Abstract
Lipopolysaccharide (LPS) is a major component of the Gram-negative outer membrane and is an important virulence determinant. The O-antigen polysaccharide of the LPS molecule provides protection from host defenses, and the length of O-antigen chains plays a pivotal role. In the Wzy-dependent O-antigen biosynthesis pathway, the integral inner membrane protein Wzz determines the O-antigen chain length. How these proteins function is currently unknown, but the hypothesis includes activities such as a “molecular ruler” or a “molecular stopwatch,” and other possibilities may exist. Wzz homologs are membrane proteins with two transmembrane helices that flank a large periplasmic domain. Recent x-ray crystallographic studies of the periplasmic portions of Wzz proteins found multiple oligomeric forms, with quaternary structures favoring the “molecular ruler” interpretation. Here, we have studied full-length Wzz proteins with the transmembrane portions embedded in lipid membranes. Using electron microscopy and image analysis we find a unique hexameric state rather than differing oligomeric forms. The data suggest that in vivo Wzz proteins determine O-antigen chain length via subtle structure-function relationships at the level of primary, secondary, or tertiary structure within the context of a hexameric complex.
Many bacteria are covered by cell surface polysaccharides, playing diverse roles in the biology of these organisms. In Gram-negative pathogens, the principle surface polymers are capsular polysaccharides (CPSs)2 and lipopolysaccharide (LPS) O-antigens. CPS and LPS are virulence determinants, typically providing protective functions. Successful protection is dependent on optimization of cell surface coverage with polymers of an appropriate chain length.
LPSs are complex glycoconjugates comprising a hydrophobic lipid A that is essential for outer membrane integrity, a core oligosaccharide, and (in many bacterial genera) a long-chain polysaccharide known as the O-antigen (1). The O-antigen extends from the cell surface to form a discrete layer (2, 3). The population of LPS molecules isolated from a growing culture is heterogeneous in size, evident in characteristic ladder patterns in SDS-PAGE profiles (4-6). Each preparation contains molecules consisting of lipid A and all or part of the core region, known as rough LPS (R-LPS), as well smooth LPS (S-LPS), where the lipid A-core is substituted with varying lengths of O-antigen. Although the sizes of the O-antigens vary, they do display a characteristic modal chain length where a substantial number of molecules fall within a discrete and strain-specific range. O-antigens often confer resistance to complement-mediated killing (7) and both the amount and length of the O-antigen are important factors (8-10). For example, in Salmonella enterica serovar Typhimurium, a minimum O-antigen chain length of 4-15 repeat units is necessary for complement resistance (11). However, O-chain length also influences the capacity of this bacterium to enter macrophages.
Many CPS and O-antigens are assembled by a so-called Wzy-dependent pathway (reviewed in Refs. 1, 12). In this pathway, polymer repeat units are assembled at the cytoplasmic membrane on a polyisoprenoid lipid carrier, using sugar nucleotide donors. The intermediates are then exported across the inner membrane by a process requiring an integral membrane protein, designated Wzx. These lipid-linked repeat units provide the substrates for block-wise polymerization in a periplasmic reaction that minimally involves an additional integral protein, Wzy, which gives the pathway its name. The final stages of biosynthesis in the Wzy-dependent pathway are more variable. For LPS, the O-antigen is linked to a preformed lipid A-core molecule prior to surface translocation of the completed molecule. Export of CPS and other polysaccharides to the cell surface is not dependent on a ligation step, and is thought to be linked directly with polymerization.
A member of the polysaccharide co-polymerase (PCP) family of proteins is involved in the Wzy-dependent biosynthesis of many polysaccharides (13). These proteins have a regulatory role in polymerization, and, in the case of CPS systems, the PCP proteins are required for both polymerization and surface export (14, 15). PCP family members (including the PCP-1, PCP-2, and PCP-3 families) are integral cytoplasmic membrane proteins. They share a conserved membrane topology consisting of two transmembrane (TM) helices that flank a large hydrophilic periplasmic domain (PD) containing amino acid sequences that are predicted to form coiled-coils (CCs) and possess a proline-rich consensus motif (PX2PX4SPKX11-GGMXGAG) located at the beginning of the C-terminal TM span (TM2) (13, 16-19). PCP-2 proteins are associated with CPS biosynthesis and are differentiated from PCP-1 and PCP-3 proteins by the possession of a tyrosine kinase domain (20). The kinase domain of PCP-2a proteins such as Wzc (required for synthesis and export of Escherichia coli group 1 CPS) is located on the cytoplasmic C-terminal region. PCP-2b proteins such as CapA (involved in Gram-positive CPS biosynthesis) lack the extended C-terminal domain but possess a cognate cytoplasmic partner with ATPase activity (CapB) (21). PCP-3 proteins belong to Wzy-independent CPS pathways (e.g. E. coli group 2 CPS) (12, 13).
Homologs of the PCP-1a protein, Wzz, regulate the polymerization of LPS O-antigens to generate the characteristic strain-specific modality evident in silver-stained SDS-PAGE profiles. Batchelor et al. (22, 23) first identified a gene that was originally called rol (regulator of O chain length) or cld (chain length determinant) (16) in E. coli. Homologs of rol/cld have now been identified in many Gram-negative bacteria, and the genes have been renamed wzz (24). The SDS-PAGE profiles of LPS isolated from wzz-null mutants are non-modal, where polymerization is terminated prematurely and a random distribution of O-antigen chain lengths favoring a low number of O repeat units in the polysaccharide is observed. In complementation experiments, the expression of heterologous wzz genes in a wzz-deficient background results in O-antigens with chain lengths characteristic of the source of the wzz gene (16, 23, 25, 26).
Hypotheses have been proposed for the mechanism of action of Wzz (16, 17, 27). The original models invoke a coordination of the polymerization step, or the relative activities of the polymerization and ligation reactions. Two possible regulatory strategies have been invoked; a “molecular stopwatch” (16) or a “molecular ruler” (17). However, it is now evident that Wzz operates on the polyisoprenoid lipid-linked polymer prior to the ligation step by WaaL, indicating regulation on the polymerization reaction (28). Structural studies could provide insights into the Wzz mechanism of action. Crystal structures have been obtained for the periplasmic domains of three Wzz homologs (WzzPD) (27). In each case, the domain formed an extended α-helical hairpin connected to an α/β base domain. The protomers assembled into complexes containing three, five, eight, or nine protomers (depending on the particular Wzz homolog), and extended ∼100 Å along the n-fold symmetry axis (27).
