Abstract
YoeB is a toxin encoded by the yefM-yoeB antitoxin-toxin operon in
the Escherichia coli genome. Here we show that YoeB, a highly potent
protein synthesis inhibitor, specifically blocks translation initiation. In
in vivo primer extension experiments using two different mRNAs, a
major band was detected after YoeB induction at three bases downstream of the
initiation codon at 2.5 min. An identical band was also detected in in
vitro toeprinting experiments after the addition of YoeB to the reaction
mixtures containing 70 S ribosomes and the same mRNAs, even in the absence of
 . Notably,
this band was not detected in the presence of YoeB alone, indicating that YoeB
by itself does not have endoribonuclease activity under the conditions used.
The 70 S ribosomes increased upon YoeB induction, and YoeB was found to be
specifically associated with 50 S subunits. Using tetracycline and hygromycin
B, we demonstrated that YoeB binds to the 50 S ribosomal subunit in 70 S
ribosomes and interacts with the A site leading to mRNA cleavage at this site.
As a result, the 3′-end portion of the mRNA was released from ribosomes,
and translation initiation was effectively inhibited. These results
demonstrate that YoeB primarily inhibits translation initiation.
. Notably,
this band was not detected in the presence of YoeB alone, indicating that YoeB
by itself does not have endoribonuclease activity under the conditions used.
The 70 S ribosomes increased upon YoeB induction, and YoeB was found to be
specifically associated with 50 S subunits. Using tetracycline and hygromycin
B, we demonstrated that YoeB binds to the 50 S ribosomal subunit in 70 S
ribosomes and interacts with the A site leading to mRNA cleavage at this site.
As a result, the 3′-end portion of the mRNA was released from ribosomes,
and translation initiation was effectively inhibited. These results
demonstrate that YoeB primarily inhibits translation initiation.
YoeB is one of the toxins encoded by the genome of Escherichia coli, which is co-expressed with YefM, its cognate antitoxin (1). It has been shown that YefM is highly unstable in the cell and purified YefM exists as a denatured form without secondary structure (2). However, when YefM is co-expressed with YoeB, a dimer of YefM forms a stable complex with a single molecule of YoeB. The x-ray structure of this heterotrimer has been determined (3). Under normal growth conditions, YoeB presumably exists as a heterotrimer and is thus unable to exert its cellular toxicity. Lon is an ATP-dependent protease that is induced under various stress conditions, and has been implicated to be responsible for the degradation of YefM and release of YoeB. This consequently leads to inhibition of protein synthesis. These authors demonstrated that the yefM-yoeB system is involved in Lon-dependent lethality (1). They also showed that induction of YoeB leads to cleavage of the lpp mRNA at a few sites and concluded that YoeB functions similar to other E. coli toxins such as RelE, MazF, and ChpBK by cleaving translated mRNAs (1). Interestingly, RelE by itself has no endoribonuclease activity (4) and has been proposed to be a ribosome-associated factor, which stimulates the endogenous ribonuclease activity of ribosomes (4, 5). On the other hand, MazF is a ribosome-independent endoribonuclease that cleaves mRNAs specifically at ACA sequences, and is thus termed an mRNA interferase (6). ChpBK was also found to be another mRNA interferase specifically cleaving mRNAs at UAC sequences (7). Therefore, it remains to be determined if YoeB is a ribosome-associating factor like RelE or if it functions as a ribosome-independent mRNA interferase like MazF and ChpBK or if it inhibits translation by a yet-unknown mechanism. Notably, purified YoeB has been shown to have an intrinsic endoribonuclease activity specific for purine residues in RNA (3).
In the present paper we demonstrate that YoeB is a 50 S
ribosome-associating factor that inhibits translation initiation using a
mechanism that is different from the mechanism proposed earlier by Christensen
et al. (1) and Kamada
and Hanaoka (3). The former
authors proposed that YoeB induces cleavage of the translated mRNA and the
latter authors concluded that YoeB is a purine-specific endoribonuclease. In
in vivo primer extension experiments carried out with two different
mRNAs, a major band was detected after YoeB induction at a position three
bases downstream of the initiation codon at 2.5 min. An identical band was
also detected in in vitro toeprinting experiments when YoeB was added
to the reaction mixtures containing 70 S ribosomes and the same mRNAs even in
the absence of initiator
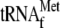 . When
. When
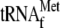 was added to
the reaction mixtures in the absence of YoeB, toeprinting sites were shifted
to a position 13 to 14 bases downstream of the initiation codon. When YoeB
forms a complex with 70 S ribosomes and mRNA in vitro, partial
cleavage of mRNAs is also observed at positions three and four bases
downstream of the initiation codon as observed in in vivo
experiments. Notably, this partial cleavage was not observed when YoeB was
incubated with mRNA in the absence of ribosomes. However, when a
nuclease-negative YoeB mutant (H83Q)
(3) was used in in
vitro toeprinting experiments, partial cleavage was observed. Using
tetracycline and hygromycin B, we demonstrated that YoeB binds to the 50 S
ribosomal subunit in 70 S ribosomes and interacts with the A site leading to
mRNA cleavage at this site. As a result, the 3′-end portion of the mRNA
was released from ribosomes. These results indicate that YoeB associates with
50 S subunits in 70 S ribosomes to effectively inhibit translation initiation
by preventing the formation of the translation initiation complex on
mRNAs.
was added to
the reaction mixtures in the absence of YoeB, toeprinting sites were shifted
to a position 13 to 14 bases downstream of the initiation codon. When YoeB
forms a complex with 70 S ribosomes and mRNA in vitro, partial
cleavage of mRNAs is also observed at positions three and four bases
downstream of the initiation codon as observed in in vivo
experiments. Notably, this partial cleavage was not observed when YoeB was
incubated with mRNA in the absence of ribosomes. However, when a
nuclease-negative YoeB mutant (H83Q)
(3) was used in in
vitro toeprinting experiments, partial cleavage was observed. Using
tetracycline and hygromycin B, we demonstrated that YoeB binds to the 50 S
ribosomal subunit in 70 S ribosomes and interacts with the A site leading to
mRNA cleavage at this site. As a result, the 3′-end portion of the mRNA
was released from ribosomes. These results indicate that YoeB associates with
50 S subunits in 70 S ribosomes to effectively inhibit translation initiation
by preventing the formation of the translation initiation complex on
mRNAs.
EXPERIMENTAL PROCEDURES
Strains and Plasmids—E. coli BL21(DE3), BW25113 (ΔaraBAD) (8), and MRE600 (9) were used. The yefM-yoeB operon was amplified by PCR using E. coli genomic DNA as template and cloned into the NdeI-XhoI sites of pET21c (Novagen). This construction created an in-frame translation fusion with a (His)6 tag at the yoeB C-terminal end. The plasmid was designated as pET21c-YefM-YoeB(His)6. The yoeB gene was cloned into pBAD creating pBAD-YoeB to tightly regulate yoeB expression by the addition of arabinose (0.2%). The yefM gene was cloned into pGEX-4T-1 (Amersham Biosciences) creating pGEX-4T-1-YefM.
Assay of Protein, DNA and RNA Synthesis in Toluene-treated Cells—A 70-ml culture of E. coli BW25113 containing pBAD-YoeB plasmid was grown at 37 °C in M9 medium with 0.5% glycerol (no glucose) and all the amino acids except for methionine and cysteine (1 mm each). When the A600 of the culture reached 0.4, arabinose was added to a final concentration of 0.2%. After incubation at 37 °C for 10 min, the cells were treated with 1% toluene (10, 11). Using toluene-treated cells, protein synthesis was carried out with [35S]methionine as described previously (10, 11). The toluene-treated cells were washed once with 0.05 m potassium phosphate buffer (pH 7.4) at room temperature, and then re-suspended into the same buffer to examine DNA synthesis using [α-32P]dCTP as described previously (12). For assaying RNA synthesis, the toluene-treated cells were washed once with 0.05 m Tris-HCl buffer (pH 7.5) at room temperature, and then re-suspended into the same buffer to measure [α-32P]UTP incorporation into RNA as described previously (13).
Assay of in Vivo Protein Synthesis—E. coli BW25113 cells containing pBAD-YoeB were grown in M9 medium with 0.5% glycerol (no glucose) and all the amino acids except for methionine and cysteine (1 mm each). When the A600 value of the culture reached 0.4, arabinose was added to a final concentration of 0.2% to induce YoeB expression. Cell cultures (0.6 ml) were taken at time intervals as indicated in Fig. 1F and mixed with 30 μCi of [35S]methionine. After a 1-min incubation at 37 °C, the rate of protein synthesis was determined as described previously (6). For SDS-PAGE analysis of the total cellular protein synthesis, samples were removed from the [35S]methionine incorporation reaction mixture (500 μl) at time intervals indicated in Fig. 1F and added into chilled test tubes containing 100 μg/ml each of non-radioactive methionine and cysteine. Cell pellets collected by centrifugation were dissolved into 50 μl of loading buffer and subjected to SDS-PAGE followed by autoradiography.
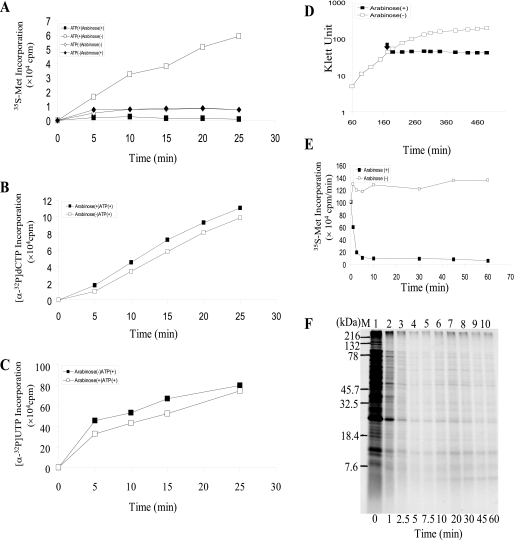
FIGURE 1.
Effect of YoeB on protein, DNA, and RNA synthesis. A, effect of YoeB on [35S]methionine incorporation in toluene-treated cells. E. coli BW25113 cells containing pBAD-YoeB were grown at 37 °C in glycerol-M9 medium. When the A600 of the culture reached 0.4, arabinose was added to a final concentration of 0.2%. After incubation at 37 °C for 10 min, the cells were treated with toluene (10, 11). Using toluene-treated cells, ATP-dependent protein synthesis was carried out with [35S]methionine as described previously (10). B, effect of YoeB on [α-32P]dCTP incorporation in toluene-treated cells (12). C, effect of YoeB on [α-32P]UTP incorporation in toluene-treated cells (13). D, growth curve of BW25113 cells containing pBAD-YoeB plasmid in LB medium. Arabinose was added at 175 min as indicated by the solid arrow. E, effect of YoeB on the rate of [35S]methionine incorporation in vivo. [35S]Methionine incorporation into E. coli BW25113 cells containing pBAD-YoeB was measured at various time points after YoeB induction as indicated. F, SDS-PAGE analysis of in vivo protein synthesis after the induction of YoeB. The same cultures in E were used.
Purification of YoeB(His)6 and GST-YefM Proteins—YoeB (His)6 tagged at the C-terminal end was purified from strain BL21(DE3) carrying pET-21cc-YefM-YoeB. The complex of YoeB(His)6 and YefM was first trapped on nickel-nitrilotriacetic acid resin. After dissociating YefM from YoeB(His)6 in 6 m guanidine HCl at 4 °C, YoeB(His)6 was re-trapped by nickel-nitrilotriacetic resin and refolded by step-by-step dialysis against buffer (50 mm NaH2PO4, 500 mm NaCl, pH 8.5) at 4 °C overnight. GST-YefM tagged at the N-terminal end was purified from strain BL21(DE3) carrying pGEX-4T-1-YefM with use of glutathione-Sepharose 4B resin (Amersham Biosciences).
