Abstract
Mitochondrial respiratory enzymes play a central role in energy production in aerobic organisms. They differentiated from the α-proteobacteria-derived ancestors by adding noncatalytic subunits. An exception is Complex II (succinate: ubiquinone reductase), which is composed of four α-proteobacteria-derived catalytic subunits (SDH1-SDH4). Complex II often plays a pivotal role in adaptation of parasites in host organisms and would be a potential target for new drugs. We purified Complex II from the parasitic protist Trypanosoma cruzi and obtained the unexpected result that it consists of six hydrophilic (SDH1, SDH2N, SDH2C, and SDH5-SDH7) and six hydrophobic (SDH3, SDH4, and SDH8-SDH11) nucleus-encoded subunits. Orthologous genes for each subunit were identified in Trypanosoma brucei and Leishmania major. Notably, the iron-sulfur subunit was heterodimeric; SDH2N and SDH2C contain the plant-type ferredoxin domain in the N-terminal half and the bacterial ferredoxin domain in the C-terminal half, respectively. Catalytic subunits (SDH1, SDH2N plus SDH2C, SDH3, and SDH4) contain all key residues for binding of dicarboxylates and quinones, but the enzyme showed the lower affinity for both substrates and inhibitors than mammalian enzymes. In addition, the enzyme binds protoheme IX, but SDH3 lacks a ligand histidine. These unusual features are unique in the Trypanosomatida and make their Complex II a target for new chemotherapeutic agents.
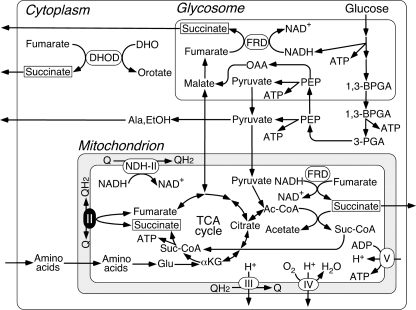
The parasitic protist Trypanosoma cruzi is the etiological agent of Chagas disease, a public health threat in Central and South America. These parasites are normally transmitted by reduviid bugs via the vector feces after a bug bite and also via transfusion of infected blood. About 16-18 million people are infected, and 100 million are at risk, but there are no definitive chemotherapeutic treatments available (1). Despite having potential pathways for oxidative phosphorylation (2), all trypanosomatids (Trypanosoma and Leishmania species) analyzed so far are characterized by incomplete oxidation of glucose with secretion of end products, such as succinate, alanine, ethanol, acetate, pyruvate, and glycerol (3, 4) (Fig. 1). Major routes for formation of succinate in Trypanosoma brucei are via NADH-dependent fumarate reductase in glycosomes and mitochondria (5, 6). In trypanosomatid mitochondria, the Krebs cycle is inefficient, and pyruvate is principally converted to acetate via acetate:succinate CoA transferase (7). A part of the Krebs cycle operates the utilization of histidine in the insect stage of T. cruzi (8).
FIGURE 1.
Metabolic pathways in T. cruzi. Incomplete oxidation of glucose takes place in glycosomes and mitochondria, and end products such as succinate, l-alanine, ethanol, and acetate are excreted from parasites (3, 4). Cytoplasmic dihydroorotate (DHO):fumarate reductase (DHOD) contributes succinate production (6).
Mitochondrial Complex II (succinate:quinone reductase (SQR)5 and succinate dehydrogenase (SDH)) serves as a membrane-bound Krebs cycle enzyme and often plays a pivotal role in adaptation of parasites to environments in their host (9, 10). In general, Complex II consists of four subunits (11). A flavoprotein subunit (SDH1, Fp) and an iron-sulfur subunit (SDH2, Ip) form a soluble heterodimer, which then binds to a membrane anchor heterodimer, SDH3 (CybL) and SDH4 (CybS). SDH1 contains a covalently bound FAD and catalyzes the oxidation of succinate to fumarate. SDH2 transfers electrons to ubiquinone via the [2Fe-2S] cluster in the N-terminal plant-type ferredoxin domain (IpN) and the [4Fe-4S] and [3Fe-4S] clusters in the C-terminal bacterial ferredoxin domain (IpC). Ubiquinone is bound and reduced in a pocket provided by SDH2, SDH3, and SDH4 (12-14). SDH3 and SDH4 contain three transmembrane helices and coordinate protoheme IX via histidine in the second helices of each subunit (11-14).
Parasitic nematodes adapted to hypoxic host environments often have modified respiratory chains. Many adult parasites perform fumarate respiration by expressing a stage-specific isoform of Complex II (9, 10). Hemonchus contortus uses an isoform for SDH2 (9), whereas Ascaris suum uses isoforms for SDH1 and SDH4 (10). To explore the adaptive strategy in a parasitic protist, we isolated mitochondria from axenic culture of T. cruzi epimastigotes and characterized the purified Complex II. Our results demonstrated for the first time that T. cruzi Complex II is an unusual supramolecular complex with a heterodimeric iron-sulfur subunit and seven novel noncatalytic subunits. Purified enzyme showed reduced binding affinities for both substrates and inhibitors. Because this novel structural organization is conserved in all trypanosomatids (2, 15, 16), parasite Complex II would be a potential target for the development of new chemotherapeutic agents for trypanosomiasis and leishmaniasis.
EXPERIMENTAL PROCEDURES
Preparation of Mitochondria—T. cruzi strain Tulahuhen was grown statically for 6-7 days at 26 °C in 300-cm2 cell culture flasks (Falcon, BD Biosciences) containing 250 ml of the modified LIT medium (17), supplemented with 0.1% (w/v) glucose, 0.001% (w/v) hemin (Sigma), and 5% (v/v) fetal bovine serum (MP Biochemicals). Mitochondria were isolated from epimastigotes by the differential centrifugation method (18) with slight modifications. Parasites grown to 6-8 × 107 cells/ml were washed with buffer A (20 mm Tris-HCl, pH 7.2, 10 mm NaH2PO4, 1 mm sodium EDTA, 1 mm dithiothreitol, 0.225 m sucrose, 20 mm KCl, and 5 mm MgCl2). Cells were disrupted by grinding with silicon carbide (Carborundum 440 mesh; Nacalai Tesque, Kyoto, Japan) in the presence of a minimum volume of buffer B (25 mm Tris-HCl, pH 7.6, 1 mm dithiothreitol, 1 mm sodium EDTA, 0.25 m sucrose, and EDTA-free Complete protease inhibitor mixture (Roche Applied Science)). The resultant cell paste was resuspended in buffer B and centrifuged at 500 × g for 5 min and 1000 × g for 15 min to remove silicon carbide and nuclear fraction, respectively. The mitochondrial fraction was recovered upon centrifugation of the last supernatant at 10,000 × g for 15 min, washed three times in buffer B, and resuspended to a protein concentration of ∼30 mg/ml and kept at -80 °C until use.
