Abstract
The genome of the Lyme disease pathogen Borrelia burgdorferi contains about a dozen linear DNA molecules that carry covalently closed hairpin telomeres as a specialized mechanism for dealing with the end-replication problem. The hairpin telomeres are generated from replicative intermediates through a two-step transesterification promoted by the telomere resolvase ResT. Although the genome of B. burgdorferi has been sequenced, the sequence of most telomeres has remained unknown because of difficulties in recovering and completely sequencing the covalently closed hairpin ends. In this study we report a new approach for the direct sequencing Borrelia telomeres and report the sequence, characterization, and in vitro reaction properties of 19 unique telomeres. Surprisingly, a variation of greater than 160-fold in the initial reaction rates of in vitro ResT-mediated telomere resolution was observed between the most active and least active telomeres. Moreover, three of the hairpin telomeres were completely inactive in vitro, but their in vivo functionality was demonstrated. Our results provide important new information on the structure and function of the B. burgdorferi telomeres and suggest the possibility that factors besides the telomere resolvase ResT may influence the reaction in vivo and rescue those telomeres that are not functional in vitro with ResT alone.
The genome of the important Lyme disease pathogen Borrelia burgdorferi is unusual in several respects. The genome is segmented and includes almost two dozen DNA molecules, including a chromosome and over 20 plasmids (1-3). The plasmids have also been referred to as mini-chromosomes (4) and essential genetic elements (5) because a number of them carry crucial information for survival in a tick vector or mammalian host (6-17). In addition to the unprecedented number of DNA molecules, the genome is also unusual in that it contains both circular and linear DNA molecules, present in roughly equal numbers (12 and 10, respectively). Linear DNA molecules are rarely found in bacteria, and those in B. burgdorferi are terminated by covalently closed hairpin ends (18, 19). These hairpin “telomeres” offer a unique solution to the end-replication problem (20) as described below.
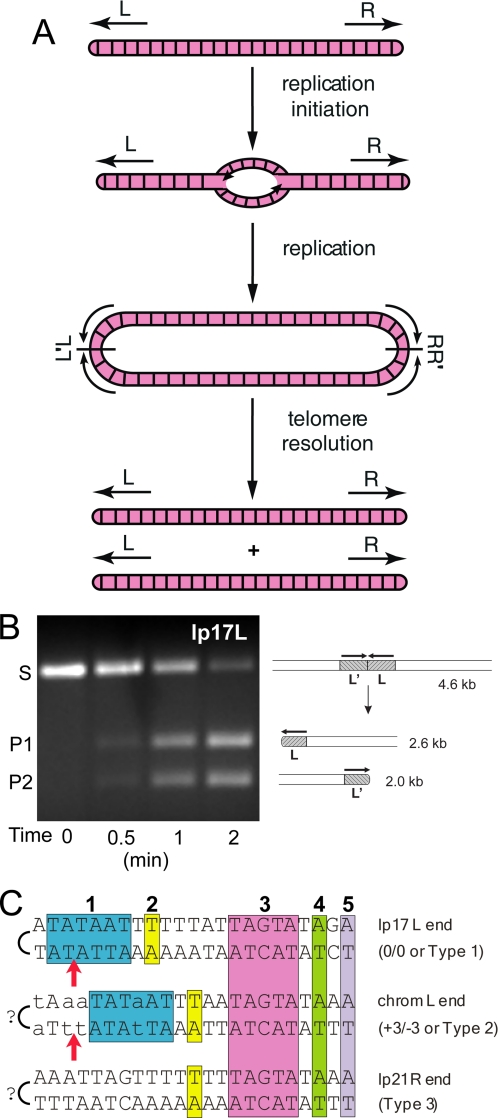
Replication of the linear DNA molecules in Borrelia species (Fig. 1A) occurs via bidirectional replication from a central origin (21, 22). Copying of the DNA on each side of the linear molecule results in the formation of a dimer junction or replicated telomere (L′L and RR′) in the replicative intermediate. These regions are subsequently processed in a DNA breakage and reunion reaction referred to as telomere resolution (23), promoted by the telomere resolvase ResT (24). This essential enzyme (6) performs a two-step transesterification to generate covalently closed hairpin ends from the replicated telomeres using a reaction mechanism similar to that of tyrosine recombinases and type IB topoisomerases (24-26). Purified ResT efficiently promotes telomere resolution in vitro when provided with a replicated telomere substrate on a linear plasmid, as shown in Fig. 1B.
FIGURE 1.
Replication pathway for linear Borrelia replicons and telomere types. A, arrows labeled L and R indicate the hairpin telomeres at the left and right ends of the linear DNA of B. burgdorferi, respectively. The lines bisecting the head-to-head (L′L) and tail-to-tail (RR′) replicated telomere junctions are axes of 180° rotational symmetry. It is not yet known whether replication is completed before telomere resolution (as shown) or whether processing can occur at either end before replication is completed at the other. See Ref. 25 for further details. B, in vitro telomere resolution of B. burgdorferi telomere lp17L by ResT. A linearized plasmid substrate (S) containing the lp17L replicated telomere (L′L) is resolved by ResT into two hairpin-containing products (P1 and P2). A schematic of the substrate and products is shown to the right of the ethidium bromide-stained 1% agarose gel used to analyze the products. C, three telomere types reported thus far in B. burgdorferi (27, 35, 37). A representative example of each telomere type is shown. The regions conserved among the sequenced telomeres are boxed and colored with numbers above the sequence. Positions of known sequence variability in or to the left of box 1 in the Type 2 telomere family are indicated by lowercase letters. The arrows indicate the position of cleavage by the telomere resolvase, ResT (27). The question marks indicate the uncertainty about the last nucleotide of the telomere ends as sequencing was not performed directly, and mung bean nuclease was used to open the hairpin ends before cloning. A and C of are reprinted from Ref. 25.
Although the ResT protein has been the subject of a number of investigations (24, 26-33), and a crystal structure has been reported for a phage telomere resolvase (34), little information has been available for the telomeres of B. burgdorferi, which is a powerful model organism for studying hairpin telomere biology. The first and only telomeres to be sequenced in their entirety were those from the small linear plasmid lp17 (19) using chemical sequencing that allowed direct sequencing around the covalently closed hairpin telomeres. Five other telomere sequences from the sequenced strain B31 have been reported by cloning the telomeres after treatment with a single strand-specific nuclease (2, 35-38). However, the precise sequence at the hairpin turnaround has remained unconfirmed because of the use of a single strand-specific nuclease in the cloning process and the possibility of nucleotide loss prior to the ligation step.