From these structural studies it was proposed that the oligomerization state of Wzz homologs defines the molecular ruler responsible for determining O-antigen chain length (27). The formation of oligomeric Wzz complexes has been described by several studies (27-33) but these reports vary greatly in terms of the number of protomers involved. It seems possible that at least part of this variability reflects non-native associations between protomers where constraints normally imposed by membrane localization have been removed. Here, we have studied various full-length Wzz homologs that display radically different oligomeric structures when periplasmic domains alone are expressed and purified (27, 32). We find that when reconstituted in a lipid bilayer, the full-length Wzz homologs display the same quaternary structure.
EXPERIMENTAL PROCEDURES
Cloning wzz Genes—Genes encoding three Wzz homologs were amplified by PCR from genomic DNA from respective strains using complementary primers incorporating overhanging restriction sites (underlined: EcoRI, forward; PstI, reverse) to facilitate cloning and encoding N-terminal polyhistidine tags. The oligonucleotide pair GATCGAATTCACCATGCATCATCATCATCATCATAGAGTAGAAA and AGCTCTGCAGTCACTTCGCGTTGTAATTACGCAGCGC was used to clone the wzzK40 gene from E. coli 2775 O8:K40 (34); the resulting protein contained an N-terminal hexahistidine tag. The wzzST and wzzFepE genes were both cloned from S. enterica serovar Typhimurium LT2 and generated derivatives with decahistidine tags. The genes encoding WzzST and WzzFepE were amplified using primer pairs GATCGAATTCACCATGCATCATCATCATCATCATCATCATCATCATACAGTGGATAGTAATACGTCTTC/GATCCTGCAGTTACAAGGCTTTTGGCTTATAGCTAC and GATCGAATTCACCATGCATCATCATCATCATCATCATCATCATCATCCATCTCTTAATGTAAAACAAG/GATCCTGCAGTCAGACTAACCGTTCATCTATCGCC, respectively. PCR fragments were digested with EcoRI and PstI, and ligated to EcoRI/PstI-digested pBAD24 (35). The resulting arabinose-inducible expression plasmid constructs, pWQ12 (encoding His6-WzzK40), pWQ14 (His10-WzzST), and pWQ15 (His10-WzzFepE) were transformed into E. coli TOP10 (F-mcrA Δ(mrr-hsdRMS-mcrBC) f80ΔlacZM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG; Novagen) by electroporation and gene constructs were verified by sequencing.
Construction of a wzz Deletion Strain of S. enterica serovar Typhimurium LT2—A double (ΔwzzST ΔwzzFepE) mutant was generated using the Lambda Red system (36) using as a parent strain S. enterica serovar Typhimurium LT2 TA262 (obtained from Dr. E. R. Vimr, University of Illinois, Urbana, IL). A chloramphenicol resistance (CmR) cassette with ends complementary to DNA sequences upstream and downstream of the wzzST gene was amplified by PCR from the pKD3 plasmid using the primers GCTTTATGGCTACACTGTCTCTCCCAGCTTCATCCTTTTTTTAGTTAGGGTATCTGTGTAGGCTGGAGCTGCTTCG and CCCGGTTTTTTAATGAGAAATTTTACCTGTCGTAGCCGACCACCATCCGGCAAAGAAGCCATATGAATATCCTCCTTAG (underlined: complementary to pKD3 priming sites). The wzzST gene was replaced in the parent strain by transforming the complementary CmR cassette into cells expressing the recombinase genes from the pKD46 plasmid. Chloramphenicol-resistant colonies were screened by PCR and silver-stained SDS-PAGE to verify introduction of the CmR cassette and loss of LPS modality, respectively. The CmR cassette was then removed by FLP-mediated recombination using the pCP20 plasmid to generate S. enterica serovar Typhimurium CWG859 ΔwzzST. The wzzFepE gene in S. enterica serovar Typhimurium CWG859 was replaced with the CmR cassette using the same approach using the primers GGATAAAGTTTTCAGGTCATACGGCGTGTAGGCTGGAGCTGCTTCG and GGATATCGCTATCCGGCTTTTCGGGTACATATGAATATCCTCCTTG (underlined: complementary to pKD3 priming sites) to amplify the CmR cassette, yielding S. enterica serovar Typhimurium CWG860 ΔwzzST ΔwzzFepE. The cat gene replacing wzzFepE was retained in Salmonella CWG860.
Mutant Complementation Analyses—To assess function of the cloned wzz genes, the respective plasmids were introduced into S. enterica serovar Typhimurium CWG860 ΔwzzSTΔwzzFepE by electroporation (37). Cells were grown in LB containing 100 μg/ml ampicillin with shaking at 37 °C until an A600 nm = 0.6 was reached. Expression of Wzz proteins was then achieved by a further 2 h of incubation following induction with 0.1% (w/v) l-arabinose. LPS profiles were then analyzed from proteinase K-digested whole cell lysates (4). The LPS molecular species were separated by SDS-PAGE using a 4-12% BisTris NuPAGE gel from Invitrogen and visualized by staining with silver (38).
Protein Overexpression and Purification—Wzz proteins were overexpressed in E. coli TOP10 strains harboring their respective pBAD-derivative recombinant expression plasmids. One liter of LB medium containing ampicillin (100 μg/ml) was inoculated with 10 ml of overnight culture and grown with shaking at 37 °C until an A600 nm = 0.6 was reached. Expression of Wzz proteins was then achieved by another 24-h incubation at 20 °C following induction with 0.1% (w/v) l-arabinose. Cells were harvested by centrifugation at 5000 × g for 15 min at 4 °C and stored at -20 °C until required. Cell pellets from 1-liter cultures were resuspended in 25 ml of 50 mm sodium phosphate buffer, pH 7.0, containing 500 mm NaCl, and the cells were disrupted by sonication. Cell-free lysate was collected by centrifugation at 15,000 × g for 30 min. Membranes were collected from the cleared supernatant by centrifugation at 75,000 × g for 1 h at 4 °C. The membrane fractions were solubilized overnight in 10 ml of 50 mm sodium phosphate buffer, pH 7.0, containing 500 mm NaCl, 20 mm imidazole, and 1% (w/v) n-dodecyl-β-d-maltoside (DDM) (Sigma) at 4 °C. Insoluble material was removed by centrifugation at 70,000 × g for 1 h at 4 °C. His6-tagged and His10-tagged proteins were purified from the solubilized membrane fraction by batch incubation with 1 ml of His-select affinity resin (Sigma) for 1 h at 4 °C. The resin-lysate mixture was loaded into 20-ml chromatography columns (Bio-Rad) and equilibrated with 50 mm sodium phosphate buffer, pH 7.0, containing 500 mm NaCl, 20 mm imidazole, and 0.008% (w/v) DDM. Proteins were eluted using an imidazole step gradient. Fractions were monitored for the presence of His-tagged proteins by SDS-PAGE and Western blots probed with anti-His5 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Protein fractions containing the His-tagged Wzz derivatives were pooled and desalted using PD-10 desalting columns (GE Healthcare, Piscataway, NJ) into storage buffer (20 mm Tris-Cl, pH 7.0, 150 mm NaCl) containing 0.008% (w/v) DDM. Samples were concentrated by filtration with 100,000 Da cutoff membranes (Sartorius, Goettingen, Germany). Protein concentrations were determined using a detergent compatible protein assay kit with BSA as a standard (Bio-Rad).