Effect of YoeB and MazF on Protein Synthesis in Prokaryotic and Eukaryotic Cell-free Systems—Prokaryotic cell-free protein synthesis was carried out with an E. coli T7 S30 extract system (Promega). The reaction mixture consisted of 10 μl of S30 premix, 7.5 μl of S30 extract, and 2.5 μl of an amino acid mixture (1 mm each of all amino acids except methionine), 1 μl of [35S]methionine, and different amounts of YoeB(His)6 and GST-YefM in a final volume of 29 μl. The different amounts of YoeB(His)6 and GST-YefM were preincubated for 10 min at 25 °C before the assay was started by adding 1 μl of pET-11a-MazG plasmid-DNA (0.16 μg/μl) (15). The reaction was performed for 1.5 h at 37 °C, and proteins were then precipitated with acetone and analyzed by SDS-PAGE followed by autoradiography. Eukaryotic cell-free protein synthesis was carried out with a rabbit reticulocyte lysates system, TnT® T7 Coupled Reticulocyte Lysate System (Promega). A luciferase T7 control DNA was used as DNA template. The reaction was performed for 1.5 h at 30 °C, and proteins were then precipitated with acetone and analyzed by SDS-PAGE.
Effect of YoeB on the yefM mRNA in Vitro—Different amounts of YoeB were incubated with 1 μg of the yefM mRNA at 37 °C for 15 min in a 10-μl reaction mixture containing 10 mm Tris-Cl (pH 7.8), 60 mm NH4Cl, 10 mm MgCl2, and 1 mm DTT2 as indicated in Fig. 2F. The reaction products were analyzed by 8% acrylamide native gel electrophoresis followed by ethidium bromide staining.
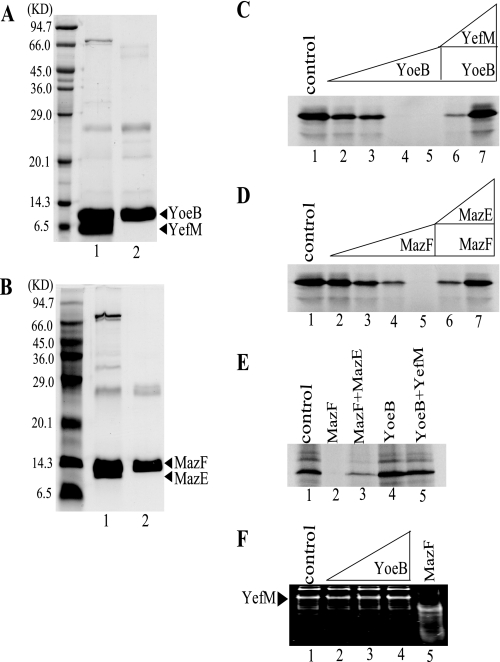
FIGURE 2.
Effect of purified YoeB(His)6 and MazF(His)6, on cell-free protein synthesis. A, purification of YoeB(His)6. Lane 1, purified YefM-YoeB(His)6 complex, and lane 2, purified YoeB(His)6. B, purification of MazF(His)6. Lane 1, purified MazE-MazF(His)6 complex, and lane 2, purified MazF(His)6. C, effect of YoeB(His)6 on MazG synthesis in a prokaryotic cell-free protein synthesis system. Lane 1, without YoeB(His)6; lanes 2–5, 30, 60, 240, and 960 nm YoeB(His)6 were added, respectively; and lanes 6 and 7, 960 nm YoeB(His)6 plus GST-YefM in its ratios to YoeB(His)6 of 2.8 and 4.2, respectively. D, effect of MazF(His)6 on MazG synthesis in a prokaryotic cell-free protein synthesis system. Lane 1, without MazF(His)6; lanes 2–5, 161, 644, 1288, and 2576 nm MazF(His)6 were added, respectively; lanes 6 and 7, 2576 nm MazF(His)6 plus MazE(His)6 in its ratios to MazF(His)6 of 1.7 and 11.9, respectively. E, effect of MazF(His)6 and YoeB(His)6 on the synthesis of luciferase in a eukaryotic cell-free protein synthesis system. Lane 1, without protein; lane 2, 2.57 μm MazF(His)6; and lane 3, 31.5 μm MazE(His)6 and 2.57 μm MazF(His)6 (the ratio of (His)6MazE to MazF(His)6, 11.9:1); lane 4, 1.92 μm YoeB(His)6; and lane 5, 8.1 μm GST-YefM and 1.92 μm YoeB(His)6 (the ratio of GST-YefM to YoeB(His)6, 4.2:1). In both cell-free systems, the reactions were carried out in the presence of [35S]methionine according to the instructions given by the manufacturer. Reaction products were analyzed by SDS-PAGE followed by autoradiography. F, effect of YoeB on the yefM mRNA in vitro. Lane 1, no protein was added; lanes 2–4, 40, 80, and 160 nm YoeB were added; lane 5, 80 nm MazF was added.
Preparation of E. coli 70 S Ribosomes—70 S ribosomes were prepared from E. coli MRE 600 as described previously (16–18) with minor modifications. Bacterial cells (2 g) were suspended in buffer A (10 mm Tris-HCl (pH 7.8) containing 10 mm MgCl2, 60 mm NH4Cl, and 6 mm 2-mercaptoethanol). Cells were lysed by French Press. Cell debris was removed by centrifugation two times at 30,000 × g for 30 min at 4 °C with a Beckman 50Ti rotor. The supernatant (three-fourths volume from the top) was then layered over an equal volume of 1.1 m sucrose in buffer B (buffer A containing 0.5 m NH4Cl) and centrifuged at 45,000 × g for 15 h at 4 °C with a Beckman 50Ti rotor. After washing with buffer A, the ribosome pellets were resuspended in buffer A and applied to a linear 5 to 40% (w/v) sucrose gradient prepared in buffer A, and centrifuged at 35,000 × g for 3 h at 4 °C with a Beckman SW41Ti rotor. Gradients were fractionated and the 70 S ribosome fractions were pooled, pelleted at 45,000 × g for 20 h at 4 °C with a Beckman 50Ti rotor. The 70 S ribosome pellets were resuspended in buffer A before they were stored at –80 °C.
Ribosome Profile Analysis—For polysome profile analysis, cells containing pBAD-YoeB plasmid were grown at 37 °C in 150 ml of LB medium and at A600 of 0.6, the arabinose was added to a final concentration of 0.2%, after 10 min induction, chloramphenicol was added to a final concentration of 200 μg/ml. Cell pellets were suspended in 1 ml of buffer A (10 mm Tris-Cl, pH 7.8, 60 mm NH4Cl, 10 mm MgCl2, and 1 mm DTT). Extracts were prepared using liquid nitrogen freezing and thawing four times. After centrifugation at 45,000 × g for 10 min at 4 °C with a Beckman 100.3 Ti rotor, about 10 A260 units of the supernatant were layered onto a 5–40% sucrose gradient in buffer A and centrifuged at 35,000 × g for 3 h at 4 °C in a Beckman SW41 rotor. Gradients were analyzed with continuous monitoring at 254 nm. To analyze a 50 S and 30 S ribosome profile, another 10 A260 units of supernatant as prepared above were dialyzed overnight against buffer B (10 mm Tris-HCl, pH 7.8, 60 mm NH4Cl, 0.5 mm MgCl2, and 1 mm DTT) (dialysis buffer was changed once) and then layered onto 10–30% sucrose gradients in buffer B and centrifuged at 35,000 × g for 3 h at 4 °C in a Beckman SW41 rotor. Gradients were analyzed with continuous monitoring at 254 nm.
Primer Extension Analysis in Vivo—For primer extension analysis of mRNA cleavage sites in vivo, total RNAs were extracted from the E. coli BW25113 cells containing pBAD-YoeB at different time points as indicated in Fig. 3, C and D. Primer extension reactions were carried out for 1 h using different primers at 42 °C using reverse transcriptase-avian myeloblastosis virus (2 units) as described previously (6).
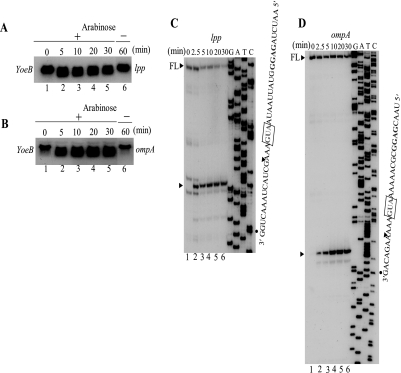
FIGURE 3.
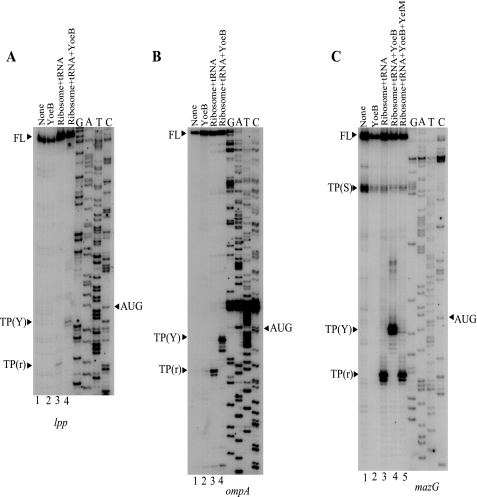
Effect of YoeB on cellular mRNAs and identification of YoeB cleavage sites in the chromosomally encoded lpp and ompA mRNAs. Total cellular RNA was extracted from E. coli BW25113 cells containing pBAD-YoeB at various time points as indicated after the addition of arabinose and subjected to Northern blot analysis using radiolabeled lpp and ompA open reading frames as probes shown in A and B, respectively. For in vivo primer extension, the lpp and ompA mRNAs were prepared from E. coli BW25113 cells containing pBAD-YoeB at various time points as indicated in C and D before and after the induction of YoeB. The sequence ladders for lpp and ompA were obtained using pCR®2.1-TOPO®-lpp and pCR®2.1-TOPO-ompA as template, respectively. The sequences around the major cleavage sites are shown at the right-hand side and the major primer extension stop sites are indicated by arrowheads. On the top of the gels, the full-length (FL) RNA bands are shown with an arrowhead and FL. C, primer extension analysis of the lpp mRNA. D, primer extension analysis of the ompA mRNA. FL, full-length.
Toeprinting Assays—Toeprinting was carried out as described
previously (19) with a minor
modification. The mixture for primer-template annealing containing mRNA and
32P-end labeled DNA primer was incubated at 70 °C for 5 min,
and then cooled slowly to room temperature. The ribosome-binding mixture
contained 2 μl of 10× buffer (100 mm Tris-HCl, pH 7.8,
containing 100 mm MgCl2, 600 mm
NH4Cl, and 60 mm 2-mercaptoethanol), different amounts
of YoeB(His)6, 0.375 mm dNTP, 0.05 μm 70 S
ribosomal subunits, 1μm
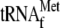 , and 2μl
of the annealing mixture in a final volume of 20 μl. The final mRNA
concentration was 0.035 μm. This ribosome-binding mixture was
incubated at 37 °C for 10 min, and then reverse transcriptase (2 units)
was added. The cDNA synthesis was carried out at 37 °C for 15 min. The
reaction was stopped by adding 12 μl of the sequencing loading buffer (95%
formamide, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene
cyanol EF). The sample was incubated at 90 °C for 5 min prior to
electrophoresis on a 6% polyacrylamide sequencing gel. The lpp and
ompA mRNAs were synthesized in vitro from their individual
DNA fragments containing a T7 promoter and part of their opening reading
frames using T7 RNA polymerase. These ∼200-bp DNA fragments for
lpp (194 bp) and ompA (248 bp), all of which had the
initiation codon at the center, were amplified by PCR using appropriate
primers using chromosome DNA as templates, respectively. The 5′-end
primers for lpp and ompA contained the T7 promoter sequence. The
mazG DNA fragment containing a T7 promoter and part of the
mazG open reading frame as described earlier
(6) was obtained by PCR
amplification using the pET-11a-MazG plasmid
(15) as DNA template. mRNA
produced from this DNA fragment was 151 bases in length.
