenin, vinexin, ponsin, actin-related protein complex (Arp 2/3), vasodilator-stimulated phosphoprotein (VASP), and paxillin. However, many of these interactions are at least partially masked in the intact, unstimulated protein due to autoinhibitory interactions between the head and tail domains (21-24).
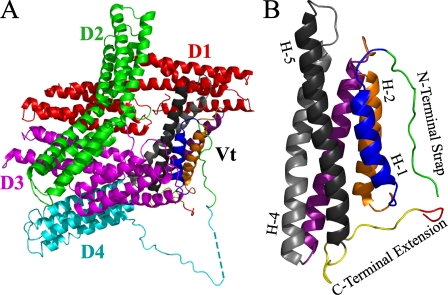
FIGURE 1.
A, a ribbon diagram illustrating the structure of full-length vinculin (PDB ID 1ST6). In the closed, autoinhibited conformation, the clamp-like head domain (Vh, D1 (red), D2 (green), D3 (magenta), and D4 (cyan)) forms a tight interaction with the tail domain (Vt, (multi-color)). Current models of vinculin activation and function require the release of the head/tail interaction to allow ligand binding. B, a ribbon diagram illustrating the isolated tail domain of vinculin (PDB ID 1ST6). The N-terminal strap and C-terminal extension are highlighted (green and yellow, respectively). The hydrophobic hairpin at the extreme C terminus is shown in red. Select helices are labeled (e.g. H-1), and for clarity, the helices are colored identically in parts A and B. A more detailed illustration, highlighting specific interactions between the N-terminal strap and C-terminal extension, is shown in Fig. 7.
Although two distinct models of vinculin activation have been proposed, one common feature of these models is that ligand binding either singly (25) or in concerted action (19) to the vinculin head and/or tail domain, cause release of the head from the tail domain to promote additional interactions. In fact, the binding of several ligands to vinculin (i.e. F-actin, acidic phospholipids, talin, and actinin) is modulated by head/tail interactions. Hence, the binding of acidic phospholipids or F-actin to the tail domain and the binding of talin or actinin to the head domain have been proposed play a role in the separation of the head and tail domains, thus activating vinculin by promoting interaction with additional ligands and/or covalent modification (25, 26). If, acidic phospholipids do participate in vinculin activation, binding of acidic phospholipids alone does not appear sufficient to disrupt the high affinity head/tail interaction (19, 27). Rather, results obtained from recent fluorescence resonance energy transfer (FRET) studies indicate that binding of both talin and actin filaments to vinculin is required for vinculin activation, suggesting an activation model involving combinatorial ligand binding to the head and tail domains (28, 29). As the binding of phospholipids and F-actin are exclusive, activation by acidic phospholipids would require a ligand distinct from F-actin, such as talin. However, given that multiple ligands have been proposed to play a role in vinculin activation, different combinations of ligands may allow vinculin activation to be spatially and temporal regulated (30). In addition to their putative role in vinculin activation, acidic phospholipids have also been proposed to facilitate PKC-phosphorylation, prevent actin binding, promote membrane association and regulate focal adhesion turnover (19, 26, 31, 32). Clearly, a better understanding of how acidic phospholipids interact with vinculin will aid in establishing its role in modulating vinculin function.
Although vinculin has been shown to bind acidic phospholipids, including phosphatidylserine (PS), phosphatidylinositol (PI), and phosphatidylinositol 4,5-bisphosphate (PIP2), the relative affinity for various acidic phospholipids under conditions that mimic physiological ionic strength and membrane composition has not been reported.
Phosphoinositides, including PIP2, have been shown to be regulated both spatially and temporally at sites of actin assembly and cytoskeletal remodeling (33-36). Although a number of structurally conserved phosphoinositides and PIP2 binding motifs have been identified (37-39), none have been found within the vinculin tail domain. The structure of Vt has been solved by x-ray crystallography, and found to be comprised of a 5-helix bundle motif, similar to that observed in the intact protein (19, 20, 26). Based on this structure, three regions of Vt were proposed to play a role in binding acidic phospholipids; a C-terminal extension containing a “hydrophobic hairpin”, a “basic collar” consisting of lysine and arginine residues from helix 1, helix 5, and the C-terminal extension which surrounding the hydrophobic hairpin, and a “basic ladder” of exposed basic residues along the length of helix 3 (26). A series of vinculin mutagenesis studies have been conducted to pinpoint the site of lipid binding within Vt. However, the data are somewhat difficult to interpret, as the number, location, and effectiveness of the mutations vary. Although some phospholipid defective variants have been characterized to determine whether the mutation(s) affect other ligand binding interactions (i.e. Vh, actin), the impact of the mutation(s) on Vt structure and stability are largely unknown. Thus, the exact site and mode of phospholipid binding is still unclear. Further complicating the field, Vt constructs of varying lengths have been used in separate studies. As the vinculin tail domain is connected to the remainder of vinculin by a flexible loop, various Vt constructs containing different N termini have been constructed. Published studies have utilized constructs containing residues 811-1066, 858-1066, 879-1066, 881-1066, and 884-1066 (26, 28, 40-42), with constructs containing residues 879-1066 and 884-1066, extensively employed for characterizing Vt-ligand interactions. For clarity, we will refer to Vt-(879-1066), the construct used to determine the crystal structure of vinculin tail domain as wild-type Vt, and the construct containing 884-1066, as VtΔN. To better characterize phospholipid binding interactions with the vinculin tail domain (Vt), we have examined the relative ability of various acidic phospholipids to associate with the vinculin tail domain using lipid co-sedimentation assays, and performed biophysical characterization and lipid binding studies on various Vt mutations that alter interactions with the N-terminal strap or within the C terminus.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—Vinculin tail (Vt) constructs containing residues 879-1066 (generously provided by Dr. Robert Liddington) and 884-1066 of chicken vinculin in a pET15b vector (Novagen), have been described previously (26, 27). Vt mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing. Vectors were transformed into Escherichia coli strain BL21(DE3), and cells were grown at 37 °C to an optical density of 0.6 (600 nm). Vt expression was initiated by addition of 0.25 mm isopropyl β-d-1-thiogalactopyranoside. Cells were grown for an additional 5 h and harvested by centrifugation. Vt was expressed at high level and purified from both the soluble and insoluble fraction. Cell pellets were resuspended in a lysis buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 5 mm imidizole, 0.1% β-mercaptoethanol (BME), and lysed by sonication. Vt, expressed in the soluble fraction, was separated from the particulate fraction by centrifugation for 1 h at 25,000 × g. The fraction containing soluble Vt, was purified by affinity separation using Ni-NTA-agarose beads (Qiagen). Vt was washed and eluted from the beads using lysis buffer containing 60 mm and 500 mm imidizole, respectively, and then dialyzed into thrombin cleavage buffer (20 mm Tris, pH 7.5, 500 mm NaCl, 2.5 mm CaCl2, 0.1% BME). The histidine (His)-tag was cleaved by thrombin (∼1 unit per 5 mg protein, Sigma) overnight at 37 °C. Cation-exchange chromatography (HiPrep 16/10 SP XL column from GE Healthcare Life Sciences) was used to further purify Vt, using a 0.05-1 m NaCl gradient in a buffer containing 20 mm Tris (pH 7.5), 2.5 mm EDTA, and 0.1% BME.
