Abstract
Catalase-peroxidase (KatG) is essential in Mycobacterium tuberculosis for oxidative stress management and activation of the antitubercular pro-drug isoniazid. The role of a unique distal side adduct found in KatG enzymes, involving linked side chains of residues Met255, Tyr229, and Trp107 (MYW), in the unusual catalase activity of KatG is addressed here and in our companion paper (Suarez, J., Ranguelova, K., Jarzecki, A. A., Manzerova, J., Krymov, V., Zhao, X., Yu, S., Metlitsky, L., Gerfen, G. J., and Magliozzo, R. S. (2009) J. Biol. Chem. 284, in press). The KatG[W107F] mutant exhibited severely reduced catalase activity yet normal peroxidase activity, and as isolated contains more abundant 6-coordinate heme in high spin and low spin forms compared with the wild-type enzyme. Most interestingly, oxyferrous heme is also found in the purified enzyme. Oxyferrous KatG[W107F] was prepared by photolysis in air of the carbonyl enzyme or was generated using hydrogen peroxide decayed with a t½ of 2 days compared with 6 min for wild-type protein. The stability of oxyenyzme was modestly enhanced in KatG[Y229F] but was not affected in KatG[M255A]. Optical stopped-flow experiments showed rapid formation of Compound I in KatG[W107F] and facile formation of oxyferrous heme in the presence of micromolar hydrogen peroxide. An analysis of the relationships between catalase activity, stability of oxyferrous enzyme, and a proposed MYW adduct radical is presented. The loss of catalase function is assigned to the loss of the MYW adduct radical and structural changes that lead to greatly enhanced stability of oxyenzyme, an intermediate of the catalase cycle of native enzyme.
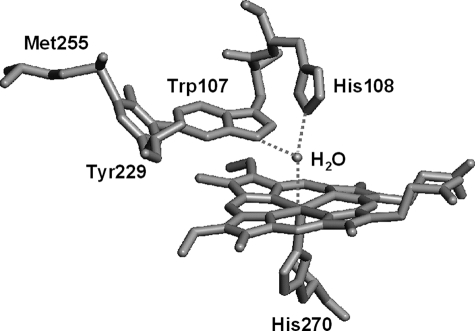
Catalase-peroxidase (KatG)4 is a dual function heme enzyme responsible in Mycobacterium tuberculosis for activation of the antitubercular pro-drug INH (1–4), and it is the sole catalase in this pathogen. KatG enzymes are homologous to class I peroxidases such as cytochrome c peroxidase and ascorbate peroxidase (5), although certain features clearly distinguish the KatG enzymes. The most unusual of these is a three-amino acid distal side adduct (MYW) in which the side chains of Met255, Tyr229, and Trp107 are covalently linked. This adduct (Fig. 1) is found in all KatG enzymes (6–8) and is a key structural element required for catalase activity. The details of post-translational modification and the exact role of the MYW adduct in the catalase reaction mechanism remain under active investigation. Our companion paper (52) provides kinetic and spectroscopic evidence that the adduct harbors a catalytically competent radical during catalase turnover.
FIGURE 1.
Structure of the distal side of M. tuberculosis KatG showing the amino acid adduct MYW. This figure was constructed from Protein Data Bank entry 2CCA using PyMOL graphics software (DeLano Scientific, Inc.). Covalent bonds occur between C-η2 of Trp107 and C-ε1 of Tyr229, and between C-ε2 of Tyr229 and S-δ of Met255 (36, 51).
Disruption of the adduct by mutation of either Trp107 or Tyr229, or changing its structure by mutation of Met255, nearly eliminates catalase activity in M. tuberculosis KatG (9, 10) and in each mutant abolishes the unique radical described in the companion paper (52). These mutations are not damaging to the peroxidase mechanism with artificial substrates in reactions where turnover is initiated with alkyl peroxides (9–14). The origin of the changes in catalatic function in Synechocystis KatG and Burkholderia pseudomallei KatG distal Trp mutants had been assigned to faulty binding of the second molecule of hydrogen peroxide that should participate in reduction of Compound (Cmpd) I in a classical catalase cycle (12, 15), because the formation of Cmpd I was undamaged and in fact was very rapid in those cases. More recently, residue Trp111 together with His112 were described as key sites in Synechocystis KatG for the steering of Cmpd I reduction by hydrogen peroxide (16). Mutations in other residues surrounding the heme pocket have also been shown to interfere with catalase activity for reasons ascribed to disruption of hydrogen bonding networks (17, 18). Other issues such as stabilization of the oxyferrous heme intermediate formed in the presence of excess H2O2 was not presented in studies of KatG from other laboratories, but in the case of M. tuberculosis KatG[Y229F] (14) and in KatG[W107F] as shown here, such stabilization is clearly a fundamental alteration in function that sets apart the behavior of these mutants. No prior work has defined a specific mechanistic role for the MYW adduct structure that could not also be fulfilled by the individual residues whether they were covalently linked or not. DFT calculations along with extensive analysis of EPR spectra allowed a reasonable assignment in the companion paper (52) of a catalytically competent radical to this adduct in KatG.
An intermediate exhibiting an optical spectrum typical of peroxidase Cmpd III (oxyperoxidase) is found when WT KatG reacts with a large excess of H2O2 (12–15, 19, 20), yet catalase function proceeds under such conditions. The formation of this species occurs in KatG[Y229F], W107F, and M255A mutants in the presence of even a few molar equivalents of peroxide, but catalase function is severely reduced. The properties of the oxy intermediates in WT KatG and three distal side mutants and a proposed mechanism for the catalase reaction are discussed here.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—INH, PAA, hydrogen peroxide, o-dianisidine, and all the other chemicals were purchased from Sigma and were of the highest purity available. INH was recrystallized from methanol and stored at 4 °C. PAA (32%) was diluted to 10 mm in potassium phosphate buffer and was incubated with 780 units/ml catalase (Roche Applied Science) for 4 h at 37 °C, followed by removal of the enzyme by ultrafiltration, to remove hydrogen peroxide that interfered with optical stopped-flow experiments. This preparation of PAA was stored at -80 °C in small aliquots.
Construction, Expression, and Purification of Distal Mutants— Overexpression of recombinant WT KatG and KatG mutant enzymes and their purification were achieved as described in the companion paper (52) and previous reports (14, 21).