Cross-linking Studies—For cross-linking experiments, Wzz samples were exchanged into 50 mm sodium phosphate buffer (pH 7.0) containing 150 mm NaCl and 0.008% (w/v) DDM using PD-10 desalting columns. Proteins were cross-linked using formaldehyde (1% (v/v), final concentration) at 37 °C. At intervals of 0, 2.5, 5, 10 15, 20, 25, and 30 min, aliquots were removed from the cross-linking mixture and quenched with 1 m Tris-Cl, pH 7.5 (to yield a final concentration of 100 mm). Samples were stored on ice until analysis by SDS-PAGE and perfluoro-octanoic acid (PFO)-PAGE. The conditions for PFO-PAGE were reported previously (39). SDS-PAGE and PFO-PAGE gels were visualized by silver staining (Pierce).
Reconstitution of Wzz in Proteoliposomes—Wzz proteins were reconstituted into lipid bilayers using the detergent adsorption method with hydrophobic beads (Bio-Beads SM) (Bio-Rad) (reviewed in Refs. 40, 41). Solubilized Wzz samples purified from E. coli crude membrane preparations were combined with 5 mg/ml 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) (Avanti Polar Lipids, Alabaster, AL) solubilized in 1% (w/v) octyl-β-d-glucopyranoside (OG) (Sigma-Aldrich) at a series of lipid-to-protein ratios lipid to protein rations ranging from 0.5:1 to 1:1 (w:w) (0.5 mg/ml final protein concentration). Protein reconstitution was initiated by the addition of hydrophobic beads (∼15 beads/100 μl) at room temperature. The formation of Wzz proteoliposomes and membrane sheets containing Wzz complexes was monitored using a transmission electron microscope (EM) (LEO 912AB). Negatively stained specimens were prepared using EM grids that had been glow-discharged for 30 s. Grids were placed carbon side down onto a 10-μl droplet from reconstitution mixture for 30 s and blotted using filter paper (Whatman). Grids coated with sample were placed carbon side down onto a 10-μl droplet of 2% (w/v) uranyl acetate (UA) for 15 s and blotted with filter paper prior to being transferred onto the EM grid holder. Micrographs were recorded using an Olympus Cantega CCD camera at magnifications of 40,000 and 50,000, corresponding to 2.70 Å/pixel and 2.18 Å/pixel, respectively. Micrographs containing two-dimensional crystals were identified and processed using CRISP software (Calidris, Sweden). Regions of 512 by 512 pixels were selected, and the reciprocal lattice parameters refined prior to generation of projection maps.
Cryo-EM—3-μl samples of Wzz proteoliposomes were removed from the 0.5:1 lipid to protein ratio reconstitution reaction, applied to Quantifoil R 2/2 holey carbon-coated EM grids and plunge frozen in liquid ethane using a Vitrobot device (FEI, Hillsboro, OR). Cryo-EM was performed using a Tecnai 200kV electron microscope operating at 200 kV with a liquid nitrogen-cooled stage. Micrographs were recorded using a Gatan 4k CCD at defocus values of 2-5 μm and at a magnification of 29,000, corresponding to a calibrated 3.84 Å/pixel.
Image Processing of Frozen Hydrated Wzz Proteoliposomes—Well-ordered areas of the two-dimensional crystals were interactively boxed out and subjected to three rounds of lattice unbending using the MRC-LMB software (42). The degree of underfocus of the microscope was determined for each crystal using the EMAN ctfit routines (43), and structure factors were then corrected for the contrast transfer function using the MRC-LMB ctfapply routine (42). Structure factors from a crystal displaying the greatest number of high quality (low IQ, 42) reflections were analyzed using the MRC-LMB routine allspace to aid the determination of the two-dimensional plane group symmetry. This identified p2, p3, p312, p321, p6, and p622 plane groups as acceptable, and of these p622 produced the lowest phase residual (10.9°) versus a theoretical target residual of 24.0°. Data for seven separate untilted crystals were merged together using phase origin refinement with a mean interimage phase residual of 27°. After averaging the merged structure factors, a projection map was calculated. The resolution of the structural data was estimated by measuring the increase in mean phase error and decrease in mean redundancy (i.e. number of observations for a given reflection) as a function of resolution shell. These data (see Table 1) implied that the crystals were sufficiently well ordered to provide data to a resolution of about 1/14 Å-1, and this cut-off was therefore used for display of the projection maps.
TABLE 1.
Summary of crystallographic image processing data
| Calibrated pixel size | 3.84 Å |
| Unit cell parameters | a = b = 93.6 Å, γ = 120° |
| Probable plane group | P622 |
| Mean interimage phase residual | 27° |
| Number of crystals analyzed | 7 |
| Resolution shell (Å−1) | Mean redundancy (range 2 to 7) | Mean phase error (range 0 to 180°) |
|---|---|---|
| 1/100-1/32 | 6 | 15° |
| 1/32-1/18 | 5 | 32° |
| 1/18-1/14 | 5 | 56° |
| 1/14-1/12 | 2 | 21° |
Some areas of the two-dimensional crystals displayed mosaicity, and were not suitable for analysis using the crystallographic software. Here, we selected small patches from the two-dimensional array (7 unit cells) and rotationally and translationally aligned the patches using the EMAN image processing package (43).
Three-dimensional Modeling—The hexameric WzzPD model was constructed using PyMOL by manually positioning a rigid model (derived from chain A of WzzEPD, PDB 3B8O; Ref. 27), while monitoring packing with other monomers related by the observed plane group symmetry. The model was optimized to maximize overall similarity with the observed electron density, while simultaneously minimizing steric clashes. The WzzST projection map includes several structural motifs (the transmembrane helices, residues 231-265 that are located at the tip of a putative coiled-coil domain, and a loop encompassing residues 86-94, of a total of 349 amino acids) not modeled in the WzzEPD x-ray structure.
