Abstract
Aminoglycoside 2″-phosphotransferases are clinically important enzymes that cause high levels of resistance to aminoglycoside antibiotics by the organisms that harbor them. These enzymes phosphorylate aminoglycosides, and the modified antibiotics show significant reduction in the binding ability to target the bacterial ribosome. This report presents a detailed characterization of the antibiotic resistance profile and the aminoglycoside and nucleotide triphosphate substrate profiles of four common aminoglycoside 2″-phosphotransferases widely distributed in clinically important Gram-positive microorganisms. Although the antibiotic resistance phenotypes exhibited by these enzymes are similar, their aminoglycoside and nucleotide triphosphate substrate profiles are distinctive. Contrary to the dogma that these enzymes use ATP as the source of phosphate in their reactions, two of the four aminoglycoside 2′-phosphotransferases utilize GTP as the phosphate donor. Of the other two enzymes, one exhibits preference for ATP, and the other can utilize either ATP or GTP as nucleotide triphosphate substrate. A new nomenclature for these enzymes is put forth that takes into account the differences among these enzymes based on their respective substrate preferences. These nucleotide triphosphate preferences should have ramifications for understanding of the evolution, selection, and dissemination of the genes for these important resistance enzymes.
Following their introduction to the clinic in the 1940s (1–3), aminoglycoside antibiotics rapidly became important chemotherapeutic agents in treatment of various microbial infections. Clinical success of aminoglycosides was in large measure the result of broad spectrum of activity (4–7), dose-dependent bactericidal activity (8–11), postantibiotic effect (ability to continue to kill bacteria after antibiotic removal following a short incubation with the microorganism) (12–14) and synergy with other antibiotics (15, 16). Another advantage was that in contrast to other antibiotic classes, such as β-lactams, emergence of resistance to aminoglycosides has occurred relatively slowly. Nonetheless, multiple mechanisms of resistance to this class of antibiotics have emerged over time (17, 18). These resistance mechanisms have resulted in reduced efficacy of several of aminoglycoside antibiotics as well as obsolescence of others, such as kanamycin (19).
Structurally, aminoglycosides are broadly classified into three classes, 4,6-disubstituted, 4,5-disubstituted, and atypical (see supplemental Fig. 1). The primary means for resistance to aminoglycosides is the production of the enzymes that chemically modify the antibiotics. The chemical modifications of aminoglycosides alter their properties both sterically and electrostatically, such that the binding to the bacterial ribosome, the target for aminoglycosides, is compromised (20, 21). Quantitative evaluations of these influences on binding to the ribosomal sites are few, but for the ones that have been investigated, the effects are substantial (22).
The antibiotic selection force has resulted in the emergence of genes that encode three families of aminoglycoside-modifying enzymes. These are aminoglycoside acetyltransferases, aminoglycoside nucleotidyltransferases, and aminoglycoside phosphotransferases (APHs).2 They are all two-substrate enzymes that facilitate transfer of functionalities from the second substrate to the aminoglycoside. The second substrate is acetyl-coenzyme A in the case of aminoglycoside acetyltransferases, which transfers the acetylate moiety to amino groups at positions 1, 3, 2′, or 6′ of the antibiotics. Aminoglycoside nucleotidyltransferases (adenylyltransferases) modify aminoglycoside antibiotics by the transfer of the AMP portion of ATP to the hydroxyl groups at positions 6, 9, 4′, 2″, or 3″ of the antibiotics. Finally, aminoglycoside phosphotransferases modify aminoglycoside antibiotics at 4-, 6-, 9-, 3′-, 2″-, 3″-, and 7″-hydroxyls of aminoglycosides by phosphorylation (see supplemental Fig. 1). Since the discovery of the APHs, it has been widely assumed that they use ATP as the second substrate (23, 24). This assumption has been based on the appreciation of the central roles that ATP plays in many biological processes but also on biochemical measurements that document that indeed ATP is a substrate for several of these enzymes.
We have undertaken an enzymological study of the four APHs that were provisionally assigned to the class of (2″)-I phosphotransferases based on good sequence identity to the APH(2″)-Ia, whose regiospecificity of phosphate transfer was experimentally determined (25). These enzymes were originally isolated from Staphylococcus aureus (26) and Enterococci (27–29) but later were also identified in Gram-negative bacterial isolates (30). They had been subsequently classified as APH(2″)-Ia, -Ib, -Ic, and -Id based on the available data on aminoglycoside resistance profiles that they produce and amino acid sequence of individual enzymes. The Roman numerals define resistance profile, and thus the assignment of identical Roman numeral I to all APH(2″) enzymes indicated that they produce the identical spectrum of resistance, whereas letters a, b, c, and d indicate that each enzyme is characterized by a unique amino acid sequence. Our analyses revealed that two of these enzymes (APH(2″)-Ia and -Ic) actually utilize GTP instead of ATP as the second substrate. This unexpected finding brings into question the dogmatic assertion that these enzymes are all ATP-dependent. Based on these findings, a new nomenclature for APH(2″) phosphotransferases is proposed herein. Future studies of APHs and also of aminoglycoside nucleotidyltransferases should bring into sharp focus the issue of selectivity of the nucleotide triphosphates for the given enzyme. These nucleotide triphosphate preferences should have ramifications for understanding of the evolution, selection, and dissemination of the genes for these important enzymes.