Optical Measurements—All standard spectrophotometric measurements were performed using an NT14 UV-visible spectrophotometer (Aviv Biomedical, Lakewood, NJ). Protein concentration, expressed as heme concentration, was determined using a heme extinction coefficient at 407 nm equal to 100 mm-1 cm-1 (22). Catalase and peroxidase activities were determined spectrophotometrically in replicate analyses using initial rates of reactions with 25 mm hydrogen peroxide (catalase activity) or t-butyl hydroperoxide plus o-dianisidine (peroxidase activity) according to published methods (22, 23).
A rapid scanning diode array stopped-flow spectrophotometer (HiTech Scientific model SF-61DX2) was used for kinetics experiments. Data acquisition and analyses were performed using the Kinet-Asyst software package (HiTech Scientific). All reactions were followed at 25 °C using freshly prepared solutions of PAA or H2O2, all in 20 mm potassium phosphate buffer, pH 7.2.
Preparation of Oxyferrous KatG[W107F]—Oxyferrous KatG[W107F] was generated through photolysis of the carbonyl enzyme dissolved in oxygen-containing buffer as described by Miller et al. (24). Briefly, enzyme solutions were placed in a sealed cuvette and purged with argon for 30 min followed by carbon monoxide for 10 min. Addition of a small excess of sodium dithionite resulted in conversion to the enzyme-CO complex according to the optical features of the product (the wavelengths of the Soret, β, and α bands were 422, 540, 570 nm, respectively). The enzyme-CO complex was then separated in air from the reaction mixture using a small ion exchange column. After extensive rinsing of the bound enzyme-CO complex, it was eluted with potassium phosphate buffer containing 0.3 m NaCl. The chromatographic purification procedure was performed in a cold room with reduced light and completed within 10 min to minimize dissociation of CO by photolysis. The enzyme-CO complex was then subjected to a 10–15-s illumination by a 75-watt xenon lamp, and the resulting protein was stored at 4 °C. For comparison, similar experiments were performed using WT KatG. In some cases, the oxyenzyme complexes were also prepared by treating the ferric enzymes with excess H2O2 followed by rapid removal of excess peroxide through ion exchange chromatography of the enzyme.
EPR/Resonance Raman Spectroscopy—X-band EPR spectra were recorded using a Bruker E500 EPR spectrometer with data acquisition and manipulation performed using XeprView and WinEPR software (Bruker). Low temperature spectra were recorded using an Oxford Spectrostat continuous flow cryostat and ITC503 temperature controller. The spectra of KatG[W107F] (100 μm) were recorded at 10 K in 20 mm potassium phosphate buffer, pH 7.2 (at room temperature). The EPR spectrum of commercial ferric horse heart Mb in 50 mm glycine/NaOH buffer at pH 10 was used as a standard to estimate the low spin concentration in samples of KatG[W107F]. Final heme concentration in the standard was based on the Soret peak at 418 nm, assuming complete conversion to the low spin alkaline form of myoglobin (ε = 95 mm-1·cm-1) (25, 26).
Resonance Raman spectra were obtained using instrumentation described previously (27). Enzyme samples (40 μm) were maintained at 6 ± 2 °C and were excited at 406.7 nm with a Kr+ laser (Coherent, I-302). The laser power at the sample was 10 milliwatts. Toluene was used to calibrate the spectra, and the spectra were corrected for a sloping base line with a polynomial function.
RESULTS AND DISCUSSION
The goal of this work was to provide characterization of resting KatG[W107F] and new mechanistic insights into the loss of catalase activity in this and two other distal side mutants to shed light on the mechanism in the WT enzyme. Spectroscopic examination of the ferric enzyme helps explain the major alteration in functional properties of the W107F mutant, which include stabilization of 6-c heme species. Prior reports have described certain properties of this mutant in the M. tuberculosis enzyme (21) and the KatG from other organisms (28–30). Here, changes in mechanism and the properties of heme and enzyme intermediates are described in the context of the unique mechanistic features afforded by the distal side MYW adduct presented in the companion paper (52).
Enzymatic and Spectroscopic Characterization of KatG[W107F]— The yield of KatG[W107F] upon overexpression and purification from Escherichia coli is similar to that of WT KatG. The catalase activity of KatG[W107F] is greatly reduced compared with WT KatG (0.5 ± 0.2 versus 3800 ± 300 units/mg for WT KatG) under assay conditions using millimolar concentrations of H2O2, whereas its peroxidase activity (with t-butyl peroxide and o-dianisidine) is only moderately reduced (0.6 ± 0.2 versus 0.9 ± 0.1 unit/mg for WT KatG).
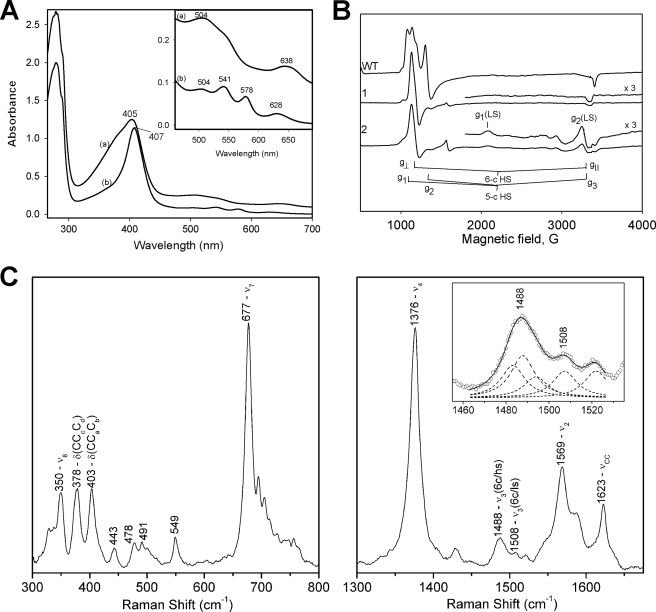
Here, it was considered important to describe the characteristics of the resting (ferric) enzyme to help explain the behavior of the oxyenzyme presented below. The optical spectrum of KatG[W107F] has a Soret peak at 407 and sharp CT2 and CT1 bands around 504 and 628 nm, respectively (Fig. 2A), characteristic of enzyme containing a high level of 6-c heme. In freshly isolated WT KatG, which contains 5-c heme as the majority species, the CT1 band is found close to 640 nm, and in “aged” WT enzyme, which contains more 6-c heme, it occurs at 629 nm (31). Interestingly, the W107F mutant also exhibits optical features at 541 and 578 nm characteristic of the β and α bands of 6-c low spin (6-c LS) ferric heme and/or oxyferrous heme, both of which are confirmed here. These low spin features decay upon storage of the enzyme, consistent with loss of an exchangeable heme ligand.