Vt was also purified from the insoluble fraction by resuspending cell pellets in 6 m guanidinium chloride (GdmCl) prior to sonication. A protocol similar to that used for soluble Vt was employed, except that purification from Ni-NTA-agarose beads was carried out under denaturing conditions. Following elution, GdmCl was removed, and Vt refolded by dialysis in a buffer containing 20 mm Tris, pH 7.5, 500 mm NaCl, and 0.1% BME. The His-tag was removed with thrombin and Vt further purified by gel filtration exchange chromatography, using methods described for the natively folded Vt protein. Proper refolding was verified by comparison of 1H-15N HSQC spectra acquired on refolded and natively folded 15N-enriched Vt.
Lipid Co-sedimentation—Lipid binding to Vt was assessed by co-sedimentation with small, unilamellar vesicles (SUV). The ability of Vt to bind PI and PS was analyzed using lipid vesicles containing 60% PE, 40% PC by weight, with either PI or PS replacing PE at the concentration indicated. PIP2 binding to Vt was characterized using vesicles containing 60% PE, 20% PC, and either 20% PS by weight or PIP2 replacing PS at the concentration indicated. For example, experiments testing the role of 10% PIP2, employed vesicles composed of 60% PC, 20% PE, 10% PS, and 10% PIP2. Vesicles were produced by combining the appropriate lipids suspended in chloroform, to produce a sample containing 250 μg of total lipid. The mixture was dried using a SpeedVac and then resuspended in 90 μl of buffer containing 40 mm 4-HEPES, 150 mm NaCl, and 2 mm dithiothreitol, pH 7.4. Resuspension and generation of the SUVs were accomplished by brief sonication with a probe tip sonicator. 10 μl of 100 μm protein (in an identical buffer) was added to each vesicle sample, producing a final volume of 100 μl and protein concentration of 10 μm. The total lipid in each sample is 250 μg or ∼3.0-3.2 mm. Samples were nutated at 4 °C for 1 h, then centrifuged at 100,000 × g for 1 h. The supernatant was removed, and the pellet resuspended in buffer containing 0.1% SDS, 25 mm glycine, and 25 mm Tris, pH 8.3. Supernatant and pellet samples were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were stained with Coomassie Blue, scanned, and protein levels quantified using Image J software (43).
NMR Samples and Spectroscopy—Bacterial-expressed 15N-labeled Vt protein was produced for nuclear magnetic resonance spectroscopy (NMR) studies by growth in M9 minimal media containing 1g/liter [15N]NH4Cl (Spectra Stable Isotopes). NMR samples were exchanged into NMR buffer (10 mm potassium phosphate, 50 mm NaCl, 2 mm dithiothreitol, 0.1% NaN3, and 10% D2O at the indicated pH) using an Amicon Ultra centrifugal filter device (10000-Da molecular weight cut-off, Millipore). 1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectra were collected on a Varian INOVA 700 MHz spectrometer at 37 °C (44). Backbone chemical shift assignments of Vt under these conditions have been reported previously (45), and have been deposited in the Biological Magnetic Resonance Data Bank (accession number 15653). NMR data processing and analysis was performed using NMRPipe (46) and NMRView (47).
Circular Dichroism—Circular dichroism (CD) spectra were collected at both near-ultraviolet (350-250 nm) and far-ultraviolet (260-190 nm) spectral regions. All spectra were acquired at 25 °C in a buffer contained 10 mm potassium phosphate, 50 mm Na2SO4, and 1 mm dithiothreitol, pH 7.5 using an Applied Photophysics Pistar-180 spectrometer. Protein concentrations were 0.45 mm and 5 μm for near-UV and far-UV CD data collection, respectively.
RESULTS
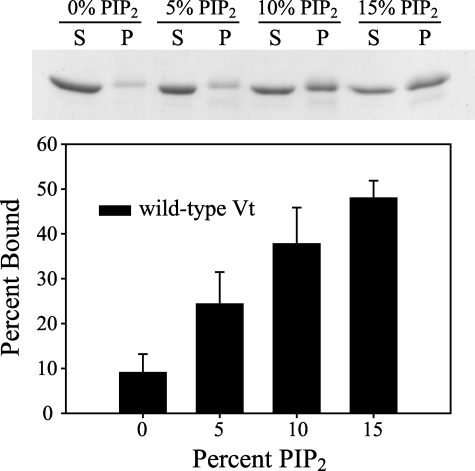
The Vinculin Tail Domain Shows Specificity for PIP2-containing Vesicles—Although the tail domain of vinculin has been reported to bind acidic phospholipids and PIP2, both the constructs used and the methods used to assess acidic phospholipid binding have varied significantly, making comparison of separate reports on lipid binding difficult (19, 26, 27, 31, 48, 49). To clarify the affinity and specificity of Vt for PIP2, we performed co-sedimentation experiments with mixed PE, PC, and PS vesicles, examining the effect of increasing PIP2 concentration. Although Vt-(884-1066) has previously been shown to bind to pure PS and PI vesicles at physiological ionic strength (27), we found that under physiological lipid and salt concentrations, no significant binding of Vt-(879-1066) to PS or PI in mixed lipid vesicles was observed. In the absence of PIP2, little co-sedimentation of Vt with mixed lipid vesicles containing 60% PC, 20% PE, and 20% PS vesicles was observed in 150 mm NaCl (Fig. 2). Upon the addition of PIP2, however, a clear concentration-dependent co-sedimentation was observed, indicating that Vt specifically recognizes PIP2.
FIGURE 2.
Lipid binding properties of wild-type Vt was examined by co-sedimentation with lipid vesicles containing 60% PE, 20% PC, and either 20% PS by weight or PIP2 at concentrations that replace PS. Soluble (S) and pellet (P) fractions were analyzed by SDS-polyacrylamide gel electrophoresis (stained with Coomassie Blue), with a representative gel shown. Gels were scanned and the amount of protein in each fraction quantified using ImageJ software. Wild-type Vt shows minimal binding to vesicles containing 60% PE, 20% PC, and 20% PS (displayed as 0% PIP2). In the presence of PIP2, a dose-dependent increase in lipid association was observed, suggesting specificity for PIP2 association. Error bars represent a S.D. of more than 3 separate measurements.