EXPERIMENTAL PROCEDURES
Cloning of the aph(2″) Genes—To ensure identical levels of expression for APH(2″)-Ia, -Ib, -Ic and -Id, we cloned the genes for these phosphotransferases under the same promoter of the aph(2″)-Ic gene in pBluescript II KS(+) vector (Stratagene). The detailed cloning procedure is described in the supplemental material and supplemental Fig. 2. For protein expression and purification, the genes for the APH(2″) enzymes were recloned into the expression vector pET22b(+) (Novagen), utilizing its NdeI site at the 5′-end and the HindIII site at the 3′-end.
Antibiotic Susceptibility Testing—Antibiotics used in this study were from the following sources: kanamycin A, kanamycin B, tobramycin, sisomicin, G418, amikacin, neomycin, paromomycin, lividomycin A, butirosin, streptomycin, spectinomycin, apramycin, and hygromycin B from Sigma, gentamicin from Fluka, netilmicin and isepamicin from Schering Plough, dibekacin and arbekacin from Meiji Seika Kaisha, and fortimicin from Kyova Hakko Kogyo. Minimum inhibitory concentrations (MICs) of various aminoglycoside antibiotics were determined in Mueller-Hinton II broth by the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (31). Escherichia coli JM83 strains harboring pBluescript::aph(2″)-Ia, -Ib, -Ic and -Id vectors were spread on agar plates supplemented with 100 μg/ml ampicillin. A parental E. coli JM83 strain harboring the plasmid pBluescript II KS(+) vector, which does not contain any genes for APH(2″) enzymes, was used as the control and was plated on antibiotic-free agar. After overnight incubation, three or four individual colonies were inoculated into 3 ml of Mueller-Hinton II broth and subgrown at 35 °C for several h until bacterial cultures reached an optical density of 0.6–0.8 at 600 nm. Optical densities of all cultures were equalized, and, after appropriate dilutions, the cultures were used to inoculate 96-well microtiter plates containing Mueller-Hinton II broth with 2-fold serial dilutions of various aminoglycoside antibiotics. The final bacterial inoculum was 5 × 105 colony-forming units/ml. The plates were incubated at 35 °C for 18 h before results were recorded. All MICs were determined in triplicate.
Protein Expression and Purification—The APH(2″)-Ib and -Ic enzymes were purified as described elsewhere (32, 33). For purification of APH(2″)-Ia and -Id, E. coli BL21(DE3) strains harboring the pET22b(+) vector with cloned aminoglycoside phosphotransferase genes were grown at 37 °C in LB medium (2 × 350 ml for each construct) supplemented with 100 μg/ml ampicillin to A590 ∼ 0.8. Protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside (0.8 mm), and cultures were incubated overnight at 20 °C (for APH(2″)-Ia) or 22 °C (for APH(2″)-Id) for the best protein yield and solubility. Bacteria were pelleted by centrifugation (3,000 × g for 15 min), resuspended in 30 ml of buffer A (25 mm HEPES, pH 7.5, 0.2 mm dithiothreitol) supplemented with 1 mm EDTA, and the cells were disrupted by repeated cycles of sonification (Bronson Sonifier 450 (VWR, West Chester, PA)), and debris was pelleted by subsequent centrifugation at 20,000 × g for 30 min. To remove nucleic acids (34), the supernatant was mixed with streptomycin sulfate (1.5% final concentration) and stirred slowly at 4 °C for 30 min. Solutions were centrifuged at 20,000 × g for 30 min, and the supernatant was dialyzed twice against 2 liters of buffer A.
The APH (2″)-Ia enzyme was purified by DEAE anion exchange chromatography. The enzyme was eluted with a gradient of NaCl (0–1 m) in buffer A, and fractions were analyzed by 12% SDS-PAGE. Enzyme-containing fractions (eluted at 0.4–0.5 m NaCl) were pooled, concentrated to 30 ml, dialyzed against buffer A, and loaded on an Affi-Gel 15-gentamicin affinity column (35). APH (2″)-Ia was eluted with gradient of NaCl (0–1 m) in buffer A. Fractions were examined by SDS-PAGE, and those that contained the purified APH (2″)-Ia protein (eluted at 0.55–0.7 m of NaCl) were pooled, concentrated to 8 mg/ml, dialyzed against 25 mm HEPES, pH 7.5, 0.5 mm dithiothreitol, and stored in liquid nitrogen.
