Abstract
Because the influenza A virus has an RNA genome, its RNA-dependent RNA polymerase, comprising the PA, PB1, and PB2 subunits, is essential for viral transcription and replication. The binding of RNA primers/promoters to the polymerases is an initiation step in viral transcription. In our current study, we reveal the 2.7 Å tertiary structure of the C-terminal RNA-binding domain of PB2 by x-ray crystallography. This domain incorporates lysine 627 of PB2, and this residue is associated with the high pathogenicity and host range restriction of influenza A virus. We found from our current analyses that this lysine is located in a unique “φ”-shaped structure consisting of a helix and an encircled loop within the PB2 domain. By electrostatic analysis, we identified a highly basic groove along with this φ loop and found that lysine 627 is located in the φ loop. A PB2 domain mutant in which glutamic acid is substituted at position 627 shows significantly lower RNA binding activity. This is the first report to show a relationship between RNA binding activity and the pathogenicity-determinant lysine 627. Using the Matras program for protein three-dimensional structural comparisons, we further found that the helix bundles in the PB2 domain are similar to that of activator 1, the 40-kDa subunit of DNA replication clamp loader (replication factor C), which is also an RNA-binding protein. This suggests a functional and structural relationship between the RNA-binding mechanisms underlying both influenza A viral transcription and cellular DNA replication. Our present results thus provide important new information for developing novel drugs that target the primer/promoter RNA binding of viral RNA polymerases.
In 1918, a pandemic expansion of influenza A virus resulted in ten million deaths worldwide (1), and strategies to prevent any future expansions of this virus are therefore an important endeavor (2, 3). Although inhibitors of neuraminidase and the M2 ion channel are widely used as anti-influenza A virus drugs (4, 5), some adverse effects of these agents and also the emergence of drug-resistant viruses have been reported (6, 7). Because the transcription and replication of influenza A virus requires the activity of its RNA-dependent RNA polymerase, this enzyme represents a very promising target for anti-viral drug development. Influenza A virus digests host mRNA via the PB1 subunit of its RNA polymerase (8). A 5′-RNA fragment of 10–15 nucleotides is used as a primer for transcription initiation (8) through primer and promoter RNA binding activities within the C-terminal region of its RNA polymerase PB2 domain (9–11). The PA subunit has protease activity and binds to PB (2). The determination of the tertiary structure of a protein is a valuable tool in the development of a targeting drug, and low resolution structures have previously been determined for the PA, PB1, and PB2 subunits of influenza A virus RNA polymerase by electron microscopy (12). In addition, the structure of PB2 nuclear localization signal domain (amino acid (aa)3 residues 687–759), the PB2 cap (7-methyl guanosine triphosphate)-binding domain (aa 318–483), and the C-terminal domain of PA with the N-terminal domain of PB1 have also been reported (13–17). The C-terminal domain of PB2 includes the Lys627 residue, which plays a role in the high pathogenicity and host range restriction of the influenza virus. The structure of PB2 has not been previously elucidated, and the structure-function relationships between this domain and Lys627 represent a significant advance in our understanding of the pathogenicity of influenza A.
EXPERIMENTAL PROCEDURES
Expression and Purification of the Influenza A Virus RNA Polymerase PB2 3/3 RNA-binding Domain—A cDNA fragment of the influenza A virus RNA polymerase PB2 3/3 domain (aa 535–759) was amplified by PCR from pBMSA-PB2 (RIKEN) using the following primers: PB2 Met 3/3, 5′-GCCGTTCATATGATGTGGGAGATTAATGGT-3′, and PB2 stop BamHI, 5′-GCCGTTGGATCCTTAATTGATGGCCATCCGAAT-3′. The amplified DNA fragment was then subcloned into the pET28a(+) plasmid (Novagen, Madison, WI) at the NdeI and BamHI restriction sites. This construct was then introduced into BL21-CodonPlus (Stratagene, La Jolla, CA) Escherichia coli. The induction of His6-tagged recombinant protein expression was achieved by the addition of isopropyl β-d-thiogalactopyranoside in TBG-M9 medium, and this was followed by purification using Ni2+-agarose. For further purification, the His-tagged proteins were cleaved by thrombin and purified using a HiTrap™ carboxymethyl (CM) FF column (GE Healthcare, Buckinghamshire, UK) with the ÄKTAprime™ plus system (GE Healthcare). For phase determination, we expressed the recombinant proteins using seleno-methionine (SeMet)-containing M9 medium (18) in E. coli B836 (DE3) cells (Novagen) to enable multiple wavelength anomalous dispersion (MAD) analysis. Site-directed mutagenesis for lysine 627 was performed by PCR. We purified PB2 3/3 Glu627 and PB2 3/3 for reference using a HiTrap™ SP XL column (GE Healthcare).