Isolation of Complex II—All steps were carried out at 4 °C. Mitochondrial fraction (∼300 mg of protein from 10 liters culture) was brought to 70 ml with buffer C (10 mm KPi, pH 7.5), 1 mm sodium EDTA, 1 mm sodium malonate, EDTA-free Complete protease inhibitor mixture (Roche Applied Science) (2 tablets/50 ml), 1% (w/v) sucrose monolaurate SM-1200 (SML) (Mitsubishi-Kagaku Foods Co., Tokyo, Japan)). The mixture was stirred for 30 min and centrifuged at 200,000 × g for 1 h. The supernatant was loaded at 1 ml/min onto a Source 15 Q column (1.6 inner diameter × 10 cm; GE Healthcare), equilibrated with buffer C containing 0.1% SML. After washing with 5 volumes of the same buffer, proteins were eluted with a 200-ml linear gradient of NaCl from 0 to 150 mm at 2 ml/min. Active fractions were concentrated to ∼250 μl by ultrafiltration with Amicon Ultra-4 (molecular weight cutoff 100,000, Millipore) and subjected to gel filtration FPLC with a Superdex 200-pg 10/300 GL column (1 cm inner diameter × 30 cm; GE Healthcare) at 0.25 ml/min in 20 mm MOPS-NaOH, pH 7.2, containing 1 mm sodium EDTA, 1 mm sodium malonate, 150 mm NaCl, and 0.1% SML. Peak fractions were rechromatographed as above, and purified enzyme was concentrated and stored at -80 °C until use.
Identification of Complex II Subunits—The purified enzyme was subjected to 12.5% SDS-PAGE, and subunits were transferred to an Immobilon-P membrane (Millipore), followed by staining with Coomassie Brilliant Blue R-250 (19, 20). Five or ten N-terminal amino acid residues were determined with a Procise 494 HT (Applied Biosystems) or an Hp G1005A (Hewlett-Packard Co.) Protein Sequencing System at the BioMedical Research Center of Juntendo University or APRO Life Science Institute, Inc. (Tokushima, Japan). When the N terminus was blocked, protein bands were digested with trypsin, and internal peptide sequences were determined (20). Genes coded for Complex II subunits were identified with BLASTP in the T. cruzi genome data base (15).
Phase Partitioning of Mitochondrial Fraction with Triton X-114—Phase partitioning by Triton X-114 was performed as described previously (21) with a slight modification. A total of 2-3 mg of mitochondrial fraction was resuspended in 1 ml of Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 2 mm sodium malonate, Complete protease inhibitors mixture (Roche Applied Science) (2 tablets/50 ml), protease inhibitors mixture for mammalian cell and tissue extracts (Sigma) (10 μl/ml), and 2% (v/v) Triton X-114. The mixture was incubated for 30 min on ice and kept at -30 °C overnight. After thawing, the insoluble material was removed by centrifugation at 4 °C, and the supernatant was incubated for 10 min at 37 °C and centrifuged at 2000 × g for 10 min to separate the aqueous and detergentrich phases. The aqueous phase was brought to 2% (v/v) Triton X-114, whereas the detergent-rich fraction was brought to 1 ml with the above buffer. After incubation on ice for 10 min, samples were incubated at 37 °C for 10 min and phases separated as before. This wash step was repeated three times. Finally, the samples were dialyzed and concentrated by Amicon Ultra-4 (Millipore) in the presence of 50 mm imidazole, 50 mm NaCl, 6 mm aminocaproic acid, 0.05% (w/v) deoxycholate, and 0.1% (w/v) SML, pH 7, and kept at -80 °C until use.
Enzyme Assay—Decylubiquinone-mediated succinate-2,4 dichlorophenolindophenol (DCIP) reductase activity was measured at 25 °C in 100 mm potassium phosphate, pH 7.4, containing 1 mm MgCl2, 2 mm KCN, 0.1 mm antimycin A (Sigma), and 0.1% SML with 63 μm decylubiquinone (Sigma) plus 60 μm DCIP. After 2 min of incubation, reduction of DCIP (ε600 = 21 mm-1 cm-1) was measured in the presence of 10 mm succinate. SQR activity was determined with 40 μm ubiquinone-2 (Q2) (Sigma, ε278 = 12.3 mm-1 cm-1). Kinetic analysis was done with KaleidaGraph version 4.0 (Synergy Software).
Miscellaneous—High resolution clear native electrophoresis (hrCNE) (22) was performed with 4-16% Novex gels (Invitrogen) using 0.02% dodecylmaltoside and 0.05% sodium deoxycholate for the cathode buffer additives, and the Complex II band was visualized by the activity staining (23) or Coomassie Brilliant Blue. Tricine-PAGE analysis was done with Novex 10-20% Tricine gels (Invitrogen), and proteins bands were sequentially stained by Sypro ruby (Invitrogen) and silver. During purification the succinate-decylubiquinone-DCIP reductase activity was monitored in a microplate spectrophotometer (Benchmark Plus, Bio-Rad). Kinetics and UV-visible absorption spectra were determined at room temperature with a V-660 UV-visible spectrophotometer (Jasco, Tokyo, Japan). Protoheme IX and protein concentrations were determined by pyridine hemochromogen method (24) and the micro BCA method (Pierce), respectively. Sequence alignment was done with ClustalX 2.0 (25).