APH(2″)-Id was purified first by affinity chromatography (Affi-Gel 15-gentamicin; 2.5 × 12 cm) and prepared as described previously (35). After elution with a NaCl gradient, fractions were analyzed by SDS-PAGE, and those that contained APH(2″)-Id (eluted at 0.3–0.5 m NaCl) were pooled, concentrated to 30 ml, and dialyzed against buffer A. The enzyme was further purified by DEAE anion exchange chromatography (2.5 × 18 cm) and eluted with a gradient of NaCl (0–1 m), and the fractions that contained the purified APH (2″)-Id (eluted at 0.25–0.35 m NaCl) were concentrated to 8 mg/ml, dialyzed against 25 mm HEPES, pH 7.5, 1 mm dithiothreitol and stored in aliquots at –80 °C. All enzymes used in this study were purified to homogeneity, as judged by SDS-PAGE.
Enzyme Kinetics—A continuous spectrophotometric assay was employed to monitor phosphorylation of kanamycin A by aminoglycoside phosphotransferases in the presence of various NTPs as the second substrates for the enzymes. In this assay, conversion of NDP released during the reaction back to NTP is coupled to the production of lactate from pyruvate and measured as a function of NADH oxidation. In a typical experiment, assays were performed in a total volume of 250 μl containing 100 mm reaction buffer (see below), 10 mm MgCl2, 20 mm KCl, 2 mm phosphoenolpyruvate, 140 μm NADH, 15 units/ml pyruvate kinase, 20 units/ml lactate dehydrogenase, kanamycin A at a fixed saturating concentration (see below) and NTP at variable concentrations. The reaction was initiated by the addition of enzyme (200 nm for APH(2″)-Ia; 4–40 nm for APH(2″)-Ib; 100–700 nm for APH(2″)-Id). To find the best conditions for kinetics assays, a set of buffers was used in pilot experiments. In the final experiments, sodium HEPES, pH 7.0, was used in assays with APH(2″)-Ia, sodium HEPES, pH 7.5, with APH(2″)-Ib and MOPS, pH 7.5, with APH(2″)-Id. Ionic strength of all buffers was adjusted to 80 mm with NaCl. Depending on the enzyme, concentrations of kanamycin A were between 20 and 100 μm (to ensure enzyme saturation and to avoid substrate inhibition, which has precedence for APHs (36).
When CTP was used in the coupled assay, the concentration of pyruvate kinase in the reaction mixture was increased to 60 units/ml due to the poor turnover of CDP by this enzyme. When UTP was used as the phosphate donor, assay conditions were modified due to the presence of 15–20% of UDP in the commercially available sample of UTP. Specifically, 5 mm phosphoenolpyruvate, 300 mm NADH, and 20 units/ml of lactate dehydrogenase were added to the reaction mixture containing all of the needed components (see above), with the exception of kanamycin A and the enzyme. The reaction was monitored spectrophotometrically to ensure complete conversion of UDP to UTP, upon which APH(2″) and kanamycin A were added to initiate the reaction. Conditions and the results for kinetics with APH(2″)-Ic were described elsewhere (32). Calculations of the steady-state kinetic parameters for each substrate were performed with GraFit 4.0 software. The values for kcat, Km, and kcat/Km were determined by fitting the kinetic data to the following equation,
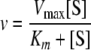 |
(Eq. 1) |
where kcat = Vmax/[E], v is the initial velocity, Vmax is the maximum reaction velocity, [S] is the substrate concentration, [E]is the enzyme concentration, and Km is the Michaelis constant of the variable substrate.
RESULTS AND DISCUSSION
Aminoglycoside Resistance Profile—Aminoglycoside resistance profiles produced by various aminoglycoside-modifying enzymes have been used as an important criterion for their classification. However, direct comparison of the reported aminoglycoside resistance profiles is complicated by the fact that the genes for these enzymes could have different promoters, might be located on different types of plasmids, or might be of chromosomal origin. These disparate locations for the requisite genes could result in expression of the genes at significantly different levels. To circumvent this problem, we fused the genes for the APH(2″)-Ia, -Ib, and -Id with the promoter of aph(2″)-Ic. Then the genes encoding the four APH(2″) enzymes were cloned in pBluescript II KS(+) vector and transformed into the same bacterial host, E. coli JM83. Levels of resistance to individual aminoglycoside antibiotics conferred by the four phosphotransferases vary significantly, with the APH(2″)-Id being the least competent (Table 1). Meanwhile, the spectrum of antibiotic resistance is similar across the strains that harbor the individual enzymes. All four APH(2″)s produced resistance to most 4,6-disubstituted aminoglycosides (kanamycin, gentamicin, tobramycin, sisomicin, netilmicin, dibekacin, and G418) with the exception of the semisynthetic isepamicin, which has a 4-amino-2-hydroxypropionyl substituent on the nitrogen at position 1 of the aminocyclitol ring (Table 1). Resistance to the semisynthetic amikacin and arbekacin, which have a 4-amino-2-hydroxybutyryl substituent at the same position, was only slightly (2-fold) increased in the strains producing APH(2″)-Ia and APH(2″)-Ib (Table 1). Of the four 4,5-disubstituted aminoglycosides tested (neomycin B, paromomycin, lividomycin A, and butirosin), resistance was increased merely 2-fold for neomycin, only for the strain expressing APH(2″)-Ia (see Table 1 for neomycin). Finally, we also tested the MICs of the so-called atypical aminoglycosides (streptomycin, spectinomycin, fortimicin, apramycin, and hygromycin) against strains producing APH(2″)s. None of these enzymes were able to elevate the MICs to atypical aminoglycoside antibiotics (data not shown).