RNA Binding Analysis by Surface Plasmon Resonance (Biacore) Assay—A Biacore 3000 instrument (Biacore AB, Uppsala, Sweden) was used to perform surface plasmon resonance assays in accordance with the manufacturer's instructions. The buffers used in these analyses were as follows: HBS-EP running buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, 0.005% surfactant P20) and sensor chip streptavidin to which 100 μl of 20 μm biotinylated RNA oligomers were immobilized. A 5′-vRNA promoter (5′-AGUAGAAACAAGGCC-3′), capped primer (5′-m7GmAAAUACUCAAG-3′), or non-capped primer (5′-AmAAUACUCAAG-3′) were synthesized by Hokkaido System Science Co. Ltd., (Hokkaido, Japan), and successful capping of the primers was confirmed by mass spectrometry (Voyager 2073 system). These molecules were then tested in the PB2 3/3 domain protein binding assays. Various concentrations (0.015, 0.044, 0.13, 0.40, 1.2, and 3.6 μm) of the purified influenza A virus RNA polymerase PB2 3/3 proteins were then applied to the RNA-immobilized sensor chip in HBS-EP buffer at a 20 μl/min flow rate. A blank control was used as a reference, and these values were subtracted from the raw data.
Estimation of the Kd (Dissociation Constant) Values of the PB2 3/3 Protein for Different RNA Substrates—For affinity analyses, the association-dissociation curves for various concentrations of PB2 3/3 or PB2 3/3 Glu627 protein were plotted using the BIAevaluation steady state affinity program (GE Healthcare) from which equilibrium dissociation constant (Kd) values (mol/liter) were calculated.
Crystallization—Purified samples were concentrated to 4–10 mg/ml by ultrafiltration. Optimal conditions for crystallization were determined initially using Crystal Screen 1™ and Crystal Screen 2™ (Hampton Research, Aliso Viejo, CA), which involved the hanging drop vapor diffusion method at 4 °C. Native crystals were obtained using a reservoir solution containing 0.1 m sodium cacodylate hydrochloric acid, pH 6.2, and 2 m sodium formate. Crystals of selenomethionine-labeled PB2 3/3 protein were obtained using a reservoir solution containing 0.1 m sodium cacodylate hydrochloric acid, pH 6.0, and 3 m sodium formate.
X-ray Diffraction Analysis—Diffraction data sets for Native and SeMet crystals were collected at 100 K, using a mixture of paratone-N (Hampton Research) and paraffin oil as a cryoprotectant, on a beamline PF-AR NW12 instrument at the Photon Factory (Tsukuba, Japan). The image data were processed using HKL2000 (19) and SCALA (20). The phase was obtained by MAD with three wavelength SeMet data sets using SOLVE and RESOLVE (21). The initial models were constructed using the XtalView program (22). Iterative refinement and model building were subsequently performed using the REFMAC5 module of the CCP4 suite (22). There were no residues found in the disallowed region of the Ramachandran plot as determined by PROCHECK (23).
RESULTS AND DISCUSSION
To perform x-ray crystallographic analysis of the RNA polymerase PB2 of influenza A virus (H1N1, A/PR/8/34), it was necessary to obtain soluble quantities of the enzyme. The PB2 subunit was therefore divided into three portions. We expressed PB2 3/3 (aa 535–759) in E. coli as a His6-tagged recombinant protein that was subsequently purified from soluble fractions using Ni2+-agarose. We further purified the PB2 3/3 domain protein by cation-exchange chromatography to near homogeneity (98%).