The sequencing of B. burgdorferi telomeres has revealed three telomere types based upon the presence, absence, or position of the box 1 and 2 region (25, 27, 37), as shown in Fig. 1C. The box 3-5 region appears to be invariant in position in all telomeres studied to date with the qualification that the position of the telomere ends (excepting lp17) remains tentative. The position of DNA cleavage is also invariant and occurs between positions 3 and 4 from the axis of symmetry in three different replicated telomere substrates (24, 27).
In this study we report the sequence of the full complement (excepting lp5) of telomeres from B. burgdorferi strain B31 and describe their similarities, differences, and the ability to serve as substrates in telomere resolution. Surprisingly, a 160-fold range in the in vitro activity of the telomeres was observed, including three telomeres with no detectable in vitro activity, although these telomeres were shown to be active in vivo.
EXPERIMENTAL PROCEDURES
B. burgdorferi Strains and Growth Conditions—B. burgdorferi strains used in this study were cultivated in liquid BSK-II + RS (BSK-II medium (39) supplemented with 6% rabbit serum (Cedarlane Laboratories)) and incubated in 1.5% CO2 at 35 °C. B31-A is a high passage noninfectious clone of B31 (40), whereas B31-5A4 and 5A10 are infectious and noninfectious clones, respectively, derived from B31 (12).
Direct Sequencing of Telomeres—B. burgdorferi plasmid DNA was prepared from 200-ml cultures using a Qiagen plasmid midi kit. Prior to lysis, bacterial pellets were washed with 20 ml of 50 mm Hepes-NaOH, pH 7.6, + 50 mm NaCl. For a single sequencing reaction, 2.3 μg of B. burgdorferi plasmid DNA was treated with 4 units of mung bean nuclease (New England Biolabs) in a 50-μl reaction using the manufacturer's buffer conditions and incubated at 30 °C for 15 min. Reactions were immediately purified by phenol chloroform extraction and ethanol precipitation. DNA pellets were resuspended in 10 μl of H2O. Sequencing was performed by the University of Calgary DNA Sequencing Facility using the following genomic sequencing conditions: 1-2 μg of template DNA, 30 pmol of PAGE purified primer (see Table 1), 4 μl of Terminator Ready Reaction Mix (Big Dye version 3.1, from ABI), 4 μl of Big Dye Terminator Sequencing Buffer, and 1 μl of ThermoFidelase (Fidelity Systems Inc.) in a total volume of 20 μl. Cycling conditions were as follows: initial denaturation for 5 min at 96 °C, followed by 80 cycles of denaturation at 96 °C for 40 s, annealing at 56 °C for 30 s, and extension at 60 °C for either 1.5 or 4 min, depending upon read length.
TABLE 1.
Oligonucleotides used for direct sequencing of telomeres
| Telomere | Primer | Sequence, 5′ to 3′ | Coordinatesa |
|---|---|---|---|
| lp17L | B694 | gatattatagctcctattaaacttgc | 240-215 |
| lp17Rb | B693 | ggggggatgtaaattaatac | 16681-16700 |
| lp21L | B867 | ggcacagtattttttaaatttttttagcg | 733-705 |
| lp21R | B866 | cgttttgtatgttgcagtaaaggggc | 18470-18495 |
| lp25L | B868 | ggtagaaaaaaagaactgagatattgc | 345-319 |
| lp25R | B869 | ctattgttgatataacactaggag | 24086-24109 |
| lp28-1L | B870 | ctaaaacaaaaccttaacgaaagg | 1003-980 |
| lp28-1L 4 | B890 | ccgttgttcaacgtataagaaaatatatc | 427-399 |
| lp28-1L 5 | B1015 | ggtgcagacaaaattaattatgtagc | 516-541 |
| lp28-1L 6 | B891 | ctctaaagattaaccctaataactac | 34-59 |
| lp28-2L | B885 | ctaaattgcaagacatcaatcc | 285-264 |
| lp28-2R | B887 | gtaagaatttcaaaagaagttataaatttg | 29749-29778 |
| lp28-3L | B883 | catttcgggttaaagttttatc | 147-126 |
| lp28-3R | B897 | ggtgtttgttgcaagtagattag | 28489-28511 |
| lp28-4L | B880 | ccaaaattataaagtttggaattttaaagttaacac | 218-183 |
| lp28-4R | B1348 | ctcagcatcttttttaaatc | 27060-27079 |
| lp36L | B879 | gattacaagacatcaatcctccc | 282-260 |
| lp36R | B878 | ctggcgaggggagccatgagtataatcg | 36344-36371 |
| lp38L | B876 | ccacttcggtttggtctagatg | 161-140 |
| lp38R | B877 | caagtgcaataattgtagttgaac | 38714-38737 |
| lp54L | B875 | gaccatttattaatggcattaagc | 264-241 |
| lp54R | B873 | gagaaattgtgcccatcgc | 53339-53357 |
| lp56L | B882 | gtaaataggtataatagaccagttcc | 507-482 |
| lp56Rb | B874 | gataatcaataaacctgatcaacg | 52821-52844 |
The coordinates used here are those of the complete plasmid sequences, including the hairpin telomeres. The sequences noted are the actual primers used and the coordinates correspond to the bottom strand for the L telomeres and the top strand for the R telomeres. The complete plasmid sequences, including telomeres, can be found at University of Calgary website.
A unique sequence for the previously sequenced lp17R and lp56R could not be found.
Sequencing of lp28-4R—A unique primer could not be found for the lp28-4 right end, because of sequence homology to the right end of lp56 and lp36. To circumvent this difficulty, the right end of lp36 was removed by targeted deletion (21, 23) in a strain lacking lp56 (B. burgdorferi 5A10 (12)). Briefly, a region of lp36 from 33,033 to 34,430 bp was PCR-amplified from B. burgdorferi plasmid DNA using primers B1260 (5′cctcataaagtattgtctggc3′) and B1261 (5′cttgtatcttcccatgatagc3′). The PCR product was then blunt end-cloned into pJET1.2 to produce pYT128. This construct was then digested with NcoI and XhoI and directionally cloned into NcoI/XhoI-digested pGCL47-4 to generate pYT129. This construct was used to transform B. burgdorferi 5A10 to delete the right end of lp36 by telomere resolution of the inserted replicated telomere. Plasmid DNA from this lp36 right end minus derivative of strain 5A10 was then mung bean nuclease-treated and used as template DNA for DNA sequencing as described above.