, and 2μl
of the annealing mixture in a final volume of 20 μl. The final mRNA
concentration was 0.035 μm. This ribosome-binding mixture was
incubated at 37 °C for 10 min, and then reverse transcriptase (2 units)
was added. The cDNA synthesis was carried out at 37 °C for 15 min. The
reaction was stopped by adding 12 μl of the sequencing loading buffer (95%
formamide, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene
cyanol EF). The sample was incubated at 90 °C for 5 min prior to
electrophoresis on a 6% polyacrylamide sequencing gel. The lpp and
ompA mRNAs were synthesized in vitro from their individual
DNA fragments containing a T7 promoter and part of their opening reading
frames using T7 RNA polymerase. These ∼200-bp DNA fragments for
lpp (194 bp) and ompA (248 bp), all of which had the
initiation codon at the center, were amplified by PCR using appropriate
primers using chromosome DNA as templates, respectively. The 5′-end
primers for lpp and ompA contained the T7 promoter sequence. The
mazG DNA fragment containing a T7 promoter and part of the
mazG open reading frame as described earlier
(6) was obtained by PCR
amplification using the pET-11a-MazG plasmid
(15) as DNA template. mRNA
produced from this DNA fragment was 151 bases in length.
RNA Isolation and Northern Blot Analysis—E. coli BW25113 containing pBAD-YoeB was grown at 37 °C in LB medium. When the A600 value reached 0.6, arabinose was added to a final concentration of 0.2%. The samples were taken at different intervals as indicated in Fig. 3, A and B. Total RNA was isolated using the hot-phenol method as described previously (20). Northern blot analysis was carried out as described previously (21).
Binding of
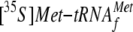 to the
mRNA-70 S Ribosome Complex—Reactions were carried out with
ompA mRNA that was used for the toeprinting experiments. The reaction
buffer used was buffer A (10 mm Tris-HCl, pH 7.8, 10 mm
MgCl2, 60 mm NH4Cl, and 6 mm
β-mercaptoethanol). 70 S ribosomes (1 pmol) were first incubated with the
ompA mRNA (0.7 pmol) and
to the
mRNA-70 S Ribosome Complex—Reactions were carried out with
ompA mRNA that was used for the toeprinting experiments. The reaction
buffer used was buffer A (10 mm Tris-HCl, pH 7.8, 10 mm
MgCl2, 60 mm NH4Cl, and 6 mm
β-mercaptoethanol). 70 S ribosomes (1 pmol) were first incubated with the
ompA mRNA (0.7 pmol) and
 (20 pmol) for 10 min at 37 °C, and then different amounts of YoeB were
added in a final reaction volume of 20 μl. The mixture was incubated for an
additional 10 min at 37 °C. Reactions mixtures were applied to
nitrocellulose filters (Millpore 0.45 μm HA) and the filters
were washed twice with 2 ml of buffer A before measuring the radioactivity.
(20 pmol) for 10 min at 37 °C, and then different amounts of YoeB were
added in a final reaction volume of 20 μl. The mixture was incubated for an
additional 10 min at 37 °C. Reactions mixtures were applied to
nitrocellulose filters (Millpore 0.45 μm HA) and the filters
were washed twice with 2 ml of buffer A before measuring the radioactivity.
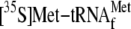 was synthesized in a buffer containing 30 mm Hepes, pH 7.6, 1
mm μm
was synthesized in a buffer containing 30 mm Hepes, pH 7.6, 1
mm μm
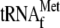 (Sigma) using
aminoacyl-tRNA synthetases, which was prepared as described previously
(25).
(Sigma) using
aminoacyl-tRNA synthetases, which was prepared as described previously
(25).
Effect of YoeB(His)6 on the 30-Base RNA in the Presence 70 S
Ribosomes and 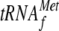 —The 30-base RNA
was 5′-end labeled with [γ-32P]ATP by T4 polynucleotide
kinase (2 units). The mixture of 0.05 μm 70 S ribosomes, 1
μm
—The 30-base RNA
was 5′-end labeled with [γ-32P]ATP by T4 polynucleotide
kinase (2 units). The mixture of 0.05 μm 70 S ribosomes, 1
μm
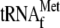 , and
different amounts of YoeB (0.16, 0.64, and 2.56 μm in lanes
6–8, respectively, in Fig.
5F) were preincubated at 37 °C for 10 min, and then
0.035 μm 32P-labeled 30-base RNA was added. The
reaction mixture was incubated at 37 °C for another 10 min and then the
reaction product was loaded on a 20% sequencing gel.
, and
different amounts of YoeB (0.16, 0.64, and 2.56 μm in lanes
6–8, respectively, in Fig.
5F) were preincubated at 37 °C for 10 min, and then
0.035 μm 32P-labeled 30-base RNA was added. The
reaction mixture was incubated at 37 °C for another 10 min and then the
reaction product was loaded on a 20% sequencing gel.
FIGURE 5.
Effect of a ribosome-binding site mutation in mazG and
ompA mRNAs on the inhibitory activity of YoeB(His)6.
A, toeprinting of the mazG mRNA after phenol extraction. The
experiment was carried out in the same way as described in the legend to
Fig. 4C except that
reaction products in lanes 4 and 6 were phenol-extracted to
remove proteins before primer extension. Lane 1, without
YoeB(His)6; lane 2, 3.4 μm
YoeB(His)6 alone; lanes 3 and 4, 0.05
μm 70 S ribosomes and 1 μm
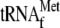 ; and
lanes 5 and 6, 0.05 μm 70 S ribosomes and 1
μm
; and
lanes 5 and 6, 0.05 μm 70 S ribosomes and 1
μm
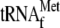 with 3.4
μm YoeB(His)6. B, effect of a mutation
(GGAG to AAUG) at the Shine-Dalgarno sequence of the
mazG mRNA on the YoeB(His)6 activity. Lanes 1 and
5, without YoeB(His)6; lanes 2 and 6,
3.4 μm YoeB(His)6; lanes 3 and 7,
0.05 μm 70 S ribosomes and 1 μm
with 3.4
μm YoeB(His)6. B, effect of a mutation
(GGAG to AAUG) at the Shine-Dalgarno sequence of the
mazG mRNA on the YoeB(His)6 activity. Lanes 1 and
5, without YoeB(His)6; lanes 2 and 6,
3.4 μm YoeB(His)6; lanes 3 and 7,
0.05 μm 70 S ribosomes and 1 μm
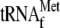 ; lanes
4 and 8, 3.4 μm YoeB(His)6, 0.05
μm 70 S ribosomes, and 1 μm
; lanes
4 and 8, 3.4 μm YoeB(His)6, 0.05
μm 70 S ribosomes, and 1 μm
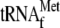 . Lanes
1–4, the wild-type mazG mRNA was used; lanes
5–8, the mutated mazG mRNA was used. C,
toeprinting of the mazG mRNA in the presence of YoeB(His)6
and 70 S ribosomes with and without
. Lanes
1–4, the wild-type mazG mRNA was used; lanes
5–8, the mutated mazG mRNA was used. C,
toeprinting of the mazG mRNA in the presence of YoeB(His)6
and 70 S ribosomes with and without
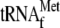 . Lane
1, without YoeB(His)6; lane 2, 3.4 μm
YoeB(His)6; lane 3, 1 μm
. Lane
1, without YoeB(His)6; lane 2, 3.4 μm
YoeB(His)6; lane 3, 1 μm
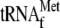 ; lane
4, 3.4 μm YoeB(His)6 and 1 μm
; lane
4, 3.4 μm YoeB(His)6 and 1 μm
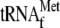 ; lane
5, 0.05 μm 70 S ribosomes; lane 6, 0.05
μm 70 S ribosomes with 3.4 μm
YoeB(His)6; lane 7, 0.05 μm 70 S ribosomes
with 1 μm
; lane
5, 0.05 μm 70 S ribosomes; lane 6, 0.05
μm 70 S ribosomes with 3.4 μm
YoeB(His)6; lane 7, 0.05 μm 70 S ribosomes
with 1 μm
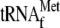 ; and lane
8, 0.05 μm 70 S ribosomes with 1 μm
; and lane
8, 0.05 μm 70 S ribosomes with 1 μm
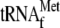 and 3.4
μm YoeB(His)6. The band at TP(S) in Fig. 5,
A–C, is likely to be formed due to a secondary
structure of the mRNA. D, toeprinting of the ompA mRNA after
phenol extraction and toeprinting of the ompA mRNA in the presence of
YoeB(His)6 and 70 S ribosomes with and without
and 3.4
μm YoeB(His)6. The band at TP(S) in Fig. 5,
A–C, is likely to be formed due to a secondary
structure of the mRNA. D, toeprinting of the ompA mRNA after
phenol extraction and toeprinting of the ompA mRNA in the presence of
YoeB(His)6 and 70 S ribosomes with and without
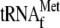 . The
experiment was carried out in the same way as described in the legend to Fig.
5, A and C. Lane 1, without YoeB(His)6;
lane 2, 3.4 μm YoeB(His)6; lane 3,
1 μm
. The
experiment was carried out in the same way as described in the legend to Fig.