TABLE 1.
MICs of aminoglycoside antibiotics for E. coli JM83 producing various APH(2″) enzymes
KanA, kanamycin A; KanB, kanamycin B; Gen, gentamicin; Tob, tobramycin; Sis, sisomicin; Ne, netilmicin; Dbk, dibekacin; Amk, amikacin; Arb, arbekacin; Ise, isepamicin; Neo, neomycin B; Par, paromomycin.
|
Enzyme
|
MIC
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KanA | KanB | Gen | Tob | Sis | Net | Dbk | G418 | Amk | Arb | Ise | Neo | Par | |||||||||||||
| μg/ml | |||||||||||||||||||||||||
| Ia | 256 | 32 | 32 | 16 | 16 | 4 | 32 | 512 | 2 | 1 | 1 | 1 | 4 | ||||||||||||
| Ib | 128 | 64 | 16 | 32 | 8 | 8 | 64 | 512 | 2 | 1 | 1 | 0.5 | 4 | ||||||||||||
| Ic | 16 | 64 | 64 | 128 | 32 | 4 | 256 | 128 | 1 | 0.5 | 1 | 0.5 | 4 | ||||||||||||
| Id | 16 | 16 | 32 | 16 | 8 | 2 | 16 | 128 | 1 | 0.5 | 1 | 0.5 | 4 | ||||||||||||
| Controla | 2 | 1 | 0.25 | 0.5 | 0.25 | 0.25 | 0.5 | 2 | 1 | 0.5 | 1 | 0.5 | 4 | ||||||||||||
Parental E. coli JM83 strain harboring plasmid pBluescript II KS(+) vector, which does not contain any genes for APH(2″) enzymes.
Aminoglycoside Substrate Profile—Although phenotypes of aminoglycoside resistance produced by various 2″-phosphotransferases are almost identical, their aminoglycoside substrate profile is more diverse. It has been reported that APH(2″)-Ia phosphorylates a broad array of aminoglycosides, including 4,6-disubstituted kanamycin A, gentamicin, tobramycin, netilmicin, dibekacin, sisomicin, isepamicin, and amikacin and 4,5-disubstituted neomycin B, paromomycin, lividomycin A, ribostamycin, and butirosin (Table 2) (37). In contrast to APH(2″)-Ia, none of the other APH(2″)-I enzymes are able to utilize 4,5-disubstituted aminoglycosides as the substrates (Table 2). APH(2″)-Ib phosphorylates a broad array of 4,6-disubstituted compounds (kanamycin A, gentamicin, tobramycin, netilmicin, dibekacin, sisomicin, isepamicin, amikacin, and arbekacin; Table 2). Among the APH(2″) enzymes, APH(2″)-Ic has the narrowest spectrum of aminoglycoside substrates, which includes 4,6-disubstituted compounds kanamycin A, gentamicin, tobramycin, netilmicin, dibekacin, and sisomicin but not 4,6-disubstituted isepamicin, amikacin, and arbekacin or any of the atypical aminoglycosides tested (Table 2). Finally, APH(2″)-Id has an aminoglycoside substrate profile identical to that of APH(2″)-Ib, which includes a wide range of 4,6-disubstituted compounds but not any of the 4,5-disubstituted or atypical aminoglycosides (Table 2).
TABLE 2.
Aminoglycoside substrate profile of APH(2″) enzymes
Note that three atypical aminoglycosides (streptomycin, spectinomycin, and hygromycin B) are not substrates for APH(2″)-Ib, -Ic, and -Id. Kinetic parameters for these substrates with APH(2″)-Ia enzyme are not reported. S, substrate; NS, not a substrate; NR, not reported. KanA, kanamycin A; Gen, gentamicin; Tob, tobramycin; Net, netilmicin; Dbk, dibekacin; Sis, sisomicin; Ise, isepamicin; Amk, amikacin; Arb, arbekacin; Neo, neomycin B; Par, paromomycin; Liv, lividomycin A; But, butirosin.