FIGURE 2.
Optical, EPR, and resonance Raman spectra of M. tuberculosis KatG[W107F]. A, optical spectra of the fresh samples of WT KatG (trace a) and KatG[W107F] (trace b) in 20 mm phosphate buffer, pH 7.2. Inset shows the enlarged spectra in the 450–700 nm range. B, low temperature EPR spectra of KatG[W107F]. Trace 1, fresh mutant enzyme “as isolated”; trace 2, mutant enzyme after storage. EPR spectra were recorded at 10 K; microwave power, 1 milliwatt; microwave frequency, 9.3869 GHz; modulation amplitude, 4 G. The EPR spectrum of WT KatG is shown for comparison. C, low and high frequency resonance Raman spectra of M. tuberculosis KatG[W107F]. The samples were excited at 406.7 nm. The ν3 band at 1488 cm-1 was deconvoluted with Lorentzian line shapes. Two bands at 1483 and 1488 cm-1 are characteristic of 6-c HS heme with a total contribution of 61%, followed by a 22% contribution from a 6-c LS heme at 1507 cm-1, and a 5-c HS heme at 1494 cm-1 with a 17% contribution. Inset, deconvolution of theν3 bands showing the individual bands as dashed lines and the combined fit as the solid line.
EPR and resonance Raman spectra also provided insight into coordination number in the W107F mutant. EPR spectra of freshly purified KatG[W107F] contain a broad axial signal (g⊥ = 5.80 and g ∼2.0) and some low intensity signals because of 6-c low spin ferric heme (g1 = 3.23, g2 = 2.05 and unresolved g3) estimated to be less than 10% of heme content in the fresh enzyme (Fig. 2B, spectrum 1). WT KatG under the same conditions instead exhibits signals assigned to 5-c heme and a rhombic signal also assigned to a 6-c heme species different from the one in the mutant (g1,2,3 = 5.94, 5.49, ∼2) (31). The 6-c complexes other than the low spin species are assigned to enzyme containing water as a sixth ligand associated with heme iron. Certain similar findings were reported earlier for the analogous mutant of Synechocystis and B. pseudomallei KatG (28, 32).
An endogenous strong field ligand is likely responsible for the 6-c LS species because the gmax value (3.21) is larger than that usually observed for O-type (oxygen and nitrogen as the strong field axial ligands) heme (33, 34). The hydroxy-form of horseradish peroxidase, for example, exhibits an EPR signal with gmax = 2.9 (35), and a similar signal is reported for the distal histidine (H123Q) mutant of Synechocystis KatG (32) arguing in favor of the hydroxide form in that histidine mutant and assignment of the 6-c LS species here to an endogenous ligand complex very likely from coordination of the distal imidazole (residue His108).
Resonance Raman spectra of KatG[W107F] contained multiple ν3 bands, including a 6-c heme band at 1488 cm-1 (along with a small contribution from a band at 1483 cm-1 also assigned to 6-c heme), and a band at 1507 cm-1 typical of 6-c LS heme (Fig. 2C), consistent with the optical and EPR spectra. The bands usually associated with out-of-plane modes in the low frequency region of the resonance Raman spectra have lower intensities in the mutant compared with WT M. tuberculosis KatG, suggesting a more planar geometry of heme. This observation is consistent with the axial EPR signal arising from the majority species at low temperature. An oxyenzyme component was not detectable most likely because of photolysis of the ligand.
In KatG[W107F], and in KatG[Y229F] as previously reported (14), elimination of the MYW adduct apparently alters the hydrogen bonding arrangements on the distal side to favor and stabilize 6-c heme. Conversely, water coordination in WT KatG could be considered to be disfavored by the presence of the MYW adduct. The heme iron-associated water molecule in the crystal structure of WT M. tuberculosis KatG from this laboratory (36) is hydrogen bonded to the indole nitrogen of Trp107 (and is also hydrogen bonded to the distal histidyl imidazole but at a longer distance). The hydrogen bond to the indole nitrogen could disfavor optimal interaction between the water ligand and iron; in fact the water iron bond length is 2.8 Å in this structure (36). The 6-c form(s) of M. tuberculosis KatG enzyme in solution displays unique spectroscopic and functional properties (37, 38). Because both the 107 and the 229 mutants favor 6-c heme species, the distal sides must have something in common. The phenolic oxygen of Tyr229 in the MYW adduct of WT KatG faces away from heme iron (36), whereas the indole nitrogen of Trp107 faces toward the sixth ligand position, yet mutation of either residue has a similar effect on coordination number that could arise due to removal of the participation of the indole group. The position of the indole is likely to be altered in the Y229F mutant because of the missing covalent bond to Trp107. Thus, disruption of the indole-water interactions in these two distal side mutants can favor water coordination to iron. There may also be a change in the position of His108 because the adjacent Trp side chain is not incorporated into the MYW adduct in the Y229F mutant, and the link between Tyr229 and Trp107 is absent in the W107F mutant, such that this region of the distal pocket can be more flexible in both cases. The histidyl imidazole of residue 108 may then become the more important hydrogen-bonding site for stabilizing sixth ligands to iron.
The W107F mutant also exhibits more 6-c LS heme because of coordination of an endogenous ligand likely to be the distal imidazole, which constitutes specific evidence for enhanced flexibility of the distal side in the heme pocket. Most importantly, the observation of enhanced stability of oxyferrous heme in these distal side mutants is consistent with the idea that the histidyl imidazole can better participate in hydrogen bonding to stabilize ligands when the indole of Trp107 is missing.