RESULTS AND DISCUSSION
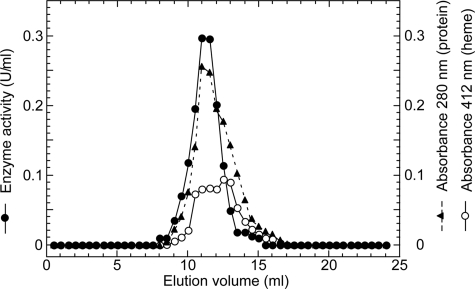
Isolation of T. cruzi Complex II—To determine the molecular organization of T. cruzi Complex II, we purified this enzyme from epimastigote mitochondria by ion-exchange and gel filtration chromatography using the nonionic detergent sucrose monolaurate (Table 1). Decylubiquinone-mediated succinate: DCIP reductase activity was eluted as a single peak at each step and co-eluted with proteins and b-type cytochrome(s) at the second Superdex 200 chromatography (Fig. 2). Specific activity was increased 34-fold to 2.9 units/mg proteins, and the yield was ∼2%. A hrCNE of the pure protein identified T. cruzi Complex II as an ∼550-kDa complex (Fig. 3, lanes 1 and 3), which is 4-fold larger than bovine and yeast Complex II (130 kDa) and potato Complex II (150 kDa) (26, 27). Upon phase partitioning of the mitochondrial fraction with Triton X-114, the Complex II of T. cruzi was found only in the detergent-rich fraction (data not shown). Analysis of the detergent-rich fraction by hrCNE showed the Complex II as a single band at the same position as the pure enzyme (∼550 kDa) (Fig. 3, lanes 2 and 4). These results indicated that the purified Complex II was obtained in its intact form. Interestingly, second dimensional analysis of both the purified Complex II and the detergent-rich fraction from phase partitioning with Triton X-114 with SDH activity showed that T. cruzi Complex II is composed of 12 subunits (Fig. 3, lanes 5 and 6). The same subunit composition was obtained by immunoaffinity purification of the partially purified enzyme (data not shown). The apparent molecular weight of the subunits ranges from 7.3 to 63 kDa (Fig. 3, lanes 7 and 8). Assuming the presence of equimolar amounts of subunits, a total molecular mass of Complex II would be 286.5 kDa, indicating that T. cruzi Complex II is a homodimer.
TABLE 1.
Purification of complex II from T. cruzi mitochondria
| Step | Protein | Succinate:DCIP reductase | Yield | Purification | |
|---|---|---|---|---|---|
| mg | units | units/mg | % | -fold | |
| Mitochondria | 314 | 27 | 0.085 | 100 | 1.0 |
| SML extract | 141 | 22 | 0.16 | 83 | 1.9 |
| Source 15Q | 7.8 | 7.6 | 0.97 | 28 | 12 |
| Superdex 200 (1st) | 1.3 | 1.4 | 1.09 | 5.3 | 13 |
| Superdex 200 (2nd) | 0.15 | 0.43 | 2.87 | 1.6 | 34 |
FIGURE 2.
Elution profile of T. cruzi Complex II on Superdex 200 chromatography. Complex II fractions from the first gel filtration chromatography with a Superdex 200-pg column were concentrated and rechromatographed at the flow rate of 0.25 ml/min. Aliquots were collected every 0.5 ml. Elution profiles for proteins and cytochromes were monitored at 280 (▴) and 412 nm (○), respectively, and the enzyme activity (•) was measured as decylquinone-mediated succinate:DCIP reductase.
FIGURE 3.
Electrophoresis analysis of T. cruzi Complex II. A, purified Complex II (2 μg; lanes 1 and 3) and the detergent-rich fraction from the phase partitioning by Triton X-114 of the mitochondrial fraction (60 μg; lines 2 and 4) were subjected to hrCNE. Proteins were stained by Coomassie Brilliant Blue (left panel), and Complex II was visualized by SDH activity staining (right panel). B, proteins of Complex II showing SDH activity in A were analyzed by 10-20% Tricine SDS-PAGE and visualized by silver stain (lane 5, pure complex; lane 6, detergent-rich fraction). C shows the subunit composition of the pure Complex II from T. cruzi stained by SYPRO ruby (lane 7) or silver stain (lane 8). Molecular weight standards used are NativeMark (Invitrogen, lane 1) and Mark 12 unstained standards (Invitrogen, lanes 5 and 7).
Identification of Genes Coded for Subunits—We determined N-terminal sequences (or internal peptide sequences in case of SDH2N and SDH8) of all subunits and identified genes coded for SDH1-1, SDH2N, SDH2C, SDH5-SDH7 (hydrophilic subunits), SDH3, SDH4, and SDH8-SDH11 (hydrophobic subunits) (Table 2). All subunits, except SDH1-1, are trypanosomatid-specific and structurally unrelated to plant-specific soluble subunits (AtSDH5-AtSDH8, 5-18 kDa) (27-29). All genes (except SDH6 with four copies) are present as two copies, which are assigned to either Esmeraldo or non-Esmeraldo haplotype (haploid genotype) in T. cruzi subgroup IIe. In contrast, only one copy each of the orthologues is present in T. brucei, Leishmania major, Leishmania infantum, and Leishmania brasiliensis (supplemental Table S1). N-terminal sequence analysis of SDH3 and SDH7 showed that yields of two isoforms are similar (i.e. SDH3-1:SDH3-2 = 63:37, and SDH7-1:SDH7-2 = 54:46), indicating that isoforms are expressed from each haplotype. Because truncated isoforms for SDH1 and SDH5 in the Esmeraldo haplotype (see below) are not assembled into the 12-subunit complex and SDH2N and SDH9 isoforms have the identical sequence, 512 (= 14 × 41 × 2(12-5)) kinds of heterogeneity may exist in the T. cruzi Complex II monomer (Table 2).
TABLE 2.
Identification of genes encoding subunits for T. cruzi complex II
| Subunita | Sequence confirmedb | Accession number or RefSeq ID at NCBI (haplotype,cMr) | Identityd | TMe |
|---|---|---|---|---|
| % | ||||
| SDH1 | Ser10-Met19 | AB031741 (NE, 66,974), XP_809281 (E, 18,231) | 59 | 0 |
| SDH5 | Ala10-Leu19 | XP_818124 (NE, 53,831), XP_810172 (E, 20,788) | 16 | 0 |
| SDH2N | Ser188-Arg196, Lys201-Ile204, Gly221-Asn223, Glu267-Ile269 | XP_814994 (merged, 32,232)f | 24 (37) | 0 |
| SDH2C | Pro2-Leu6 | XP_803796 (NE, 21,352), XP_806126 (E, 21,379) | 25 (43) | 0 |
| SDH6a | Val19-Val28 | XP_809065 (NA, 36,077), XP_812789 (NA, 36,035) | 15 | 0 |
| SDH6b | Val19-Val28 | XP_813603 (NA, 36,133), XP_813645 (NA, 36,039) | 14 | 0 |
| SDH7 | Ile26-Leu35 | XP_813318 (NE, 28,218), XP_820239 (E, 28,202) | 22 | 0 |
| SDH3 | Val2-Phe11 | XP_809410 (NE, 12,176), XP_810064 (E, 12,204) | 29 | 1 |
| SDH4 | Phe39-Thr48 | XP_808211 (E, 13,957), XP_816430 (NE, 13,975) | 27 | 2 |
| SDH8 | Gly5-Met16 | XP_809192 (NE, 16,199), XP_817545 (E, 16,143) | NDg | 2 |
| SDH9 | Ile10-Pro19 | XP_807105 (merged, 15,736) | ND | 1 |
| SDH10 | Pro25-Val33 | XP_808894 (NE, 15,565), XP_808903 (E, 15,554) | ND | 1 |
| SDH11 | Phe20-Cys29 | XP_814088 (E, 10,346), XP_814509 (NE, 10,337) | ND | 1 |
Alleles were named as SDH3-1 (XP_809410) and SDH3-2 (XP_810064) in the order of the accession numbers, except for SDH5.