RESULTS
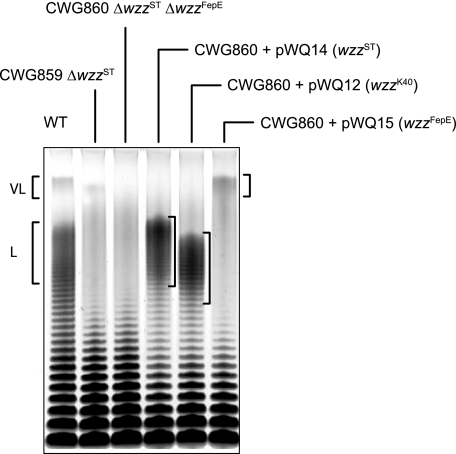
Confirmation of Chain-length Specificity of Wzz Proteins—Two genes encoding Wzz proteins have been identified in the genome of S. enterica serovar Typhimurium; the first is located adjacent to the O-antigen biosynthesis locus (hereafter designated wzzST) (16, 23), and the second is within a cluster of genes associated with ferric enterochelin synthesis and transport (wzzFepE) (44). WzzST directs the formation of “long” (L) O-chain lengths comprising 16-35 repeat units (29) whereas WzzFepE is responsible for “very long” (VL >100 repeat units) chains (44). Modal clusters of LPS molecules containing the L and VL forms are evident in SDS-PAGE of LPS isolated from the wildtype S. enterica serovar Typhimurium (Fig. 1). A double mutant (CWG860) lacking both wzzST and wzzFepE was constructed, and the LPS from this strain displays an SDS-PAGE profile typical of Wzz-deficient mutants, where lower molecular weight forms (<15 repeat units) predominate and modal clusters are absent. The expected modality was restored when the appropriate Wzz derivative was expressed from a plasmid carrying only the cloned gene (Fig. 1). The WzzK40 protein is involved in the biosynthesis of the K40 antigen in E. coli. This polymer exists as an LPS-linked polysaccharide, as well as an LPS-independent polymer known as a group 4 capsule or “O-antigen capsule”. Both forms are synthesized by the same biosynthesis proteins (34). The WzzK40 protein controls modality of the LPS-linked form of K40 polysaccharide (34, 45). When expressed in S. enterica serovar Typhimurium CWG860, WzzK40 determines a modality of 15-28 repeat-units, slightly shorter than those formed by overexpressed WzzST in this background (Fig. 1).
FIGURE 1.
O-antigen chain length is regulated by Wzz. LPS preparations from S. enterica serovar Typhimurium LT2 (WT) and its derivative wzz-null strains (CWG859 and CWG860) were analyzed by SDS-PAGE on a 4-12% Bis-Tris gradient gel and silver-stained. wzz genes were expressed in trans from pBAD-based plasmids in CWG860 and result in the production of long (L) and very long (VL) mode LPS as previously reported (16, 23, 29, 34, 44).
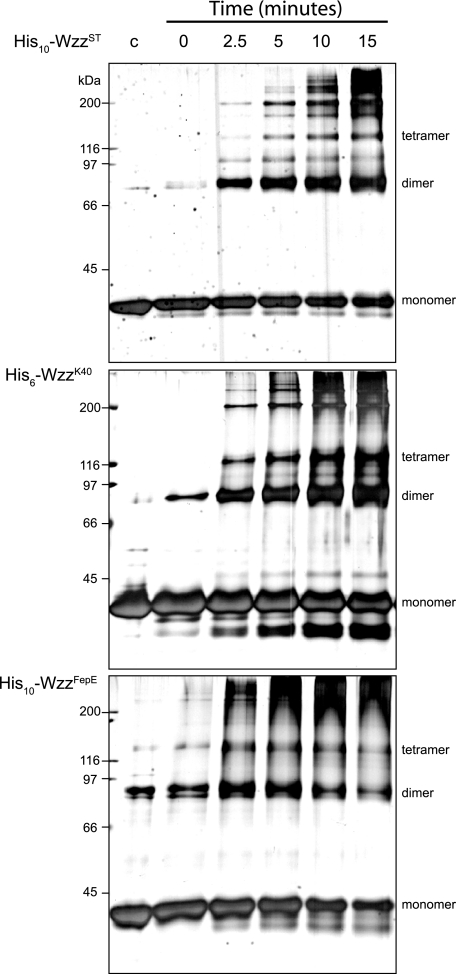
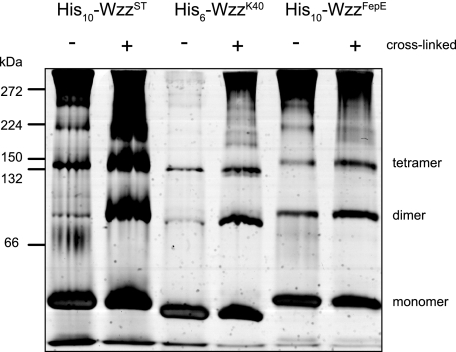
Oligomeric Analysis of Solubilized Wzz—Previous independent reports have used formaldehyde cross-linked cells to show that Wzz proteins form a variety of oligomeric structures (28-30). His6-WzzK40, His10-WzzST, and His10-WzzFepE were overexpressed in E. coli Top10 from their respective pBAD-based plasmids and purified from solubilized crude membrane preparations by Ni2+-NTA affinity chromatography. SDS-PAGE and Western blot analysis showed that the proteins exist in the monomeric forms under denaturing conditions, consistent with the predicted molecular masses of His6-WzzK40 (37.2 kDa), His10-WzzST (37.6 kDa), and His10-WzzFepE (43.7 kDa) (data not shown). A small amount of material consistent with an SDS stable dimer was evident in the His10-WzzST and His6-WzzK40 samples. His10-WzzFepE preparations also revealed a band consistent with a dimer and a band equivalent to a trimer or tetramer. To investigate oligomerization further, the purified Wzz homologs were subjected to formaldehyde cross-linking. Analysis of time course in cross-linking by SDS-PAGE showed a prominent band that is consistent with a dimer, as well as higher molecular weight species (Fig. 2). There were no obvious differences in the cross-linking patterns between the various Wzz homologs. In addition to SDS-PAGE, we took advantage of the relatively non-denaturing conditions of PFO-PAGE (28-30, 46). This approach was used with Wzc to show its tendency to form tetramers (39). PFO-PAGE analysis of His6-WzzK40 His10-WzzST, and His10-WzzFepE revealed bands equivalent to dimeric and tetrameric species as well as some higher mass bands. As for the formaldehyde cross-linked samples, there were no major differences in the PFO-PAGE profiles of the three Wzz homologs (Fig. 3). The cross-linking and PFO-PAGE studies suggest that the detergent-solubilized Wzz proteins form dimeric and tetrameric complexes that can assemble into even higher molecular weight oligomers, as reported elsewhere (28-31, 33). However, neither study is consistent with the hypothesis that the Wzz homologs display radically different oligomeric forms (27).