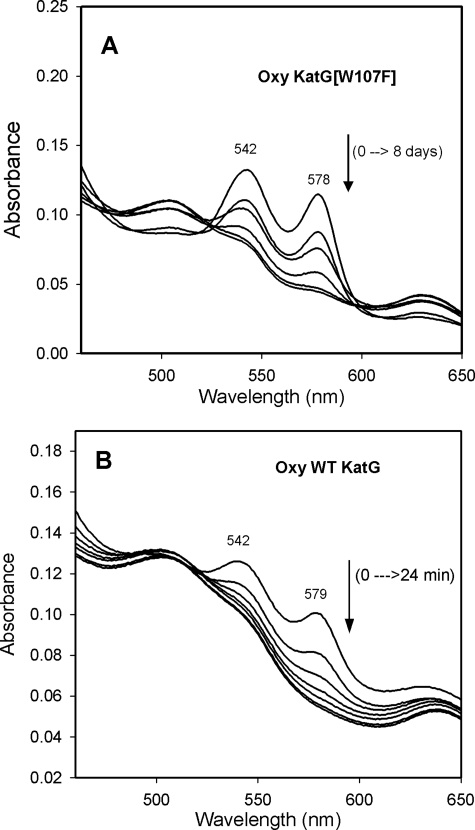
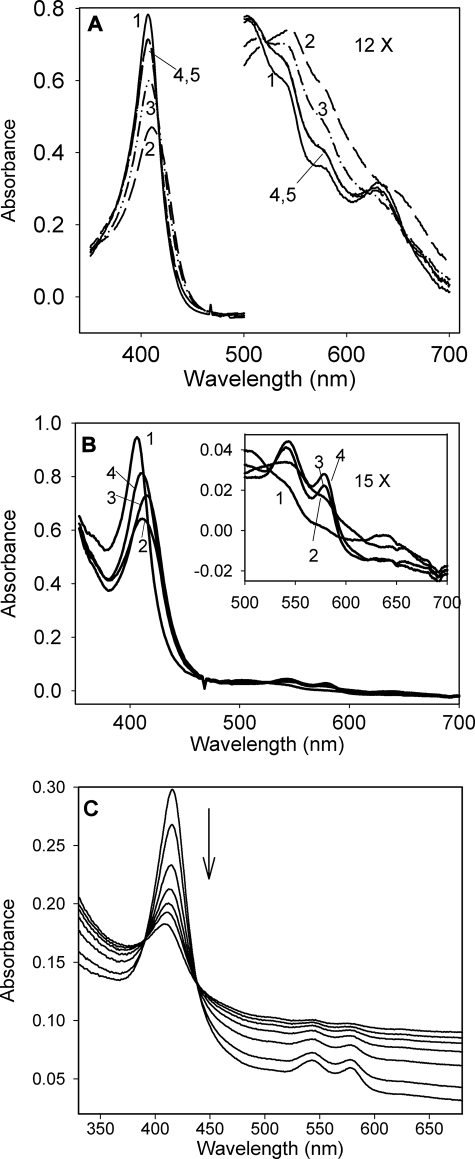
Oxyferrous KatG[W107F]—The presence of some oxyferrous enzyme in samples of purified KatG[W107F] was suspected because of the broad shape of the features around 540 and 580 nm in the optical spectrum of the enzyme as isolated. However, oxyferrous heme has not been reported for purified KatG or other “native” peroxidases, either because there is no route to its formation in situ or because this intermediate may be formed but is unstable. Thus, the presence of oxyferrous heme in preparations of KatG[W107F] would indicate first that it can be formed during cell growth and that it is stable enough to persist during purification and storage. To help confirm these ideas, authentic oxyferrous KatG[W107F] was prepared by photolysis in aerated buffer of the carbonyl complex formed from reduced (ferrous) enzyme and CO. The prepared oxy-KatG[W107F] shows characteristic absorption bands at 413, 542, and 578 nm. Like oxy-horseradish peroxidase (39), the oxyenzyme undergoes auto-oxidation to ferric enzyme with an isosbestic point consistent with direct conversion to the ferric enzyme (Fig. 3A). Oxyferrous KatG[W107F] decays very slowly, and the reaction follows single exponential kinetics. The t½ is estimated to be at least 2 days at 4 °C, as compared with 6 min for the similarly prepared oxy-WT KatG (Fig. 3B).
FIGURE 3.
Decay to ferric protein of oxyferrous M. tuberculosis KatG and KatG[W107F]. Authentic oxyferrous KatG samples were prepared through photolysis of carbon monoxide-bound ferrous protein as described under “Experimental Procedures.” A, absorbance traces of oxy-KatG[W107F] were acquired at 0, 1, 2, 4, 6, and 8 days after photolysis of the enzyme-CO complex in aerobic conditions. B, absorbance traces of oxy-WT KatG were acquired at 0, 4, 8, 12, 16, 20, and 24 min after photolysis of the enzyme-CO complex in aerobic conditions.
In similar experiments, the half-lives of oxyferrous Y229F and M255A mutants were found to be 25 and 7 min, respectively. The oxy-WT KatG and KatG[Y229F] prepared here are more stable than the corresponding enzyme forms from Synechocystis (40). Most interestingly, the stability of oxyferrous M. tuberculosis KatG was hardly affected by the Met255 mutation, was improved by the Tyr299 mutation, and was greatly enhanced by Trp107 mutation. The Met255 mutation still allows formation of a Tyr-Trp cross-link on the distal side (10). These observations lead to the conclusion that the MYW adduct in WT KatG disfavors ligand binding to the sixth coordination position of heme iron in confirmation of the observations on the ferric enzyme above.
The very slow rate of decay of the W107F oxyenzyme is consistent with the formation of this species during cell culture and its persistence during the 3–4-day purification in the cold. Oxyenzyme stability is notably affected by its handling; at room temperature and especially with strong light irradiation, it was found to decay more quickly than in the dark at 4 °C.