These are N-terminal sequences except for SDH2N and SDH8, where the N-terminal residues were blocked.
Homozygous alleles located in a merged assembly of Esmeraldo (E) and non-Esmeraldo (NE) homologous sequences whose different copies were merged genes during the genome assembly are indicated by “merged.” Haplotypes for gene with more than two copies in the genome that does not belong to a merged region are not assigned (NA).
Identity % to counterparts in human were as follows: SDH1 (D30648), SDH2 (P21912), SDH3 (Q99643), or SDH4 (O14521). In parentheses, the identity % of SDH2N and SDH2C that correspond to either Met1-Pro155 (IpN domain) or Tyr156-Val280 (IpC domain), respectively, of human SDH2 is shown. Identity % for truncated forms of SDH1 and SDH5 (SDH1-2 and SDH5-2) in the Esmeraldo haplotype was 66 and 20%, respectively.
SDH2N from other trypanosomatids lack Met1 to Arg42 of TcSDH2N.
ND indicates not determined because these hydrophobic sequences are a highly divergent form of mammalian sequences.
Flavoprotein Subunit—SDH1-1 (63-kDa band in Tricine-PAGE) cross-reacted with the antiserum against bovine SDH1 (data not shown) and is highly homologous to counterparts in T. brucei (93% identity), L. major (90%), Homo sapiens (59%), Arabidopsis thaliana (62%), Saccharomyces cerevisiae (61%), and Escherichia coli (48%, SdhA). Amino acid residues proposed for dicarboxylate binding and a FAD ligand histidine (12-14) are all conserved in SDH1-1. SDH1-1 and SDH5-1 of the non-Esmeraldo haplotype share a weak sequence similarity in the entire region, but the latter lacks amino acid residues responsible for FAD and dicarboxylate binding. In the Esmeraldo haplotype, SDH1-2 and SDH5-2 are truncated and contain only Met1 to Gly167 of TcSDH1-1 and Ile305 to Met486 of TcSDH5-1, respectively (Table 2). These findings suggest that TcSDH1-1, TcSDH1-2, TcSDH5-1, and TcSDH5-2 might have evolved by gene duplication and subsequent degeneration.
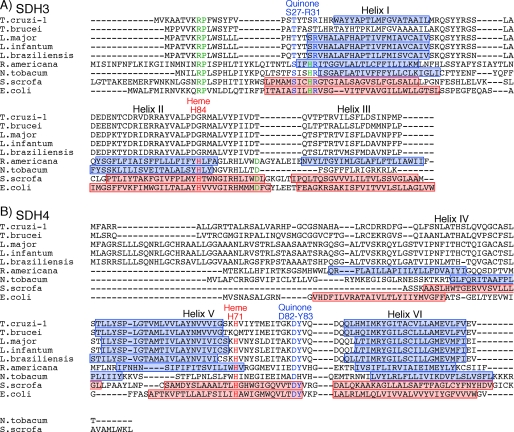
Iron-Sulfur Subunit—Sequence analysis of the 25- and 21-kDa band proteins revealed that they contain the plant ferredoxin domain (IpN) and bacterial ferredoxin domain (IpC) of canonical SDH2 (Ip) in the N- and C-terminal half, respectively (Fig. 4). Sequence identities of IpN and IpC are 37 and 43%, respectively, to those of human SDH2 (Table 2), and the IpN and IpC domains contain all amino acid residues responsible for binding of iron-sulfur clusters and ubiquinone (12, 13, 30) (Fig. 4). Such a heterodimeric Ip subunit can be found in T. brucei (31), T. cruzi, L. major, L. infantum, and L. brasiliensis (Tables 2), which belong to the order Trypanosomatida. Thus we named these subunits as SDH2N and SDH2C, respectively.
FIGURE 4.
Alignment of heterodimeric SDH2 sequences. Amino acid residues proposed for binding of the iron-sulfur clusters are shown in red and those for the quinone binding in blue. Residue numbers refer to the E. coli SDH2 (SdhB) sequence. GenBank™ accession numbers for SDH2N and SDH2C sequences used are T. cruzi (XP_814994 and XP_803796), T. brucei (XP_847169 and XP_826981), and L. major (XP_001683488 and XP_001682013). Other SDH2 sequences used are E. coli (NP_415252), Rickettsia prowazekii (Q9ZEA1), Reclinomonas americana (NP_044798), Dictyostelium discoideum (XP_646559), Plasmodium falciparum (D86574), Tetrahymena thermophila (XP_001024894), S. cerevisiae (NP_012774), Caenorhabditis elegans (NP_495992), H. sapiens (NP_002991), and A. thaliana (NP_189374).
Splitting of mitochondrial membrane proteins has been reported for cytochrome c oxidase CoxII in Apicomplexa and Chlorophyceae (32, 33), and ATP synthase α subunit in Leishmania tarentolae and T. brucei (34, 35). The former occurs at the gene level and the latter by post-translational cleavage. Sequence analysis indicates that heterodimeric SDH2 and CoxII have emerged from gene duplication followed by degeneration of the N- or C-terminal half of the duplication products. Conserved domains in degenerated duplicons, which have arisen from mitochondrion-to-nucleus transfer of the duplicated genes (32, 33, 36), must retain the potential for protein-protein interactions and constitute a heterodimeric functional subunit by trans-complementation.