|
Enzyme
|
4,6-Disubstituted aminoglycosides
|
4,5-Disubstituted aminoglycosides
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kan | Gen | Tob | Net | Dbk | Sis | Ise | Amk | Arb | Neo | Par | Liv | But | |
| Ia | S | S | S | S | S | S | S | S | NR | S | S | S | S |
| Ib | S | S | S | S | S | S | S | S | S | NS | NS | NS | NS |
| Ic | S | S | S | S | S | S | NS | NS | NS | NS | NS | NS | NS |
| Id | S | S | S | S | S | S | S | S | S | NS | NS | NS | NS |
Nucleotide Triphosphate Profile—We recently performed the detailed kinetic analysis of APH(2″)-Ic and for the first time demonstrated that it utilizes not ATP but rather GTP as the phosphate donor (32). Based on this unexpected finding, we studied five nucleotide triphosphates (NTPs), ATP, GTP, ITP, UTP, and CTP as phosphate donors for the APH(2″)-catalyzed reaction with kanamycin A for the APH(2″)-Ia, -Ib, and -Id. For these experiments, we purified the three additional APH(2″)s to homogeneity. NTPs containing pyrimidine bases in their structures are very poor substrates for APH(2″)-Ia, -Ib, -Ic, and -Id. Catalytic efficiencies (kcat/Km) for phosphorylation of kanamycin A using UTP as the second substrate are between 60 and 900 m–1 s–1. The reaction with CTP could not even be attempted, because it was an exceedingly poor substrate. We estimate that the turnover number (kobs) for the case of CTP was <0.1 s–1 (Table 3). Although it is widely accepted that aminoglycoside phosphotransferases utilize ATP as the phosphate donor in their catalytic reaction, our data demonstrate that it is not broadly the case with APH(2″)s. Only one, APH(2″)-Ib, showed significant preference for ATP (Km = 16 ± 2 μm and kcat/Km = (2.7 ± 0.4) × 106 m–1 s–1) over GTP (Km = 70 ± 2 μm and kcat/Km = (1.3 ± 0.1) × 105 m–1 s–1) and ITP (Km = 660 ± 40 μm and kcat/Km = (2.5 ± 0.2) × 104 m–1 s–1) (Table 3). APH(2″)-Ia, and -Ic demonstrate strong preference for GTP over ATP, as judged by both smaller Km values and larger kcat/Km. For APH(2″)-Ia, kcat/Km of phosphate transfer from GTP is (1.4 ± 0.1) × 105 m–1 s–1, which is more than 800-fold larger than for ATP. This significantly diminished catalytic efficiency with ATP results from both larger Km (870 μm for ATP versus 3.5 μm for GTP) and slightly smaller kcat (3-fold lower for ATP, compared with GTP) values. ITP is also a much better substrate than ATP for APH(2″)-Ia (more than 400-fold difference in kcat/Km). As is the case with GTP, improved catalytic efficiency with ITP versus ATP is the result of a significantly smaller Km value. It would appear that the differences in structures within the purine bases of ATP and GTP are crucial for the efficiency of NTP binding by APH(2″)-Ia (Fig. 1). The active sites would have evolved differentially for selective recognition of these structural differences in the substrates. Very similar Km values and identical turnover numbers for APH(2″)-Ia with GTP and ITP as the substrates argue against significant contribution of an amino group at position C2 for binding of NTP substrate (Fig. 1).
TABLE 3.
NTP substrate profile of APH(2″) enzymes
|
Enzyme
|
ATP
|
GTP
|
ITP
|
UTP
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| μm | s-1 | m-1 s-1 | μm | s-1 | m-1 s-1 | μm | s-1 | m-1 s-1 | μm | s-1 | m-1 s-1 | |
| APH(2″)-Ia | 870 ± 85 | 0.17 ± 0.01 | (1.9 ± 0.2) × 102 | 3.5 ± 0.2 | 0.51 ± 0.02 | (1.4 ± 0.1) × 105 | 5.9 ± 0.3 | 0.49 ± 0.02 | (8.3 ± 0.6) × 104 | 720 ± 20 | 0.47 ± 0.01 | (6.6 ± 0.3) × 102 |
| APH(2″)-Ib | 16 ± 2 | 43 ± 2 | (2.7 ± 0.4) × 106 | 70 ± 2 | 9.4 ± 0.1 | (1.3 ± 0.1) × 105 | 660 ± 40 | 16.5 ± 0.5 | (2.5 ± 0.2) × 104 | 1860 ± 200 | 1.7 ± 0.1 | (9.1 ± 1.1) × 102 |
| APH(2″)-Ic | 1600 ± 200 | 0.1 ± 0.02 | (6.2 ± 1.4) × 101 | 4 ± 0.5 | 0.8 ± 0.1 | (2.0 ± 0.3) × 105 | 39 ± 5 | 2.1 ± 0.2 | (5.4 ± 0.8) × 104 | 1300 ± 200 | 0.14 ± 0.02 | (1.1 ± 0.2) × 102 |
| APH(2″)-Id | 101 ± 8 | 0.83 ± 0.02 | (8.2 ± 0.7) × 103 | 137 ± 5 | 0.83 ± 0.01 | (6.1 ± 0.2) × 103 | 235 ± 5 | 0.9 ± 0.01 | (3.9 ± 0.1) × 103 | 2200 ± 250 | 0.13 ± 0.01 | (6.1 ± 0.7) × 101 |
FIGURE 1.
The chemical structures of nucleoside triphosphates used in kinetics studies.