Construction of Replicated Telomere Plasmids—Each telomere resolution substrate was constructed from a pair of synthetic oligonucleotides (see Table 2), corresponding to the elucidated sequence of each B. burgdorferi telomere. Each oligonucleotide pair was phosphorylated, annealed, and ligated. The ligation was subsequently digested with BamHI and then ligated to BamHI-cut pKK81, as described previously (23). Ligations were precipitated and used to transform an Escherichia coli sbc mutant strain. Plasmid DNA from this strain was prepared from 50-ml cultures according to the GeneElute HP plasmid midiprep protocol (Sigma) or from 5-ml cultures using the Qiagen miniprep protocol. The replicated telomere inserts were sequenced after plasmid preparation to confirm the integrity of the constructs, and subsequently used in both in vitro and in vivo telomere resolution assays, as described below.
TABLE 2.
Oligonucleotides (Oligo) used for replicated telomere plasmid substrate construction in E. coli
| Telomere | Plasmid | Oligo 1 | Oligo 2 | Oligo 1 sequence | Oligo 2 sequence | Strain |
|---|---|---|---|---|---|---|
| ChromL | pYT92 | OGCB253 | OGCB254 | gatcctttttatactattaaattatatt | tataaatataatttaatagtataaaaag | GCE1708 |
| ChromR | pYT94 | OGCB287 | OGCB288 | gatccattctatactaattaaaaattat | atatataatttttaattagtatagaatg | GCE1710 |
| lp17L | pYT1 | OGCB17 | OGCB18 | gatccactctatactaataaaaaattat | atatataattttttattagtatagagtg | GCE203 |
| lp17Ra | pYT95 | OGCB289 | OGCB290 | gatccactttgtactaaataaatattat | atatataatatttatttagtacaaagtg | GCE1711 |
| lp21Lb | pYT96 | OGCB291 | OGCB292 | gatccactctatactaataaaaaattat | atatataattttttattagtatagagtg | GCE1712 |
| lp21R | pYT97 | OGCB293 | OGCB294 | gatccgctttatactaaaaaaaactaat | ttaaattagttttttttagtataaagcg | GCE1713 |
| lp25L | pYT91 | OGCB285 | OGCB286 | gatcctaactatactatttaaaaattta | tatataaatttttaaatagtatagttag | GCE1707 |
| lp25R | pYT105 | OGCB309 | OGCB310 | gatcctctctatactaataaaaaaatat | ttaaatatttttttattagtatagagag | GCE1721 |
| lp28-1L | pYT98 | OGCB295 | OGCB296 | gatcctatctatactaattaaaaattat | atatataatttttaattagtatagatag | GCE1714 |
| lp28-1R | pYT93 | OGCB255 | OGCB256 | gatcctatatatactaaaagataataaa | tatatttattatcttttagtatatatag | GCE1709 |
| lp28-2L | pYT109 | OGCB317 | OGCB318 | gatcctatttatactaattaataataaa | ttaatttattattaattagtataaatag | GCE1725 |
| lp28-2R | pYT102 | OGCB303 | OGCB304 | gatcctttctatactaattaataattgt | atatacaattattaattagtatagaaag | GCE1718 |
| lp28-3L | pYT104 | OGCB307 | OGCB308 | gatccactctatactaataaaaaattat | atatataattttttattagtatagagtg | GCE1720 |
| lp28-3R | pYT103 | OGCB305 | OGCB306 | gatccaatttatactaagtaataataat | atatattattattacttagtataaattg | GCE1719 |
| lp28-4Lb | pYT96 | OGCB291 | OGCB292 | gatccactctatactaataaaaaattat | atatataattttttattagtatagagtg | GCE1712 |
| lp28-4R | pYT139 | OGCB356 | OGCB357 | gatcctcattatactaaaagataataaa | ttaatttattatcttttagtataatgag | GCE1753 |
| lp36L | pYT90 | OGCB283 | OGCB284 | gatcctatttatactaattaataataaa | ttaatttattattaattagtataaatag | GCE1706 |
| lp36R | pYT107 | OGCB313 | OGCB314 | gatcctcattatactaaaagataataaa | tatatttattatcttttagtataatgag | GCE1723 |
| lp38L | pYT131 | OGCB339 | OGCB340 | gatcctttttatactaaaaaattgtaaa | ttaatttacaattttttagtataaaaag | GCE1747 |
| lp38R | pYT100 | OGCB299 | OGCB300 | gatccctattatactaaaagataataaa | ttaatttattatcttttagtataatagg | GCE1716 |
| lp54L | pYT134 | OGCB345 | OGCB346 | gatccttagtatactaatagattatctt | catgaagataatctattagtatactaag | GCE1750 |
| lp54R | pYT132 | OGCB341 | OGCB342 | gatccttagtatactaataaactcttat | tataataagagtttattagtatactaag | GCE1748 |
| lp56L | pYT99 | OGCB297 | OGCB298 | gatccgcattatactaaaagataataaa | ttaatttattatcttttagtataatgcg | GCE1715 |
| lp56Ra | pYT95 | OGCB289 | OGCB290 | gatccactttgtactaaataaatattat | atatataatatttatttagtacaaagtg | GCE1711 |
| lp17L + 1 | pYT9 | OGCB33 | OGCB34 | gatccactctatactaataaaaaattata | ttaatataattttttattagtatagagtg | GCE257 |
| lp17L − 1 | pYT17 | OGCB49 | OGCB50 | gatccactctatactaataaaaaatta | tatataattttttattagtatagagtg | GCE300 |
| lp25L + 1 | pYT117 | OGCB333 | OGCB334 | gatcctaactatactatttaaaaatttat | atatataaatttttaaatagtatagttag | GCE1733 |
| lp25L − 1 | pYT118 | OGCB335 | OGCB336 | gatcctaactatactatttaaaaattt | atataaatttttaaatagtatagttag | GCE1734 |
| lp25R + 1 | pYT113 | OGCB325 | OGCB326 | gatcctctctatactaataaaaaaatatt | ttaaaatatttttttattagtatagagag | GCE1729 |
| lp25R − 1 | pYT114 | OGCB327 | OGCB328 | gatcctctctatactaataaaaaaata | ttaatatttttttattagtatagagag | GCE1730 |
| lp28-1R + 1 | pYT111 | OGCB321 | OGCB322 | gatcctatatatactaaaagataataaat | atatatttattatcttttagtatatatag | GCE1727 |
| lp28-1R − 1 | pYT112 | OGCB323 | OGCB324 | gatcctatatatactaaaagataataa | atatttattatcttttagtatatatag | GCE1728 |
| lp56L + 1 | pYT115 | OGCB329 | OGCB330 | gatccgcattatactaaaagataataaat | ttaaatttattatcttttagtataatgcg | GCE1731 |
| lp56L − 1 | pYT116 | OGCB331 | OGCB332 | gatccgcattatactaaaagataataa | atatttattatcttttagtataatgcg | GCE1732 |
Telomere sequences for lp17R and lp56R are identical and therefore represented by the same replicated telomere plasmid construct, pYT95.