Membrane Anchor Subunits—Membrane anchor subunits in protist enzymes are highly divergent from bacterial and mammalian counterparts and difficult to find with conventional BLAST programs. We identified candidates for T. cruzi SDH3 and SDH4 by the presence of the quinone/heme-binding motifs “RPX16SX2HR (SDH3 helix I)” and “HX10DY (SDH4 helix V),” respectively, present in membrane anchor subunits. In Complex II, Trp164 in SDH2 (Fig. 4) and Tyr83 in the SDH4 HX10DY motif (Fig. 5B) (E. coli numbering) could hydrogen bond to the O-1 atom of ubiquinone and contribute to the binding affinity (12, 37). Arg31 in the SDH3 SX2HR motif (Fig. 5A) and Asp82 in the SDH4 HX10DY motif are in close proximity to ubiquinone and could interact with Tyr83 (37). Ser27 in the SDH3 SX2HR motif has been shown to be essential for quinone binding (38) and is a candidate for hydrogen bonding to the O-4 atom of ubiquinone (30). The first arginine (Arg9 in E. coli SDH3) in the RPX16SX3R motif is in the vicinity of Glu186 in SDH1 and Asp106 in SDH2 and may play a structural role by making a hydrogen bond network.
FIGURE 5.
Alignments of SDH3 (A) and SDH4 (B) sequences. Amino acid residues proposed for binding of protoheme IX are shown in red and those for the quinone binding in blue. Other conserved residues are indicated by green. Transmembrane helices found in E. coli (Protein Data Bank code 1NEK) and porcine (Protein Data Bank code 1ZOY) Complex II are shown by red rectangles, and transmembrane helices predicted by TMHMM are indicated by blue rectangles. TMHMM failed to predict transmembrane helices in T. brucei SDH3. Residue numbers refer to E. coli SDH3 (SdhC) and SDH4 (SdhD). GenBank™ accession numbers for SDH3 and SDH4 sequences used are T. cruzi (XP_809410, XP_808211), T. brucei (XP_845531, XP_823384), L. major (XP_001684890, XP_001685874), L. infantum (XP_001467132), L. brasiliensis (XP_001566908, XP_001567905), R. americana (NP_044796, NP_044797), Nicotiana tobacum (YP_173376, YP_173457), Sus scrofa (1ZOY_C, 1ZOY_D), and E. coli (NP_415249, NP_415250).
In T. cruzi, SDH3 has the “RPX11SX2HR motif in front of the predicted transmembrane helix I and lacks transmembrane helices II and III. However, sequence alignment suggests the presence of the alternative motif “TX2SR/(T)” in the Trypanosomatida (Fig. 5A). In mitochondrial Complex II, protoheme IX is ligated by two His residues in the second transmembrane helix of SDH3 (“HX10D” motif) and SDH4 (“HX10DY” motif). A heme ligand in helix II (His84 in E. coli SDH3) may be substituted by a nearby histidine in the quinone-binding motif “SX2HR” (39). In contrast, SDH4 lacks helix IV and appears to interact with heme and ubiquinone with the HX10DY motif. As in rice SDH4 (GenBank™ accession number NP_001045324), the heme ligand His is substituted by Gln in T. brucei SDH4. The presence of a bound heme or an alternative ligand in T. brucei SDH4 needs to be tested in future studies. It is also possible that trypanosomatid-specific subunits could be assembled as a jigsaw puzzle-like membrane anchor.
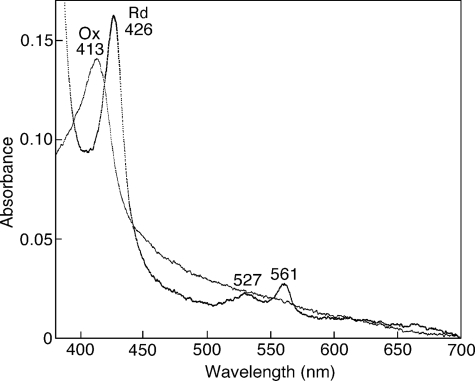
Spectroscopic Properties of T. cruzi Complex II—Pyridine ferrohemochrome analysis showed that T. cruzi Complex II binds a stoichiometric amount of protoheme IX (0.85 heme/monomer of enzyme) indicating that monomer enzyme complex contains one heme. At room temperature, the air-oxidized and fully reduced forms of the purified enzyme showed peaks at 413 and 426, 527, and 561 nm, respectively (Fig. 6). Peak positions are similar to those reported for Complex II from E. coli (40), adult A. suum (41), and bovine (42, 43), where heme is ligated via histidine in the second helices of SDH3 and SDH4. Although heme has an important role in the assembly of Complex II, it is not essential for the reduction of ubiquinones (43, 44).
FIGURE 6.
Visible absorption spectra of T. cruzi Complex II. Purified Complex II was desalted by ultrafiltration and diluted with 0.1 m sodium phosphate, pH 7.2, containing 0.1% SML at a final concentration of 0.06 mg/ml. Absorption spectra of the air-oxidized (Ox, thin line) and dithionite-reduced (Rd, thick line) forms were recorded at room temperature with UV-2400 spectrophotometer (Shimadzu Corp., Kyoto, Japan).
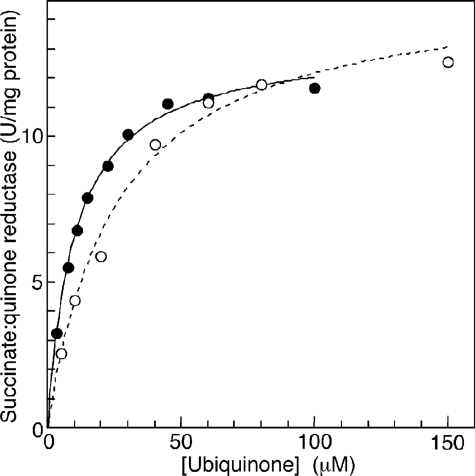
Enzymatic Properties of T. cruzi Complex II—We examined SQR activity of the purified enzyme and found the difference in apparent Km values between Q1 (33.9 ± 3.6 μm) and Q2 (18.8 ± 6.4 μm) (Fig. 7), indicating that the 6-polyprenyl group of ubiquinone contributes to the binding affinity. The apparent Vmax value of the T. cruzi Complex II was rather constant, 11.9 ± 2.2 for Q1 and 11.5 ± 0.4 Q2 units/mg proteins, respectively, and one-fourth of those reported for bovine and E. coli enzymes (45, 46). This is not surprising because T. cruzi complex II has about 2-3 times more proteins than the other enzymes. Km values for ubiquinone and succinate (18.8 ± 6.4 μm (Q2) and 1.48 ± 0.17 mm, respectively) were higher than 0.3 and 130 μm, respectively, of bovine enzyme (45), and 2 and 277 μm, respectively, of the E. coli enzyme (46, 47). Notably, the Km value for succinate was comparable with 610 μm in adult A. suum (10), which expresses the stage-specific Complex II as quinol:fumarate reductase under hypoxic habitats in host organisms.