FIGURE 2.
Wzz oligomerization in time course cross-linking experiments. Silver-stained SDS-PAGE gels of polyhistidine-tagged Wzz proteins incubated at 37 °C with 1% formaldehyde. Aliquots were removed from cross-linking reactions at time intervals and quenched in 100 mm Tris-Cl, pH 7.5. An untreated sample of each homolog was included as a control (lane c).
FIGURE 3.
Membrane-extracted Wzz exists in several oligomeric states. PFO-PAGE analysis, visualized with silver-staining, shows that DDM-solubilized Wzz preparations contain a heterogeneous population of oligomers. Formaldehyde cross-linking does not produce any novel protein bands that are not evident in untreated samples.
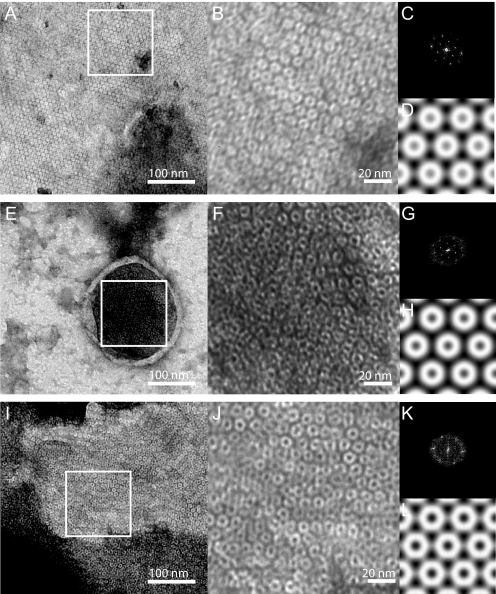
EM of Negatively Stained Wzz Proteoliposomes—To investigate the organization of Wzz under conditions closer to the physiological ones, reconstitution experiments were carried out using purified protein and lipid (DMPC) at lipid to protein ratios ranging from 0.25:1 to 1:1 (w/w). Reconstitution reactions were monitored by EM using negatively stained samples. The reconstitution of His10-WzzST and His6-WzzK40 was successful at lipid to protein ratios of 0.25:1 to 0.75:1. These conditions produced proteoliposomes and large membrane sheets containing a homogeneous population of densely packed protein complexes organized into two-dimensional crystalline arrays. Reconstitution at a lipid to protein ratio of 0.5:1 resulted in the most homogenous population of proteoliposomes with respect to size and protein content (Fig. 4, A and I). His10-WzzFepE did not produce large proteoliposomes with DMPC. However, small membrane patches and small vesicles containing crystalline arrays were observed (Fig. 4E). A comparison of membrane embedded His6-WzzK40, His10-WzzST, and His10-WzzFepE complexes showed that the Wzz homologs all form a ring-like structure with an outside diameter of ∼100 Å. Crystallographic image processing indicated that the unit cells had the dimensions a = b = 95.2 Å (WzzST), a = b = 98.8 Å (WzzK40) and a = b = 107.0 Å (WzzFepE), with γ = 120° for all three. Preliminary analysis on the basis of symmetry-related reflections indicated a p622 plane group as most likely.
FIGURE 4.
Transmission electron microscopy of negatively stained Wzz proteoliposomes. A, reconstituted WzzST forms large two-dimensional crystalline arrays. The inset area (white box) is a 512 × 512 pixel region that is shown at higher magnification in panel B, and was used to determine the reciprocal lattice parameters (panel C) and for generation of a projection map (panel D). The dimensions of the unit cell are a = b = 95.2 Å and γ = 120°. E, WzzFepE proteoliposome and the 512 × 512 pixel region (white box) shown at higher magnification in panel F that was used for reciprocal lattice refinement (panel G) and for generation of a projection map (panel H). The dimensions of the unit cell are a = b = 107.0 Å and γ = 120°. I, WzzK40 proteoliposomes and the 512 × 512 pixel region (white box) shown at higher magnification in panel J that was used for reciprocal lattice refinement (panel K) and for generation of a projection map (panel L). The dimensions of the unit cell are a = b = 98.8 Å and γ = 120°.
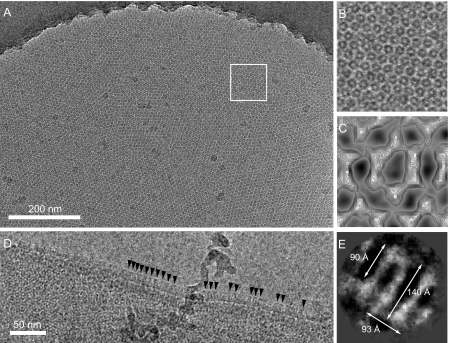
Cryo EM of WzzST and WzzK40 Proteoliposomes in Vitreous Ice—WzzST and WzzK40 proteoliposomes generated in reconstitution experiments at a lipid to protein ratio of 0.5:1 were applied to holey-carbon grids and rapidly frozen for cryo-EM. Fig. 5A shows a representative micrograph of a two-dimensional WzzST crystal present in a large proteoliposome (∼2-μm diameter). The dimensions of the molecules present in the frozen-hydrated WzzST two-dimensional crystal are consistent with those observed in negatively stained proteoliposomes (∼100 Å diameter). Side views of WzzST complexes were also visible at the folded edge of many proteoliposomes (Fig. 5D). Single particle averaging of such side views (Fig. 5E) implies that a large density extending from (∼90 Å × 93Å) the lipid bilayer that has dimensions consistent with the WzzPD crystal structures (FepEPD = 105 Å × 120 Å; WzzEPD = 95 Å × 113 Å; WzzSTPD = 92 Å × 71 Å) (27). Well ordered arrays were not observed for frozen hydrated samples of WzzK40, but these proteoliposomes also contained complexes (data not shown) of dimensions consistent with WzzST.
FIGURE 5.
Cryo-electron microscopy of WzzST proteoliposomes embedded in vitreous buffer. A, large ∼2-μm diameter vesicle containing a two-dimensional Wzz crystalline array. The inset area (white box) is shown at higher magnification in panel B. C, projection map of the Wzz complex after patch averaging without the application of symmetry (p1). D, magnified few of the edge of a flattened vesicle with side views of Wzz molecules emerging from the membrane (arrowheads). E, average of side views generated with EMAN (43), showing the membrane-embedded region (band of density) at the bottom.
