Abstract
Cytoplasmic linker protein 170 (CLIP-170) is the prototype microtubule (MT) plus-end tracking protein (+TIP) and is involved in regulating MT dynamics. A comprehensive understanding of the process by which CLIP-170 tracks MT plus ends would provide insight into its function. However, the precise molecular mechanism of CLIP-170 +TIP behavior is unknown, and many potential models have been presented. Here, by separating the two CLIP-170 CAP-Gly domains and their adjacent serine-rich regions into fragments of varied size, we have characterized the minimal plus-end tracking unit of CLIP-170 in vivo. Each CLIP-170 fragment was also characterized for its tubulin polymerization activity in vitro. We found that the two CAP-Gly domains have different activities, whereas CAP-Gly-1 appears incompetent to mediate either +TIP behavior or MT nucleation, a CLIP-170 fragment consisting of the second CAP-Gly domain and its adjacent serine-rich region can both track MT plus ends in vivo and induce tubulin polymerization in vitro. These observations complement recent work on CLIP-170 fragments, demonstrate that CAP-Gly motifs do not require dimerization for +TIP and polymerization-promoting activities, and provide insight into CLIP-170 function and mechanism.
Microtubules (MTs)2 play essential roles in vital cellular processes, including intracellular transport, cell division, and signal transduction (1–4). They are highly dynamic polymers, constantly shifting between phases of growing and shortening in a behavior termed “dynamic instability” (1, 2, 5–7). MT dynamics are regulated spatially and temporally, but the mechanisms of this regulation remain poorly understood. Because molecular events at the MT plus end govern whether the MT grows or shrinks, proteins that localize specifically to the plus end are expected to play a significant role in the control of MT dynamics. These proteins have been classified as MT plus-end tracking proteins (+TIPs), because they dynamically track the tips of growing MTs in vivo.
As expected, +TIPs contribute to the regulation of MT dynamics (5–9). For example, the +TIPs CLIP-170, EB1, p150Glued, and CLIP-associated proteins promote MT polymerization (5–14). Conversely, the +TIPs KLP10A and KLP59C increase MT depolymerization; KLP10A stimulates catastrophe, whereas KLP59C suppresses rescue (15). Many +TIPs interact with each other and/or other proteins, forming a loose network of protein-protein interactions known as the “MT plus-end complex” (8, 9, 16). One way to better understand the significance and activity of this complex is to elucidate the activities and mechanisms of the individual proteins in this complex. We have chosen to focus on CLIP-170 because of its history as the first identified +TIP and because of its interactions with other +TIPs (14, 17–19).
CLIP-170 (cytoplasmic linker protein 170) was originally identified as a linker between MTs and endocytic carrier vesicles (20), before later being identified as the prototype +TIP (21). CLIP-170 has also been implicated in interactions between the cell cortex and MTs (22). The CLIP-170 structure is composed of an N-terminal MT-binding domain, a central coiled-coil domain that allows homodimerization, and a C-terminal metal-binding domain (Fig. 1A) (23, 24). Previous work has shown that the N-terminal head domain (H1-(1–350)) is sufficient for MT plus-end tracking activity (10, 25).
FIGURE 1.
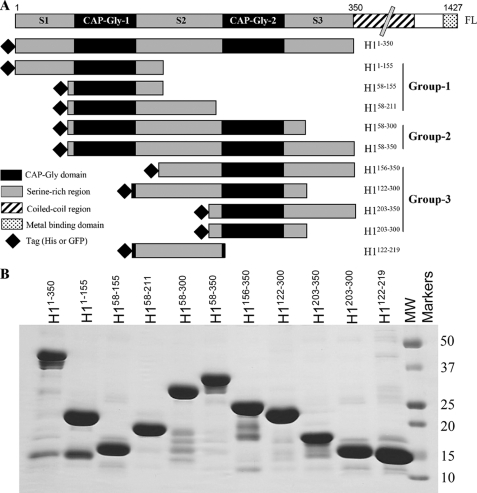
Schematic diagram and purification of recombinant CLIP-170 fragments. A, schematic representation of the CLIP-170 constructs used in this study. Numbers represent the amino acid numbers of the corresponding constructs. FL, full length. B, SDS-PAGE of purified His-tagged proteins (∼5.0 μg). Molecular weight markers and their molecular masses are indicated at the right side of the figure in kDa. The additional bands are degradation products commonly observed in bacterially expressed CLIP-170 preparations (24, 25).