We then subjected the recombinant PB2 3/3 protein to crystallization as described under “Experimental Procedures.” Native crystals were found to belong to the space group P3221 with the properties a = b = 52.5 Å, c = 156.3 Å. The Matthew's value (Vm) was 2.7, assuming one molecule in an asymmetric unit. SeMet crystals were found in the same space group with the properties a = b = 52.6 Å, c = 156.4 Å. The crystal structure of PB2 3/3 was then determined by MAD phasing guided by the six SeMet positions out of the 10 methionine sites using SeMet crystals (24). The model structure for PB2 3/3 was constructed from 208 residues (aa 535–742), except for the C-terminal 18 residues, and 86 water molecules (R value, 23.7%, and Rfree, 29.0%) using reflection data of up to 2.7 Å. There were no residues found in the disallowed region of the Ramachandran plot.
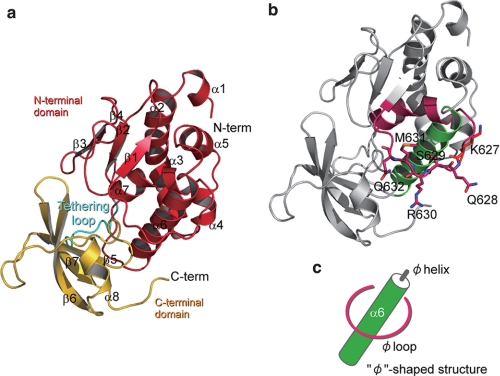
The determined structure for PB2 3/3 is shown in Fig. 1 and consists of eight helices and seven strands (Fig. 1, a and b). The N-terminal region (aa 535–684) is rich in α-helices, whereas the C-terminal portion is predominantly composed of β-strands (Fig. 1, a and b). The structure of the PB2 3/3 C terminus (aa 685–742) consists of three strands and one helix, which is quite similar to the already reported structure of the importin-binding domain of PB2 (Fig. 1b) (13). The N-terminal region, which has not previously been reported, consists of seven helix bundles and four β-strands (Fig. 1a). The C-terminal importin-binding region and the N terminus are linked by a “tethering loop” (aa 676–692) (Fig. 1a). Because the N terminus of PB2 3/3 is located in front of the importin-binding surface of its C-terminal region in the tertiary structure, the binding of the C-terminal region of PB2 to importin α-protein (13) requires its N-terminal portion to move or rotate away (domain-domain movement).
FIGURE 1.
Tertiary structure of the RNA-binding domain in the influenza A virus RNA polymerase PB2 subunit. a, tertiary structure of the RNA-binding domain of the PB2 subunit. The N-terminal domain (N-term), C-terminal domain (C-term), and tethering loop are shown in red, yellow, and cyan, respectively. b, φ-shaped structure in the PB2 subunit. The φ loop contains α7 and φ helices indicated in purple and green, respectively. Lys627 is shown in red, and other flexible residues 628QSRMQ632 are shown in purple. c, schematic illustration of the φ structure.
Recently, the other structure of the domain was reported by Tarendeau et al. (25). There are eight amino acid differences among the two structures outside of the N-terminal regions (former: this structure (Protein Data Bank accession code 3cw4); latter: Protein Data Bank accession code 2vy6) A613T, F636L, A661T, V667I, T676I, G682S, A684S, and K702R. A661T, G682S, A684S, and K702R are host range mutations from (avian to avian) to (human to human). Unless these changes occur, the tertiary structure is basically identical to the root mean square deviation 1.103 Å. These host range mutations never affect the tertiary structure, suggesting that they directly interact with other host or virus factors. Their structures also do not include the residues Asp678–Gly685 in the tethering loop.
We identified a characteristic structure in PB2 3/3 in which one helix (aa 589–605) is encircled by a loop (aa 623–635) (Fig. 1, b–d). We denoted this the “φ structure” because its shape resembles this Greek letter (Fig. 1, b and c). In the encircled “φ loop,” two prolines and one phenylalanine (PFXXXXP) are conserved among the influenza A, B, and C virus PB2 subunits (supplemental Fig. 1), suggesting important roles, possibly including structural maintenance, for this region. The residue at position 627 in PB2 has been shown to be a determinant of pathogenicity and host range restriction for the influenza A virus (26–28). Interestingly, lysine 627 is located in the φ loop (Fig. 1, b and c). In the φ helix,“ there is also a conserved sequence GXXYSXhhR (“h” denotes a hydrophobic amino acid), which would be expected to contribute to the essential functions of the overall φ structure in the life cycle of the influenza A virus. Because the side chains of Lys (627: red) QSRMQ (purple) are toward the outside on the flexible φ loop, they would be expected to function in interactions with other molecules. The inherent flexibility of this structure further suggests important roles for the PB2 domain, which include the pathogenicity of this virus, as discussed in the next section.