Crystallographic Image Processing of WzzST Proteoliposomes—Projection data for the WzzST two-dimensional crystals is compatible with the hypothesis that the arrays are formed from two crystalline layers of Wzz complexes stacked head-to-head on top of each other. This may arise due to the flattening of vesicles within the thin ice layer on the microscope grid, which occurs during specimen preparation, or it may reflect a pre-existing interaction in the specimen. The suggested p622 plane group is also compatible with the appearance of hexagonally shaped complexes with some bilateral symmetry, and a central void (or deficit) of ∼25-Å diameter that are revealed in projection maps calculated with no (P1) symmetry imposed (data not shown). The same hexagonal complexes displaying bilateral symmetry were also observed after averaging small crystal patches (Fig. 5C). The overall dimensions of the unit cell, the hexagonal shape, and the suggested plane group symmetry, would specifically argue against the hypothesis that the crystals were composed of pentameric, octameric, or nonameric complexes as observed for the periplasmic domains in Wzz x-ray structures (27). By far the most likely interpretation is that the two-dimensional Wzz crystals are composed of hexameric complexes stacked on top of each other.
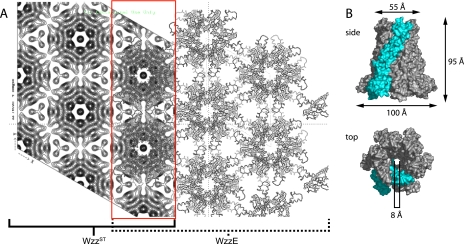
Three-dimensional Model of a WzzPD Hexamer and a Crystal Lattice Model—The WzzPD structures from three homologs solved by x-ray diffraction show that individual protomers share a conserved secondary structure and assemble into complexes with different oligomeric states and external diameters (FepEPD: n = 9, D = 120 Å; WzzEPD: n = 8, D = 113 Å; WzzSTPD: n = 5, D =∼72 Å) (27). A Wzz hexamer was modeled using the WzzEPD protomer (PDB: 3B8O) with application of 6-fold rotational symmetry and energy minimization (Fig. 6B). While the crystal structure of the WzzSTPD protomer is available, we chose the WzzEPD protomer because the WzzSTPD structure lacks information with respect to protomer-protomer contacts that is available in both the FepEPD and WzzEPD structures. Modeling the WzzEPD protomer into a hexamer required less rotational manipulation than generating a model with the FepEPD protomer, producing a more accurate approximation of intermolecular contacts within a hexameric Wzz complex. The model has a base diameter of 100 Å and height of 95 Å. These dimensions match the data obtained from two-dimensional WzzST crystals and molecule side views (Fig. 5, D and E). In addition, the close agreement between the WzzST projection map superimposed with the hexameric model along the 6-fold symmetry axis provides evidence for the interaction between adjacent complexes, whereby helices in the α/β base domain of the Wzz protomer extend laterally and may form the hexagonal contact points in the two-dimensional lattice (Fig. 6A).
FIGURE 6.
The projection map of WzzST is consistent with a hexameric quaternary structure. A, left, projection map of the WzzST crystalline array from cryo-EM after image processing with the application of p622 symmetry. The unit cell dimensions are a = b = 93 Å. Right, model of a p622 crystalline array of WzzEPD hexamers containing two stacked layers. A superimposed region of WzzST and WzzEPD is highlighted in red. B, model of a hexameric Wzz complex generated using the WzzEPD protomer (PDB: 3B8O) shown as molecular surface representations from side and top views. A single protomer is shown in cyan.
DISCUSSION
We used an EM approach to characterize three full-length Wzz homologs reconstituted in lipid bilayers. Micrographs of Wzz proteoliposomes and membrane sheets show that WzzST, WzzFepE, and WzzK40 form two-dimensional crystalline arrays. Crystallographic image processing shows that the dimensions of the unit cell are nearly identical for all three homologs, where the diameter of a Wzz complex is ∼100 Å. To obtain detailed structural information of the Wzz complex, we performed cryo-EM using WzzST and WzzK40 proteoliposomes and generated a projection map of WzzST at 14-Å resolution. We modeled the periplasmic domain of a Wzz hexamer using the Wzz-EPD x-ray structure (27). A two-dimensional crystal lattice generated using this model with the application of p622 symmetry is highly consistent with the WzzST projection map. Side views of WzzST and WzzK40 show that the hexameric complexes extend ∼100 Å from the membrane and has an approximate diameter of ∼90 Å. These dimensions correspond well with the model of hexameric Wzz based on the x-ray data (27).
Formaldehyde cross-linking experiments have shown that Wzz proteins are capable of forming a variety of oligomeric structures, including dimers, tetramers, and hexamers (28, 29, 33). SAXS analysis of purified WzzO86 suggest that solubilized samples form a tetrameric complex with an extended conformation (31) that is consistent with data from size exclusion chromatography and cross-linking experiments using the same homolog (30). Formaldehyde cross-linking experiments using detergent-solubilized Wzz samples showed that the oligomerization of solubilized Wzz is variable. These data suggest that high molecular weight Wzz oligomers represent aggregates of a stable lower molecular weight species, where a dimer appears to be the most likely candidate. PFO-PAGE supports this interpretation, showing that the Wzz oligomers identified in cross-linking reactions are detected under mildly denaturing conditions.
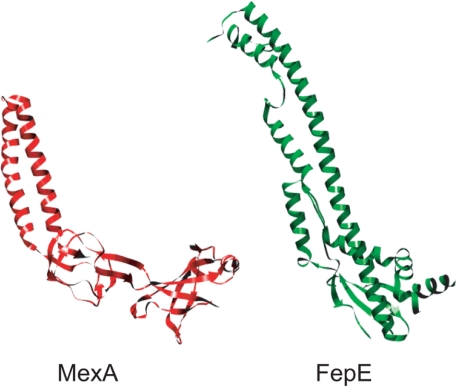
The WzzPD protomers are some-what similar to the protomer structures of AcrA and MexA, two homologs that form the periplasmic adaptor components of the tripartite multidrug efflux pumps in E. coli and Pseudomonas aeruginosa, respectively (Fig. 7). They are required for the assembly of an active channel between a cognate inner membrane antiporter (AcrB and MexB) and the outer membrane channel-forming protein, TolC (47). The MexA and AcrA crystal structures share a conserved architecture consisting of a 50 Å α-helical hairpin domain and basal region consisting of lipoyl and β-barrel domains (48-50). It is proposed that MexA assembles into a ring containing nine molecules that interacts with TolC and MexB (49, 50) and the same is likely true for AcrA. The crystal structures of nonameric and octameric Wzz rings are nearly identical to the proposed model for MexA, suggesting that the self-assembly of periplasmic proteins with extended helical structures into ring-like scaffolds is a wider structural theme.
FIGURE 7.