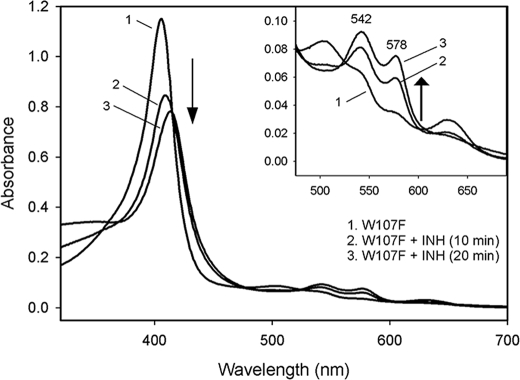
Interaction of KatG[W107] with INH—Binding of INH to WT KatG has been reported to convert 6-c heme to 5-c species (37, 41), and residue Trp107 has been ascribed as a unique hydrogen bonding role in a proposed INH-binding site in KatG (42, 43). These observations suggested that the W107F mutant would exhibit poor or no binding of the drug. No obvious optical changes were detected in anaerobic titrations of KatG[W107F] with INH. In the presence of oxygen, however, ferric KatG[W107F] was converted to the oxyenzyme upon addition of INH (Fig. 4). This process was inhibited by the metal-chelating reagent EDTA and was accelerated by the addition of micromolar Mn2+, suggesting that this conversion was mediated by trace metal ion-initiated redox chemistry (44). Oxyenzyme formation was inhibited by superoxide dismutase, which indicates that it was formed through the ligation of superoxide by ferric enzyme. One source of superoxide here is INH radicals, formed upon drug oxidation either by KatG peroxidase activity or in other oxidations of the drug, reacting with dioxygen. Additionally, hydrogen peroxide formed in situ could also produce oxyenzyme. Given the stability of oxy mutant enzyme, this species would accumulate in the presence of INH because oxy-KatG does not react with the drug (not shown). Oxyenzyme was not detected when WT KatG was treated with INH under similar conditions.
FIGURE 4.
INH induced oxyferrous KatG[W107F] formation. KatG[W107F] (10 μm) was aerobically mixed with INH (2 mm) and MnCl2 (5 μm) in 20 mm phosphate buffer, pH 7.2, and spectra were taken at 10 and 20 min after mixing.
Reaction of KatG[W107F] with Peroxides—The kinetics of the reaction of KatG[W107F] with various peroxides was monitored using stopped-flow optical measurements to gain insights into the origins of the poor catalase activity and other functions of the mutant. Very rapid turnover of KatG[W107F] to Cmpd I was found upon addition of PAA or m-chloroperoxybenzoic acid, whereas in the presence of hydrogen peroxide, formation of oxyenzyme rapidly followed Cmpd I formation. Fig. 5A shows an example of the spectral changes for KatG[W107F] reacted with a 3-fold molar excess of PAA. The decrease in intensity of the Soret peak and concomitant changes in the visible region are consistent with generation of an oxoferryl porphyrin π-cation radical, peroxidase Cmpd I (19). The kobs values for Cmpd I formation, which were linearly dependent on the concentration of peroxides collected as a function of PAA, m-chloroperoxybenzoic acid, or H2O2 concentration, gave second-order rate constants for Cmpd I formation equal to 1.4 × 106 m-1 s-1 (PAA), 1.7 × 106 m-1 s-1 (m-chloroperoxybenzoic acid), or 3.2 × 106 m-1 s-1 (H2O2). The rate using PAA was more than 100-fold more rapid than the rate for WT KatG (1.2 × 104 m-1 s-1) (19). Rapid formation of Cmpd I was also reported for the analogous mutant (W105F) of E. coli KatG (15) and of Synechocystis KatG[W122F] (12), and for Synechocystis KatG[Y249F] (11) and M. tuberculosis KatG[Y229F] (14). Thus, the presence of the distal side adduct may actually inhibit turnover of peroxides by ferric heme in WT KatG or the adduct has some other impact on the mechanism in the pre-steady state. A recent analysis of ligand binding to WT KatG and Ser315 mutants (38) demonstrated that distal side water facilitates ligand binding and the presence of abundant 6-c heme in these mutants may contribute to the enhanced rate of peroxide turnover, although other features of the distal side may also be important. The elimination of the potential to form the distal side MYW adduct radical after Cmpd I is formed (see companion paper (52)) is expected to have a major impact on initial redox processes in the distal side mutants. For example, the increased apparent rate of Cmpd I formation found for KatG[W107F] may actually reflect the loss of electron transfer pathways that rapidly deplete (reduce) this intermediate in WT KatG; repeated turnovers with peroxide may be required before a detectable yield of Cmpd I accumulates, whereas in the mutants, Cmpd I is rapidly produced and depleted only slowly because the MYW adduct is unavailable for an immediate electron transfer step.
FIGURE 5.
Reaction of KatG[W107F] with peroxides. A, spectral changes upon addition of 30 μm PAA to 10 μm KatG[W107F] in 20 mm phosphate buffer, pH 7.2. 1, resting enzyme; 2, t = 1 s; 3, t = 22 s; 4, t = 100 s; 5, t = 200 s. B, spectral changes of 10 μm KatG[W107F] upon addition of 50 μm H2O2 in 20 mm phosphate buffer, pH 7.2, at 25 °C. 1, resting enzyme; 2, t = 0.5 s; 3, t = 5 s; 4, t = 105 s. C, heme breakdown during long term incubation of KatG[W107F] with excess H2O2. M. tuberculosis KatG[W107F] (4 μm) was mixed with 1 mm H2O2 in 20 mm phosphate buffer, pH 7.2, at 25 °C. Spectra were recorded at 1, 6, 12, 24, 36, 48, 60, and 84 min after mixing.
Catalase Pathway and Oxyenzyme Intermediate—The poor catalase activity of KatG[W107F] was further probed using stopped-flow experiments. A species with an optical spectrum typical of peroxidase Cmpd III (Fig. 5B, spectrum 3), with peaks at 415, 542, and 578 nm, rapidly develops using even a small molar excess of H2O2. The optical features here are very similar to those described above for the oxyenzyme prepared from the ferrous form. Examination of the time course at 578 nm allowed evaluation of kobs for formation of oxyenzyme, which was linearly dependent on the concentration of H2O2. A single second-order rate constant equal to 2.0 × 106 m-1 s-1 was estimated from the data. The W107F mutant was susceptible to decomposition in the presence of excess H2O2 because it is unable to decompose hydrogen peroxide efficiently. As shown in Fig. 5C, incubating 4 μm enzyme with 1 mm H2O2 caused a continuous decrease of the Soret peak and gradual loss of features in the visible region. This indicates secondary reactions likely involving hydrogen peroxide reacting with Cmpd I and more complex behavior than the decomposition of the oxyenzyme demonstrated above when it was prepared without H2O2 or when excess peroxide was removed, as follows.