Even a greater difference is observed for APH(2″)-Ic, whose catalytic efficiency with GTP and kanamycin A as substrates is more than 3,000-fold above that with ATP as a phosphate donor. As in the case with the APH(2″)-Ia, the decrease of kcat and significant increase of Km contribute to the observed drop in catalytic efficiency (Table 3). The comparison of kcat and Km values for APH(2″)-Ic with kanamycin and ATP, GTP, or ITP indicates that the nature of the functional groups at both C6 and C2 positions of the purine are important for NTP binding by this enzyme (see Table 3 and Fig. 1). Finally, APH(2″)-Id demonstrates similar catalytic efficiencies with three purine-containing NTPs, ATP (kcat/Km = (8.2 ± 0.7) × 103 m–1s–1), GTP (kcat/Km = (6.1 ± 0.2) × 103 m–1 s–1), or ITP (kcat/Km = (3.9 ± 0.1) × 103 m–1 s–1) as phosphate donors (Table 3). Overall, among APH(2″) enzymes, APH(2″)-Id exhibits the lowest catalytic efficiency for the preferred NTP substrate that correlates well with the lowest MICs values produced by this enzyme.
The recently determined estimates of the concentrations of various NTPs in E. coli in midlog phase of culture growth are 3,560 μm for ATP, 1,160 μm for GTP, 667 μm for UTP, and 325 μm for CTP (38). Given that the Km values for GTP for APH(2″)-Ia and APH(2″)-Ic are 3.5 and 4.0 μm, respectively, whereas the corresponding values for ATP are 870 and 1,600 μm, it is reasonable to assume that GTP and not ATP is the preferred second substrate for these enzymes in vivo.
New Nomenclature for APH(2″)s—According to the existing nomenclature for aminoglycoside-modifying enzymes (39), the abbreviations APH (aminoglycoside phosphotransferase), AAC (aminoglycoside acetyltransferase), and ANT (aminoglycoside nucleotidyl- or adenylyltransferase) in the enzyme names define the type of enzymatic modification and signify the enzyme family. The site of antibiotic modification is used to further designate members of the three families into classes. This is indicated in parenthesis after the family name; for example, (3), (3′), and (2″), indicate modifications of functionalities at positions 3, 3′, and 2″ of the antibiotic, respectively. Roman numerals I, II, III, etc., are used for subdividing the enzymes into unique antibiotic resistance profiles. Finally, individual enzymes of the same type that differ by amino acid sequence are distinguished by lowercase letters a, b, c, etc. According to this nomenclature, APH(2″)-Ia, -Ib, Ic, and -Id are four unique proteins conferring an identical spectrum of antibiotic resistance. Aminoglycoside phosphotransferase of the bifunctional aminoglycoside acetyltransferase(6′)-Ie-APH(2″)-Ia enzyme from S. aureus was the first characterized enzyme of the APH(2″) class. Regiospecificity of phosphate transfer to 4,6-disubstituted aminoglycosides by this enzyme was experimentally confirmed (25). This enzyme is able to phosphorylate the hydroxyl groups at different positions other than 2″ in various aminoglycoside antibiotics (37). Although 4,6-disubstituted aminoglycosides were phosphorylated exclusively at 2″-OH, the 4,5-disubstituted aminoglycoside lividomycin was modified at the 5″-OH, whereas another 4,5-disubstituted aminoglycoside neomycin was phosphorylated at two positions, the 3′-OH (primarily) and the 3‴-OH. Subsequently, the three additional enzymes APH(2″)-Ib, -Ic, and -Id were provisionally assigned to the class of (2″)-I phosphotransferases based on the sequence identity (from 22 to 33%) with the APH(2″)-Ia enzyme (Table 4) (27–29). Although amino acid sequence identity among the enzymes assigned to the APH(2″) class of phosphotransferases is relatively low (e.g. enzymes of the APH(3′) family exhibit up to 44% sequence identity at the maximum), they are even less homologous to other aminoglycoside phosphotransferases (amino acid sequence identity between APH(2″) and APH(3′), for example, is between 14 and 20%).
TABLE 4.
Amino acid sequence identity between APH(2″) phosphotransferases
|
Enzyme
|
Percentage identity
|
|||
|---|---|---|---|---|
| APH(2″)-Ia | APH(2″)-Ib | APH(2″)-Ic | APH(2″)-Id | |
| % | % | % | % | |
| APH(2″)-Ia | 100 | |||
| APH(2″)-Ib | 33 | 100 | ||
| APH(2″)-Ic | 22 | 24 | 100 | |
| APH(2″)-Id | 26 | 30 | 27 | 100 |
As an important step in confirming their assignment as members of the APH(2″) class of phosphotransferases, we were able to confirm that APH(2″)-Ib and -Ic transfer phosphate to the 2″-hydroxyl of kanamycin A (32, 33). The nature of the 2″-phosphorylation of aminoglycosides by APH(2″)-Ia was documented earlier (25). Furthermore, we have performed the same for APH(2″)-Id and compared the phosphorylated kanamycin A with an authentic sample. Indeed, APH(2″)-Id also phosphorylates the 2″-hydroxyl. Hence, all of these are bona fide APH(2″) enzymes. We note that when we cloned the genes for these APH(2″)s under the same promoter in the pBluescript II KS(+) vector, the antibiotic resistance profiles produced by the individual enzymes were similar but not identical (Table 1). Therefore, one cannot assign the same Roman numeral to all four enzymes. Even greater differences were observed when kinetic analyses were performed. Kinetics studies reported for APH(2″)-Ia (37) and our studies of APH(2″)-Ib, -Ic, and -Id phosphotransferases demonstrate that they have unique aminoglycoside substrate profiles (Table 2). The APH(2″)-Ia phosphotransferase domain of the bifunctional enzyme has a very broad spectrum of antibiotic substrates, including both 4,5- and 4,6-disubstituted aminoglycosides, whereas 4,5-disubstituted aminoglycosides are not substrates for APH(2″)-Ib, -Ic, and -Id enzymes. Moreover, the APH(2″)-Ib and -Id enzymes are able to utilize as substrates all 4,6-disubstituted aminoglycosides tested, but some of these antibiotics (amikacin and isepamicin) are not substrates for APH(2″)-Ic.