The periplasmic region of PCP proteins and the periplasmic component of the TolC multi-drug efflux pump share a similar secondary structure. Ribbon representations of the FepEPD protomer (PDB 3B8N) from E. coli O157 and the MexA protomer (PDB 1T5E) from P. aeruginosa. Both proteins possess extended α-helical domains that are proposed to self-assemble into ring-shaped periplasmic complexes (27, 48, 49).
The crystallographic evidence that Wzz protomers interface via a narrow strip along the length of each protomer (27) is comparable to the molecular contacts observed in AcrA and MexA crystals (48, 49, 51). Although the antiparallel interaction observed between AcrA monomers in the crystal structure are not thought to be physiologically relevant, it is possible that similar parallel and antiparallel interactions occur in Wzz proteins extracted from the cytoplasmic membrane. Like Wzz, the analysis of AcrA oligomerization yielded variable results depending on the experimental approach; cross-linking identifies an AcrA monomer, dimer, and trimer (52), while analytical ultracentrifugation and dynamic light scattering indicate that AcrA is monomeric in solution (53). From the collective information available for AcrA and MexA, we speculate that the formation of Wzz tetramers and higher ordered oligomers in solubilized samples is non-physiological, where these structures likely represent a variety of homotypic interactions occurring between α-helical hairpin domains. Data reported by Tocilj et al. (27) implied that the oligomeric status of PCP proteins varies between homologs. The report of multiple FepEPD crystal forms (27) may suggest that the high protein concentration environment required for crystallization mediates the assembly non-physiological oligomers. Our EM data for reconstituted WzzST, WzzFepE, and WzzK40 rather indicates that complexes of the three Wzz homologs have a conserved quaternary structure.
The side views reported in this work provide the first structural information for a full length PCP protein that includes its TM helices interacting with a lipid bilayer. The classification of Wzz proteins into the PCP family of proteins is based on conserved primary sequence characteristics and their involvement in bacterial polysaccharides biosynthesis (13). The common structural features of PCP members are perhaps indicative of a broader functional significance linking these proteins in polysaccharide biosynthesis. In wzz-deficient strains, O polysaccharides are assembled, ligated to lipid A-core and successfully exported to the cell surface, implying that Wzz proteins are not required for polysaccharide translocation. Genetic evidence exists for an interaction between Wzz and a putative Wzy/Wzx complex (54), which would allow Wzz to regulate O-antigen biosynthesis at the polymerization step (16, 17, 27). Bacterial polysaccharide biosynthesis pathways also share a conserved priming reaction that initiates repeat unit assembly. Enzymes belonging to the N-acetylhexosame-1-phosphate transferase (PNPT) and polyisoprenyl-phosphate hexose-1-phosphate transferase (PHPT) families initiate the assembly of all known Wzy-dependent polysaccharides (1, 12, 55-60). Recently, the PHPT from S. enterica serovar Typhimurium (called WbaP) was shown to influence O-antigen chain length determination. The deletion of a putative 146-amino acid periplasmic loop from the central portion of WbaP produced a wzz-like phenotype (61). Thus, Wzz could modulate O-antigen chain length by regulating the flow of repeat units into the polymerization reaction. The exact role will only be resolved by a detailed understanding of the molecular interactions between Wzz and other components in the system. Identification of conserved Wzz hexameric complexes does not resolve these questions but does argue against a role for the precise oligomeric state in determination of modal chain length.
This work was supported by Canadian Institutes of Health Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CPS, capsular polysaccharides; PCP, polysaccharide co-polymerase; TM, transmembrane; LPS, lipopolysaccharide; PD, periplasmic domain; DDM, n-dodecyl-β-d-maltoside; PFO, perfluoro-octanoic acid; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; OG, octyl-β-d-glucopyranoside; EM, electron microscope; PDB, Protein Data Bank.
References
- 1.Raetz, C. R., and Whitfield, C. (2002) Annu. Rev. Biochem. 71 635-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam, J. S., Graham, L. L., Lightfoot, J., Dasgupta, T., and Beveridge, T. J. (1992) J. Bacteriol. 174 7159-7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kastowsky, M., Gutberlet, T., and Bradaczek, H. (1992) J. Bacteriol. 174 4798-4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hitchcock, P. J., and Brown, T. M. (1983) J. Bacteriol. 154 269-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munford, R. S., Hall, C. L., and Rick, P. D. (1980) J. Bacteriol. 144 630-640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palva, E. T., and Makela, P. H. (1980) Eur. J. Biochem. 107 137-143 [DOI] [PubMed] [Google Scholar]
- 7.Joiner, K. A. (1988) Annu. Rev. Microbiol. 42 201-230 [DOI] [PubMed] [Google Scholar]
- 8.Pangburn, M. K. (1989) J. Immunol. 142 2766-2770 [PubMed] [Google Scholar]
- 9.Pangburn, M. K. (1989) J. Immunol. 142 2759-2765 [PubMed] [Google Scholar]
- 10.Grossman, N., Schmetz, M. A., Foulds, J., Klima, E. N., Jimenez-Lucho, V. E., Leive, L. L., and Joiner, K. A. (1987) J. Bacteriol. 169 856-863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray, G. L., Attridge, S. R., and Morona, R. (2006) J. Bacteriol. 188 2735-2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield, C. (2006) Annu. Rev. Biochem. 75 39-68 [DOI] [PubMed] [Google Scholar]
- 13.Morona, R., Van Den Bosch, L., and Daniels, C. (2000) Microbiology 146 1-4 [DOI] [PubMed] [Google Scholar]
- 14.Wugeditsch, T., Paiment, A., Hocking, J., Drummelsmith, J., Forrester, C., and Whitfield, C. (2001) J. Biol. Chem. 276 2361-2371 [DOI] [PubMed] [Google Scholar]
- 15.Collins, R. F., Beis, K., Dong, C., Botting, C. H., McDonnell, C., Ford, R. C., Clarke, B. R., Whitfield, C., and Naismith, J. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 2390-2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastin, D. A., Stevenson, G., Brown, P. K., Haase, A., and Reeves, P. R. (1993) Mol. Microbiol. 7 725-734 [DOI] [PubMed] [Google Scholar]