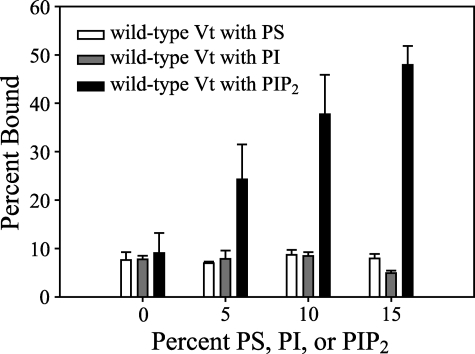
The vinculin tail domain has also been reported to interact with pure PI vesicles (using a 30-kDa V8 proteolytic fragment) (27) and pure PS vesicles (using Vt-(879-1066)) (26). To ascertain the relative affinity of Vt for PI, PS, and PIP2, Vt association was observed in mixed lipid vesicles containing each of these lipids. As shown in Fig. 3, Vt does not bind PI or PS significantly in the context of mixed lipid vesicles, and demonstrated a marked preference for PIP2 over either PI or PS.
FIGURE 3.
To compare the relative affinity of Vt for PS, PI, and PIP2, the ability of Vt to co-sediment with mixed lipid vesicles containing PE, PC, and either PS or PI was assessed. These results indicate that Vt specifically recognizes PIP2 relative to PS and PI, as concentration-dependent binding to PIP2 is observed, with minimal binding of PS and PI observed at concentrations up to 15%. Error bars represent the S.D. of >3 separate measurements.
Vt Demonstrates Loss of Tertiary in Lipid Micelles—As shown in Figs. 2 and 3, the vinculin tail domain shows enhanced association with PIP2 in lipid co-sedimentation relative to PS and PI. In an effort to determine the site(s) of interaction between Vt with PIP2, we employed CD and solution NMR spectroscopy. Studies of phospholipid interactions were initiated using micelles as opposed to larger vesicles. However, introduction of dodecylphosphocholine (DPC) at concentrations that promote micelle formation causes a collapse in Vt structure, as determined by both near-UV (ultraviolet) CD and 1H-15N heteronuclear two-dimensional NMR.
Near-UV CD is sensitive to the tertiary packing of aromatic residues and therefore the tertiary structure of proteins, whereas far-UV CD is sensitive to the conformation of the peptide backbone and therefore the secondary structure of proteins. The far-UV CD of Vt exhibits only minor changes in the presence of 100 mm DPC indicating there is no significant change in the secondary structure whereas the near-UV CD spectra exhibits a significant loss of signal, indicating a loss of tertiary structure (Fig. 4A).
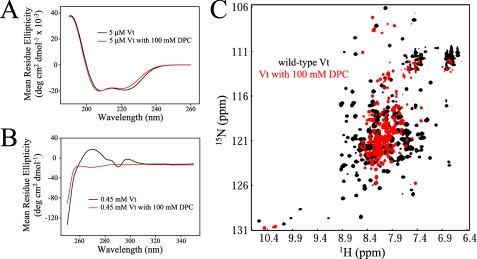
FIGURE 4.
CD and NMR analysis of Vt in the presence of DPC micelles. Comparison of far (A) and near (B) UV CD spectra of Vt in the presence and absence of 100 mm DPC. While far UV CD spectra show similar spectral profiles, significant alterations are observed in the near UV CD spectra. Results from these analyses indicate that DPC micelles cause a loss in Vt tertiary structure but maintain helical secondary structure. The data are further supported by NMR studies. 1H-15N HSQC NMR spectra were collected on 0.15 mm Vt at pH 5.5, both alone and in the presence of 100 mm DPC (C). The loss of spectral dispersion and the increase in resonances with chemical shifts close to the random coil values (∼8.3 ppm), suggest a loss in Vt tertiary structure upon association with DPC micelles. As vinculin does not appear to specifically bind PC, interactions with DPC micelles are likely to be nonspecific. In addition, similar structural perturbations were observed with all lipid micelles tested (i.e. LPPG and short-chain PIP2), suggesting that lipid micelles may act as a detergent, unfolding Vt.
The 1H-15N HSQC NMR spectra detect signals for protons attached to 15N nuclei. The backbone NH of each amino acid (with the exception of proline) providing a residue specific probe sensitive to changes in its electrochemical environment. 1H-15N HSQC spectral dispersion arises from the unique environment of each distinct NH pair. Loss of a distinct, folded structure results in convergence of resonances toward random coil chemical shifts (centered at ∼8.3 ppm). The significant loss of spectral dispersion observed for Vt in the presence of 100 mm DPC (Fig. 4B) is indicative of a significant loss in tertiary structure, in agreement with the near-UV CD. The residual dispersion observed by NMR, also agrees with the persistence of helices as detected by far-UV CD.
Although Vt has been proposed to undergo a conformational change upon association with acidic phospholipids (26), DPC micelles may act as a detergent causing unfolding of Vt. As Vt does not bind to PC containing vesicles, the interaction with DPC micelles may be nonspecific. Adding to this possibility, a similar collapse in NMR chemical shift dispersion was observed in 1H-15N HSQC spectra of Vt in the presence of 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LPPG) (data not shown).
Attempts to map the PIP2 interaction site by NMR using the PIP2 headgroup, d-myo-inositol-1,4,5-triphosphate or a short chain (C8) derivative of PIP2 were also unsuccessful (data not shown), as a clear, specific binding site was not observed. These results indicate that the head group alone does not have sufficient affinity for Vt to either bind specifically or promote a conformational change in Vt, necessary for high affinity binding.