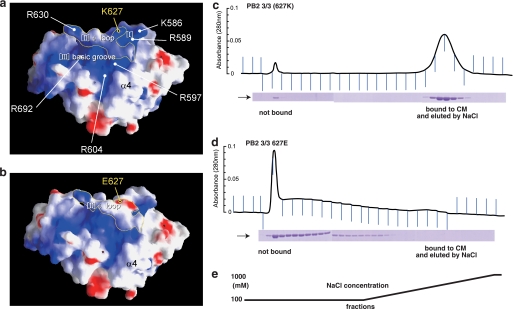
To predict the function of the PB2 3/3 domain, we calculated the electrostatic potentials on its surface using the GRASP program (Fig. 2). One surface of PB2 3/3 (Fig. 2a) is highly basic when compared with the opposite side, suggesting that it may play an important role in the association with RNA primers and/or promoters as described in a later section (Fig. 2a). The φ loop generates a boundary along with a basic groove (Fig. 2a). Thus, the basic surface of PB2 consists of three parts: (I) a small portion separated by the φ loop (Lys586, Arg589), (II) a boundary of the φ loop (Lys627, Arg630), and (III) a basic groove separated by the φ loop (Fig. 2a). This basic surface is also separated from non-basic surfaces by helix 4 (Fig. 2a).
FIGURE 2.
Electrostatic potential of the RNA-binding domain surface of the influenza A virus RNA polymerase PB2 subunit. a, electrostatic potential of PB2 3/3 calculated using the GRASP program. Blue and red indicate basic and acidic surfaces, respectively. Basic regions (I) and (III) are separated by the φ loop (II). b, electrostatic potential of PB2 3/3 Glu627 calculated using the GRASP program. c–e, alteration of the surface electrostatic potentials of PB2 3/3 by substitution of lysine 627 with glutamic acid. The blue line in the upper panel shows the optical density at 280 nm. The lower panel shows PB2 3/3 or PB2 3/3 Glu627 proteins in each fraction analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. PB2 3/3 or PB2 3/3 Glu627 proteins were applied to a CM column and eluted using a gradient concentration of NaCl. PB2 3/3 bound successfully to the CM column and could be subsequently eluted with NaCl (panel c), whereas PB2 3/3 Glu627 did not bind to this resin (panel d). Panel e shows the NaCl concentration in each fraction.
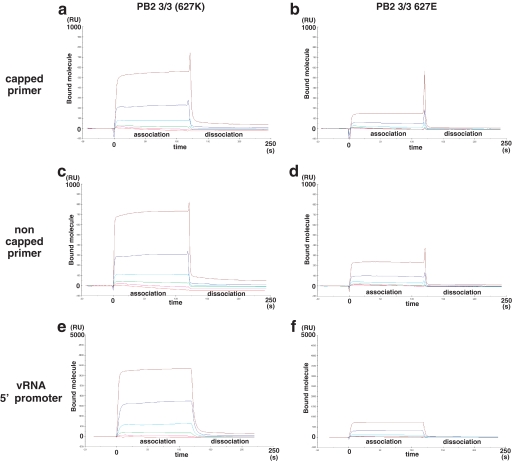
Based upon our prediction that the basic surface would function in RNA binding, we tested whether the recombinant PB2 3/3 product had RNA primer and/or promoter binding activities using surface plasmon resonance analysis. This assay method is also suitable for performing kinetic analysis. The interactions of various concentrations (0.015, 0.044, 0.13, 0.40, 1.2, and 3.6 μm) of the PB2 3/3 protein with synthetic RNA oligomers (capped and non-capped primer RNAs and a 5′-vRNA promoter) were then evaluated (Fig. 3). Capped primer (5′-m7GpppAmAAUACUCAAG-3′), non-capped primer (5′-AmAAUACUCAAG-3′), or vRNA 5′ promoter (5′-AGUAGAAACAAGGCC-3′) were tested in a PB2 3/3 domain protein binding assay (11, 29). An increase in the resonance units indicates an association. The PB2 3/3 protein was found to associate with all of these RNA oligomers in a dose-dependent manner (Fig. 3), indicating that it possesses RNA binding activity. Significantly, this is the first report to show that this domain of PB2 has RNA binding activity.