- 17.Morona, R., van den Bosch, L., and Manning, P. A. (1995) J. Bacteriol. 177 1059-1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker, A., and Puhler, A. (1998) J. Bacteriol. 180 395-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doublet, P., Grangeasse, C., Obadia, B., Vaganay, E., and Cozzone, A. J. (2002) J. Biol. Chem. 277 37339-37348 [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. C., Zheng, J., She, Y. M., and Jia, Z. (2008) EMBO J. 27 1758-1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olivares-Illana, V., Meyer, P., Bechet, E., Gueguen-Chaignon, V., Soulat, D., Lazereg-Riquier, S., Mijakovic, I., Deutscher, J., Cozzone, A. J., Laprevote, O., Morera, S., Grangeasse, C., and Nessler, S. (2008) PLoS Biol. 6 e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batchelor, R. A., Haraguchi, G. E., Hull, R. A., and Hull, S. I. (1991) J. Bacteriol. 173 5699-5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batchelor, R. A., Alifano, P., Biffali, E., Hull, S. I., and Hull, R. A. (1992) J. Bacteriol. 174 5228-5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves, P. R., Hobbs, M., Valvano, M. A., Skurnik, M., Whitfield, C., Coplin, D., Kido, N., Klena, J., Maskell, D., Raetz, C. R., and Rick, P. D. (1996) Trends Microbiol. 4 495-503 [DOI] [PubMed] [Google Scholar]
- 25.Franco, A. V., Liu, D., and Reeves, P. R. (1998) J. Bacteriol. 180 2670-2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klee, S. R., Tzschaschel, B. D., Timmis, K. N., and Guzman, C. A. (1997) J. Bacteriol. 179 2421-2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tocilj, A., Munger, C., Proteau, A., Morona, R., Purins, L., Ajamian, E., Wagner, J., Papadopoulos, M., Van Den Bosch, L., Rubinstein, J. L., Fethiere, J., Matte, A., and Cygler, M. (2008) Nat. Struct. Mol. Biol. 15 130-138 [DOI] [PubMed] [Google Scholar]
- 28.Daniels, C., Griffiths, C., Cowles, B., and Lam, J. S. (2002) Environ. Microbiol. 4 883-897 [DOI] [PubMed] [Google Scholar]
- 29.Daniels, C., and Morona, R. (1999) Mol. Microbiol. 34 181-194 [DOI] [PubMed] [Google Scholar]
- 30.Guo, H., Lokko, K., Zhang, Y., Yi, W., Wu, Z., and Wang, P. G. (2006) Protein Expr. Purif. 48 49-55 [DOI] [PubMed] [Google Scholar]
- 31.Tang, K. H., Guo, H., Yi, W., Tsai, M. D., and Wang, P. G. (2007) Biochemistry 46 11744-11752 [DOI] [PubMed] [Google Scholar]
- 32.Marolda, C. L., Haggerty, E. R., Lung, M., and Valvano, M. A. (2008) J. Bacteriol. 190 2128-2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purins, L., Van Den Bosch, L., Richardson, V., and Morona, R. (2008) Microbiology 154 1104-1116 [DOI] [PubMed] [Google Scholar]
- 34.Amor, P. A., and Whitfield, C. (1997) Mol. Microbiol. 26 145-161 [DOI] [PubMed] [Google Scholar]
- 35.Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995) J. Bacteriol. 177 4121-4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datsenko, K. A., and Wanner, B. L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6640-6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binotto, J., MacLachlan, P. R., and Sanderson, K. E. (1991) Can J. Microbiol. 37 474-477 [DOI] [PubMed] [Google Scholar]
- 38.Tsai, C. M., and Frasch, C. E. (1982) Anal. Biochem. 119 115-119 [DOI] [PubMed] [Google Scholar]
- 39.Collins, R. F., Beis, K., Clarke, B. R., Ford, R. C., Hulley, M., Naismith, J. H., and Whitfield, C. (2006) J. Biol. Chem. 281 2144-2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosser, G. (2001) Micron 32 517-540 [DOI] [PubMed] [Google Scholar]
- 41.Yeager, M., Unger, V. M., and Mitra, A. K. (1999) Methods Enzymol. 294 135-180 [DOI] [PubMed] [Google Scholar]
- 42.Amos, L. A., Henderson, R., and Unwin, P. N. (1982) Prog. Biophys. Mol. Biol. 39 183-231 [DOI] [PubMed] [Google Scholar]
- 43.Ludtke, S. J., Jakana, J., Song, J. L., Chuang, D. T., and Chiu, W. (2001) J. Mol. Biol. 314 253-262 [DOI] [PubMed] [Google Scholar]
- 44.Murray, G. L., Attridge, S. R., and Morona, R. (2003) Mol. Microbiol. 47 1395-1406 [DOI] [PubMed] [Google Scholar]
- 45.Dodgson, C., Amor, P., and Whitfield, C. (1996) J. Bacteriol. 178 1895-1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramjeesingh, M., Huan, L. J., Garami, E., and Bear, C. E. (1999) Biochem. J. 342 119-123 [PMC free article] [PubMed] [Google Scholar]
- 47.Thanabalu, T., Koronakis, E., Hughes, C., and Koronakis, V. (1998) EMBO J. 17 6487-6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akama, H., Matsuura, T., Kashiwagi, S., Yoneyama, H., Narita, S., Tsukihara, T., Nakagawa, A., and Nakae, T. (2004) J. Biol. Chem. 279 25939-25942 [DOI] [PubMed] [Google Scholar]
- 49.Higgins, M. K., Bokma, E., Koronakis, E., Hughes, C., and Koronakis, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9994-9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lobedanz, S., Bokma, E., Symmons, M. F., Koronakis, E., Hughes, C., and Koronakis, V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4612-4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikolosko, J., Bobyk, K., Zgurskaya, H. I., and Ghosh, P. (2006) Structure 14 577-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zgurskaya, H. I., and Nikaido, H. (2000) J. Bacteriol. 182 4264-4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zgurskaya, H. I., and Nikaido, H. (1999) J. Mol. Biol. 285 409-420 [DOI] [PubMed] [Google Scholar]
- 54.Marolda, C. L., Tatar, L. D., Alaimo, C., Aebi, M., and Valvano, M. A. (2006) J. Bacteriol. 188 5124-5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price, N. P., and Momany, F. A. (2005) Glycobiology 15 29R-42R [DOI] [PubMed] [Google Scholar]
- 56.Anderson, M. S., Eveland, S. S., and Price, N. P. (2000) FEMS Microbiol. Lett. 191 169-175 [DOI] [PubMed] [Google Scholar]
- 57.Amer, A. O., and Valvano, M. A. (2001) Microbiology 147 3015-3025 [DOI] [PubMed] [Google Scholar]
- 58.Wang, L., Liu, D., and Reeves, P. R. (1996) J. Bacteriol. 178 2598-2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, L., and Reeves, P. R. (1994) J. Bacteriol. 176 4348-4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valvano, M. A. (2003) Front Biosci. 8 s452-471 [DOI] [PubMed] [Google Scholar]
- 61.Saldias, M. S., Patel, K., Marolda, C. L., Bittner, M., Contreras, I., and Valvano, M. A. (2008) Microbiology 154 440-453 [DOI] [PubMed] [Google Scholar]