FIGURE 7.
Kinetic analysis of succinate-quinone reductase activity. Succinate:ubiquinone reductase activity of the purified Complex II was determined with Q1 (○) and Q2 (•) at a protein concentration of 1.25 μg/ml in the presence of 10 mm sodium succinate. Data were fitted with the Michaelis-Menten equation using KaleidaGraph, and apparent Km and Vmax values were 30.3 ± 4.3 μm and 14.0 ± 1.2 units/mg protein, respectively, for Q1 and 12.4 ± 0.7 μm and 11.9 ± 0.3 units/mg protein, respectively, for Q2.
Then we examined effects of inhibitors for binding sites of quinones and dicarboxylates on SQR activity. Atpenin A5, a potent inhibitor for Complex II, inhibited the T. cruzi enzyme with the IC50 value of 6.4 ± 2.4 μm, which is 3 orders of magnitude higher than that of bovine Complex II (4 nm) (48). Furthermore, carboxin, 2-theonyltrifluoroacetone, plumbagin, and 2-heptyl-4-hydroxyquinoline N-oxide were ineffective (100 μm <IC50). Structural divergence in trypanosomatid SDH3 and SDH4 could be the cause for lower binding affinities for both quinones and inhibitors. In addition, we found for the dicarboxylate-binding site that the IC50 value for malonate (40 μm) was much higher than the Ki value for bovine Complex II (1.3 μm) (45).
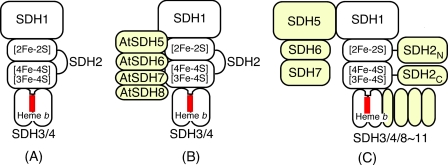
Structure of Trypanosomatid Complex II—To the best of our knowledge, this is the first report on the isolation of protist Complex II. T. cruzi Complex II has unusual subunit organization with six each of hydrophilic and hydrophobic subunits. Such a supramolecular structure and heterodimeric SDH2 (SDH2N and SDH2C) are conserved in the Trypanosomatida. Furthermore, SDH1, SDH2N, SDH2C, SDH3, SDH4, and SDH8-SDH10 can be identified in the ongoing genome projects on the evolutionary relatives, the photosynthetic free-living Euglena gracilis, and the nonphotosynthetic euglenoid Astasia longa in the Euglenida. Thus a part of these features are common in the Euglenozoa, a divergent lineage of eukaryotes (Fig. 8).
FIGURE 8.
Subunit organization of Complex II. A, common four-subunit Complex II (e.g. mammals, E. coli); B, eight-subunit Complex II in plants (e.g. A. thaliana); and C, 12-subunit Complex II in the Trypanosomatida. Noncatalytic subunits and domains are shown in yellow and heme in red.
Accumulation of noncatalytic subunits through expanding the protein interaction network could be a driving force for protein evolution. Structural and catalytic features are unique, and thus this enzyme could be a potential target for novel chemotherapeutic agents for trypanosomiasis and leishmaniasis.
Conclusion—The parasitic protist T. brucei is a gold mine where unprecedented biological phenomena like RNA editing and trans-splicing in mitochondria were originally discovered. It was found recently in Diplonema papillatum, a free-living evolutionary cousin, that mature mRNA for cytochrome c oxidase CoxI was assembled from nine gene fragments by a jigsaw puzzle mechanism (49). From a characterization of Complex II from T. cruzi, we revealed a novel supramolecular organization, which is conserved in the Trypanosomatida.
Parasites have exploited unique energy metabolic pathways as adaptations to their natural habitats within their hosts (50, 51). In fact, the respiratory systems of parasites typically show greater diversity in electron transfer pathways than those of host animals. As shown in this study, such is also the case with Complex II, which is a well known marker enzyme of mitochondria. Studies on the role of supramolecular Complex II in adaptation of trypanosomatids is now underway in our laboratory.
Supplementary Material
Acknowledgments
We thank Drs. J. L. Concepcion (Universidad de Los Andes, Merida-Venezuela) and T. Nara (Juntendo University) for kind advice; and Drs. M. Matsuzaki (University of Tokyo), T. Hashimoto (University of Tsukuba), G. Cecchini (University of California San Francisco), and M. Müller (Rockefeller University) for critical reading of the manuscript.
This work was supported in part by Grant-in-aid for Scientific Research 20570124 (to T. M.), Creative Scientific Research Grant 18GS0314 (to K. K.), Grant-in-aid for Scientific Research on Priority Areas 18073004 (to K. K.) from the Japanese Society for the Promotion of Science, and Targeted Proteins Research Program (to K. K.) from the Japanese Ministry of Education, Science, Culture, Sports and Technology (MEXT). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: SQR, succinate:quinone reductase; hrCNE, high resolution clear native electrophoresis; IC50, the 50% inhibitory concentration; IpN, the N-terminal plant-type ferredoxin domain; IpC, the C-terminal bacterial ferredoxin domain; DCIP, 2,4-dichlorophenolindophenol; SML, sucrose monolaurate; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]-glycine; MOPS, 3-(N-morpholino)propanesulfonic acid; Qn, ubiquinone-n.