5, A and C. Lane 1, without YoeB(His)6;
lane 2, 3.4 μm YoeB(His)6; lane 3,
1 μm
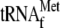 ; lane
4, 3.4 μm YoeB(His)6 and 1 μm
; lane
4, 3.4 μm YoeB(His)6 and 1 μm
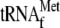 ; lane
5, 0.05 μm 70 S ribosomes; lane 6, 0.05
μm 70 S ribosomes with 3.4 μm
YoeB(His)6; lane 7, 0.05 μm 70 S ribosomes
with 1 μm
; lane
5, 0.05 μm 70 S ribosomes; lane 6, 0.05
μm 70 S ribosomes with 3.4 μm
YoeB(His)6; lane 7, 0.05 μm 70 S ribosomes
with 1 μm
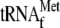 ; and lane
8, 0.05 μm 70 S ribosomes with 1 μm
; and lane
8, 0.05 μm 70 S ribosomes with 1 μm
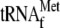 and 3.4
μm YoeB(His)6; lane 9, 0.05 μm
70 S ribosomes and 1 μm
and 3.4
μm YoeB(His)6; lane 9, 0.05 μm
70 S ribosomes and 1 μm
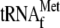 with 3.4
μm YoeB(His)6; lane 10, 0.05
μm 70 S ribosomes and 1 μm
with 3.4
μm YoeB(His)6; lane 10, 0.05
μm 70 S ribosomes and 1 μm
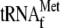 . Reaction
products in lanes 9 and 10 were phenol-extracted before
primer extension. E, effect of the mutation (GGAG to
GAAA) at the Shine-Dalgarno sequence of the ompA mRNA on
the YoeB(His)6 activity. Lanes 1 and 5, without
YoeB(His)6; lanes 2 and 6, 3.4 μm
YoeB(His)6; lanes 3 and 7, 0.05 μm
70 S ribosomes and 1 μm
. Reaction
products in lanes 9 and 10 were phenol-extracted before
primer extension. E, effect of the mutation (GGAG to
GAAA) at the Shine-Dalgarno sequence of the ompA mRNA on
the YoeB(His)6 activity. Lanes 1 and 5, without
YoeB(His)6; lanes 2 and 6, 3.4 μm
YoeB(His)6; lanes 3 and 7, 0.05 μm
70 S ribosomes and 1 μm
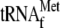 ; lanes
4 and 8, 0.05 μm 70 S ribosomes and 1
μm
; lanes
4 and 8, 0.05 μm 70 S ribosomes and 1
μm
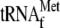 together with
3.4 μm YoeB(His)6. Lanes 1–4, the
wild-type ompA mRNA; lanes 5–8, the mutated
ompA mRNA. F, the RNA sequence of a 30-base synthetic RNA
and the effect of YoeB(His)6 on the 30-base RNA in the presence 70
S ribosomes and
together with
3.4 μm YoeB(His)6. Lanes 1–4, the
wild-type ompA mRNA; lanes 5–8, the mutated
ompA mRNA. F, the RNA sequence of a 30-base synthetic RNA
and the effect of YoeB(His)6 on the 30-base RNA in the presence 70
S ribosomes and
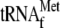 . Lane
1, without YoeB(His)6; lane 2, 2.56 μm
YoeB(His)6; lane 3, 1 μm
. Lane
1, without YoeB(His)6; lane 2, 2.56 μm
YoeB(His)6; lane 3, 1 μm
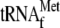 ; lane
4, 0.05 μm 70 S ribosomes; lane 5, 0.05
μm 70 S ribosomes and 1 μm
; lane
4, 0.05 μm 70 S ribosomes; lane 5, 0.05
μm 70 S ribosomes and 1 μm
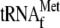 ; and
lanes 6–8, 0.05 μm 70 S ribosomes, 1
μm
; and
lanes 6–8, 0.05 μm 70 S ribosomes, 1
μm
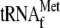 , and 0.16,
0.64, and 2.56 μm YoeB(His)6, respectively. An
arrowhead indicates the cleavage product, which migrated at the
position seven bases shorter than the 30-base RNA on the basis of the ladder
(a partial alkaline hydrolysate of the 30-base RNA). SD,
Shine-Dalgarno; FL, full-length.
, and 0.16,
0.64, and 2.56 μm YoeB(His)6, respectively. An
arrowhead indicates the cleavage product, which migrated at the
position seven bases shorter than the 30-base RNA on the basis of the ladder
(a partial alkaline hydrolysate of the 30-base RNA). SD,
Shine-Dalgarno; FL, full-length.
RESULTS
YoeB Inhibits Protein Synthesis Immediately After Its Induction—To identify the cellular functions inhibited by YoeB, a cell-free system prepared from E. coli BW25113 cells carrying arabinose-inducible pBAD-YoeB vector was used. The cells were permeabilized by toluene treatment (10, 11). ATP-dependent [35S]methionine incorporation was completely inhibited when cells were preincubated for 10 min in the presence of arabinose before toluene treatment (Fig. 1A). However, the incorporation of [α-32P]dCTP (Fig. 1B) and [α-32P]UTP (Fig. 1C) was not significantly affected under similar conditions (12, 13). These results demonstrate that YoeB inhibits protein synthesis, but not DNA replication or RNA synthesis. Notably, cell growth was inhibited almost immediately after the addition of arabinose (Fig. 1D). The in vivo incorporation of [35S]methionine was also dramatically inhibited within 5 min after YoeB induction (Fig. 1, E and F). MazF is another potent E. coli toxin that functions as an mRNA interferase that specifically cleaves mRNAs at ACA triplet sequences and consequently leads to inhibition of protein synthesis (6). However, in contrast to YoeB, complete inhibition of protein synthesis by MazF requires significantly longer time (15–20 min) after its induction (6).
Inhibitory Effect of Purified YoeB on Cell-free Protein Synthesis—Next, we examined the effect of purified YoeB on E. coli cell-free protein synthesis. YoeB was purified from cells co-expressing both YefM and YoeB as described under “Experimental Procedures” (Fig. 2A). MazF was purified (Fig. 2B) as described previously (6). The synthesis of MazG protein (30 kDa) (15) from plasmid pET-11a-MazG was tested at 37 °C for 1 h in the absence and presence of YoeB using an E. coli T7 S30 extract system (Promega) (Fig. 2C). MazG synthesis was almost completely blocked at YoeB concentrations of 240 nm or above. Similar inhibition was observed when purified MazF was added (Fig. 2D) consistent with the previous result (6).
We then tested the effect of YefM antitoxin on the YoeB-mediated inhibition of MazG synthesis. The addition of GST-YefM rescued MazG synthesis in a dose-dependent manner (Fig. 2C). Importantly, YoeB did not inhibit eukaryotic cell-free protein synthesis (Fig. 2E, lane 4) in contrast to MazF (Fig. 2E, lane 2), which is known to inhibit both prokaryotic, and eukaryotic protein synthesis (6). These results indicate that YoeB is a protein synthesis inhibitor specific to prokaryotes. This is also consistent with the observation that YoeB induction in yeast cells had no effect on cell growth, whereas MazF induction blocked yeast cell growth (data not shown). It should be noted that YoeB alone could not degrade YefM mRNA under our experimental condition (Fig. 2F).
YoeB Induction Does Not Affect the Stability of Cellular mRNAs—Despite the abrupt inhibition of protein synthesis by YoeB, its induction did not severely affect the stability of cellular mRNAs. On the other hand, the induction of MazF significantly decreased stability of cellular mRNAs (6). Most significantly, full-length lpp and ompA mRNAs very quickly disappeared after MazF induction (10 min) (6), whereas all these mRNAs appeared to be more stable after YoeB induction (Fig. 3, A and B).
In Vivo Cleavage of the lpp and ompA mRNAs—As shown in Fig. 3, A and B, cellular mRNAs were partially degraded upon induction of YoeB. Therefore we next attempted to identify the cleavage sites of the chromosomally encoded lpp and ompA mRNAs by primer extension experiments. For this purpose, total RNA was extracted from E. coli BW25113 cells harboring pBAD-YoeB at different time intervals following induction of YoeB. The primer extension analyses of lpp (Fig. 3C) and ompA (Fig. 3D) mRNAs demonstrated that the distinct major bands exhibiting the specific cleavage sites in each mRNA appeared as early as 2.5 min after YoeB induction (Fig. 3, C and D, lane 2). The band intensities for ompA (Fig. 3D) mRNA further increased from 2.5 to 10–20 min after YoeB induction. Interestingly, in both cases, the major bands resulted from cleavage of the mRNAs at three bases downstream of AUG, and most notably no other bands were observed in the regions between 5′-ends of the mRNAs and the initiation codon suggesting that YoeB may function only when it associates with the ribosomal translation machine. It is interesting to note that the substantial amounts of the full-length mRNAs for lpp (Fig. 3C) and ompA (Fig. 3D) mRNAs remained uncleaved even 30 min after induction of YoeB. This observation is again consistent with the fact that unlike MazF, YoeB does not significantly affect the stability of cellular mRNAs.
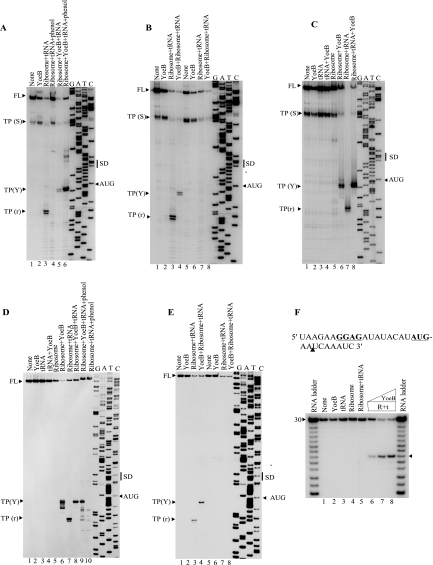
YoeB Binds to the Translation Initiation Complex in
Vitro—Next, we examined if similar cleavage of lpp and
ompA mRNAs was observed in vitro by toeprinting (TP)
(19). In this experiment, we
also used the mazG mRNA (Fig.
4C), which we previously used in our experiments with
MazF (6) in addition to
lpp (Fig. 4A)
and ompA (Fig.
4B) mRNAs. Primer extension analysis of the mRNAs alone
yielded the full-length bands (Fig. 4,
A–C, lane 1). Importantly, the addition of
YoeB alone did not result in formation of new bands indicating that YoeB by
itself has no endoribonuclease activity under the conditions used
(Fig. 4,
A–C, lane 2). When 70 S ribosomes
and initiator 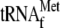 were added to the reaction mixtures, typical toeprinting band (TP(r))
downstream of the initiation codon appeared
(Fig. 4,
A–C, lane 3). On the other hand,
when YoeB was added together with 70 S ribosomes and initiator
were added to the reaction mixtures, typical toeprinting band (TP(r))
downstream of the initiation codon appeared
(Fig. 4,
A–C, lane 3). On the other hand,
when YoeB was added together with 70 S ribosomes and initiator
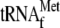 , a new band
TP(Y) (Fig. 4,
A–C, lane 4) appeared with
concomitant disappearance of the TP(r) band in all cases. This new band,
TP(Y), is 10 bases upstream of the normal toeprinting band (13–14 bases
downstream of the initiation codon) as judged from the sequence ladder at the
right-hand side. It should be noted that this band is observed only in the
presence of 70 S ribosomes, initiator
, a new band
TP(Y) (Fig. 4,
A–C, lane 4) appeared with
concomitant disappearance of the TP(r) band in all cases. This new band,
TP(Y), is 10 bases upstream of the normal toeprinting band (13–14 bases
downstream of the initiation codon) as judged from the sequence ladder at the
right-hand side. It should be noted that this band is observed only in the
presence of 70 S ribosomes, initiator
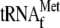 , and YoeB
(Fig. 4,
A–C, lane 4). Interestingly, this
TP(Y) band disappeared with concomitant appearance of the TP(r) band when
YefM, the YoeB antitoxin, was added together with YoeB
(Fig. 4C, lane
5). The band below the full-length band shown by TP(S) was probably due
to the secondary structure present in the mazG mRNA because this band
was detected in the absence of any factors
(Fig. 4C, lane
1).
, and YoeB
(Fig. 4,
A–C, lane 4). Interestingly, this
TP(Y) band disappeared with concomitant appearance of the TP(r) band when
YefM, the YoeB antitoxin, was added together with YoeB
(Fig. 4C, lane
5). The band below the full-length band shown by TP(S) was probably due
to the secondary structure present in the mazG mRNA because this band
was detected in the absence of any factors
(Fig. 4C, lane
1).
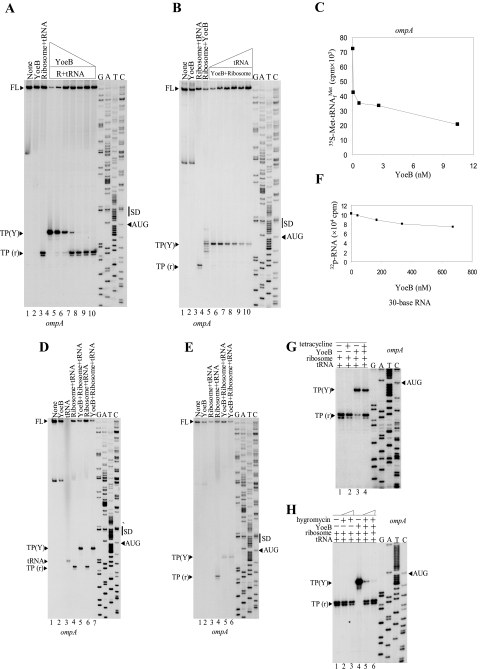
FIGURE 4.