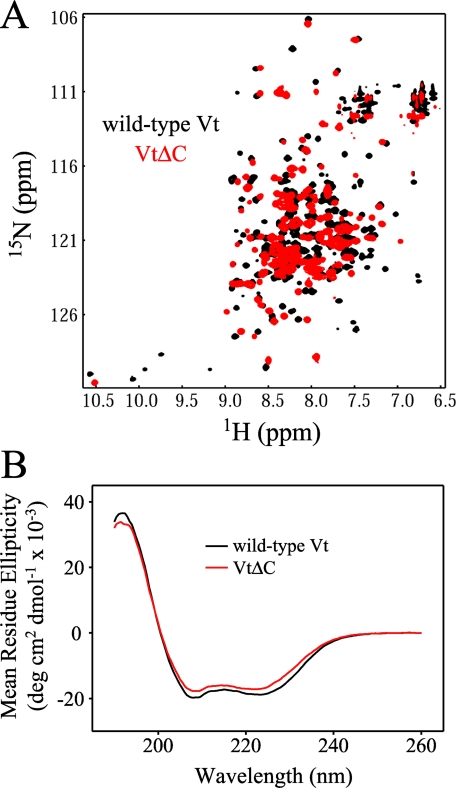
Certain C-terminal Residues Stabilize the Tertiary Fold of Vt—A C-terminal deletion variant of Vt reported to be deficient in lipid binding has been utilized in multiple studies (19, 26, 31, 32). This mutant, VtΔC, lacks 15 C-terminal amino acids (1052-1066). The deletion of this fragment has been reported to decrease PIP2 binding but does not significantly affect interaction with either actin or the head domain of vinculin (26, 32). However, VtΔC demonstrates an increased susceptibility to protease degradation (26, 32) as well as alterations in one dimensional 1H NMR spectra, indicating loss of structure (32). As residues in the C terminus of Vt form tertiary interactions with other residues in the tail domain, the loss of these interactions upon deletion could alter the structure and stability of Vt. In particular, the deletion results in removal of tryptophan 1058 (Trp-1058), which packs against tryptophan 912 (Trp-912) in the loop between helix 1 and 2. Interestingly, both tryptophan residues are conserved in all vinculins as well as α-catenin, a cell adhesion protein with high homology to vinculin (32, 50). Along with Trp-1058, VtΔC removes leucine 1056 (Leu-1056), which also packs into the base of the hydrophobic core. Loss of these packing interactions could alter Vt structure resulting in destabilization. In full-length vinculin, there is also an interaction between the C terminus and the N-terminal strap of Vt, with Asp-882 of the N-terminal strap making polar interactions with C-terminal residues, Lys-1061 and Tyr-1065 (19). To better understand the role of the Vt C terminus in lipid binding, we conducted NMR and CD analyses of the VtΔC variant. As reported previously, we found VtΔC to be significantly more susceptible to proteolytic cleavage than wild-type Vt (26, 32). Moreover, two-dimensional 1H-15N (NMR) spectra of 15N-enriched wild-type and VtΔC, indicate that the C-terminal deletion results in significant loss of tertiary structure. As shown in Fig. 5A, the 1H-15N HSQC spectra of wild-type Vt displays spectral dispersion consistent with that of a well folded α-helical protein. In contrast, the 1H-15N spectrum of VtΔC exhibits increased spectral overlap, resulting from a collapse in chemical shift dispersion and an increase in the number of resonances with random coil chemical shifts (Fig. 5A). Moreover, the majority of resonances exhibit chemical shift changes. These NMR data support a loss of structural stability upon deletion of the C-terminal residues.
FIGURE 5.
NMR and CD analysis of the lipid-defective C-terminal deletion mutant, VtΔC. Compared with wild-type Vt, the 1H-15N 2D NMR spectra of VtΔC(A) shows increased overlap resulting from a collapse in chemical shift dispersion and an increase of resonances with random coil chemical shifts (∼8.3 ppm), consistent with a loss of structural stability due to the C-terminal deletion (VtΔC in red overlaid on wild-type Vt in black). In contrast, far UV CD spectra of wild-type Vt and VtΔC are similar (B), suggesting retention in the overall helical content.
While the near UV CD (350-250 nm) of VtΔC could not be directly compared with wild-type Vt due to the removal two of the three tryptophan residues in the protein (Trp residues dominate absorption in the near UV CD spectrum), far UV CD (260-190 nm) which is sensitive to the secondary structure of proteins, suggests that the helical content of wild-type Vt and VtΔC are nearly identical (Fig. 5B). Together, this data suggest that while the secondary structure of VtΔC is largely unaltered relative to wild-type Vt, its tertiary conformation and stability may be significantly altered.
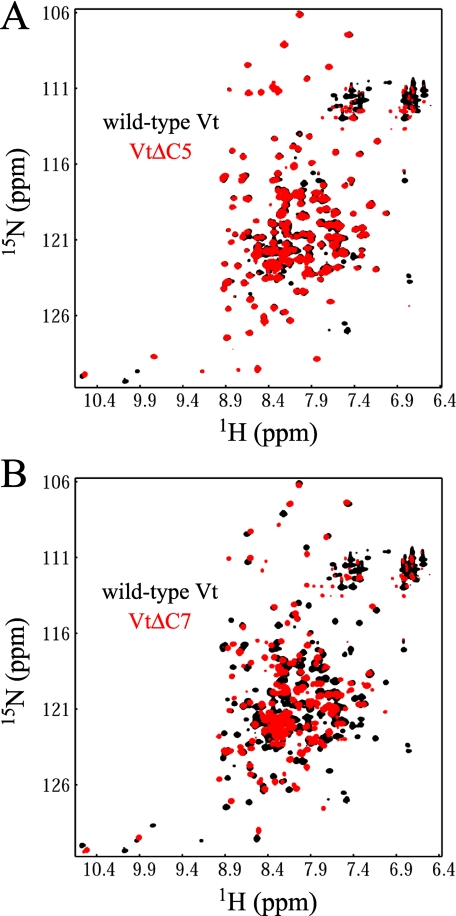
Structural changes in VtΔC have been attributed, at least in part, to loss of Trp-1058 (32). However, other residues associated with the C-terminal deletion may be involved in lipid binding. The “hydrophobic hairpin” (TPWYQ at the extreme C terminus) has been postulated to be important for vinculin insertion into the membrane, and mutagenesis studies indicate that Arg-1057, Arg-1060, and Lys-1061 (part of the “basic collar”) may be involved in binding acidic lipid head groups (26, 31). To elucidate the role of these residues in Vt structural integrity, we generated two new C-terminal deletion mutants, VtΔC5 and VtΔC7. VtΔC5 is a deletion which removes 5 amino acids from Vt that make up the hydrophobic hairpin (TPWYQ), whereas in VtΔC7, two additional amino acids are deleted, Arg-1060 and Lys-1061, which are part of the basic collar. As shown in Fig. 6A, the 1H-15N HSQC spectra of VtΔC5 is nearly identical to that of wild-type Vt, suggesting that removal of the hydrophobic hairpin has a minimal effect on the structure of the protein. In contrast, the 1H-15N HSQC spectra of VtΔC7 exhibits significant spectral changes, showing some similarity to that observed in VtΔC (chemical shift changes, increase in overlap, increase in resonances with random coil chemical shifts), although the spectra changes observed for VtΔC7 are not as extensive as those observed for VtΔC (Figs. 5A and 6B). Thus, our NMR data indicate that Arg-1060 and Lys-1061 may play a role in maintaining the tertiary fold of Vt. These observations are consistent with observations that these residues form tertiary contacts with other residues in the crystal structure of full-length vinculin. Located in the C terminus, Arg-1060 and Lys-1061 make contacts with residues in the N terminus and in the helix 1-2 loop, including polar contacts with Asp-882 (N terminus) and Lys-911 (helix 1-2 loop). Although the effect of deleting these residues may differ significantly from that of point mutations, it remains possible that mutation of these residues could affect the structure or stability of Vt.