The evidence for Cmpd III formation in these experiments permitted additional insight into the properties of this species formed during catalytic turnover rather than by photolysis of the carbonyl enzyme. The KatG[W107F] oxyenzyme intermediate generated by addition of a 10-fold molar excess of H2O2 was isolated by immediate removal of residual peroxide. This species is also very stable in the absence of peroxide and decays at a rate similar to the oxyferrous enzyme prepared from the aerobic photolysis of carbonyl enzyme. The oxyferrous forms of KatG[Y229F] or KatG[M255A] prepared by brief H2O2 treatment also decay at rates similar to the decay of the oxyferrous enzymes prepared from the carbonyl forms (data not shown).
For the WT enzyme, Cmpd III is formed in the presence of large excesses of H2O2 (19, 52) under catalase turnover conditions. Under such conditions, this intermediate must be unstable as the kcat for catalase turnover is on the order of 5100 s-1. Thus, its half-life must be in the millisecond range, in contrast to the half-life of 6 min for oxyferrous WT KatG prepared from the carbonyl enzyme. In other words, the peroxide-induced Cmpd III species decays to ferric enzyme thousands of times faster than does the oxyferrous protein prepared under non-catalytic conditions in the absence of hydrogen peroxide. The main difference between the two routes to oxyferrous protein is that H2O2 treatment generates Cmpd I during the initial turnover and potentiates formation of a protein-based radical, whereas formation of oxyenzyme from carbonyl enzyme bypasses the hypervalent heme intermediate and does not produce the radical. The absence of the radical was confirmed using EPR spectroscopy (data not shown). These observations support the proposal in the companion paper (52) that the MYW adduct radical or some other structural feature of the enzyme in the state containing oxyferrous heme and this radical accelerates turnover and the return to the ferric state. In the case of the W107F, Y229F, and M255A mutants, because of the lack of intact adduct and/or adduct radical, the H2O2-induced Cmpd III intermediates return to the ferric state at rates similar to the oxyferrous enzymes prepared from the carbonyl forms without peroxide.
Conclusions—KatG[W107F] forms a highly stable oxyferrous heme species, and the amino acid replacement eliminates the unique structure responsible for robust catalase activity that can only occur when the oxyenzyme formed in the presence of H2O2 decays very rapidly.
The finding here that M. tuberculosis KatG[W107F] as isolated contains a fraction of heme in the oxyferrous form is unusual as it has not been observed for WT M. tuberculosis KatG nor has it been reported for the analogous mutants of KatG from other organisms (12, 13, 15, 45). The oxyferrous enzyme could result either from turnover induced by H2O2 or by direct binding of superoxide to ferric heme in situ. Micromolar hydrogen peroxide is sufficient to induce rapid formation of oxyenzyme in purified M. tuberculosis KatG[W107F] in vitro, and this level of H2O2 is easily attained in aerobic cultures (46, 47). H2O2 can accumulate upon dismutation of superoxide, whereas superoxide addition to the ferric enzyme can also directly occur; either pathway produces oxyenzyme that will persist during isolation and purification. We have demonstrated both routes to a stable oxyenzyme in this study.
The enhanced stability of oxyferrous enzyme in the W107F mutant may depend directly on good hydrogen bonding of the dioxygen ligand with His108 (48, 49). Furthermore, the possibility that the indole group of Trp107 may specifically destabilize sixth ligands ensures that oxy-WT KatG is unstable enough to avoid being a dead end intermediate in the catalase reaction pathway.
A recently published article addressing the catalase mechanism in Synechocystis KatG and the distal tryptophan (W122F) mutant of that enzyme demonstrates that dioxygen is formed in a nonscrambling mechanism during turnover of millimolar H2O2 (30). The mechanism of this reaction has not been explained. In our proposed catalatic pathway for WT KatG, which is consistent with previously published ideas (50), the final step involves electron transfer from superoxide to the MYW adduct radical and release of dioxygen from the enzyme. Thus, the conversion of oxyenzyme to ferric protein should be among the rate-limiting steps in the catalase pathway in WT KatG.
The relationships between catalase activity, stability of oxyenzyme alone, and adduct radical formation are summarized in Table 1. Of the three distal side adduct mutants, the decay rates of oxyferrous enzyme are in the order M255A > Y229F > W107F, which is also the order of their catalase activities. As reported in our companion paper (52), none of these mutants forms the radical assigned to the distal adduct in WT KatG under catalase turnover conditions. The inherent decay rate of oxyferrous heme in WT KatG (t½ ≈ 6 min or kdecay = 2.8 × 10-3 s-1) is far slower than the turnover rate of intermediates in the catalase reaction (7). Thus, the MYW radical itself or some unique structural feature of this species, or both, contribute to the rapid decay of the WT oxyenzyme. Note that the KatG[M255A] mutant, whereas its oxy-form decays at the same rate as WT enzyme, does not form a radical in the presence of mm H2O2 and only exhibits 0.3% of the WT catalase activity. DFT calculations in the companion paper (52) show that an MYW radical species should contain a hydrogen on the indole nitrogen, producing partial cationic character in the adduct radical and favoring electron removal from superoxide dissociating from the oxyenzyme. Therefore, both a direct redox role provided by the MYW radical and a structural effect in the enzyme harboring both oxyferrous heme and the MYW-adduct radical likely contribute to the rapid decay of oxyenzyme in WT KatG.
TABLE 1.
Comparison of Mtb KatG and distal side adduct mutants
| Enzyme | Oxyferrous enzyme,astability t½ | H2O2-induced adduct radical | Catalase activity |
|---|---|---|---|
| min | units/mg | ||
| WT KatG | 6 | + | 3800 |
| KatG[M255A] | 7 | – | 12.3 |
| KatG[Y229F] | 25 | – | 2.7 (14) |
| KatG[W107F] | 2800 | – | 0.5 |
Oxyferrous enzymes were prepared through photolysis of carbonyl enzymes in air
The unique physiological roles of WT KatG, which is the sole catalase expressed in M. tuberculosis, and its sustained role in antibiotic (INH) activation is only possible because of the unique architecture governed by the distal Trp107 and the MYW adduct in this enzyme.