Toeprinting of the lpp, ompA, and mazG mRNA.
A, toeprinting of the lpp mRNA in the presence of
YoeB(His)6. B, toeprinting of the ompA mRNA in
the presence of YoeB(His)6. C, toeprinting of the
mazG mRNA in the presence of YoeB(His)6. For each panel,
lane 1, without YoeB(His)6, 70 S ribosomes, and
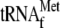 ; lane
2, 3.4 μm YoeB(His)6 alone; lane 3,
0.05 μm 70 S ribosomes and 1 μm
; lane
2, 3.4 μm YoeB(His)6 alone; lane 3,
0.05 μm 70 S ribosomes and 1 μm
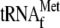 ; lane
4, 0.05 μm 70 S ribosomes and 1 μm
; lane
4, 0.05 μm 70 S ribosomes and 1 μm
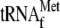 with 3.4
μm YoeB(His)6; lane 5, 0.05 μm
70 S ribosomes and 1 μm
with 3.4
μm YoeB(His)6; lane 5, 0.05 μm
70 S ribosomes and 1 μm
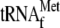 with 3.4
μm YoeB(His)6 and 13.6 μm glutathione
S-transferase-YefM. The sequence ladders shown at the
right-hand side in all cases were obtained using the same primers
used for toeprinting with their corresponding genes cloned in pCR 2.1-TOPO as
template. All mRNA sequences shown are complementary to the sequencing
ladders. The initiation codon, AUG, is indicated with an arrowhead.
TP(Y) is the band where toeprinting was stopped in the
presence of YoeB(His)6. FL, the full-length of the mRNA;
and TP(r), the toeprinting site due to normal ribosome
binding to mRNA in the absence of YoeB(His)6. The band at TP(S) is
likely to be formed due to a secondary structure of the mRNA.
with 3.4
μm YoeB(His)6 and 13.6 μm glutathione
S-transferase-YefM. The sequence ladders shown at the
right-hand side in all cases were obtained using the same primers
used for toeprinting with their corresponding genes cloned in pCR 2.1-TOPO as
template. All mRNA sequences shown are complementary to the sequencing
ladders. The initiation codon, AUG, is indicated with an arrowhead.
TP(Y) is the band where toeprinting was stopped in the
presence of YoeB(His)6. FL, the full-length of the mRNA;
and TP(r), the toeprinting site due to normal ribosome
binding to mRNA in the absence of YoeB(His)6. The band at TP(S) is
likely to be formed due to a secondary structure of the mRNA.
Next, we tested whether the TP(Y) and TP(r) bands resulted from the
cleavage of mRNAs using mazG and ompA mRNAs. These mRNAs
were first preincubated without (Fig. 5,
A, lane 3, and D, lane 7) or with YoeB
(Fig. 5, A, lane
5 for the mazG mRNA, and D, lane 8 for the
ompA mRNA) in the presence of 70 S ribosomes and initiator
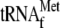 . RNAs were
then phenol extracted and used for primer extension shown in
Fig. 5A (lanes
4 and 6) for the mazG mRNA and
Fig. 5D (lanes
9 and 10) for the ompA mRNA. The TP(Y) band was
observed even after phenol extraction (compare
Fig. 5, A, lanes
5 and 6 for the mazG mRNA, and D, lanes 8 and
9 for the ompA mRNA), indicating that the TP(Y) band
resulted from at least partial cleavage of the mazG and ompA
mRNAs. In contrast, the TP(r) band disappeared after phenol extraction,
indicating that this band was not due to mRNA cleavage, but was caused by
binding of ribosomes to the mRNAs. It is important to note that the mRNAs were
cleaved three and four bases downstream of the translation initiation codon
only in the presence of ribosomes, initiator
. RNAs were
then phenol extracted and used for primer extension shown in
Fig. 5A (lanes
4 and 6) for the mazG mRNA and
Fig. 5D (lanes
9 and 10) for the ompA mRNA. The TP(Y) band was
observed even after phenol extraction (compare
Fig. 5, A, lanes
5 and 6 for the mazG mRNA, and D, lanes 8 and
9 for the ompA mRNA), indicating that the TP(Y) band
resulted from at least partial cleavage of the mazG and ompA
mRNAs. In contrast, the TP(r) band disappeared after phenol extraction,
indicating that this band was not due to mRNA cleavage, but was caused by
binding of ribosomes to the mRNAs. It is important to note that the mRNAs were
cleaved three and four bases downstream of the translation initiation codon
only in the presence of ribosomes, initiator
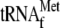 , and YoeB,
but not in the presence of YoeB alone. Therefore, the latent endogenous
endoribonuclease activity of 70 S ribosomes
(5) may be enhanced only when
YoeB binds to the ribosome initiation complex. The ribosome-dependent
endoribonuclease activity induced by RelE preferentially cleaves mRNA at stop
codons (UAG > UAA > UGA) and at certain sense codons (UCG and CAG) in
in vitro experiments
(4). Notably, the primary
sequence of YoeB has only 15% identity with that of RelE. Comparison of the
x-ray structures of the YoeB-YefM complex
(3) and the RelE-RelB complex
(26) demonstrates little
homology between these complexes, although they share a microbial RNase fold
(3). The possibility that YoeB
may function as an endoribonuclease when it binds to the ribosome initiation
complex is unlikely because a nuclease-negative YoeB mutant H83Q
(3) was still able to block
protein synthesis as shown later.
, and YoeB,
but not in the presence of YoeB alone. Therefore, the latent endogenous
endoribonuclease activity of 70 S ribosomes
(5) may be enhanced only when
YoeB binds to the ribosome initiation complex. The ribosome-dependent
endoribonuclease activity induced by RelE preferentially cleaves mRNA at stop
codons (UAG > UAA > UGA) and at certain sense codons (UCG and CAG) in
in vitro experiments
(4). Notably, the primary
sequence of YoeB has only 15% identity with that of RelE. Comparison of the
x-ray structures of the YoeB-YefM complex
(3) and the RelE-RelB complex
(26) demonstrates little
homology between these complexes, although they share a microbial RNase fold
(3). The possibility that YoeB
may function as an endoribonuclease when it binds to the ribosome initiation
complex is unlikely because a nuclease-negative YoeB mutant H83Q
(3) was still able to block
protein synthesis as shown later.
When the Shine-Dalgarno (SD) sequences, GGAG in the mazG mRNA (Fig. 5B, lanes 1–4) and GGAG in the ompA mRNA (Fig. 5E, lanes 1–4) were mutated to AAUG (Fig. 5B, lanes 5-8) and GAAA (Fig. 5E, lanes 5–8), respectively, both TP(Y) and TP(r) completely disappeared (compare Fig. 5, B, lanes 4 and 8, for the mazG mRNA, and E, for the ompA mRNA). These results indicate that the binding of 70 S ribosomes to the mRNA is absolutely required for the YoeB-mediated cleavage of mRNAs.
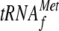 Is not Required for YoeB-Mediated
A Site Cleavage—Next we examined if the initiator
Is not Required for YoeB-Mediated
A Site Cleavage—Next we examined if the initiator
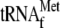 is required
for YoeB-mediated mRNA cleavage in the presence of YoeB. Intriguingly, the
mazG mRNA was cleaved even in the absence of
is required
for YoeB-mediated mRNA cleavage in the presence of YoeB. Intriguingly, the
mazG mRNA was cleaved even in the absence of
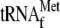 if YoeB and
70 S ribosomes were included in the reaction, yielding identical toeprinting
bands (compare Fig.
5C, lane 6 with 8). It should be noted
that in the absence of YoeB, the addition of
if YoeB and
70 S ribosomes were included in the reaction, yielding identical toeprinting
bands (compare Fig.
5C, lane 6 with 8). It should be noted
that in the absence of YoeB, the addition of
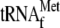 was required
for the detection of the toeprinting band at the TP(r) position (compare
lane 7 with 5, where toeprinting in lane 7 was
carried out in the presence of
was required
for the detection of the toeprinting band at the TP(r) position (compare
lane 7 with 5, where toeprinting in lane 7 was
carried out in the presence of
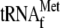 , whereas that
in lane 5 was performed in the absence of
, whereas that
in lane 5 was performed in the absence of
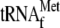 ). The
addition of YoeB alone (lane 2),
). The
addition of YoeB alone (lane 2),
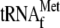 alone
(lane 3), or YoeB plus
alone
(lane 3), or YoeB plus
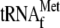 (lane
4) did not yield any bands either at TP(Y) or TP(r) positions. The
identical cleavage pattern was observed with the ompA mRNA
(Fig. 5D). These
results suggest that
(lane
4) did not yield any bands either at TP(Y) or TP(r) positions. The
identical cleavage pattern was observed with the ompA mRNA
(Fig. 5D). These
results suggest that
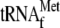 was not
required for YoeB-mediated A site cleavage. Note that both TP(Y) bands for
mazG and ompA (Fig. 5,
C and D, lane 6, respectively) were
still detectable after phenol extraction (data not shown).
was not
required for YoeB-mediated A site cleavage. Note that both TP(Y) bands for
mazG and ompA (Fig. 5,
C and D, lane 6, respectively) were
still detectable after phenol extraction (data not shown).
Cleavage of a Synthetic 30-Base RNA—To further unambiguously
demonstrate that the mRNA cleavage occurring downstream of the initiation
codon requires both ribosomes and YoeB, we repeated a similar experiment as
above using a 30-base synthetic RNA that contains an SD sequence and AUG (see
Fig. 5F). When YoeB
was incubated with 70 S ribosomes,
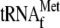 , and the
5′-end 32P-labeled 30-base RNA at 37 °C for 10 min, a new
band appeared (Fig.
5F, lanes 6–8) and its intensity increased
with the increasing amounts of YoeB. This band was not formed when only YoeB
(lane 2), or only
, and the
5′-end 32P-labeled 30-base RNA at 37 °C for 10 min, a new
band appeared (Fig.
5F, lanes 6–8) and its intensity increased
with the increasing amounts of YoeB. This band was not formed when only YoeB
(lane 2), or only
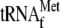 (lane 3), or only 70 S ribosomes (lane 4) or only
70 S ribosomes plus
(lane 3), or only 70 S ribosomes (lane 4) or only
70 S ribosomes plus
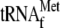 (lane
5) were incubated with the 30-base RNA. As judged by using an RNA ladder,
the new band is 7 bases shorter than the 30-base RNA. This result is
consistent with the results seen with mazG
(Fig. 5,
A–C) and ompA
(Fig. 5D) mRNAs,
because the 30-base RNA was cleaved only when YoeB,
(lane
5) were incubated with the 30-base RNA. As judged by using an RNA ladder,
the new band is 7 bases shorter than the 30-base RNA. This result is
consistent with the results seen with mazG
(Fig. 5,
A–C) and ompA
(Fig. 5D) mRNAs,
because the 30-base RNA was cleaved only when YoeB,
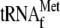 , and 70 S
ribosomes were added together. Judged from the RNA ladder, the cleavage of the
30-base RNA occurred at two bases downstream of AUG. The result confirmed the
notion that the RNA cleavage occurs only in the presence of both 70 S
ribosomes and YoeB.