FIGURE 3.
The role of lysine 627 in the binding of PB2 3/3 to RNA oligomers. RNA binding analyses of PB2 3/3 and PB2 3/3 Glu627 proteins were conducted using a surface plasmon resonance assay. Various concentrations (0.015, 0.044, 0.13, 0.40, 1.2, and 3.6 μm) of recombinant PB2 3/3 and PB2 3/3 Glu627 proteins were incubated with the RNA oligonucleotides that had been immobilized on a sensor chip. The 5′-vRNA promoter (5′-AGUAGAAACAAGGCC-3′), a capped primer (5′-m7GmAAAUACUCAAG-3′), or a non-capped primer (5′-AmAAUACUCAAG-3′) was tested for PB2 3/3 domain protein binding. The vertical axis in each panel indicates the resonance units (RU), which reflect the number of attached molecules observed in real time (horizontal axis, full scale is 250 s). The red, magenta, green, cyan, blue, and brown lines indicate the binding curves for the 0.015, 0.044, 0.15, 0.44, 1.2, and 3.6 μm protein concentrations, respectively. The data shown indicate the resonance units after subtraction of the background values (no RNA measurements). The labels association and dissociation represent the buffer in the presence or absence of the input molecules, respectively. Affinity analysis of the binding of oligomer RNA to recombinant PB2 3/3 protein was performed, and the calculated Kd and Rmax values are shown in supplemental Table 1.
To determine whether any preferences exist for the binding of RNA to the PB2 3/3 domain protein, the Kd values were determined for various RNA molecules using the BIAevaluation program (Fig. 3). The Kd value for 5′-vRNA promoter was calculated as 3.2 μm, whereas those for capped and non-capped primers were found to be 12 and 8.8 μm, respectively. This suggests that PB2 3/3 has 2–3-fold higher binding affinity for vRNA promoter in comparison with capped and non-capped RNA oligomers. Honda and Ishihama's review (9) has suggested that this domain has primer binding activity but did not show any data in support of this. Jung and Brownlee (11) have shown that the PB2 subunit binds to the vRNA promoter but did not show which domain contributes to this activity. We show in our current study that the PB2 3/3 domain has RNA binding activity with a Kd in the μm range. This affinity was found not to be dependent on capping, but our current data do not exclude the possibility that the whole PB2 subunit or trimer RNA polymerase complex binds to capped primers through this domain. The affinity of binding to the vRNA promoter is higher than that found for capped primer RNA, suggesting that this domain facilitates promoter binding with the PB1 subunit (11).
We next focused on the lysine residue at position 627, which is located in the φ loop (Figs. 1, b and c, and 2, a and b), because this residue has been shown to be a determinant of the pathogenicity and host range restriction of the influenza A virus (4–6). When the residue at position 627 is glutamic acid, influenza A virus shows low temperature sensitivity in mammalian cells (30). When this residue is lysine, the virus shows high pathogenicity in mammals (26, 28). Lysine is almost exclusively found in human influenza virus isolates, whereas glutamic acid is almost exclusively found in avian influenza virus isolates at this position. This amino acid was also previously reported to be involved in the interaction with NP (nucleoprotein) protein (30, 31). Based on our data regarding the basic surface of this domain, we predicted that the residue would also contribute to the binding of acidic molecules such as RNA. Thus, to investigate whether lysine 627 contributes to RNA binding activity, we replaced it with glutamic acid at this position in the PB2 3/3 protein (PB2 3/3 Glu627). During the purification of the PB2 3/3 and PB2 3/3 Glu627 proteins using CM column chromatography, we found that they had quite different properties. PB2 3/3 bound to CM resin, whereas PB2 3/3 Glu627 did not (Fig. 2, c–e). This suggests that the substitution of only one lysine at position 627 with glutamic acid has profound effects upon the surface electric charges of this domain, although there are 13 basic residues on the basic surface of the PB2 3/3 domain. Because the lysine at position 627 is located in the center of the basic surface and within the φ loop (Fig. 2a), it may have strong effects upon surface electric charges.