Acknowledgments
We thank Jun He for purification of KatG[W107F].
This work was supported, in whole or in part, by National Institutes of Health Grants AI-43582 and AI-060014 (NIAID) (to R. S. M.). This work was also supported by National Science Foundation grant (to J. P. M. S.) and the Heiser Program for Research in Leprosy and Tuberculosis (to S. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: KatG, catalase-peroxidase; KatG[W107F], W107F mutant of KatG; KatG[Y229F], Y229F mutant of KatG; KatG[M255A], M255A mutant of KatG; INH, isoniazid (isonicotinic acid hydrazide); Cmpd, compound; PAA, peroxyacetic acid; 5-c, five coordinate; 6-c, six coordinate; LS, low spin; MYW, Met-Tyr-Trp; WT, wild type.
References
- 1.Zhang, Y. (1993) Res. Microbiol. 144 143-149 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, Y., Heym, B., Allen, B., Young, D., and Cole, S. (1992) Nature 358 591-593 [DOI] [PubMed] [Google Scholar]
- 3.Rozwarski, D. A., Grant, G. A., Barton, D. H. R., Jacobs, W. R., Jr., and Sacchettini, J. C. (1998) Science 279 98-102 [DOI] [PubMed] [Google Scholar]
- 4.Johnsson, K., and Schultz, P. G. (1994) J. Am. Chem. Soc. 116 7425-7426 [Google Scholar]
- 5.Dunford, H. B. (1999) Heme Peroxidases, pp. 276-278, Wiley-VCH, New York
- 6.Yamada, Y., Fujiwara, T., Sato, T., Igarashi, N., and Tanaka, N. (2002) Nat. Struct. Biol. 9 691-695 [DOI] [PubMed] [Google Scholar]
- 7.Bertrand, T., Eady, N. A. J., Jones, J. N., Jesmin, Nagy, J. M., Jamart-Gregoire, B., Raven, E. L., and Brown, K. A. (2004) J. Biol. Chem. 279 38991-38999 [DOI] [PubMed] [Google Scholar]
- 8.Carpena, X., Loprasert, S., Mongkolsuk, S., Switala, J., Loewen, P. C., and Fita, I. (2003) J. Mol. Biol. 327 475-489 [DOI] [PubMed] [Google Scholar]
- 9.Ghiladi, R. A., Knudsen, G. M., Medzihradszky, K. F., and Ortiz de Montellano, P. R. (2005) J. Biol. Chem. 280 22651-22663 [DOI] [PubMed] [Google Scholar]
- 10.Ghiladi, R. A., Medzihradszky, K. F., and Ortiz de Montellano, P. R. (2005) Biochemistry 44 15093-15105 [DOI] [PubMed] [Google Scholar]
- 11.Jakopitsch, C., Auer, M., Ivancich, A., Ruker, F., Furtmuller, P. G., and Obinger, C. (2003) J. Biol. Chem. 278 20185-20191 [DOI] [PubMed] [Google Scholar]
- 12.Regelsberger, G., Jakopitsch, C., Ruker, F., Krois, D., Peschek, G. A., and Obinger, C. (2000) J. Biol. Chem. 275 22854-22861 [DOI] [PubMed] [Google Scholar]
- 13.Regelsberger, G., Jakopitsch, C., Furtmuller, P. G., Rueker, F., Switala, J., Loewen, P. C., and Obinger, C. (2001) Biochem. Soc. Trans. 29 99-105 [DOI] [PubMed] [Google Scholar]
- 14.Yu, S., Girotto, S., Zhao, X., and Magliozzo, R. S. (2003) J. Biol. Chem. 278 44121-44127 [DOI] [PubMed] [Google Scholar]
- 15.Hillar, A., Peters, B., Pauls, R., Loboda, A., Zhang, H., Mauk, A. G., and Loewen, P. C. (2000) Biochemistry 39 5868-5875 [DOI] [PubMed] [Google Scholar]
- 16.Deemagarn, T., Wiseman, B., Carpena, X., Ivancich, A., Fita, I., and Loewen, P. C. (2007) Proteins 66 219-228 [DOI] [PubMed] [Google Scholar]
- 17.Jakopitsch, C., Droghetti, E., Schmuckenschlager, F., Furtmuller, P. G., Smulevich, G., and Obinger, C. (2005) J. Biol. Chem. 280 42411-42422 [DOI] [PubMed] [Google Scholar]
- 18.Jakopitsch, C., Auer, M., Regelsberger, G., Jantschko, W., Furtmuller, G. P., Ruker, F., and Obinger, C. (2003) Eur. J. Biochem. 270 1006-1013 [DOI] [PubMed] [Google Scholar]
- 19.Chouchane, S., Lippai, I., and Magliozzo, R. S. (2000) Biochemistry 39 9975-9983 [DOI] [PubMed] [Google Scholar]
- 20.Jakopitsch, C., Ivancich, A., Schmuckenschlager, F., Wanasinghe, A., Poltl, G., Furtmuller, P. G., Ruker, F., and Obinger, C. (2004) J. Biol. Chem. 279 46082-46095 [DOI] [PubMed] [Google Scholar]
- 21.Ranguelova, K., Girotto, S., Gerfen, G. J., Yu, S., Suarez, J., Metlitsky, L., and Magliozzo, R. S. (2007) J. Biol. Chem. 282 6255-6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcinkeviciene, J. A., Magliozzo, R. S., and Blanchard, J. S. (1995) J. Biol. Chem. 270 22290-22295 [DOI] [PubMed] [Google Scholar]
- 23.Saint-Joanis, B., Souchon, H., Wilming, M., Johnsson, K., Alzari, P. M., and Cole, S. T. (1999) Biochem. J. 338 753-760 [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, M. A., Bandyopadhyay, D., Mauro, J. M., Traylor, T. G., and Kraut, J. (1992) Biochemistry 31 2789-2797 [DOI] [PubMed] [Google Scholar]