FIGURE 6.
To assess the role the hydrophobic hairpin and C-terminal basic collar residues on Vt structure, two deletion mutants, VtΔC5 and VtΔC7, were created. VtΔC5 removes only the hydrophobic hairpin (the final five C-terminal residues), while VtΔC7 removes an additional two residues that are part of the basic collar (R1060 and K1061). As observed in A, the 1H-15N HSQC spectra of VtΔC5 is nearly identical to that of wild-type Vt, indicating that deletion of the hydrophobic hairpin does not alter the structure of Vt. In contrast, the 1H-15N HSQC spectrum of VtΔC7 (B) shows large scale spectral changes compared with wild-type Vt, with both intensity and chemical shift changes observed for the majority of NH resonances. Moreover, several of the NH resonances show higher intensity, random coil chemical shifts, suggesting some loss of structure. However, the spectral changes observed for VtΔC7 are not as extensive as those observed for VtΔC (Fig. 5). These data indicate that deletion of R1060 and K1061 in the C terminus of Vt causes a loss in structure, suggesting that these residues play a role in the tertiary stability of Vt.
The Role of the N-terminal Strap in PIP2 Binding—A significant number of mutations have been made in Vt to assess their affect on lipid binding. In addition to the VtΔC deletion mutant, a variety of basic residues have been mutated, targeted predominately to residues in the “basic collar” and “basic ladder” (19, 31, 32). To elucidate lipid binding determinants in Vt, we characterized a series of mutants in both Vt-(879-1066) as well as a shorter construct of Vt containing residues 884-1066, referred to here as VtΔN (27).
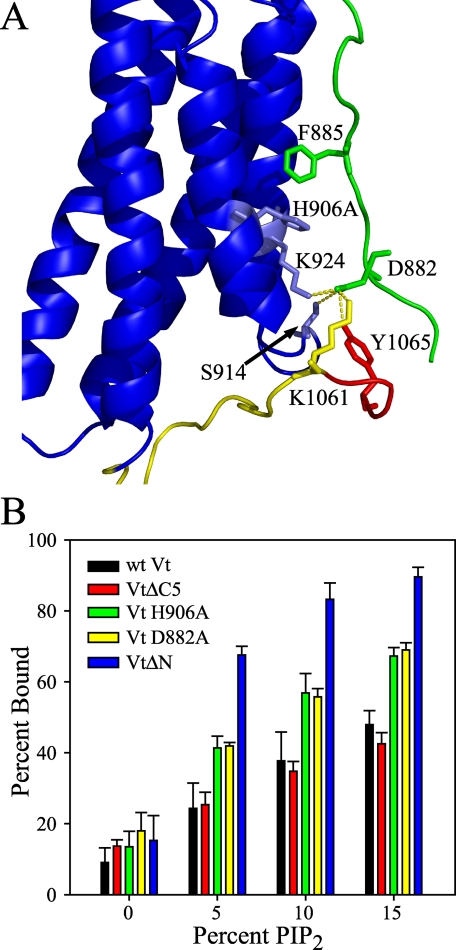
The N terminus of Vt contains a “strap”, residues 879-892, which is found in an extended conformation and packs against the interface of helices 1 and 2 in the crystal structure of full-length vinculin (19). The N-terminal strap is found in multiple conformations in the crystal structure of Vt, suggesting conformational flexibility (26). In the full-length crystal structure, the N-terminal strap forms contacts with the Vt helix bundle via contacts with both phenylalanine 885 (Phe-885) and aspartic acid 882 (Asp-882). The side chain of Phe-885 packs against the side chain of histidine 906 (His-906) in helix 1, while Asp-882 makes a number of polar contacts with other residues in Vt, including Ser-914 (helix 1-2 loop), Lys-924 (helix 2), and Lys-1061 and Tyr-1065 in the C terminus (Fig. 7A). Thus, removal of 5 amino acids from the N terminus of Vt (i.e, VtΔN) results in the loss of Asp-882, and multiple interactions between Asp-882 and the Vt helix bundle and C terminus. Comparison of 1H-15N HSQC spectra of 15N-enriched wild-type Vt and VtΔN, show differences in NH chemical shifts associated predominately with residues in the N-terminal strap, the helix 1-2 interface, and the C terminus, consistent with the loss of contacts between these regions and the strap, due to the deletion of 5 N-terminal residues. The lack of chemical shift changes for most residues in the helix core, suggest that the helix bundle remains intact. Molecular dynamics (MD) simulations of VtΔN were also conducted using the x-ray crystal structure of Vt (taken from PDB ID 1ST6) as a starting point and removing the 5 N-terminal amino acids. Consistent with our NMR data, results from the MD simulations of VtΔN show an intact helical bundle but flexible N and C termini (data not shown). Hence, our computational and NMR data suggest that deletion of the N terminus of Vt perturbs interactions between the strap, helix 1-2 interface, and C terminus, but does not cause a large scale structural changes in the 5-helix bundle fold. This is consistent with previous observations, showing that VtΔN can interact with acidic phospholipids and Vh, and can bind/bundle F-actin (21, 27, 29, 30, 51).
FIGURE 7.
A, structure of Vt (from 1ST6), highlighting the interactions of the N-terminal strap of Vt with the helix 1-2 interface and the C terminus. The N-terminal strap of Vt is depicted in green, while the C terminus is shown in yellow (with the hydrophobic hairpin shown in red). The N-terminal strap of Vt packs in an extended conformation with the helix 1-2 interface. Two residues within the strap, F885 and D882, form multiple interactions within Vt that are likely to stabilize the extended conformation of the strap. F885 of the N-terminal strap packs against H906 of helix 1, while D882 forms multiple polar interactions with S914 (helix 1-2 loop), K924 (helix 2), and K1061 and Y1065 in the C terminus of Vt. B, while wild-type Vt exhibits a PIP2-dependent association with lipid vesicles, a number of Vt mutants show a marked increase in PIP2 affinity relative to wild-type Vt. Of these, VtΔN, Vt D882A, and Vt H906A are likely to perturb interactions between the Vt-helix bundle and the strap, thereby increasing conformational mobility of the N-terminal strap. The increase in PIP2 binding observed with these mutants is consistent with the hypothesis that a conformational change or removal of the N-terminal strap of Vt facilitates higher affinity PIP2 association. Error bars represent the S.D. of >3 separate measurements.