To investigate whether lysine 627 contributes to RNA binding, we examined the PB2 3/3 and PB2 3/3 Glu627 proteins for this activity using a surface plasmon resonance assay for four types of oligonucleotide RNAs, such as primer/promoter RNA (Fig. 3). The RNA binding activity of PB2 3/3 Glu627 was significantly decreased for all RNA oligomers tested when compared with that of PB2 3/3 (Fig. 3 and supplemental Table 1). We thus concluded that residue 627 is indeed involved in RNA binding. The differences in the RNA binding activity of PB2 might therefore contribute to the pathogenicity of influenza in humans. Our current results are also not necessarily inconsistent with previous reports for NP as it is not unusual for a domain to have multiple functions and bind to several molecules. Taken together, our present findings thus indicate that the PB2 surface is a potential candidate for novel anti-influenza A virus drugs from the viewpoint of pathogenicity and host range restriction levels.
To further investigate the functional and structural relationships of the recombinant PB2 3/3 domain, we screened for proteins with a similar tertiary structure using the Matras program (32). The results of these searches indicated that the helix bundles in the N-terminal portion of PB2 3/3 have similarities with that of subunit C from DNA clamp loader (replication factor C). The tertiary structure of DNA clamp loader was reported previously by Kuriyan and colleagues (33) (supplemental Fig. 2a, blue line). The red and blue lines overlap at the helix as shown by the stereo structures (supplemental Fig. 2a). Kuriyan's model illustrates that DNA clamp loader binds to RNA primers, proliferating cell nuclear autoantigen (PCNA), DNA clamp), and DNA polymerase at the initiation of DNA replication (supplemental Fig. 2c). Taken together, our analysis outlines the structural and functional relationship between influenza A virus transcription and cellular DNA replication from the view point of RNA primer binding. Because each of the two domains, influenza RNA polymerase PB2 3/3 and DNA clamp loader, have RNA binding activity, it is probable that these helix bundles are involved with an RNA binding function.
In conclusion, the unique φ-shaped structure within the influenza A virus RNA-dependent RNA polymerase and the associated structure-function relationships between RNA binding and pathogenicity for this enzyme have the potential to lead to novel anti-influenza A virus drugs. The development of such agents may be achieved by performing docking simulations in silico in which the surfaces of these unique domains can be comprehensively tested using small molecule databases.
Supplementary Material
Acknowledgments
We thank N. Igarashi, N. Matsugaki, Y. Yamada, and S. Wakatsuki for data collection using the PF AR-NW12. The influenza (A/PR/8/34) RNA polymerase PB2 plasmid, pBMSA-PB2, was provided by the DNA Bank, RIKEN BioResource Center (Tsukuba, Japan; originally deposited by Dr. Susumu Nakada) with the support of National Bio-Resources Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT).
The atomic coordinates and structure factors (code 3CW4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental figures and a supplemental table.
Footnotes
The abbreviations used are: aa, amino acids; SeMet, seleno-methionine; MAD, multiple anomalous dispersion; vRNA, viral RNA; CM, carboxymethyl.
References
- 1.Taubenberger, J. K., Reid, A. H., Lourens, R. M., Wang, R., Jin, G., and Fanning, T. G. (2005) Nature 437 889-893 [DOI] [PubMed] [Google Scholar]
- 2.Morse, S. S. (2007) Nat. Med. 13 681-684 [DOI] [PubMed] [Google Scholar]
- 3.Horimoto, T., and Kawaoka, Y. (2005) Nat. Rev. Microbiol. 3 591-600 [DOI] [PubMed] [Google Scholar]