- 25.Antonini, E., and Brunori, M. (1971) in Hemoglobin and Myoglobin in Their Reactions with Ligands (Neuberger, A., and Tatum, E. L., eds) p. 44, North-Holland Publishing Co., Amsterdam
- 26.Ikeda-Saito, M., Hori, H., Andersson, L. A., Prince, R. C., Pickering, I. J., George, G. N., Sanders, C. R., II, Lutz, R. S., McKelvey, E. J., and Mattera, R. (1992) J. Biol. Chem. 267 22843-22852 [PubMed] [Google Scholar]
- 27.Kapetanaki, S., Chouchane, S., Girotto, S., Yu, S., Magliozzo, R. S., and Schelvis, J. P. (2003) Biochemistry 42 3835-3845 [DOI] [PubMed] [Google Scholar]
- 28.Carpena, X., Wiseman, B., Deemagarn, T., Herguedas, B., Ivancich, A., Singh, R., Loewen, P. C., and Fita, I. (2006) Biochemistry 45 5171-5179 [DOI] [PubMed] [Google Scholar]
- 29.Jakopitsch, C., Regelsberger, G., Furtmuller, P. G., Ruker, F., Peschek, G. A., and Obinger, C. (2001) Biochem. Biophys. Res. Commun. 287 682-687 [DOI] [PubMed] [Google Scholar]
- 30.Vlasits, J., Jakopitsch, C., Schwanninger, M., Holubar, P., and Obinger, C. (2007) FEBS Lett. 581 320-324 [DOI] [PubMed] [Google Scholar]
- 31.Chouchane, S., Girotto, S., Kapetanaki, S., Schelvis, J. P., Yu, S., and Magliozzo, R. S. (2003) J. Biol. Chem. 278 8154-8162 [DOI] [PubMed] [Google Scholar]
- 32.Ivancich, A., Jakopitsch, C., Auer, M., Un, S., and Obinger, C. (2003) J. Am. Chem. Soc. 125 14093-14102 [DOI] [PubMed] [Google Scholar]
- 33.Peisach, J., and Blumberg, W. E. (1971) Proceedings of the 1st Inter-American Symposium on Hemoglobins, Caracas, 1969 (Arends, T., Bemski, G., and Nagel, R. L., eds.) p. 199, Karger, Basel
- 34.Svistunenko, D. A., Sharpe, M. A., Nicholls, P., Blenkinsop, C., Davies, N. A., Dunne, J., Wilson, M. T., and Cooper, C. E. (2000) Biochem. J. 351 595-605 [PMC free article] [PubMed] [Google Scholar]
- 35.Blumberg, W. E., Peisach, J., Wittenberg, B. A., and Wittenberg, J. B. (1968) J. Biol. Chem. 243 1854-1862 [PubMed] [Google Scholar]
- 36.Zhao, X., Yu, H., Yu, S., Wang, F., Sacchettini, J. C., and Magliozzo, R. S. (2006) Biochemistry 45 4131-4140 [DOI] [PubMed] [Google Scholar]
- 37.Zhao, X., Yu, S., and Magliozzo, R. S. (2007) Biochemistry 46 3161-3170 [DOI] [PubMed] [Google Scholar]
- 38.Ranguelova, K., Suarez, J., Metlitsky, L., Yu, S., Brejt, S. Z., Zhao, L., Schelvis, J. P., and Magliozzo, R. S. (2008) Biochemistry 47 12583-12592 [DOI] [PubMed] [Google Scholar]
- 39.Wittenberg, B. A., Antonini, E., Brunori, M., Noble, R. W., Wittenberg, J. B., and Wymann, J. (1967) Biochemistry 6 1970-1974 [DOI] [PubMed] [Google Scholar]
- 40.Jakopitsch, C., Wanasinghe, A., Jantschko, W., Furtmuller, P. G., and Obinger, C. (2005) J. Biol. Chem. 280 9037-9042 [DOI] [PubMed] [Google Scholar]
- 41.Wengenack, N. L., Todorovic, S., Yu, L., and Rusnak, F. (1998) Biochemistry 37 15825-15834 [DOI] [PubMed] [Google Scholar]
- 42.Pierattelli, R., Banci, L., Eady, N. A. J., Bodiguel, J., Jones, J. N., Moody, P. C. E., Raven, E. L., Jamart-Gregoire, B., and Brown, K. A. (2004) J. Biol. Chem. 279 39000-39009 [DOI] [PubMed] [Google Scholar]
- 43.Metcalfe, C., Macdonald, I. K., Murphy, E. J., Brown, K. A., Raven, E. L., and Moody, P. C. (2008) J. Biol. Chem. 283 6193-6200 [DOI] [PubMed] [Google Scholar]
- 44.Magliozzo, R. S., and Marcinkeviciene, J. A. (1996) J. Am. Chem. Soc. 118 11303-11304 [Google Scholar]
- 45.Singh, R., Wiseman, B., Deemagarn, T., Donald, L. J., Duckworth, H. W., Carpena, X., Fita, I., and Loewen, P. C. (2004) J. Biol. Chem. 279 43098-44106 [DOI] [PubMed] [Google Scholar]
- 46.Seaver, L. C., and Imlay, J. A. (2001) J. Bacteriol. 183 7173-7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez-Flecha, B., and Demple, B. (1997) J. Bacteriol. 179 382-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller, M. A., Shaw, A., and Kraut, J. (1994) Nat. Struct. Biol. 1 524-531 [DOI] [PubMed] [Google Scholar]
- 49.Berglund, G. I., Carlsson, G. H., Smith, A. T., Szoke, H., Henriksen, A., and Hajdu, J. (2002) Nature 417 463-468 [DOI] [PubMed] [Google Scholar]
- 50.Jakopitsch, C., Vlasits, J., Wiseman, B., Loewen, P. C., and Obinger, C. (2007) Biochemistry 46 1183-1193 [DOI] [PubMed] [Google Scholar]
- 51.Delano, W. L. (2004) The PyMol Molecular Graphics System, DeLano Scientific, San Carlos, CA
- 52.Suarez, J., Ranguelova, K., Jarzecki, A. A., Manzerova, J., Krymov, V., Zhao, X., Yu, S., Metlitsky, L., Gerfen, G. J., and Magliozzo, R. S. (2009) J. Biol. Chem. 284 7017-7029 [DOI] [PMC free article] [PubMed] [Google Scholar]