Intriguingly, we found the lipid binding capacity of VtΔN to be significantly different from wild-type Vt. While the lipid binding specificity of VtΔN was nearly identical to Vt (minimal binding to PS and PI, data not shown), the affinity for PIP2 was considerably higher (Fig. 7B). As one of the points of interaction between the N-terminal strap and the helical bundle (Asp-882) has been removed in VtΔN, we hypothesized that the enhancement observed in PIP2 binding may be due to release or partial release of strap. To test this hypothesis, we conducted lipid co-sedimentation assays on two mutants, Vt D882A and Vt H906A. Mutation of either of these residues should disrupt interactions with the N-terminal strap. In particular, mutation of His-906 to alanine should disrupt packing with Phe-885 while the aspartic acid to alanine mutation at 882 should disrupt polar contacts Ser-914, Lys-924, Lys-1061, and Tyr-1065. Results shown in Fig. 7B, indicate that both mutants significantly increase the co-sedimentation of Vt with PIP2 containing lipids, consistent with our hypothesis that release of the N-terminal strap increases PIP2 binding. As the base lipid vesicle composition in the 0% PIP2 samples contain 20% PS, these results indicate that the Vt variants (VtΔN, VtD882, and VtH906A) retain specificity for PIP2 over PS.
As previously mentioned, we found that changes in the 1H-15N HSQC spectra of VtΔC are indicative of loss in structure relative to Vt, while VtΔC5, which removes only the hydrophobic hairpin at the C terminus of Vt, shows similar NMR spectra to wild-type Vt, indicating a similar fold. As VtΔC5 does not appear to perturb the tertiary fold of Vt and is postulated to play a role in membrane insertion, we conducted lipid co-sedimentation experiments with VtΔC5. As shown in Fig. 7B, we find that VtΔC5 exhibits similar binding to PIP2 containing vesicles, indicating that the hydrophobic hairpin is not critical for lipid binding.
DISCUSSION
The regulation of the actin cytoskeleton, its connections to the cell membrane, and the linkage to neighboring cells or extra cellular matrix, play an integral part in many physiological and pathological processes. Cell processes including migration, differentiation, proliferation, and survival, along with larger scale processes such as tissue organization, wound healing, and tumorigenesis are regulated in part by dynamic regulation of cell adhesions and the actin cytoskeleton (52-55). Motility changes required for many of theses processes involve dynamic creation, stabilization, and turnover of sites of adhesion (56, 57), with vinculin playing an important role (5, 31, 32, 40, 58). The activation and function of vinculin has been shown to be spatially and temporally regulated in cells, and vinculin is believed to play an integral role in strengthening of adhesions (59, 60). Intriguingly, interactions with lipids play a role in the regulation of adhesion site turnover (31, 32). The lipid binding function of vinculin is localized to the tail domain (Vt), which has previously been shown to associate with acidic phospholipids, including PS, PI, and PIP2 (22, 27). Of these, PIP2 is of particular interest, as it is known to be an important regulator of the actin cytoskeleton (39). Many of the reports of Vt lipid binding were in the context of pure lipid vesicles. Hence, we were interested in assessing phospholipid binding using mixed lipid vesicles that better mimic cellular membranes. We found that Vt does not bind significantly to vesicles containing PE, PC, and PS, while demonstrating a significant concentration dependent binding to PIP2 containing vesicles (Fig. 2). Although Vt has been shown to bind pure PI and PS vesicles, we observed only minimal co-sedimentation with mixed lipid vesicles containing PI or PS (Fig. 3). Local concentrations of PIP2 are controlled by both synthesis and sequestration, and regulated by signaling pathways known to affect the actin cytoskeleton (38). The selective affinity of vinculin for PIP2 suggests that this interaction may provide an important link in the regulation of actin cytoskeletal dynamics.
Our lipid co-sedimentation assays indicate that Vt exhibits specificity for binding to PIP2, but does not specifically associate with vesicles containing mixtures of PE, PC, and PS. It is of interest to note that while vinculin does not appear to specifically bind PC, at the 100 mm concentration used for NMR we see interactions with dodecylphosphocholine (DPC) micelles. In fact our CD and NMR data suggest that, while secondary structure may be retained, there is a loss of tertiary structure and lipid specificity upon association with micelles (Fig. 4). These observations suggest that lipid micelles may function as a detergent and unfold Vt. Therefore, micelles may not mimic physiological lipid interactions and caution should be used in interpreting data on Vt in micelles.
How PIP2 interacts with Vt to modulate vinculin function remains unclear. Vinculin does not contain known PIP2 binding motifs (37), and mutation and deletions reported to block lipid binding have not identified a clear site of binding (19, 22, 26, 27, 31, 32). Supporting the observations of Saunders et al. (32), our data support a loss in structural stability associated with the C-terminal deletion mutant VtΔC (Fig. 5). Hence, loss of lipid binding may result from an altered tertiary structure. In contrast, removal of the hydrophobic hairpin does not significantly alter PIP2 association and does not appear to alter Vt structure. Thus, although the hydrophobic hairpin has previously proposed to be important for insertion of vinculin into membranes (26), our results indicate that removal of the hydrophobic hairpin (TPWYQ) is not critical for lipid insertion. Removal of an additional two amino acids from the C terminus, VtΔC7, causes spectral perturbations that may be indicative of a structural change, albeit not as extensive as those observed for VtΔC (Fig. 6). These results indicate that perturbations of contacts between the basic collar residues, Arg-1060 and Lys-1061, with the helix 1-2 loop and the N-terminal strap, may alter Vt structure, which should be noted when interpreting the lipid binding data of Vt variants containing mutations at these positions.
Intriguingly, analysis of lipid binding data on multiple Vt mutants (VtΔN, D882A, and H906A), indicates that perturbation of interactions with the N-terminal strap of Vt enhances PIP2 association (Fig. 7). It is possible that the release of this strap may expose a surface important for lipid binding, allowing for the formation of a lipid binding surface not present in the closed conformation, or more readily allow a conformational change required for lipid binding. It is intriguing to speculate that residues in the basic collar (Lys-911 and Lys-924) become more accessible for interaction with PIP2 upon release of the N-terminal strap, as these residues have been proposed to be important for PIP2 association (19, 32). Additionally, as the N-terminal strap makes interactions with the C terminus, it is possible that rearrangement of the strap could cause perturbations or increased conformational mobility in the C terminus, which could facilitate PIP2 interactions. It has also been reported that a rearrangement in the N-terminal strap of Vt may be required for F-actin bundling (61), and that PIP2 binding inhibits interactions of vinculin with F-actin (30, 48). Rearrangement of the N-terminal strap of Vt may be a common requirement for the binding of either PIP2 or F-actin, with both ligands sharing a mutually exclusive, overlapping site of interaction. This would be consistent with the hypothesis that PIP2 binding may displace vinculin from F-actin, allowing focal adhesion turnover as has been proposed (31, 32).