Telomere sequences for lp21L and lp28-4L are identical and therefore represented by the same replicated telomere plasmid construct, pYT96.
In Vitro Telomere Resolution Reaction Conditions—B. burgdorferi ResT was expressed in E. coli and purified as described previously (24). Telomere resolution reactions contained 25 mm Tris-HCl (pH 8.5), 100 mm NaCl, 1 mm EDTA, 100 μg/ml bovine serum albumin, 5 mm spermidine, and 5-10 μg/ml PstI-linearized substrate DNA. Time course reactions contained 200 nm ResT and were incubated at 30 °C with 20-μl aliquots removed at the time intervals indicated in the figures. Reactions were performed in duplicate or triplicate and stopped by the addition of SDS to a final concentration of 0.2-0.5%. Samples were resolved on 20-cm 0.8-1% agarose gels in 1× TAE buffer at 80-100 V for 2 h. The gels were stained with ethidium bromide, and fluorescence of DNA bands was quantified using AlphaInnotech software. The percentage of telomere resolution was determined by dividing the fluorescence of the reaction products by the total fluorescence (products plus substrate).
In Vivo Telomere Resolution Assay—Plasmid constructs (50 μg) carrying the telomeres to be tested (see Fig. 6) were used to transform electrocompetent B. burgdorferi B31-A, as reported previously (23). The electroporated cells were resuspended in 1 ml of BSK-II + RS and then transferred to 9 ml of BSK-II + RS. Following a 24-h recovery at 35 °C, the 10-ml culture was added to 100 ml of BSK-II + RS supplemented with 200 μg/ml kanamycin and then distributed into 96-well plates in 250-μl aliquots. Plates were incubated at 35 °C for 10 days, at which time yellow wells containing live B. burgdorferi were diluted 1 to 100 in 200 μl of BSK-II + RS and grown to mid-log phase. Diluted cultures were screened for the kanamycin resistance gene in a GeneAmp PCR System 9700 (Applied Biosystems) using 1 μl of BSK-II culture and 30 pmol of each primer (B70, CATATGAGCCATATTCAACGGGAAACG; B71, AAAGCCGTTTCTGTAATGAAGGAG) in a final reaction volume of 20 μl. The PCR conditions were 94 °C for 15 min, followed by 25 cycles of 94 °C for 30 s, 50 °C for 30 s, and 68 °C for 1 min. Total reactions were analyzed on 1.5% agarose gels in TAE buffer and visualized by ethidium bromide staining. Plasmid DNA from kanamycin-positive B. burgdorferi clones was prepared and analyzed by field inversion gel electrophoresis. Using an MJ Research PPI-200 programmable power inverter, ∼250 ng of plasmid DNA was separated on a 0.65% Seakem agarose gel at 80 V for 40 min, followed by 21 h with Program 0 and buffer recirculation.
FIGURE 6.
B. burgdorferi telomere sequence alignment. The telomere Type (1, 2, or 3) is shown in the left column. Telomere sequences are arranged in descending order, according to the initial rate of resolution, as determined in Fig. 4. The initial rate, expressed in femtomoles/min is shown in the right column and telomere sequences are aligned with the hairpins (or symmetry axis in the replicated telomeres) to the left. The telomeres shown are half of the actual replicated telomere substrates used in the telomere resolution reactions. The colored boxes labeled 1 and 3 refer to previously identified regions of sequence homology, with some modifications. The original box 1 sequence, TATAAT is indicated by a light blue box, whereas the newly identified box 1 sequence, TATTAT is shown in dark blue. The homology box 3 region has been expanded from the five-nucleotide sequence TAGTA to the eight-nucleotide sequence TTAGTATA. The telomere sequences of lp17L, lp17R, lp21R, lp28-1R, lp56R ChromL, and ChromR have been reported previously (2, 19, 35-38).
RESULTS
Direct Genomic Sequencing of the B. burgdorferi Telomeres—To sequence the telomeres of B. burgdorferi, we utilized the previously sequenced B31 strain (1, 2). This allowed for design of sequencing primers for the telomeric regions based upon an adjacent previously reported sequence. Our sequencing approach is shown in Fig. 2A. Total plasmid DNA was first treated with mung bean nuclease to open the hairpin ends. The DNA was subsequently used directly as a template in sequencing reactions using genomic sequencing conditions as described under “Experimental Procedures.” The use of Thermofidelase, a thermostable topoisomerase (Fidelity Systems, Inc.), was essential for the generation of high quality sequence in the genomic sequencing reactions and greatly increased read quality and length. Fig. 2B shows primary data for the telomeres of lp17, lp21R, and lp21L. The hairpin ends of lp17 are the only B. burgdorferi telomeres that have been sequenced in their entirety by chemical sequencing (19). The sequence generated by our genomic sequencing approach corresponds to the previously reported lp17 sequence. At the end of the sequence the addition of two nontemplated A residues followed by the termination of sequence output was observed.
FIGURE 2.