H1-(1–350) is characterized by two conserved CAP-Gly (cytoskeleton-associated protein glycine-rich) domains (23, 24, 26) surrounded by three basic serine-rich regions (Fig. 1A). In addition to CLIP-170, CAP-Gly domains are present in many other proteins, including p150Glued, CLIP-115, CLIPR-59, and tubulin folding cofactors B and E (17, 27–31). The CLIP-170 CAP-Gly domains are well conserved from yeast to humans (27, 30–33), whereas the surrounding serine-rich regions are poorly conserved at the primary sequence level (34).
Previous reports have shown that the CAP-Gly domains are largely responsible for mediating interactions between CLIP-170 (or its close relative CLIP-115) and MTs (17, 27, 28, 32, 33). The CAP-Gly domains of CLIP-170 have also been found to mediate binding to other proteins, including EB1 (18, 35). However, very little is known about the role of the serine-rich regions except in the case of CLIP-115, where they have been shown to contribute to MT interactions (28).
Here we have characterized the activities of the individual CAP-Gly domains of CLIP-170 and their surrounding serinerich regions by dividing the CLIP-170 head domain (H1-(1– 350)) into different subdomains. Specifically, the two conserved CAP-Gly domains have been separated to determine whether one or both structures are essential for MT plus-end tracking. In addition, the roles of the different serine-rich regions are examined either alone or in combination with their adjacent CAP-Gly domain to better define the role of these regions in MT assembly and plus-end tracking.
Using a combination of live-cell imaging and standard biochemical analysis, we demonstrate that there are significant differences in the plus-end tracking and tubulin polymerization activities of CAP-Gly-1 and CAP-Gly-2. Moreover, contrary to a previous report (36), we provide evidence that a fragment consisting of a single CAP-Gly domain (CAP-Gly-2 in combination with its adjacent serine-rich domain) is sufficient for both tracking MT plus ends in vivo and for inducing MT nucleation and elongation in vitro.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant CLIP-170 Fragments—The plasmids for expression of GFP-CLIP-170 fragments in mammalian cells were constructed using directional TOPO cloning and Gateway technology (Invitrogen) (25). Briefly, varying lengths of coding sequence were PCR-amplified from a pET15b vector (Novagen) already containing the CLIP-170 H1-(1–350) sequence (24, 25). The resulting PCR products were subcloned into the pENTR D-TOPO vector following the manufacturer's protocol. These entry clones were then further used in a Gateway LR recombination reaction to insert them into the pcDNA-DEST53 vector (Invitrogen), which contains an N-terminal GFP tag.
To obtain the bacterially expressed His-tagged CLIP-170 fragments, the PCR products from above were subcloned back into empty pET15b vectors using the unique NdeI and BamHI restriction sites to maintain the N-terminal His tag. All constructs were verified by restriction digestion and DNA sequencing analysis.
The recombinant His-tagged CLIP-170 fragments were expressed in Escherichia coli BL21 (DE3) and purified using the standard Novagen His tag purification protocol with the following modifications. Briefly, after induction with isopropyl 1-thio-β-d-galactopyranoside for 4 h, cells were pelleted and resuspended in lysis buffer (20 mm Tris-HCl, pH 7.9, 300 mm NaCl, and 5 mm imidazole). The cell lysate was then purified by Ni2+ affinity chromatography. All the fragments were eluted at 200 mm imidazole. Finally, salts were removed by using Bio-Gel P-6 desalting resin. All constructs expressed proteins of the expected molecular weights and were found in the soluble fraction. The purity of the recovered proteins was assayed by SDS-PAGE (Fig. 1B) (the smaller bands observed in this gel are CLIP-170 fragments typical of bacterially expressed CLIP-170 preparations (Refs. 24, 25 and data not shown)). Protein concentration was determined by the method of Bradford using bovine serum albumin as the standard (37). All proteins were centrifuged at 4 °C for 15 min at 165,000 × g before all experiments to remove any aggregated protein.