In summary, results from our studies indicate that Vt shows specificity for PIP2 in mixed lipid vesicles that better mimic membrane lipid composition than those used in earlier studies. In particular, Vt binds to lipid vesicles in a PIP2-dependent manner, with minimal binding in the absence of PIP2. In contrast, no significant binding was observed with lipid vesicles containing up to 15% PS or PI. However, in the presence of lipid micelles, Vt loses tertiary structure while retaining secondary structure. Additionally, while Vt does not specifically interact with PC in larger lipid vesicles, it does interact with DPC in micelles, suggesting lipid specificity may be lost in the context of lipid micelles. Thus, Vt-micelle interactions may not adequately reflect Vt-membrane interactions. Furthermore, our CD and NMR data indicate that the mutant, VtΔC, reported to be deficient in lipid binding, appears to have significantly altered tertiary structure, likely due to deletion of packed hydrophobic residues (Leu-1056 and Trp-1058). A loss of tertiary structure is also observed for the shorter C-terminal deletion mutant, VtΔC7, suggesting that the basic collar residues Arg-1060 and Lys-1061, may be important for the structural integrity of Vt. In contrast, removal of the hydrophobic hairpin (i.e. VtΔC5) shows no evidence of structural perturbation and retains PIP2 binding, suggesting the hydrophobic hairpin is not critical for lipid interactions. Conversely, mutations likely to alter the interaction of the N-terminal strap of Vt (VtΔN, VtD882, and VtH906A) exhibit significant increases in PIP2 binding and retain PIP2 specificity, suggesting that release of the N terminus strap promotes PIP2 binding. Thus, PIP2 binding may require a conformational change in the N-terminal strap to promote a higher affinity PIP2 association.
Acknowledgments
We thank Dr. Greg Young, the NMR facility manager at UNC-CH, for assistance in NMR data collection, Dr. Ashutosh Tripathy, Director of the UNC Macromolecular Interactions Facility, for assistance with CD, Dr. Francesca Marassi for help with the lipid micelle experiments, and Dr. Brenda Temple, director of the R. L. Juliano Structural Bioinformatics Core Facility, for assistance with molecular dynamics simulations.
This work was authored, in whole or in part, by NIH staff. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Vt, Vinculin tail domain; PIP2, phosphatidylinositol 4,5-bisphosphate; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine; PI, phosphatidylinositol; DPC, dodecylphosphocholine.
References
- 1.Geiger, B., Tokuyasu, K. T., Dutton, A. H., and Singer, S. J. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 4127-4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkin, A. M., Ornatsky, O. I., Kabakov, A. E., Glukhova, M. A., and Koteliansky, V. E. (1988) J. Biol. Chem. 263 6631-6635 [PubMed] [Google Scholar]
- 3.Pardo, J. V., Siliciano, J. D., and Craig, S. W. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 1008-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coll, J. L., Ben-Ze'ev, A., Ezzell, R. M., Rodriguez Fernandez, J. L., Baribault, H., Oshima, R. G., and Adamson, E. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9161-9165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subauste, M. C., Pertz, O., Adamson, E. D., Turner, C. E., Junger, S., and Hahn, K. M. (2004) J. Cell Biol. 165 371-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu, W., Baribault, H., and Adamson, E. D. (1998) Development 125 327-337 [DOI] [PubMed] [Google Scholar]
- 7.Xu, W., Coll, J. L., and Adamson, E. D. (1998) J. Cell Sci. 111 1535-1544 [DOI] [PubMed] [Google Scholar]
- 8.Barstead, R. J., and Waterston, R. H. (1991) J. Cell Biol. 114 715-724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemljic-Harpf, A. E., Miller, J. C., Henderson, S. A., Wright, A. T., Manso, A. M., Elsherif, L., Dalton, N. D., Thor, A. K., Perkins, G. A., McCulloch, A. D., and Ross, R. S. (2007) Mol. Cell. Biol. 27 7522-7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zemljic-Harpf, A. E., Ponrartana, S., Avalos, R. T., Jordan, M. C., Roos, K. P., Dalton, N. D., Phan, V. Q., Adamson, E. D., and Ross, R. S. (2004) Am. J. Pathol. 165 1033-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez Fernandez, J. L., Geiger, B., Salomon, D., and Ben-Ze'ev, A. (1993) J. Cell Biol. 122 1285-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez Fernandez, J. L., Geiger, B., Salomon, D., Sabanay, I., Zoller, M., and Ben-Ze'ev, A. (1992) J. Cell Biol. 119 427-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasile, V. C., Will, M. L., Ommen, S. R., Edwards, W. D., Olson, T. M., and Ackerman, M. J. (2006) Mol. Genet. Metab. 87 169-174 [DOI] [PubMed] [Google Scholar]
- 14.Vasile, V. C., Edwards, W. D., Ommen, S. R., and Ackerman, M. J. (2006) Biochem. Biophys. Res. Commun. 349 709-715 [DOI] [PubMed] [Google Scholar]
- 15.Vasile, V. C., Ommen, S. R., Edwards, W. D., and Ackerman, M. J. (2006) Biochem. Biophys. Res. Commun. 345 998-1003 [DOI] [PubMed] [Google Scholar]
- 16.Groesch, M. E., and Otto, J. J. (1990) Cell Motil. Cytoskeleton 15 41-50 [DOI] [PubMed] [Google Scholar]
- 17.Molony, L., and Burridge, K. (1985) J. Cell. Biochem. 29 31-36 [DOI] [PubMed] [Google Scholar]
- 18.Winkler, J., Lunsdorf, H., and Jockusch, B. M. (1996) J. Struct. Biol. 116 270-277 [DOI] [PubMed] [Google Scholar]
- 19.Bakolitsa, C., Cohen, D. M., Bankston, L. A., Bobkov, A. A., Cadwell, G. W., Jennings, L., Critchley, D. R., Craig, S. W., and Liddington, R. C. (2004) Nature 430 583-586 [DOI] [PubMed] [Google Scholar]