Direct sequencing of B. burgdorferi telomeres. A, schematic of the sequencing strategy. B. burgdorferi linear plasmid DNA treated with mung bean nuclease to open the covalently closed hairpin ends was used as template in a DNA sequencing reaction with primers specific for the left or right end of each plasmid. B, electropherograms of telomere sequencing reactions from lp17 left end, lp21 left end, and lp21 right end. The positions of sequence homology boxes are indicated above each sequence, whereas the end of the telomere is marked by a dotted line. In some instances, one or two nontemplated A residues were added by the polymerase during the cycle sequencing.
The middle panel of Fig. 2B shows the sequence output for lp21R, a telomere also previously sequenced (37). Again, there were nontemplated residues followed by a cessation in sequence output. At the end of the lp21R telomere, we observed the addition of nontemplated A residues at both the last nucleotide and at the position beyond. The sequence we obtained was identical to that previously reported by mung bean nuclease treatment followed by cloning and sequencing (37). The reason for the presence of a mixed read (A and T) for the terminal nucleotide probably results from mung bean cleavage to remove the terminal nucleotide from some hairpins, followed by addition of a nontemplated A at this position. Based upon our observation of the addition of nontemplated A residues beyond and sometimes at the terminal nucleotide, in cases where a unique A was the output for the terminal nucleotide the sequence was read as an A. However, when mixed reads were observed at the terminal nucleotide, A residues were ignored. The assignment of the telomere ends in our study was made by the sequence output. The end point of each telomere agreed with the previous tentative observation of a spacing of 14 nucleotides between the telomere end and the start of box 3 (see Fig. 1B), as reported for all known B. burgdorferi telomeres (2, 19, 35-38). The validity of the 14-nucleotide spacing assignment is further addressed below. The bottom panel of Fig. 2B shows the sequence output for the left end of lp21, a previously unsequenced telomere.
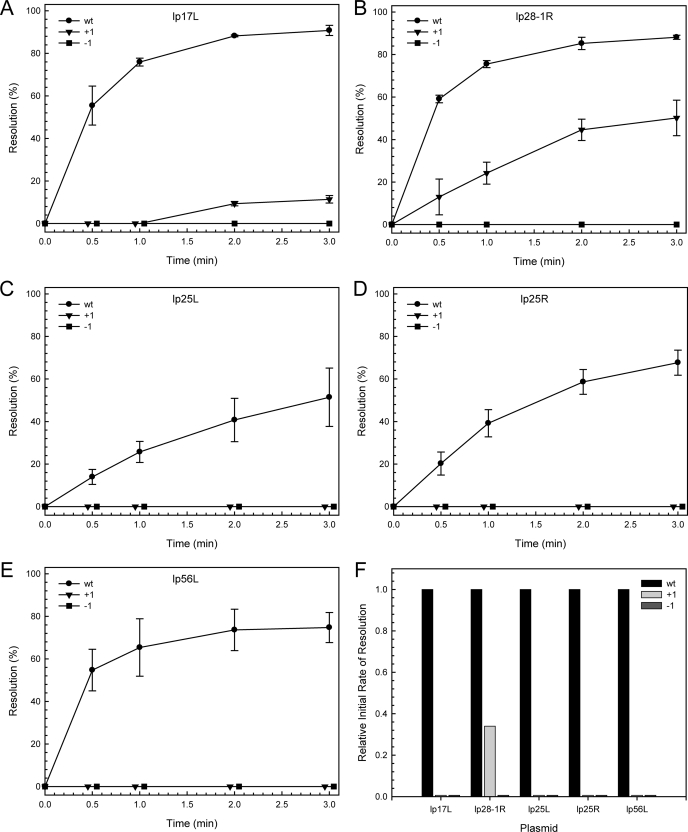
To further address the issue of the 14-nucleotide spacing between box 3 and the terminal nucleotide of the telomeres, we generated plasmids carrying each of five sequenced replicated telomeres (lp17L, lp25L, lp25R, lp28-1R, and lp56L) in their wild-type configuration or with a single nucleotide addition (+1) or deletion (-1) adjacent to the axis of symmetry. Plasmids carrying the replicated telomeres were linearized and used as substrates for in vitro telomere resolution reactions as shown in Fig. 3, A-E. In comparison with the wild-type telomeres, the +1 and -1 derivatives showed little or no activity, except for the +1 derivative of lp28-1R, which showed an initial rate of about 30% that of the wild-type telomere. A summary of the relative initial rates of resolution for each of the telomeres is shown in Fig. 3F. The reason for the residual activity in the +1 telomere from lp28-1R may be related to the fact that lp28-1R is a highly active telomere and displays one of the highest initial rates of all the B. burgdorferi telomeres, as discussed below.
FIGURE 3.
In vitro telomere resolution of wild type, +1, and -1 telomere substrates. The kinetics of in vitro telomere resolution on wild-type, +1, and -1 versions of lp17L (A), lp28-1R (B), lp25L (C), lp25R (D), and lp56L (E) are shown. The ± notation indicates the insertion or deletion of a nucleotide on each side at the axis of symmetry (to the left of box 1). The data were derived from quantification of ethidium bromide stained agarose gels similar to that shown in Fig. 1B (see “Experimental Procedures”). F illustrates the relative initial rate of resolution for each substrate as compared with the corresponding wild-type telomere. Experiments were performed in triplicate. The error bars indicate the standard deviation.
Our conclusion from Fig. 3 and our sequencing data is that B. burgdorferi telomeres display an invariant spacing between the terminal nucleotide and box 3 and that this spacing is an important feature for substrate utilization by ResT. The previously reported sequences of B. burgdorferi telomeres cloned with a mung bean nuclease digestion step therefore appear to be complete sequences. Using the approach outlined above, we determined the sequence of all unknown telomeres in the prototype strain B31, with the exception of lp5 for which no unique sequencing primer could be found. The sequences have been deposited in Gen-Bank™ under the accession numbers FJ472325-FJ472342. In addition, the complete linear B31 plasmid sequences with their telomeres can be obtained from the website of G. Chaconas at the University of Calgary.
There are several other points worthy of note from our sequencing studies. A unique primer could not be found to the right end of lp28-4 because of sequence homology with lp56 and lp36. Therefore, to sequence lp28-4, we made use of a B31 clone lacking lp56 (12) from which we subsequently removed the right end of lp36 through targeted deletion (21, 23) (see under “Experimental Procedures”). A unique primer was generated for lp28-44 in the resulting strain, and a good quality sequence was obtained. In addition, our sequencing studies revealed an additional 675 bp of previously unreported (41) DNA sequence with an open reading frame at the left end of lp28-1. Finally, a unique set of 19 telomeres was sequenced with several telomere duplications. The lp17L telomere sequence was identical for lp28-3L, lp28-4L, and lp21L. The lp28-2L telomere was shared by lp36L, and the match between lp17R and lp56R has been reported previously (19).