References
- 1.World Health Organization (2007) Report of the First Meeting of WHO Strategic and Technical Advisory Group on Neglected Tropical Diseases, pp. 1-26, Geneva, Switzerland
- 2.Berriman, M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D. C., Lennard, N. J., Caler, E., Hamlin, N. E., Haas, B., Bohme, U., Hannick, L., Aslett, M. A., Shallom, J., Marcello, L., Hou, L., Wickstead, B., Alsmark, U. C., Arrowsmith, C., Atkin, R. J., Barron, A. J., Bringaud, F., Brooks, K., Carrington, M., Cherevach, I., Chillingworth, T. J., Churcher, C., Clark, L. N., Corton, C. H., Cronin, A., Davies, R. M., Doggett, J., Djikeng, A., Feldblyum, T., Field, M. C., Fraser, A., Goodhead, I., Hance, Z., Harper, D., Harris, B. R., Hauser, H., Hostetler, J., Ivens, A., Jagels, K., Johnson, D., Johnson, J., Jones, K., Kerhornou, A. X., Koo, H., Larke, N., Landfear, S., Larkin, C., Leech, V., Line, A., Lord, A., Macleod, A., Mooney, P. J., Moule, S., Martin, D. M., Morgan, G. W., Mungall, K., Norbertczak, H., Ormond, D., Pai, G., Peacock, C. S., Peterson, J., Quail, M. A., Rabbinowitsch, E., Rajandream, M. A., Reitter, C., Salzberg, S. L., Sanders, M., Schobel, S., Sharp, S., Simmonds, M., Simpson, A. J., Tallon, L., Turner, C. M., Tait, A., Tivey, A. R., Van Aken, S., Walker, D., Wanless, D., Wang, S., White, B., White, O., Whitehead, S., Woodward, J., Wortman, J., Adams, M. D., Embley, T. M., Gull, K., Ullu, E., Barry, J. D., Fairlamb, A. H., Opperdoes, F., Barrell, B. G., Donelson, J. E., Hall, N., Fraser, C. M., Melville, S. E., and El-Sayed, N. M. (2005) Science 309 416-42216020726 [Google Scholar]
- 3.Cazzulo, J. J. (1994) J. Bioenerg. Biomembr. 26 157-165 [DOI] [PubMed] [Google Scholar]
- 4.Besteiro, S., Barrett, M. P., Riviere, L., and Bringaud, F. (2005) Trends Parasitol. 21 185-191 [DOI] [PubMed] [Google Scholar]
- 5.Bringaud, F., Riviere, L., and Coustou, V. (2006) Mol. Biochem. Parasitol. 149 1-9 [DOI] [PubMed] [Google Scholar]
- 6.Takashima, E., Inaoka, D. K., Osanai, A., Nara, T., Odaka, M., Aoki, T., Inaka, K., Harada, S., and Kita, K. (2002) Mol. Biochem. Parasitol. 122 189-200 [DOI] [PubMed] [Google Scholar]
- 7.Van Hellemond, J. J., Opperdoes, F. R., and Tielens, A. G. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3036-3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harington, J. S. (1961) Parasitology 51 309-318 [DOI] [PubMed] [Google Scholar]
- 9.Roos, M. H., and Tielens, A. G. (1994) Mol. Biochem. Parasitol. 66 273-281 [DOI] [PubMed] [Google Scholar]
- 10.Saruta, F., Kuramochi, T., Nakamura, K., Takamiya, S., Yu, Y., Aoki, T., Sekimizu, K., Kojima, S., and Kita, K. (1995) J. Biol. Chem. 270 928-932 [DOI] [PubMed] [Google Scholar]
- 11.Cecchini, G. (2003) Annu. Rev. Biochem. 72 77-109 [DOI] [PubMed] [Google Scholar]
- 12.Yankovskaya, V., Horsefield, R., Tornroth, S., Luna-Chavez, C., Miyoshi, H., Leger, C., Byrne, B., Cecchini, G., and Iwata, S. (2003) Science 299 700-704 [DOI] [PubMed] [Google Scholar]
- 13.Sun, F., Huo, X., Zhai, Y., Wang, A., Xu, J., Su, D., Bartlam, M., and Rao, Z. (2005) Cell 121 1043-1057 [DOI] [PubMed] [Google Scholar]
- 14.Huang, L. S., Sun, G., Cobessi, D., Wang, A. C., Shen, J. T., Tung, E. Y., Anderson, V. E., and Berry, E. A. (2006) J. Biol. Chem. 281 5965-5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Sayed, N. M., Myler, P. J., Bartholomeu, D. C., Nilsson, D., Aggarwal, G., Tran, A. N., Ghedin, E., Worthey, E. A., Delcher, A. L., Blandin, G., Westenberger, S. J., Caler, E., Cerqueira, G. C., Branche, C., Haas, B., Anupama, A., Arner, E., Aslund, L., Attipoe, P., Bontempi, E., Bringaud, F., Burton, P., Cadag, E., Campbell, D. A., Carrington, M., Crabtree, J., Darban, H., da Silveira, J. F., de Jong, P., Edwards, K., Englund, P. T., Fazelina, G., Feldblyum, T., Ferella, M., Frasch, A. C., Gull, K., Horn, D., Hou, L., Huang, Y., Kindlund, E., Klingbeil, M., Kluge, S., Koo, H., Lacerda, D., Levin, M. J., Lorenzi, H., Louie, T., Machado, C. R., McCulloch, R., McKenna, A., Mizuno, Y., Mottram, J. C., Nelson, S., Ochaya, S., Osoegawa, K., Pai, G., Parsons, M., Pentony, M., Pettersson, U., Pop, M., Ramirez, J. L., Rinta, J., Robertson, L., Salzberg, S. L., Sanchez, D. O., Seyler, A., Sharma, R., Shetty, J., Simpson, A. J., Sisk, E., Tammi, M. T., Tarleton, R., Teixeira, S., Van Aken, S., Vogt, C., Ward, P. N., Wickstead, B., Wortman, J., White, O., Fraser, C. M., Stuart, K. D., and Andersson, B. (2005) Science 309 409-41516020725 [Google Scholar]
- 16.Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., Sisk, E., Rajandream, M. A., Adlem, E., Aert, R., Anupama, A., Apostolou, Z., Attipoe, P., Bason, N., Bauser, C., Beck, A., Beverley, S. M., Bianchettin, G., Borzym, K., Bothe, G., Bruschi, C. V., Collins, M., Cadag, E., Ciarloni, L., Clayton, C., Coulson, R. M., Cronin, A., Cruz, A. K., Davies, R. M., De Gaudenzi, J., Dobson, D. E., Duesterhoeft, A., Fazelina, G., Fosker, N., Frasch, A. C., Fraser, A., Fuchs, M., Gabel, C., Goble, A., Goffeau, A., Harris, D., Hertz-Fowler, C., Hilbert, H., Horn, D., Huang, Y., Klages, S., Knights, A., Kube, M., Larke, N., Litvin, L., Lord, A., Louie, T., Marra, M., Masuy, D., Matthews, K., Michaeli, S., Mottram, J. C., Muller-Auer, S., Munden, H., Nelson, S., Norbertczak, H., Oliver, K., O'Neil, S., Pentony, M., Pohl, T. M., Price, C., Purnelle, B., Quail, M. A., Rabbinowitsch, E., Reinhardt, R., Rieger, M., Rinta, J., Robben, J., Robertson, L., Ruiz, J. C., Rutter, S., Saunders, D., Schafer, M., Schein, J., Schwartz, D. C., Seeger, K., Seyler, A., Sharp, S., Shin, H., Sivam, D., Squares, R., Squares, S., Tosato, V., Vogt, C., Volckaert, G., Wambutt, R., Warren, T., Wedler, H., Woodward, J., Zhou, S., Zimmermann, W., Smith, D. F., Blackwell, J. M., Stuart, K. D., Barrell, B., and Myler, P. J. (2005) Science 309 436-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourguignon, S. C., Mello, C. B., Santos, D. O., Gonzalez, M. S., and Souto-Padron, T. (2006) Acta Trop. 98 103-109 [DOI] [PubMed] [Google Scholar]