- 4.De Clercq, E. (2006) Nat. Rev. Drug Discov. 5 1015-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden, F. G., Atmar, R. L., Schilling, M., Johnson, C., Poretz, D., Paar, D., Huson, L., Ward, P., and Mills, R. G. (1999) N. Engl. J. Med. 341 1336-1343 [DOI] [PubMed] [Google Scholar]
- 6.Reece, P. A. (2007) J. Med. Virol. 79 1577-1586 [DOI] [PubMed] [Google Scholar]
- 7.Izumi, Y., Tokuda, K., O'dell, K. A., Zorumski, C. F., and Narahashi, T. (2007) Neurosci. Lett. 426 54-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, L., Summers, D. F., Peng, Q., and Galarz, J. M. (1995) Virology 208 38-47 [DOI] [PubMed] [Google Scholar]
- 9.Honda, A., and Ishihama, A. (1997) Biol. Chem. 378 483-488 [PubMed] [Google Scholar]
- 10.Crescenzo-Chaigne, B., van der Werf, S., and Naffakh, N. (2002) Virology 303 240-252 [DOI] [PubMed] [Google Scholar]
- 11.Jung, T. E., and Brownlee, G. G. (2006) J. Gen. Virol. 87 679-688 [DOI] [PubMed] [Google Scholar]
- 12.Torreira, E., Schoehn, G., Fernández, Y., Jorba, N., Ruigrok, R. W., Cusack, S., Ortín, J., and Llorca, O. (2007) Nucleic Acids Res. 35 3774-3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarendeau, F., Boudet, J., Guilligay, D., Mas, P. J., Bougault, C. M., Boulo, S., Baudin, F., Ruigrok, R. W., Daigle, N., Ellenberg, J., Cusack, S., Simorre, J. P., and Hart, D. J. (2007) Nat. Struct. Mol. Biol. 14 229-233 [DOI] [PubMed] [Google Scholar]
- 14.Guilligay, D., Tarendeau, F., Resa-Infante, P., Coloma, R., Crepin, T., Sehr, P., Lewis, J., Ruigrok, R. W., Ortin, J., Hart, D. J., and Cusack, S. (2008) Nat. Struct. Mol. Biol. 15 500-506 [DOI] [PubMed] [Google Scholar]
- 15.Obayashi, E., Yoshida, H., Kawai, F., Shibayama, N., Kawaguchi, A., Nagata, K., Tame, J. R., and Park, S. Y. (2008) Nature 454 1127-1131 [DOI] [PubMed] [Google Scholar]
- 16.He, X., Zhou, J., Bartlam, M., Zhang, R., Ma, J., Lou, Z., Li, X., Li, J., Joachimiak, A., Zeng, Z., Ge, R., Rao, Z., and Liu, Y. (2008) Nature 454 1123-1126 [DOI] [PubMed] [Google Scholar]
- 17.Collins, P. J., Haire, L. F., Lin, Y. P., Liu, J., Russell, R. J., Walker, P. A., Skehel, J. J., Martin, S. R., Hay, A. J., and Gamblin, S. J. (2008) Nature 453 1258-1261 [DOI] [PubMed] [Google Scholar]
- 18.Doublié, S. (1997) Methods Enzymol. 276 523-530 [PubMed] [Google Scholar]
- 19.Otowinovski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, Number 4. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 21.Terwilliger, T. C. (2003) Methods Enzymol. 374 22-37 [DOI] [PubMed] [Google Scholar]
- 22.McRee, D. E. (1999) J. Struct. Biol. 125 156-165 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 24.Hendrickson, W. A., and Ogata, C. M. (1997) Methods Enzymol. 276 494-523 [DOI] [PubMed] [Google Scholar]
- 25.Tarendeau, F., Crepin, T., Guilligay, D., Ruigrok, R. W., Cusack, S., and Hart, D. J. (2008) PLoS Pathog. 4 e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subbarao, E. K., London, W., and Murphy, B. R. (1993) J. Virol. 67 1761-1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shinya, K., Hamm, S., Hatta, M., Ito, H., Ito, T., and Kawaoka, Y. (2004) Virology 320 258-266 [DOI] [PubMed] [Google Scholar]
- 28.Hatta, M., Gao, P., Halfmann, P., and Kawaoka, Y. (2001) Science 293 1840-1842 [DOI] [PubMed] [Google Scholar]
- 29.Fechter, P., Mingay, L., Sharps, J., Chambers, A., Fodor, E., and Brownlee, G. G. (2003) J. Biol. Chem. 278 20381-20388 [DOI] [PubMed] [Google Scholar]
- 30.Labadie, K., Dos Santos Afonso, E., Rameix-Welti, M. A., van der Werf, S., and Naffakh, N. (2007) Virology 362 271-282 [DOI] [PubMed] [Google Scholar]
- 31.Ye, Q., Krug, R. M., and Tao, Y. J. (2006) Nature 444 1078-1082 [DOI] [PubMed] [Google Scholar]
- 32.Kawabata, T. (2003) Nucleic Acids Res. 31 3367-3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowman, G. D., O'Donnell, M., and Kuriyan, J. (2004) Nature 429 724-730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.