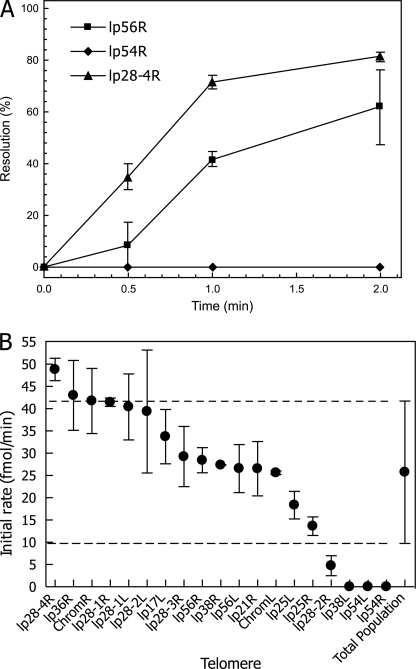
Activity of the B. burgdorferi Telomeres—To investigate the requirements for telomere function, we constructed plasmids (Table 2) carrying each of the 19 unique B31 telomeres in their replicated form (L′L or RR′, see Fig. 1) for use in telomere resolution assays. The plasmids were linearized with PstI and used in standard telomere resolution reactions with ResT as described under “Experimental Procedures.” The results were surprising in that a large variability in initial rates was observed for the different telomeres. Fig. 4A shows a kinetic profile of three replicated telomere substrates in in vitro telomere resolution reactions as follows: the highly active lp28-4R, the intermediate lp56R, and the inactive lp54R. Initial reaction rates were determined from kinetic profiles of the complete set of unique telomeres, and the data are presented in Fig. 4B. A range of 160-fold or greater was observed in the initial rate between the most active (lp28-4) and the three inactive telomeres. The three inactive telomeres displayed no detectable activity under reaction conditions where the stimulatory agents bovine serum albumin and spermidine were present.
FIGURE 4.
In vitro telomere resolution of all B. burgdorferi telomeres by ResT. A, kinetics of telomere resolution are shown for three representative plasmid substrates containing the replicated telomeres from lp28-4R (high activity), lp56R (intermediate activity), and lp54R (no activity). The average initial rates of ResT-mediated in vitro resolution are shown for all B. burgdorferi telomeres in B. Error bars indicate the standard deviation observed when assays were performed with two independently prepared DNA substrates. The average initial rate of resolution for the entire population of telomeres is shown at the far right. Dotted lines indicate the upper and lower limits of the average activity range. The lp17L telomere is identical to lp28-3L, lp28-4L, and lp21L, and the lp28-2L telomere matches lp36L. The lp17R telomere is also found at lp56R (19).
The lack of activity of three telomeres as substrates in in vitro telomere resolution was surprising and prompted us to test whether these telomeres were active in vivo. Therefore, we performed an in vivo telomere resolution assay (21, 23). Each inactive telomere was inserted, in its replicated form, into a plasmid carrying a recombination target region from lp17 (Fig. 5A). The constructs were used to transform B. burgdorferi B31-A, resulting in integration of the transforming plasmids into lp17. The resulting integrants with active telomeres would serve as substrates for telomere resolution at the integrated telomeres, resulting in the deletion in the left end of lp17. Plasmid DNA was prepared from transformants and analyzed by pulse field gel electrophoresis as shown in Fig. 5B. The position of lp17 in the B31-A parent DNA sample is shown in Fig. 5B, lane 1. Fig. 5B, lane 2, contains DNA from B31-A transformed with pKK81, a control plasmid lacking a telomere, and provides a marker for an unresolved plasmid integrant. Fig. 5B, lane 3, shows DNA from a positive control plasmid carrying the lp17L telomere (23) where telomere resolution has occurred, and the left end of lp17 has been deleted. Fig. 5B, lanes 3-6, contain DNA from B31-A transformed with plasmids carrying the three inactive telomeres, lp38L, lp54L, and lp54R, respectively. In all three cases telomere resolution occurred in vivo with the telomeres that were completely inactive in the in vitro assay. We therefore conclude that although these three telomeres are inactive in the in vitro assay, they nonetheless represent bona fide Borrelia telomeres that function in vivo.
FIGURE 5.
In vivo telomere resolution of telomere substrates inactive in vitro. A, schematic of in vivo telomere resolution assay. A plasmid (pGCL10-2) with a replicated telomere (red) also carries a recombination target sequence from near the left end of lp17 (green) and a kanamycin resistance gene (blue). Following transformation of B. burgdorferi B31-A, the plasmid recombines with the lp17 target, generating a plasmid integrant. When the replicated telomere present on the construct is processed by the B. burgdorferi telomere resolution machinery, the left end of lp17 is deleted to generate a resolved plasmid. The ability of different replicated telomeres to be resolved in vivo is assayed by insertion of the desired replicated telomere sequence into lp17, by recombination as shown in the schematic, reproduced from Ref. 23. B, field inversion gel electrophoresis of DNA from B. burgdorferi B31-A transformed with plasmids containing different replicated telomeres in the assay plasmid shown in A. pKK81 is the parental vector and does not contain a telomere. pGCL10-2 contains the left end telomere from lp17. The migration positions of wild type lp17, unresolved lp17 integrant, and resolved lp17 integrants are indicated to the left of the gel.
DISCUSSION
In the work described here we used a direct genomic sequencing approach coupled with prior mung bean nuclease treatment of B. burgdorferi plasmids to sequence the hairpin telomeres of the linear plasmids. We also investigated the spacing requirement between the hairpin ends and the box 3 sequence recognized by ResT, in order to establish a fixed 14-nucleotide distance and to define precisely the position of the terminal nucleotide. These results allowed the confirmation of previously reported telomere sequences that were tentative in the hairpin turnaround region. The approach used here will be useful for sequencing hairpin telomeres from other Borrelia species and other organisms where they exist.