Cell Culture, Transfections, and Live Cell Imaging—COS-7 cells were routinely grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% l-glutamine under standard tissue culture conditions (17). The cells were plated on 18-mm2 coverslips and the following day transiently transfected with ∼5 μg of plasmid DNA from each of the various GFP-CLIP-170 constructs using calcium phosphate (17). 18–24 h post-transfection the coverslips were inverted onto slides and sealed with VaLaP. Imaging was performed on a Nikon inverted microscope (TE2000, Melville, NY) fitted for immunofluorescence with HighQ filter sets from Chroma (Brattleboro, VT). Cells were visualized using a 60× objective and a 1.5× optivar. Metamorph software (Molecular Devices, Sunnyvale, CA) controlled the data acquisition from a cooled backthinned CCD camera (Cascade 512B Photometrics, Tucson, AZ). Images were further processed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Preparation of Tubulin and Microtubules—Porcine brain MT protein was isolated by two cycles of polymerization and depolymerization as described previously (25). MT-associated protein-free tubulin was purified from the MT protein by phosphocellulose (P-11) chromatography. Taxol (paclitaxel, Sigma) MTs were prepared from P-11-purified porcine brain tubulin by the stepwise addition of taxol as described previously (25).
Microtubule Cosedimentation Assay—The affinity of each CLIP-170 fragment for MTs was measured by cosedimentation assay (25). 2–3 μm of a CLIP-170 fragment was incubated with varying concentrations of taxol-MTs in PEM buffer (80 mm PIPES, pH 6.8, 2 mm MgSO4, and 1 mm EGTA). The samples were incubated for 20 min at 37 °C and then centrifuged at 165,000 × g for 15 min. The supernatants and pellets were separated, and equal fractions were analyzed by SDS-PAGE. Gels were scanned, and band intensities of CLIP-170 fragments were quantified with ImageJ (National Institutes of Health). The fraction of CLIP-170 fragment in the pellet was assumed to be the fraction bound because each experiment included a control without MTs in which the CLIP-170 fragment remained entirely in the supernatant. The binding affinities of CLIP-170 fragments with MTs were calculated as described previously (25). Data were fit to a bimolecular binding equation Y = Bmax·X/(Kd + X), where Y is the fraction of CLIP-170 in the pellet; X is the concentration of MTs, and Bmax is the maximal achievable binding. All analysis was performed under the assumption of 1:1 stoichiometry for CLIP-170 fragment:tubulin dimer binding in MTs.
Tubulin Polymerization Assay and Electron Microscopy—Tubulin polymerization was monitored using a light scattering assay at 350 nm as described previously (38). Briefly, tubulin (12 μm) was mixed with CLIP-170 fragments at 0 °C in PEM buffer (described above) and 1 mm GTP. Polymerization was initiated by transferring the samples to a PerkinElmer Life Sciences Lambda 2 spectrophotometer connected to a 37 °C water bath. Because 12 μm tubulin is below the critical concentration for nucleation, only CLIP-170 fragments with nucleation activity will produce a signal in this assay. To observe the effects of the CLIP-170 fragments on MT elongation, seeded assembly was performed using taxol-MT seeds (11). Seeds were prepared by polymerizing the tubulin with taxol. Polymers were sedimented, washed, and resuspended in PEM buffer without taxol. Polymers were sheared by passage five times through a 25-gauge needle. For electron microscopy, tubulin was polymerized as described above. Samples were fixed with prewarmed 0.5% glutaraldehyde for 5 min and then diluted 3-fold in PEM buffer before being applied to carbon-coated electron microscope grids (300 mesh). The grids were subsequently negatively stained with 1% uranyl acetate solution and observed in a H-600 transmission electron microscope (Hitachi).