- 20.Borgon, R. A., Vonrhein, C., Bricogne, G., Bois, P. R., and Izard, T. (2004) Structure (Camb) 12 1189-1197 [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. P., and Craig, S. W. (1994) J. Biol. Chem. 269 12611-12619 [PubMed] [Google Scholar]
- 22.Johnson, R. P., and Craig, S. W. (1995) Biochem. Biophys. Res. Commun. 210 159-164 [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R. P., and Craig, S. W. (1995) Nature 373 261-264 [DOI] [PubMed] [Google Scholar]
- 24.Ziegler, W. H., Liddington, R. C., and Critchley, D. R. (2006) Trends Cell Biol. 16 453-460 [DOI] [PubMed] [Google Scholar]
- 25.Izard, T., Evans, G., Borgon, R. A., Rush, C. L., Bricogne, G., and Bois, P. R. (2004) Nature 427 171-175 [DOI] [PubMed] [Google Scholar]
- 26.Bakolitsa, C., de Pereda, J. M., Bagshaw, C. R., Critchley, D. R., and Liddington, R. C. (1999) Cell 99 603-613 [DOI] [PubMed] [Google Scholar]
- 27.Johnson, R. P., Niggli, V., Durrer, P., and Craig, S. W. (1998) Biochemistry 37 10211-10222 [DOI] [PubMed] [Google Scholar]
- 28.Chen, H., Choudhury, D. M., and Craig, S. W. (2006) J. Biol. Chem. 281 40389-40398 [DOI] [PubMed] [Google Scholar]
- 29.Cohen, D. M., Chen, H., Johnson, R. P., Choudhury, B., and Craig, S. W. (2005) J. Biol. Chem. 280 17109-17117 [DOI] [PubMed] [Google Scholar]
- 30.Steimle, P. A., Hoffert, J. D., Adey, N. B., and Craig, S. W. (1999) J. Biol. Chem. 274 18414-18420 [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekar, I., Stradal, T. E., Holt, M. R., Entschladen, F., Jockusch, B. M., and Ziegler, W. H. (2005) J. Cell Sci. 118 1461-1472 [DOI] [PubMed] [Google Scholar]
- 32.Saunders, R. M., Holt, M. R., Jennings, L., Sutton, D. H., Barsukov, I. L., Bobkov, A., Liddington, R. C., Adamson, E. A., Dunn, G. A., and Critchley, D. R. (2006) Eur. J. Cell Biol. 85 487-500 [DOI] [PubMed] [Google Scholar]
- 33.Botelho, R. J., Teruel, M., Dierckman, R., Anderson, R., Wells, A., York, J. D., Meyer, T., and Grinstein, S. (2000) J. Cell Biol. 151 1353-1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holz, R. W., Hlubek, M. D., Sorensen, S. D., Fisher, S. K., Balla, T., Ozaki, S., Prestwich, G. D., Stuenkel, E. L., and Bittner, M. A. (2000) J. Biol. Chem. 275 17878-17885 [DOI] [PubMed] [Google Scholar]
- 35.Honda, A., Nogami, M., Yokozeki, T., Yamazaki, M., Nakamura, H., Watanabe, H., Kawamoto, K., Nakayama, K., Morris, A. J., Frohman, M. A., and Kanaho, Y. (1999) Cell 99 521-532 [DOI] [PubMed] [Google Scholar]
- 36.Stauffer, T. P., Ahn, S., and Meyer, T. (1998) Curr. Biol. 8 343-346 [DOI] [PubMed] [Google Scholar]
- 37.Cullen, P. J., Cozier, G. E., Banting, G., and Mellor, H. (2001) Curr. Biol. 11 R882-893 [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin, S., Wang, J., Gambhir, A., and Murray, D. (2002) Annu. Rev. Biophys. Biomol. Struct. 31 151-175 [DOI] [PubMed] [Google Scholar]
- 39.Yin, H. L., and Janmey, P. A. (2003) Annu. Rev. Physiol. 65 761-789 [DOI] [PubMed] [Google Scholar]
- 40.Goldmann, W. H., and Ingber, D. E. (2002) Biochem. Biophys. Res. Commun. 290 749-755 [DOI] [PubMed] [Google Scholar]
- 41.Weekes, J., Barry, S. T., and Critchley, D. R. (1996) Biochem. J. 314 827-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziegler, W. H., Tigges, U., Zieseniss, A., and Jockusch, B. M. (2002) J. Biol. Chem. 277 7396-7404 [DOI] [PubMed] [Google Scholar]
- 43.Rasband, W. S. (2006) Image J, Version 1.37c, National Institutes of Health, Bethesda, MD
- 44.Kay, L., Keifer, P., and Saarinen, T. (1992) J. Am. Chem. Soc. 114 10663-10665 [Google Scholar]
- 45.Palmer, S. M., and Campbell, S. L. (2008) Biomolecular NMR Assignments 2 69-71 [DOI] [PubMed] [Google Scholar]
- 46.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 47.Johnson, B. A., and Blevins, R. A. (1994) J. Biomol. NMR 4 603-614 [DOI] [PubMed] [Google Scholar]
- 48.Gilmore, A. P., and Burridge, K. (1996) Nature 381 531-535 [DOI] [PubMed] [Google Scholar]
- 49.Miller, G. J., and Ball, E. H. (2001) J. Biol. Chem. 276 28829-28834 [DOI] [PubMed] [Google Scholar]
- 50.Pokutta, S., and Weis, W. I. (2000) Mol. Cell 5 533-543 [DOI] [PubMed] [Google Scholar]
- 51.Johnson, R. P., and Craig, S. W. (2000) J. Biol. Chem. 275 95-105 [DOI] [PubMed] [Google Scholar]
- 52.Berrier, A. L., and Yamada, K. M. (2007) J. Cell. Physiol. 213 565-573 [DOI] [PubMed] [Google Scholar]
- 53.Danen, E. H., and Sonnenberg, A. (2003) J. Pathol. 201 632-641 [DOI] [PubMed] [Google Scholar]
- 54.Delon, I., and Brown, N. H. (2007) Curr. Opin. Cell Biol. 19 43-50 [DOI] [PubMed] [Google Scholar]
- 55.Kedrin, D., van Rheenen, J., Hernandez, L., Condeelis, J., and Segall, J. E. (2007) J. Mammary Gland Biol. Neoplasia 12 143-152 [DOI] [PubMed] [Google Scholar]
- 56.Moissoglu, K., and Schwartz, M. A. (2006) Biol. Cell 98 547-555 [DOI] [PubMed] [Google Scholar]
- 57.Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T., and Horwitz, A. R. (2003) Science 302 1704-1709 [DOI] [PubMed] [Google Scholar]
- 58.Humphries, J. D., Wang, P., Streuli, C., Geiger, B., Humphries, M. J., and Ballestrem, C. (2007) J. Cell Biol. 179 1043-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen, H., Cohen, D. M., Choudhury, D. M., Kioka, N., and Craig, S. W. (2005) J. Cell Biol. 169 459-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galbraith, C. G., Yamada, K. M., and Sheetz, M. P. (2002) J. Cell Biol. 159 695-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Janssen, M. E., Kim, E., Liu, H., Fujimoto, L. M., Bobkov, A., Volkmann, N., and Hanein, D. (2006) Mol. Cell 21 271-281 [DOI] [PubMed] [Google Scholar]