Telomere Features—The sequence of the 19 unique telomeres is aligned in Fig. 6 in order of descending reactivity from the top. The sequences of several telomeres are identical to one another (lp17L = lp28-3L = lp28-4L = lp21L; lp282L = lp36L; lp17R = lp56R) as indicated in the figure. Several interesting points emerge from analysis of the telomere alignment. The first is that the original box 1 sequence (Fig. 6, TATAAT, light blue) is present in only 5 of the 19 unique telomeres. In all cases except one (ChromL), it is positioned to start with the penultimate nucleotide of the telomere. The second is that a modified box 1 (Fig. 6, TATTAT, dark blue) is found in 7 of the 19 unique telomeres, and its start is at position 4 in all cases. The third is that the 5-nucleotide box 3 sequence (TAGTA) can be expanded to an 8-nucleotide sequence (TTAGTATA). This exact sequence is present in 16 of the 19 telomeres with a single base pair mismatch in the 3 remaining telomeres. The universality of this sequence is in agreement with its importance as the recognition site for ResT binding as determined by mutagenesis of the site as well as DNase footprinting (27, 31).
Three Telomere Types—The set of 24 hairpin telomeres from the B. burgdorferi type strain B31 now allows for further analysis of the previous telomere classification scheme shown in Fig. 1C. The new data support the presence of three telomere classes with an approximately equal number of telomeres in each, but allow for more stringent descriptive criteria. All telomere types possess the now expanded box 3 sequence (TTAGTATA) at positions 14-21. The Type 1 telomere carries the original box 1 sequence (TATAAT) at positions 2-7, and the Type 2 telomere exhibits the new modified box 1 sequence (TATTAT) at positions 5-10, and the Type 3 telomere does not have either box 1. The only telomere that does not fit neatly into this classification is the chromosome left end (ChromL), which has the original box 1 motif of a Type 1 telomere located at positions 5-10, the characteristic location for Type 2 telomeres. This telomere is likely a Type 2 telomere that has acquired a single base pair mutation within box 1, and it seems most logical to group this telomere within the Type 2 category because of the location of its box 1.
Telomere Reactivity—Surprisingly, a very large range in initial reaction rates, at least 160-fold between the most active and the least active telomeres, was observed for the set of 19 unique telomeres. No apparent difference existed between the Type 1 and Type 2 telomeres, with a less than 2-fold variance between members of these two classes and an interspersed rank order in terms of their reactivity (Fig. 6). In contrast, the Type 3 telomeres were clustered at the lower end of the reactivity scale, and all three inactive telomeres were Type 3 telomeres and did not contain a clearly defined box 1. Therefore, the box 1 sequence appears to play a significant role in telomere reactivity as indicated by an earlier mutagenic study (27). Nonetheless, four of the Type 3 telomeres displayed activity in vitro, whereas three were completely inactive. The more active Type 3 telomeres (lp21R, lp25L, and lp25R) all carried an uninterrupted stretch of 5-7 T residues to the left of box 3, which could conceivably play a role in stimulating substrate activity; however, further studies will be required to test this possibility. Finally, we could not correlate telomere reactivity with other sequence features, including the nucleotides flanking the cleavage site (positions 3 and 4), or the sequence of the sticky ends resulting from DNA cleavage.
Plasmid Stability, Telomere Exchanges, and in Vivo Rescue—The linear B. burgdorferi plasmids display widely variable stability properties during in vitro propagation of the organism. Some plasmids are easily lost, such as lp28-1, lp56, and lp25 (12). Interestingly, of these, lp28-1 carries two highly active telomeres. The plasmids lp56 and lp25, which are also easily lost upon in vitro passage, carry two intermediate level and low reactivity telomeres, respectively. In contrast, lp54, which carries two telomeres that are completely unreactive in vitro, is rarely, if ever, lost upon continuous passage (12, 42). Therefore, there appears to be no correlation between plasmid instability and low in vitro reactivity of its telomeres.
Another fascinating aspect of the B. burgdorferi genome is the extensive mosaic distribution of DNA sequences near the telomeres of almost every linear plasmid (1, 35, 37). These telomere exchanges are believed to be promoted by reversal of the telomere resolution reaction to fuse two unrelated linear plasmids, followed by mutation of the dimer junction to block resolution of the fused structure (25, 29). The collection of telomere sequences from B. burgdorferi B31 now establishes a set of rules for any subsequent telomere exchanges based upon the complementarity of the sticky ends generated upon telomere cleavage and predicts which linear plasmids may exchange genetic information with each other through reversal of the telomere resolution reaction by ResT.
Finally, the intriguing question remaining to be answered is why three telomeres that are functional in vivo are completely inactive under standard in vitro reaction conditions. It is tempting to speculate that additional host factors may be involved in the telomere resolution reaction in vivo for some or all of the hairpin telomeres. Studies are currently underway in an attempt to identify such factors.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ472325-FJ472342.