The data presented in this report demonstrate yet another important distinction among enzymes of the APH(2″) family, namely their preference for the second substrate, the nucleotide triphosphate (Table 3). Remarkably, and contrary to the established dogma, only one of the four APH(2″) enzymes, APH(2″)-Ib, has a clear preference for ATP. APH(2″)-Ia and APH(2″)-Ic strongly prefer GTP as the second substrate, whereas APH(2″)-Id demonstrates similar catalytic efficiencies with either of these NTPs. Taken together, these data indicate that classification of aminoglycoside-modifying enzymes based solely on their antibiotic resistance profile would not be adequate, and a more detailed characterization of enzymes that includes both the antibiotic and NTP substrate profiling is required. Our kinetic studies of APH(2″) phosphotransferases clearly demonstrated that each of these enzymes has its unique substrate profile. We thus propose a new nomenclature for the class of APH(2″)s. According to this new nomenclature, the Roman numeral would indicate the unique substrate profile, which would include both the aminoglycoside and the NTP substrates rather than an antibiotic resistance profile of bacterial isolates. Based on this nomenclature, the APH(2″) enzymes formerly known as APH(2″)-Ib, -Ic, and -Id are now named APH(2″)-IIa, -IIIa, and IVa, respectively (Table 5), thus accommodating our finding that each enzyme of the APH(2″) family has a unique substrate profile.
TABLE 5.
New nomenclature for APH(2″) enzymes
Kan, kanamycin A; Gen, gentamicin; Tob, tobramycin; Net, netilmicin; Dbk, dibekacin; Sis, sisomicin; Ise, isepamicin; Amk, amikacin; Neo, neomycin B; Par, paromomycin; Liv, lividomycin A; Rib, ribostamycin; But, butirosin; Arb, arbekacin.
|
Substrate profile
|
NTP substrate
|
Old nomenclature
|
New nomenclature
|
||
|---|---|---|---|---|---|
| Enzyme name | Gene name | Enzyme name | Gene name | ||
| Kan, Gen, Tob, Net, Dbk, Sis, Ise, Amk, Neo, Par, Liv, Rib, But | GTP | APH(2″)-Ia | aph(2″)-Ia | APH(2″)-Ia | aph(2″)-Ia |
| Kan, Gen, Tob, Net, Dbk, Sis, Ise, Amk, Arb, | ATP | APH(2″)-Ib | aph(2″)-Ib | APH(2″)-IIa | aph(2″)-IIa |
| Kan, Gen, Tob, Net, Dbk, Sis | GTP | APH(2″)-Ic | aph(2″)-Ic | APH(2″)-IIIa | aph(2″)-IIIa |
| Kan, Gen, Tob, Net, Dbk, Sis, Ise, Amk, Arb, | ATP/GTP | APH(2″)-Id | aph(2″)-Id | APH(2″)-IVa | aph(2″)-IVa |
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI 057393. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: APH, aminoglycoside phosphotransferase; MIC, minimal inhibitory concentration; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Schatz, A., and Waksman, S. A. (1944) Proc. Soc. Exp. Biol. Med. 57 244–248 [Google Scholar]
- 2.Waksman, S. A., and Fenner, F. (1949) Ann. N. Y. Acad. Sci. 52 750–787 [DOI] [PubMed] [Google Scholar]
- 3.Waksman, S. A., and Lechevalier, H. A. (1949) Science 109 305–307 [DOI] [PubMed] [Google Scholar]
- 4.Akova, M., Gur, D., Livermore, D. M., Kocagoz, T., and Akalin, H. E. (1999) Antimicrob. Agents Chemother. 43 1298–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, C. N., Hollis, D. G., and Thornsberry, C. (1985) J. Clin. Microbiol. 22 212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie, S. H. (2002) Antimicrob. Agents Chemother. 46 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinzl, B., Eber, E., Oberwaldner, B., Haas, G., and Zach, M. S. (2002) Pediatr. Pulmonol. 33 32–37 [DOI] [PubMed] [Google Scholar]
- 8.Craig, W. A., and Ebert, S. C. (1990) Scand. J. Infect. Dis. 74 (suppl.) 63–70 [PubMed] [Google Scholar]
- 9.Drusano, G. L. (1988) Antimicrob. Agents Chemother. 32 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert, S. C., and Craig, W. A. (1990) Infect. Control Hospital Epidemiol. 11 319–326 [DOI] [PubMed] [Google Scholar]
- 11.Moore, R. D., Lietman, P. S., and Smith, C. R. (1987) J. Infect. Dis. 155 93–99 [DOI] [PubMed] [Google Scholar]