, and 70 S
ribosomes were added together. Judged from the RNA ladder, the cleavage of the
30-base RNA occurred at two bases downstream of AUG. The result confirmed the
notion that the RNA cleavage occurs only in the presence of both 70 S
ribosomes and YoeB.
YoeB Competes with
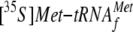 for 70 S
Ribosome Binding—Next, we tested if YoeB is able to displace
for 70 S
Ribosome Binding—Next, we tested if YoeB is able to displace
 from the ribosome in vitro. For this purpose, YoeB was added at the
minimal concentration that is just enough to produce the TP(Y) band without
forming the TP(r) band. As the amounts of YoeB added in the reaction mixture
were reduced (Fig. 6A,
from lanes 4 to 10), the intensities of the TP(Y) band were
progressively reduced, whereas the intensities of the TP(r) band increased
gradually to reach the normal level.
from the ribosome in vitro. For this purpose, YoeB was added at the
minimal concentration that is just enough to produce the TP(Y) band without
forming the TP(r) band. As the amounts of YoeB added in the reaction mixture
were reduced (Fig. 6A,
from lanes 4 to 10), the intensities of the TP(Y) band were
progressively reduced, whereas the intensities of the TP(r) band increased
gradually to reach the normal level.
FIGURE 6.
Competition of
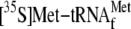 ,
tetracycline, and hygromycin B binding to the mRNA-70 S ribosome complex with
YoeB, and release of the 3′-end portion of the ompA mRNA
cleaved by the addition of YoeB from ribosomes. A, effect of YoeB
on the formation of the ompA mRNA-70 S
,
tetracycline, and hygromycin B binding to the mRNA-70 S ribosome complex with
YoeB, and release of the 3′-end portion of the ompA mRNA
cleaved by the addition of YoeB from ribosomes. A, effect of YoeB
on the formation of the ompA mRNA-70 S
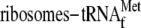 complex. Lane 1, without YoeB(His)6; lane 2, 20.7
nm YoeB(His)6; lane 3, 1 μm
complex. Lane 1, without YoeB(His)6; lane 2, 20.7
nm YoeB(His)6; lane 3, 1 μm
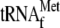 and 0.05
μm 70 S ribosomes; lanes 4–10, 1 μm
and 0.05
μm 70 S ribosomes; lanes 4–10, 1 μm
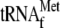 and 0.05
μm 70 S ribosomes and 20.7, 10.4, 2.58, 0.64, 0.16, 0.04, and
0.01 nm YoeB(His)6, respectively. B, effect of
and 0.05
μm 70 S ribosomes and 20.7, 10.4, 2.58, 0.64, 0.16, 0.04, and
0.01 nm YoeB(His)6, respectively. B, effect of
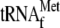 on the
activity of YoeB. Lane 1, without YoeB(His)6; lane
2, 20.7 nm YoeB(His)6; lane 3, 1
μm
on the
activity of YoeB. Lane 1, without YoeB(His)6; lane
2, 20.7 nm YoeB(His)6; lane 3, 1
μm
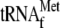 and 0.05
μm 70 S ribosomes; lane 4, 0.05 μm 70 S
ribosomes and 20.7 nm YoeB(His)6; lanes
5–10, 0.05 μm 70 S ribosomes and 20.7 nm
YoeB with 1, 4, 8, 16, 32, and 64 μm
and 0.05
μm 70 S ribosomes; lane 4, 0.05 μm 70 S
ribosomes and 20.7 nm YoeB(His)6; lanes
5–10, 0.05 μm 70 S ribosomes and 20.7 nm
YoeB with 1, 4, 8, 16, 32, and 64 μm
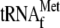 ,
respectively. C, binding of
,
respectively. C, binding of
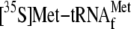 to 70 S ribosomes at 37 °C in the absence and presence of different
amounts of YoeB(His)6. 70 S ribosomes (0.05 μm) were
first incubated with
to 70 S ribosomes at 37 °C in the absence and presence of different
amounts of YoeB(His)6. 70 S ribosomes (0.05 μm) were
first incubated with
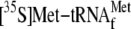 (1 μm) and the ompA mRNA (0.035 μm) for
10 min at 37 °C, then 0, 0.04, 0.64, 2.58, and 10.4 nm YoeB was
added. The reaction mixtures were incubated for an additional 10 min at 37
°C and then applied to nitrocellulose filters (Millpore 0.45
μm HA), which were washed twice with 2 ml of buffer A before
measuring the radioactivity. D, toeprinting experiment with the
ompA mRNA and
(1 μm) and the ompA mRNA (0.035 μm) for
10 min at 37 °C, then 0, 0.04, 0.64, 2.58, and 10.4 nm YoeB was
added. The reaction mixtures were incubated for an additional 10 min at 37
°C and then applied to nitrocellulose filters (Millpore 0.45
μm HA), which were washed twice with 2 ml of buffer A before
measuring the radioactivity. D, toeprinting experiment with the
ompA mRNA and
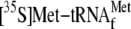 .
Lane 1, without YoeB(His)6; lane 2, 20.7
nm YoeB(His)6; lane 3,
.
Lane 1, without YoeB(His)6; lane 2, 20.7
nm YoeB(His)6; lane 3,
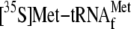 ;
lane 4, 1 μm
;
lane 4, 1 μm
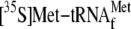 and 0.05 μm 70 S ribosomes; lane 5, 1 μm
and 0.05 μm 70 S ribosomes; lane 5, 1 μm
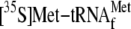 ,
0.05 μm 70 S ribosomes, and 20.7 nm
YoeB(His)6; lane 6, 1 μm
,
0.05 μm 70 S ribosomes, and 20.7 nm
YoeB(His)6; lane 6, 1 μm
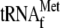 and 0.05
μm 70 S ribosomes; lane 7, 1 μm
and 0.05
μm 70 S ribosomes; lane 7, 1 μm
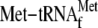 ,
0.05μm 70 S ribosomes, and 20.7 nm
YoeB(His)6. E, release of the 3′-end portion of the
ompA mRNA cleaved by the addition of YoeB from ribosomes. Lane
1, without YoeB(His)6; lane 2, 20.7 nm
YoeB(His)6; lanes 3 and 4, with 1
μm
,
0.05μm 70 S ribosomes, and 20.7 nm
YoeB(His)6. E, release of the 3′-end portion of the
ompA mRNA cleaved by the addition of YoeB from ribosomes. Lane
1, without YoeB(His)6; lane 2, 20.7 nm
YoeB(His)6; lanes 3 and 4, with 1
μm
 and 0.05
μm 70 S ribosomes; lanes 5 and 6, 1
μm
and 0.05
μm 70 S ribosomes; lanes 5 and 6, 1
μm
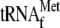 and 0.05
μm 70 S ribosomes and 20.7 nm YoeB(His)6.
Lanes 3 and 5, the samples were centrifuged at 90,000
× g for 1 h at 4 °C, and the supernatant was taken for the
primer extension. F, effect of YoeB on the 30-base RNA binding to the
and 0.05
μm 70 S ribosomes and 20.7 nm YoeB(His)6.
Lanes 3 and 5, the samples were centrifuged at 90,000
× g for 1 h at 4 °C, and the supernatant was taken for the
primer extension. F, effect of YoeB on the 30-base RNA binding to the
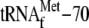 S
ribosome complex. 70 S ribosomes (0.05 μm) were first incubated
with 0, 42, 168, 336, and 672 nm YoeB(His)6 and
S
ribosome complex. 70 S ribosomes (0.05 μm) were first incubated
with 0, 42, 168, 336, and 672 nm YoeB(His)6 and
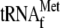 (1
μm) for 10 min at 37 °C. The 5′-end labeled 30-base
RNA (35 nm) was then added. The reaction mixture was incubated for
an additional 10 min at 37 °C, and then applied to nitrocellulose filters
(Millpore 0.45 μm HA), which were washed twice with 2 ml of
buffer A before measuring the radioactivity. G, effect of
tetracycline on the activity of YoeB-mediated mRNA cleavage. Lane 1,
1 μm
(1
μm) for 10 min at 37 °C. The 5′-end labeled 30-base
RNA (35 nm) was then added. The reaction mixture was incubated for
an additional 10 min at 37 °C, and then applied to nitrocellulose filters
(Millpore 0.45 μm HA), which were washed twice with 2 ml of
buffer A before measuring the radioactivity. G, effect of
tetracycline on the activity of YoeB-mediated mRNA cleavage. Lane 1,
1 μm
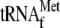 and 0.05
μm 70 S ribosomes; lane 2, 1 μm
and 0.05
μm 70 S ribosomes; lane 2, 1 μm
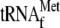 , 0.05
μm 70 S ribosomes, and 160 μm tetracycline;
lane 3, 1 μm
, 0.05
μm 70 S ribosomes, and 160 μm tetracycline;
lane 3, 1 μm
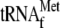 , 0.05
μm 70 S ribosomes, and 10.4 nm YoeB(His)6;
lane 4, 1 μm
, 0.05
μm 70 S ribosomes, and 10.4 nm YoeB(His)6;
lane 4, 1 μm
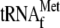 , 0.05
μm 70 S ribosomes, 10.4 nm YoeB(His)6, and
160 μm tetracycline. H, effect of hygromycin B on the
activity of YoeB-mediated mRNA cleavage. Lane 1, 1 μm
, 0.05
μm 70 S ribosomes, 10.4 nm YoeB(His)6, and
160 μm tetracycline. H, effect of hygromycin B on the
activity of YoeB-mediated mRNA cleavage. Lane 1, 1 μm
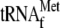 and 0.05
μm 70 S ribosomes; lanes 2 and 3, 1
μm
and 0.05
μm 70 S ribosomes; lanes 2 and 3, 1
μm
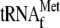 , 0.05
μm 70 S ribosomes, and 0.64 and 1.6 mm hygromycin B,
respectively; lane 4, 1 μm
, 0.05
μm 70 S ribosomes, and 0.64 and 1.6 mm hygromycin B,
respectively; lane 4, 1 μm
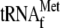 , 0.05
μm 70 S ribosomes, and 20.7 nm YoeB(His)6;
lanes 5 and 6, 1 μm
, 0.05
μm 70 S ribosomes, and 20.7 nm YoeB(His)6;
lanes 5 and 6, 1 μm
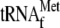 , 0.05
μm 70 S ribosomes, 20.7 nm YoeB(His)6, and
0.64 and 1.6 mm hygromycin B, respectively. SD,
Shine-Dalgarno; FL, full-length.
, 0.05
μm 70 S ribosomes, 20.7 nm YoeB(His)6, and
0.64 and 1.6 mm hygromycin B, respectively. SD,
Shine-Dalgarno; FL, full-length.
We also tested if
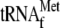 bound to the
ribosomes is displaced with YoeB under the condition as that used in
Fig. 6A. For this
purpose,
bound to the
ribosomes is displaced with YoeB under the condition as that used in
Fig. 6A. For this
purpose,
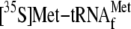 was first incubated with 70 S ribosomes and mRNA at 37 °C for 10 min, and
then different amounts of YoeB as used in
Fig. 6A were added.
The reaction mixtures were incubated at 37 °C for another 10 min. The
reaction mixtures were then applied to nitrocellulose filters (Millpore 0.45
μm HA) and the filters were washed twice with 2 ml of buffer A
before measuring the radioactivity retained on the filters. As shown in
Fig. 6C,
was first incubated with 70 S ribosomes and mRNA at 37 °C for 10 min, and
then different amounts of YoeB as used in
Fig. 6A were added.