This research was supported in part by the Canadian Institutes of Health Research Grant MOP-53086, the Canada Research Chairs Program, and the Alberta Heritage Fund for Medical Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Casjens, S., Palmer, N., Van Vugt, R., Huang, W. H., Stevenson, B., Rosa, P., Lathigra, R., Sutton, G., Peterson, J., Dodson, R. J., Haft, D., Hickey, E., Gwinn, M., White, O., and Fraser, C. M. (2000) Mol. Microbiol. 35 490-516 [DOI] [PubMed] [Google Scholar]
- 2.Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., White, O., Ketchum, K. A., Dodson, R., Hickey, E. K., Gwinn, M., Dougherty, B., Tomb, J. F., Fleischmann, R. D., Richardson, D., Peterson, J., Kerlavage, A. R., Quackenbush, J., Salzberg, S., Hanson, M., van Vugt, R., Palmer, N., Adams, M. D., Gocayne, J., Weidman, J., Utterback, T., Watthey, L., McDonald, L., Artiach, P., Bowman, C., Garland, S., Fujii, C., Cotton, M. D., Horst, K., Roberts, K., Hatch, B., Smith, H. O., and Venter, J. C. (1997) Nature 390 580-586 [DOI] [PubMed] [Google Scholar]
- 3.Miller, J. C., Bono, J. L., Babb, K., El-Hage, N., Casjens, S., and Stevenson, B. (2000) J. Bacteriol. 182 6254-6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G., and Zuckert, W. R. (1997) Nature 390 553, 555 [DOI] [PubMed] [Google Scholar]
- 5.Stewart, P. E., Byram, R., Grimm, D., Tilly, K., and Rosa, P. A. (2005) Plasmid 53 1-13 [DOI] [PubMed] [Google Scholar]
- 6.Byram, R., Stewart, P. E., and Rosa, P. (2004) J. Bacteriol. 186 3561-3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimm, D., Eggers, C. H., Caimano, M. J., Tilly, K., Stewart, P. E., Elias, A. F., Radolf, J. D., and Rosa, P. A. (2004) Infect. Immun. 72 5938-5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm, D., Tilly, K., Bueschel, D. M., Fisher, M. A., Policastro, P. F., Gherardini, F. C., Schwan, T. G., and Rosa, P. A. (2005) J. Med. Entomol. 42 676-684 [DOI] [PubMed] [Google Scholar]
- 9.Labandeira-Rey, M., and Skare, J. T. (2001) Infect. Immun. 69 446-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal, U., Yang, X., Chen, M., Bockenstedt, L. K., Anderson, J. F., Flavell, R. A., Norgard, M. V., and Fikrig, E. (2004) J. Clin. Investig. 113 220-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purser, J. E., Lawrenz, M. B., Caimano, M. J., Howell, J. K., Radolf, J. D., and Norris, S. J. (2003) Mol. Microbiol. 48 753-764 [DOI] [PubMed] [Google Scholar]
- 12.Purser, J. E., and Norris, S. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13865-13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, X. F., Pal, U., Alani, S. M., Fikrig, E., and Norgard, M. V. (2004) J. Exp. Med. 199 641-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jewett, M. W., Lawrence, K., Bestor, A. C., Tilly, K., Grimm, D., Shaw, P., VanRaden, M., Gherardini, F., and Rosa, P. A. (2007) Mol. Microbiol. 64 1358-1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Revel, A. T., Blevins, J. S., Almazan, C., Neil, L., Kocan, K. M., de la Fuente, J., Hagman, K. E., and Norgard, M. V. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6972-6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strother, K. O., Broadwater, A., and De Silva, A. (2005) Vector Borne Zoonotic Dis. 5 237-245 [DOI] [PubMed] [Google Scholar]
- 17.Jewett, M. W., Byram, R., Bestor, A., Tilly, K., Lawrence, K., Burtnick, M. N., Gherardini, F., and Rosa, P. A. (2007) Mol. Microbiol. 66 975-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbour, A. G., and Garon, C. F. (1987) Science 237 409-411 [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch, J., and Barbour, A. G. (1991) J. Bacteriol. 173 7233-7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson, J. D. (1972) Nat. New Biol. 239 197-201 [DOI] [PubMed] [Google Scholar]
- 21.Beaurepaire, C., and Chaconas, G. (2005) Mol. Microbiol. 57 132-142 [DOI] [PubMed] [Google Scholar]
- 22.Picardeau, M., Le Dantec, C., and Vincent, V. (2000) Microbiology 146 305-313 [DOI] [PubMed] [Google Scholar]
- 23.Chaconas, G., Stewart, P. E., Tilly, K., Bono, J. L., and Rosa, P. (2001) EMBO J. 20 3229-3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobryn, K., and Chaconas, G. (2002) Mol. Cell 9 195-201 [DOI] [PubMed] [Google Scholar]
- 25.Chaconas, G. (2005) Mol. Microbiol. 58 625-635 [DOI] [PubMed] [Google Scholar]
- 26.Deneke, J., Burgin, A. B., Wilson, S. L., and Chaconas, G. (2004) J. Biol. Chem. 279 53699-53706 [DOI] [PubMed] [Google Scholar]
- 27.Tourand, Y., Kobryn, K., and Chaconas, G. (2003) Mol. Microbiol. 48 901-911 [DOI] [PubMed] [Google Scholar]
- 28.Bankhead, T., and Chaconas, G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 13768-13773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobryn, K., and Chaconas, G. (2005) Mol. Cell 17 783-791 [DOI] [PubMed] [Google Scholar]
- 30.Kobryn, K., Burgin, A. B., and Chaconas, G. (2005) J. Biol. Chem. 280 26788-26795 [DOI] [PubMed] [Google Scholar]
- 31.Tourand, Y., Lee, L., and Chaconas, G. (2007) Mol. Microbiol. 64 580-590 [DOI] [PubMed] [Google Scholar]
- 32.Bankhead, T., Kobryn, K., and Chaconas, G. (2006) Mol. Microbiol. 62 895-905 [DOI] [PubMed] [Google Scholar]
- 33.Tourand, Y., Bankhead, T., Wilson, S. L., Putteet-Driver, A. D., Barbour, A. G., Byram, R., Rosa, P. A., and Chaconas, G. (2006) J. Bacteriol. 188 7378-7386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aihara, H., Huang, W. M., and Ellenberger, T. (2007) Mol. Cell 27 901-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casjens, S., Murphy, M., DeLange, M., Sampson, L., van Vugt, R., and Huang, W. M. (1997) Mol. Microbiol. 26 581-596 [DOI] [PubMed] [Google Scholar]
- 36.Hinnebusch, J., Bergstrom, S., and Barbour, A. G. (1990) Mol. Microbiol. 4 811-820 [DOI] [PubMed] [Google Scholar]
- 37.Huang, W. M., Robertson, M., Aron, J., and Casjens, S. (2004) J. Bacteriol. 186 4134-4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, J. R., Hardham, J. M., Barbour, A. G., and Norris, S. J. (1997) Cell 89 275-285 [DOI] [PubMed] [Google Scholar]
- 39.Barbour, A. G. (1984) Yale J. Biol. Med. 57 521-525 [PMC free article] [PubMed] [Google Scholar]
- 40.Bono, J. L., Elias, A. F., Kupko, J. J., III, Stevenson, B., Tilly, K., and Rosa, P. (2000) J. Bacteriol. 182 2445-2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casjens, S. (2000) J. Mol. Microbiol. Biotechnol. 2 401-410 [PubMed] [Google Scholar]
- 42.Lawrenz, M. B., Kawabata, H., Purser, J. E., and Norris, S. J. (2002) Infect. Immun. 70 4798-4804 [DOI] [PMC free article] [PubMed] [Google Scholar]