- 12.Craig, W. A., and Gudmundsson, S. (1996) in Antibiotics in Laboratory Medicine, 4th Ed. (Lorian, V., ed) pp. 296–329, Williams and Wilkins, Baltimore
- 13.Craig, W. A., and Vogelman, B. (1987) Ann. Intern. Med. 106 900–902 [DOI] [PubMed] [Google Scholar]
- 14.McDonald, P. J., Craig, W. A., and Kunin, C. M. (1977) J. Infect. Dis. 135 217–223 [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, D. N. (2005) in Principles and Practice of Infectious Diseases (Mandell, G. E., Bennett, J. E., and Dolin, R., eds) pp. 328–356, Churchill Livingstone, New York
- 16.Nicolau, D. P., and Quintiliani, R., Jr. (1999) in Antimicrobial Therapy and Vaccines (Yu, V. L., Merigan, T. C., Jr., and Barriere, S. L., eds) pp. 621–638, Williams and Wilkins, Baltimore
- 17.Shaw, K. J., Munayyer, H., Rather, P. N., Hare, R. S., and Miller, G. H. (1993) Antimicrob. Agents Chemother. 37 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakulenko, S. B., and Mobashery, S. (2003) Clin. Microbiol. Rev. 16 430–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright, G. D., Berghuis, A. M., and Mobashery, S. (1998) Adv. Exp. Med. Biol. 456 27–69 [PubMed] [Google Scholar]
- 20.Moazed, D., and Noller, H. F. (1987) Nature 327 389–394 [DOI] [PubMed] [Google Scholar]
- 21.Woodcock, J., Moazed, D., Cannon, M., Davies, J., and Noller, H. F. (1991) EMBO J. 10 3099–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llano-Sotelo, B., Azucena, E. F., Jr., Kotra, L. P., Mobashery, S., and Chow, C. S. (2002) Chem. Biol. 9 455–463 [DOI] [PubMed] [Google Scholar]
- 23.Benveniste, R., and Davies, J. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 2276–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster, T. J. (1983) Microbiol. Rev. 47 361–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azucena, E., Grapsas, I., and Mobashery, S. (1997) J. Am. Chem. Soc. 119 2317–2318 [Google Scholar]
- 26.Ferretti, J. J., Gilmore, K. S., and Courvalin, P. (1986) J. Bacteriol. 167 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow, J. W., Zervos, M. J., Lerner, S. A., Thal, L. A., Donabedian, S. M., Jaworski, D. D., Tsai, S., Shaw, K. J., and Clewell, D. B. (1997) Antimicrob. Agents Chemother. 41 511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, S. J., You, I., Clewell, D. B., Donabedian, S. M., Zervos, M. J., Petrin, J., Shaw, K. J., and Chow, J. W. (2000) Antimicrob. Agents Chemother. 44 2876–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, S. F., Zervos, M. J., Clewell, D. B., Donabedian, S. M., Sahm, D. F., and Chow, J. W. (1998) Antimicrob. Agents Chemother. 42 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow, J. W., Kak, V., You, I., Kao, S. J., Petrin, J., Clewell, D. B., Lerner, S. A., Miller, G. H., and Shaw, K. J. (2001) Antimicrob. Agents Chemother. 45 2691–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute (2006) Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard, 7th Ed., Clinical and Laboratory Standards Institute, Wayne, PA
- 32.Badarau, A., Shi, Q., Chow, J. W., Zajicek, J., Mobashery, S., and Vakulenko, S. (2008) J. Biol. Chem. 283 7638–7647 [DOI] [PubMed] [Google Scholar]
- 33.Toth, M., Zajicek, J., Kim, C., Chow, J. W., Smith, C., Mobashery, S., and Vakulenko, S. (2007) Biochemistry 46 5570–5578 [DOI] [PubMed] [Google Scholar]
- 34.Van Pelt, J. E., and Northrop, D. B. (1984) Arch. Biochem. Biophys. 230 250–263 [DOI] [PubMed] [Google Scholar]
- 35.Kim, C., Haddad, J., Vakulenko, S. B., Meroueh, S. O., Wu, Y., Yan, H., and Mobashery, S. (2004) Biochemistry 43 2373–2383 [DOI] [PubMed] [Google Scholar]
- 36.McKay, G. A., and Wright, G. D. (1995) J. Biol. Chem. 270 24686–24692 [DOI] [PubMed] [Google Scholar]
- 37.Daigle, D. M., Hughes, D. W., and Wright, G. D. (1999) Chem. Biol. 6 99–110 [DOI] [PubMed] [Google Scholar]
- 38.Buckstein, M. H., He, J., and Rubin, H. (2008) J. Bacteriol. 190 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993) Microbiol. Rev. 57 138–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