The reaction mixtures were incubated at 37 °C for another 10 min. The
reaction mixtures were then applied to nitrocellulose filters (Millpore 0.45
μm HA) and the filters were washed twice with 2 ml of buffer A
before measuring the radioactivity retained on the filters. As shown in
Fig. 6C,
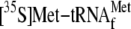 was displaced in a YoeB concentration-dependent manner. Over 70% of
was displaced in a YoeB concentration-dependent manner. Over 70% of
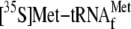 was released at 10 nm YoeB (the ratio of YoeB to ribosome is 1:5).
It should be noted that ribosome-bound initiator tRNA drops from 72 to about
42, upon addition of 0.04 nm YoeB and further to about 30 upon
addition of 3 nm YoeB. Because the major part of the decrease in
ribosome bound initiator tRNA occurred at substoichiometric additions of YoeB,
this result is not due to direct competition between initiator tRNA and YoeB.
It is more likely due to indirect competition caused by mRNA cleavage. The
binding stability of initiator tRNA to 70 S ribosomes is known to be coupled
to that of mRNA, and therefore if mRNA is cleaved, the binding of mRNA to 70 S
ribosomes should be reduced, which in turn, likely destabilizes initiator tRNA
binding (Fig. 6C).
was released at 10 nm YoeB (the ratio of YoeB to ribosome is 1:5).
It should be noted that ribosome-bound initiator tRNA drops from 72 to about
42, upon addition of 0.04 nm YoeB and further to about 30 upon
addition of 3 nm YoeB. Because the major part of the decrease in
ribosome bound initiator tRNA occurred at substoichiometric additions of YoeB,
this result is not due to direct competition between initiator tRNA and YoeB.
It is more likely due to indirect competition caused by mRNA cleavage. The
binding stability of initiator tRNA to 70 S ribosomes is known to be coupled
to that of mRNA, and therefore if mRNA is cleaved, the binding of mRNA to 70 S
ribosomes should be reduced, which in turn, likely destabilizes initiator tRNA
binding (Fig. 6C).
In a reciprocal experiment, where increasing amounts of
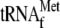 were added in
the reaction mixture, the YoeB-mediated mRNA cleavage activity was diminished
(Fig. 6B), However,
the TP(r) band did not appear, further suggesting that YoeB and initiator tRNA
appear to indirectly compete for binding to 70 S ribosomes. It should be noted
that the use of
were added in
the reaction mixture, the YoeB-mediated mRNA cleavage activity was diminished
(Fig. 6B), However,
the TP(r) band did not appear, further suggesting that YoeB and initiator tRNA
appear to indirectly compete for binding to 70 S ribosomes. It should be noted
that the use of
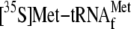 instead of uncharged
instead of uncharged
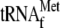 did not
affect the toeprinting results (Fig.
6D, compare lane 4 with 6, and lane
5 with 7).
did not
affect the toeprinting results (Fig.
6D, compare lane 4 with 6, and lane
5 with 7).
Tetracycline and Hygromycin B Inhibit YoeB-mediated mRNA
Cleavage—To identify the exact YoeB-acting site, we next examined
the effect of tetracycline and hygromycin B on YoeB-mediated mRNA cleavage.
Both tetracycline and hygromycin B are known to bind to the A site in
ribosomes (23).
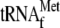 was first
incubated with 70 S ribosomes and mRNA at 37 °C for 10 min to form the
translation initial complex, and then tetracycline
(Fig. 6G, lanes
2 and 4) and different amounts of hygromycin B
(Fig. 6H, lanes 2,
3, 5, and 6) were added. The reaction mixtures were incubated at
37 °C for another 10 min. YoeB was then added to the reaction mixtures for
another 10 min (Fig. 6,
G, lanes 3 and 4, and H, lanes
4–6). Reverse transcriptase was finally added to initiate cDNA
synthesis. As shown in Fig.
6G, 160 μm tetracycline (the ratio of
tetracycline to 70 S ribosomes is 3250 to 1) inhibited YoeB-mediated mRNA
cleavage at the A site with concomitant appearance of the TP(r) band
(Fig. 6G, lane
4). With hygromycin B, two different concentrations (0.64 mm
for lanes 2 and 5 and 1.6 mm for lanes 3
and 6) were used. As shown in Fig.
6H, 1.6 mm hygromycin B had little inhibitory
effect on the translation initiation complex formation (compare lane
1 with lane 3 in Fig.
6H). However, 1.6 mm hygromycin B (the ratio
of hygromycin B to 70 S ribosomes is 32500) almost completely blocked
YoeB-mediated mRNA cleavage activity (Fig.
6H, compare lane 4 with 6). At 0.64
mm hygromycin B, the mRNA cleavage was partially inhibited with
concomitant appearance of the TP(r) band
(Fig. 6H, lane
5). Note that at 1.6 mm hygromycin B, the intensity of the
TP(r) band was almost identical to that in the absence of both hygromycin and
YoeB (Fig. 6H, compare
lane 6 with 1). These results are consistent with the result
obtained with tetracycline in Fig.
6G, suggesting that hygromycin B binding to A sites
interferes with YoeB-mediated mRNA cleavage at the A site.
was first
incubated with 70 S ribosomes and mRNA at 37 °C for 10 min to form the
translation initial complex, and then tetracycline
(Fig. 6G, lanes
2 and 4) and different amounts of hygromycin B
(Fig. 6H, lanes 2,
3, 5, and 6) were added. The reaction mixtures were incubated at
37 °C for another 10 min. YoeB was then added to the reaction mixtures for
another 10 min (Fig. 6,
G, lanes 3 and 4, and H, lanes
4–6). Reverse transcriptase was finally added to initiate cDNA
synthesis. As shown in Fig.
6G, 160 μm tetracycline (the ratio of
tetracycline to 70 S ribosomes is 3250 to 1) inhibited YoeB-mediated mRNA
cleavage at the A site with concomitant appearance of the TP(r) band
(Fig. 6G, lane
4). With hygromycin B, two different concentrations (0.64 mm
for lanes 2 and 5 and 1.6 mm for lanes 3
and 6) were used. As shown in Fig.
6H, 1.6 mm hygromycin B had little inhibitory
effect on the translation initiation complex formation (compare lane
1 with lane 3 in Fig.
6H). However, 1.6 mm hygromycin B (the ratio
of hygromycin B to 70 S ribosomes is 32500) almost completely blocked
YoeB-mediated mRNA cleavage activity (Fig.
6H, compare lane 4 with 6). At 0.64
mm hygromycin B, the mRNA cleavage was partially inhibited with
concomitant appearance of the TP(r) band
(Fig. 6H, lane
5). Note that at 1.6 mm hygromycin B, the intensity of the
TP(r) band was almost identical to that in the absence of both hygromycin and
YoeB (Fig. 6H, compare
lane 6 with 1). These results are consistent with the result
obtained with tetracycline in Fig.
6G, suggesting that hygromycin B binding to A sites
interferes with YoeB-mediated mRNA cleavage at the A site.
Cleavage of mRNA at the A Site Codons Releases the 3′-End Portion of the mRNA from 70 S Ribosomes—To examine if the cleavage of mRNA results in the release of the 3′-end portion of the mRNA from 70 S ribosomes, the reaction mixture after complex formation with the use of the ompA mRNA was centrifuged at 90,000 × g at 4°C for 1 h to pellet ribosomes. Subsequently, we determined if the 3′-end portion of the mRNA was released into the supernatant by primer extension reaction. As shown in Fig. 6E, the TP(r) band was not detectable in the supernatant fraction (lane 3), whereas the TP(Y) band was almost fully recovered in the supernatant fraction (lane 5). These results indicate that the TP(Y) band detected in the toeprinting experiments was derived from the 3′-end portion of the ompA mRNA, subsequently released into the soluble fraction (or into the cytoplasmic fraction in the cells). Using the same 30-base synthetic RNA as used in Fig. 5F, we examined if the 5′-end portion of the RNA dissociates from ribosomes in the presence of YoeB. Although the RNA is cleaved at 7 bases from the 3′ end (see Fig. 5F), the remaining 23-base RNA (labeled at its 5′ end with 32P) was bound to the ribosomes even at a very high concentration of YoeB (70 times higher concentration than that used in Fig. 6C; see Fig. 6F), indicating that the 5′-end portion of mRNA stays bound to 70 S ribosomes even after cleavage of the mRNA.
Effect of a Nuclease-negative Mutation (H83Q) in YoeB on the Cell Growth and Protein Synthesis—H83Q, a nuclease negative YoeB mutant, previously reported by Kamada and Hannoka (3) was also constructed to examine its effect on cell growth. Induction of the YoeB(H83Q) mutant inhibited cell growth and the addition of YoeB(H83Q) also inhibited MazG protein synthesis in vitro (data not shown). When the toeprinting experiment was carried out using the purified YoeB(H83Q), toeprinting bands (TP(Y)) three and four bases downstream of the initiation codon were detected in the manner identical to that of the wild-type YoeB (data not shown). Thus, YoeB(H83Q), a nuclease negative mutant, does not affect YoeB-mediated mRNA cleavage at the A site. Notably a higher concentration of the mutant YoeB than the wild-type YoeB was required to result in the same toeprinting effect as caused with the wild-type YoeB, indicating the mutant YoeB seems to have a weaker affinity to 70 S ribosomes than the wild-type YoeB.
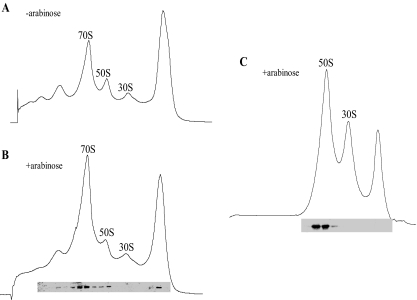
YoeB Associates with 50 S Ribosomal Subunits—Because YoeB binds to the translation initiation complex (Fig. 4), we examined whether YoeB affects the polysome profiles and also specifically associates with one of the ribosomal subunits. When the polysome pattern of E. coli BW25113 cells carrying the pBAD-YoeB plasmid was analyzed by sucrose density gradient at 10 min after induction of YoeB by arabinose, a significant increase in the 70 S ribosomal fraction was observed without change in the 30 S and 50 S ribosomal profiles (compare Fig. 7B with Fig. 7A). Western blotting analysis showed that YoeB was associated with both 70 S ribosome and 50 S ribosome fractions, but not with 30 S ribosome (Fig. 7B). Cell lysates were prepared in 0.5 mm Mg2+ to dissociate 70 S ribosomes to 50 S and 30 S ribosomes and analyzed by sucrose density gradient centrifugation (Fig. 7C), where YoeB was detected only in the 50 S ribosome fraction, demonstrating that YoeB is a 50 S ribosome associating protein.
FIGURE 7.
YoeB associates with the large 50 S ribosomal subunits. Ribosome profiles were analyzed by sucrose density gradient centrifugation as described under “Experimental Procedures.” A, polysome profiles of E. coli BW25113 containing pBAD-YoeB without YoeB induction. B, polysome profiles of E. coli BW25113 containing pBAD-YoeB at 10 min after YoeB induction. C, 70 S ribosomes of the same lysate used in B were disassociated into 50 S and 30 S ribosomal subunits in 0.5 mm Mg2+. In B and C, Western blot analysis was carried out to detect YoeB in each gradient fraction.