- 18.Concepcion, J. L., Chataing, B., and Dubourdieu, M. (1999) Comp. Biochem. Physiol. 122 211-222 [DOI] [PubMed] [Google Scholar]
- 19.Matsudaira, P. (1987) J. Biol. Chem. 262 10035-10038 [PubMed] [Google Scholar]
- 20.Rosenfeld, J., Capdevielle, J., Guillemot, J. C., and Ferrara, P. (1992) Anal. Biochem. 203 173-179 [DOI] [PubMed] [Google Scholar]
- 21.Brusca, J. S., and Radolf, J. D. (1994) Methods Enzymol. 228 182-193 [DOI] [PubMed] [Google Scholar]
- 22.Wittig, I., Karas, M., and Schagger, H. (2007) Mol. Cell. Proteomics 6 1215-1225 [DOI] [PubMed] [Google Scholar]
- 23.Sabar, M., Balk, J., and Leaver, C. J. (2005) Plant J. 44 893-901 [DOI] [PubMed] [Google Scholar]
- 24.Berry, E. A., and Trumpower, B. L. (1987) Anal. Biochem. 161 1-15 [DOI] [PubMed] [Google Scholar]
- 25.Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007) Bioinformatics (Oxf.) 23 2947-2948 [DOI] [PubMed] [Google Scholar]
- 26.Schagger, H., and Pfeiffer, K. (2000) EMBO J. 19 1777-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millar, A. H., Eubel, H., Jansch, L., Kruft, V., Heazlewood, J. L., and Braun, H. P. (2004) Plant Mol. Biol. 56 77-90 [DOI] [PubMed] [Google Scholar]
- 28.Eubel, H., Heinemeyer, J., and Braun, H. P. (2004) Plant Physiol. 134 1450-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eubel, H., Heinemeyer, J., Sunderhaus, S., and Braun, H. P. (2004) Plant Physiol. Biochem. 42 937-942 [DOI] [PubMed] [Google Scholar]
- 30.Horsefield, R., Yankovskaya, V., Sexton, G., Whittingham, W., Shiomi, K., Omura, S., Byrne, B., Cecchini, G., and Iwata, S. (2006) J. Biol. Chem. 281 7309-7316 [DOI] [PubMed] [Google Scholar]
- 31.Allen, J. W., Ginger, M. L., and Ferguson, S. J. (2004) Biochem. J. 383 537-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funes, S., Davidson, E., Reyes-Prieto, A., Magallon, S., Herion, P., King, M. P., and Gonzalez-Halphen, D. (2002) Science 298 2155. [DOI] [PubMed] [Google Scholar]
- 33.Waller, R. F., and Keeling, P. J. (2006) Gene (Amst.) 383 33-37 [DOI] [PubMed] [Google Scholar]
- 34.Williams, N., and Frank, P. H. (1990) Mol. Biochem. Parasitol. 43 125-132 [DOI] [PubMed] [Google Scholar]
- 35.Nelson, R. E., Aphasizheva, I., Falick, A. M., Nebohacova, M., and Simpson, L. (2004) Mol. Biochem. Parasitol. 135 221-224 [DOI] [PubMed] [Google Scholar]
- 36.Adams, K. L., Rosenblueth, M., Qiu, Y. L., and Palmer, J. D. (2001) Genetics 158 1289-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2006) J. Biol. Chem. 281 32310-32317 [DOI] [PubMed] [Google Scholar]
- 38.Yang, X., Yu, L., He, D., and Yu, C. A. (1998) J. Biol. Chem. 273 31916-31923 [DOI] [PubMed] [Google Scholar]
- 39.Maklashina, E., Rothery, R. A., Weiner, J. H., and Cecchini, G. (2001) J. Biol. Chem. 276 18968-18976 [DOI] [PubMed] [Google Scholar]
- 40.Kita, K., Vibat, C. R., Meinhardt, S., Guest, J. R., and Gennis, R. B. (1989) J. Biol. Chem. 264 2672-2677 [PubMed] [Google Scholar]
- 41.Takamiya, S., Furushima, R., and Oya, H. (1986) Biochim. Biophys. Acta 848 99-107 [DOI] [PubMed] [Google Scholar]
- 42.Tushurashvili, P. R., Gavrikova, E. V., Ledenev, A. N., and Vinogradov, A. D. (1985) Biochim. Biophys. Acta 809 145-159 [DOI] [PubMed] [Google Scholar]
- 43.Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18007-18012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyedotun, K. S., Sit, C. S., and Lemire, B. D. (2007) Biochim. Biophys. Acta 1767 1436-1445 [DOI] [PubMed] [Google Scholar]
- 45.Grivennikova, V. G., Gavrikova, E. V., Timoshin, A. A., and Vinogradov, A. D. (1993) Biochim. Biophys. Acta 1140 282-292 [DOI] [PubMed] [Google Scholar]
- 46.Maklashina, E., and Cecchini, G. (1999) Arch. Biochem. Biophys. 369 223-232 [DOI] [PubMed] [Google Scholar]
- 47.Miyadera, H., Hiraishi, A., Miyoshi, H., Sakamoto, K., Mineki, R., Murayama, K., Nagashima, K. V., Matsuura, K., Kojima, S., and Kita, K. (2003) Eur. J. Biochem. 270 1863-1874 [DOI] [PubMed] [Google Scholar]
- 48.Miyadera, H., Shiomi, K., Ui, H., Yamaguchi, Y., Masuma, R., Tomoda, H., Miyoshi, H., Osanai, A., Kita, K., and Omura, S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 473-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marande, W., and Burger, G. (2007) Science 318 415. [DOI] [PubMed] [Google Scholar]
- 50.Kita, K., and Takamiya, S. (2002) Adv. Parasitol. 51 95-131 [DOI] [PubMed] [Google Scholar]
- 51.Tielens, A. G., Rotte, C., van Hellemond, J. J., and Martin, W. (2002) Trends Biochem. Sci. 27 564-572 [DOI] [PubMed] [Google Scholar]
- 52.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001) J. Mol. Biol. 305 567-580 [DOI] [PubMed] [Google Scholar]
- 53.Mitaku, S., Hirokawa, T., and Tsuji, T. (2002) Bioinformatics 18 608-616 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.