DISCUSSION
In the present paper, we demonstrated that YoeB is a potent inhibitor of protein synthesis, which functions in a manner that is distinctly different from that of MazF, an mRNA interferase (6). Upon YoeB induction, [35S]methionine incorporation into cellular proteins was abruptly stopped within less than 5 min (Fig. 1, E and F). This is in sharp contrast to MazF induction, which stops [35S]methionine incorporation only after 15–20 min of its induction (6). Another important difference between YoeB and MazF is that significant amounts of full-length cellular mRNAs remain in the YoeB-induced cells even 30 min after its induction (Fig. 3, A and B), whereas they rapidly disappear after 10 min upon MazF induction (6). In addition, MazF inhibits protein synthesis in both prokaryotes and eukaryotes as expected from its function as an mRNA interferase (6), whereas YoeB was found to be a prokaryote-specific inhibitor (Fig. 2, C and E).
On the basis of these results, we speculated that YoeB does not primarily function as a sequence-specific endoribonuclease or an mRNA interferase as originally proposed by Christensen et al. (1) and Kamada and Hanaoka (3). To substantiate our hypothesis, we carried out in vivo primer extension experiments using chromosomally encoded mRNAs as targets rather than mRNAs encoded by genes cloned in a multicopy plasmid to avoid artifacts (Fig. 3, C and D). Interestingly, both mRNAs tested were detected either as the corresponding full-length mRNAs or as a product cleaved at three bases downstream of the initiation codon (Fig. 3, C and D). Notably, in the ∼200-nucleotide 5′ leader sequences for both mRNAs, no other cleavage bands were detected. These mRNAs encoded outer membrane proteins and were used in our experiment because of their abundance in the cell. However, the mRNA from the glnH gene that encodes a cytoplasmic protein was also cleaved at three bases downstream of AUG (data not shown), indicating that the cleavage of mRNAs by YoeB does not depend on the type of these mRNAs.
Importantly, this in vivo observation was consistent with the
in vitro toeprinting experiments using 70 S ribosomes,
 , purified
YoeB, and the same mRNAs (Fig.
4). We demonstrated that the addition of YoeB caused the upstream
shift of the stop site for primer extension (toeprinting site) from normal 13
and 14 bases downstream of AUG to three and four bases from AUG. This apparent
10-base upward shift resulted from the cleavage of the mRNA at the A site
mediated by YoeB bound to 70 S ribosomes
(Fig. 5, A, lane 6, and
D, lane 9, and
Fig. 6, E, lane
5), although the exact mechanism for this cleavage remains to be
determined. YoeB specifically binds to 50 S ribosomes
(Fig. 7, B and
C), whereas hygromycin B and tetracycline bind to 30 S
ribosomes. Therefore it is reasonable to speculate that YoeB locates at the
interface between 50 S and 30 S ribosomes to interact with the A site to
cleave mRNA at that site. This interface is known to play important roles in
translation initiation
(22).
, purified
YoeB, and the same mRNAs (Fig.
4). We demonstrated that the addition of YoeB caused the upstream
shift of the stop site for primer extension (toeprinting site) from normal 13
and 14 bases downstream of AUG to three and four bases from AUG. This apparent
10-base upward shift resulted from the cleavage of the mRNA at the A site
mediated by YoeB bound to 70 S ribosomes
(Fig. 5, A, lane 6, and
D, lane 9, and
Fig. 6, E, lane
5), although the exact mechanism for this cleavage remains to be
determined. YoeB specifically binds to 50 S ribosomes
(Fig. 7, B and
C), whereas hygromycin B and tetracycline bind to 30 S
ribosomes. Therefore it is reasonable to speculate that YoeB locates at the
interface between 50 S and 30 S ribosomes to interact with the A site to
cleave mRNA at that site. This interface is known to play important roles in
translation initiation
(22).
Importantly, these antibiotics (tetracycline and hygromycin B), which are
known to inhibit protein synthesis by binding to the A site
(23), block YoeB-mediated mRNA
cleavage at the A site (Fig. 6, G
and H). This supports the notion that YoeB binds to the
50 S ribosomal subunit in 70 S ribosomes and interacts with the A site leading
to mRNA cleavage. It is also apparent that mRNA cleavage upon YoeB binding
makes the translational initial complex unstable as
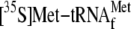 was released (Fig.
6C). On the other hand, it should be noted the mRNA is
able to form a complex with 70 S ribosomes even in the absence of
was released (Fig.
6C). On the other hand, it should be noted the mRNA is
able to form a complex with 70 S ribosomes even in the absence of
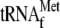 if YoeB is
added (Fig. 5, C and
D, lane 6).
if YoeB is
added (Fig. 5, C and
D, lane 6).
It is important to note that YoeB alone did not show any endoribonuclease activity to several mRNAs under the conditions used in the toeprinting experiments (Fig. 4, lane 2). Only when it was added along with 70 S ribosomes, mRNAs were cleaved at three and four bases downstream of AUG (Fig. 5, C and D, lane 6), suggesting that YoeB is a ribosome-associating factor (Fig. 7, B and C), which may stimulate the intrinsic endogenous endoribonuclease activity of ribosomes or YoeB itself. Although purified YoeB has been shown to have a purine-specific endoribonuclease activity, albeit very weak (3), the fact that the nuclease-negative YoeB(H83Q) was able to inhibit cell growth and protein synthesis supports the notion that the endoribonuclease activity of YoeB is not primarily required for its inhibitory function for protein synthesis. It should be noted that even though protein synthesis was inhibited in vivo in 5–10 min after YoeB induction (1), purified YoeB only partially cleaved mRNA even 60 min after its incubation with the mRNA substrate (3). Notably, YoeB stimulates only partial ribosome-mediated mRNA cleavage at three bases downstream of AUG as seen in the in vivo primer extension experiments for all mRNAs tested (Fig. 3, C and D), whereas protein synthesis is completely inhibited (Fig. 1, E and F). This further suggests that the primary inhibitory function of YoeB for protein synthesis is not its endoribonuclease activity, but is to prevent the formation of the initiation complex. However, the binding of YoeB to 70 S ribosomes seems to stimulate their latent endoribonuclease activity, resulting in partial cleavage of mRNAs at three bases downstream of the initiation codon (Fig. 5, A, lane 6, and D, lane 9). As a result, the 3′-end portion of the mRNA is dissociated from the ribosomes and released to the cytoplasm (Fig. 6E, lane 5).
In summary, YoeB binds to 50 S ribosomal subunit in 70 S ribosomes and interacts with the A site (Fig. 6, G and H) to very effectively prevent the translational initiation complex formation resulting in the inhibition of protein synthesis almost immediately after YoeB induction. This apparently leads to activation of the latent endoribonuclease activity of either ribosomes or YoeB resulting in the cleavage of mRNA at the A site, and release of the 3′-end portion of mRNA from ribosomes (Fig. 6E).
Recently, it has been reported that E. coli cold-shock protein Y
(22) and an antibiotic GE82221
(24) cause specific inhibition
of translation initiation. Both of these, however, bind to 30 S ribosomes to
directly prevent
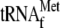 binding to
the P site, which is quite different from the mechanism of YoeB-mediated
inhibition of translation. It is quite interesting to point out that E.
coli uses different strategies to regulate translation; at the level of
initiation (YoeB and protein Y), or at the level of elongation by mRNA
interferases (MazF and ChpBK) and termination (RelE).
binding to
the P site, which is quite different from the mechanism of YoeB-mediated
inhibition of translation. It is quite interesting to point out that E.
coli uses different strategies to regulate translation; at the level of
initiation (YoeB and protein Y), or at the level of elongation by mRNA
interferases (MazF and ChpBK) and termination (RelE).
Acknowledgments
We thank Dr. Marilyn Kozak and Sangita Phadtare for critical reading of this manuscript. We also thank Guangying Sun and Ling Zhu for assistance during the course of experiments.
This work was supported by National Institutes of Health Grant RO1GM081567 and by a grant from Takara Bio Inc. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DTT, dithiothreitol; TP, toeprinting.
References
- 1.Christensen, S. K., Maenhaut-Michel, G., Mine, N., Gottesman, S., Gerdes, K., and Van Melderen, L. (2004) Mol. Microbiol. 51 1705–1717 [DOI] [PubMed] [Google Scholar]
- 2.Cherny, I., and Gazit, E. (2004) J. Biol. Chem. 279 8252–8261 [DOI] [PubMed] [Google Scholar]
- 3.Kamada, K., and Hanaoka, F. (2005) Mol. Cell 19 497–509 [DOI] [PubMed] [Google Scholar]
- 4.Pedersen, K., Zavialov, A. V., Pavlov, M. Y., Elf, J., Gerdes, K., and Ehrenberg, M. (2003) Cell 112 131–140 [DOI] [PubMed] [Google Scholar]
- 5.Hayes, C. S., and Sauer, R. T. (2003) Mol. Cell 12 903–911 [DOI] [PubMed] [Google Scholar]
- 6.Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003) Mol. Cell 12 913–923 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Y., Zhu, L., Zhang, J., and Inouye, M. (2005) J. Biol. Chem. 280 26080–26088 [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swaney, S. M., Aoki, H., Ganoza, M. C., and Shinabarger, D. L. (1998) Antimicrob. Agents Chemother. 42 3251–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halegoua, S., Hirashima, A., Sekizawa, J., and Inouye, M. (1976) Eur. J. Biochem. 69 163–167 [DOI] [PubMed] [Google Scholar]
- 11.Halegoua, S., Hirashima, A., and Inouye, M. (1976) J. Bacteriol. 126 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moses, R. E., and Richardson, C. C. (1970) Proc. Natl. Acad. Sci. U. S. A. 67 674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson, R. L., Radcliffe, C. W., and Pace, N. R. (1971) J. Bacteriol. 107 585–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleted in proof
- 15.Zhang, J., and Inouye, M. (2002) J. Bacteriol. 184 5323–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki, H., Ke, L., Poppe, S. M., Poel, T. J., Weaver, E. A., Gadwood, R. C., Thomas, R. C., Shinabarger, D. L., and Ganoza, M. C. (2002) Antimicrob. Agents Chemother. 46 1080–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, H., and Babitzke, P. (1998) J. Biol. Chem. 273 20494–20503 [DOI] [PubMed] [Google Scholar]
- 18.Hesterkamp, T., Deuerling, E., and Bukau, B. (1997) J. Biol. Chem. 272 21865–21871 [DOI] [PubMed] [Google Scholar]
- 19.Moll, I., and Blasi, U. (2002) Biochem. Biophys. Res. Commun. 297 1021–1026 [DOI] [PubMed] [Google Scholar]
- 20.Sarmientos, P., Sylvester, J. E., Contente, S., and Cashel, M. (1983) Cell 32 1337–1346 [DOI] [PubMed] [Google Scholar]
- 21.Baker, K. E., and Mackie, G. A. (2003) Mol. Microbiol. 47 75–88 [DOI] [PubMed] [Google Scholar]
- 22.Vila-Sanjurjo, A., Schuwirth, B. S., Hau, C. W., and Cate, J. H. (2004) Nat. Struct. Mol. Biol. 11 1054–1059 [DOI] [PubMed] [Google Scholar]
- 23.Brodersen, D. E., Clemons, W. M., Jr., Carter, A. P., Morgan-Warren, R. J., Wimberly, B. T., and Ramakrishnan, V. (2000) Cell 103 1143–1154 [DOI] [PubMed] [Google Scholar]
- 24.Brandi, L., Fabbretti, A., La Teana, A., Abbondi, M., Losi, D., Donadio, S., and Gualerzi, C. O. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajbhandary U., and Ghosh, H. (1969) J. Biol. Chem. 244 1104–1113 [PubMed] [Google Scholar]
- 26.Takagi, H., Kakuta, Y., Okada, T., Yoo, M., Tanaka, I., and Kimuya, M. (2005) Nat. Struct. Mol. Biol. 12 327–331 [DOI] [PubMed] [Google Scholar]