RESULTS
Dissection of the CLIP-170 Head Domain—To determine whether one or both of the CAP-Gly domains and surrounding serine-rich regions are necessary for plus-end tracking behavior and tubulin polymerization, the H1-(1–350) head domain of CLIP-170 was separated into 10 smaller constructs of varied size. Two versions for each construct were generated, one with an N-terminal GFP tag and the other with an N-terminal His tag (Fig. 1A). The constructs can be grouped into three categories as follows: Group 1 (constructs containing CAP-Gly-1: H1-(1–155), H1-(58–155), and H1-(58–211)), Group 2 (constructs with both CAP-Gly motifs: H1-(58–300) and H1-(58–350)), and Group 3 (constructs containing CAP-Gly-2: H1-(156–350), H1-(122–300), H1-(203–350), and H1-(203–300)).
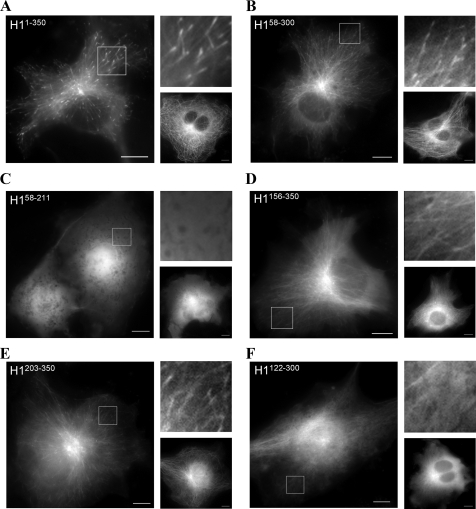
Minimal Unit for CLIP-170 Plus-end Tracking—To examine the behavior of GFP-tagged CLIP-170 fragments in vivo, we transiently transfected each construct into COS-7 cells and examined the localization of these proteins. For reference, Fig. 2A and supplemental movie 1 show the head domain of CLIP-170 (H1-(1–350)) labeling the plus end of the MTs as reported previously (25). At higher expression levels, H1-(1–350) localizes along the MT lattice (Fig. 2A, right lower panel) (10, 25).
FIGURE 2.
Plus-end tracking behavior of CLIP-170 fragments in vivo. COS-7 cells were transfected with the GFP-tagged plasmid constructs described in Fig. 1A. Single frames from movies have been extracted for presentation here. Right upper panels have been magnified 2.5× from each boxed region. Right lower panels represent the same construct at a high level of expression as measured by fluorescence intensity. A, full-length head domain H1-(1–350) tracks microtubule plus ends. B, H1-(58–300) tracks plus ends in most transfected cells and decorates the MTs in overexpressing cells to similar levels as H1-(1–350). C, H1-(58–211) does not track microtubule plus ends and remains diffuse throughout the cytoplasm at all levels of expression. D, H1-(156–350) has some ability to localize to the MT lattice but no real preference for the plus end. E, H1-(203–350) tracks microtubule plus ends, although with less robust tips compared with either construct containing two CAP-Gly domains; this construct does bind the lattice at higher levels of expression. F, H1-(122–300) is largely diffuse, localizing only weakly to MT fibers, and was not observed to track MT plus ends. (Scale bar, 10 μm.)
Consistent with previous efforts to dissect CLIP-170 +TIP activity (25, 36), the constructs containing both CAP-Gly motifs (H1-(58–300) and H1-(58–350)) track MT plus ends at low expression and at higher expression visibly decorate the MT lattice (Fig. 2B, supplemental movie 2, and data not shown). Removal of the first serine-rich region (H1-(58–350)) or both first and third serine-rich regions together (H1-(58–300)) did not abolish the plus-end tracking activity or the MT labeling, although it did make the plus-end labeling less distinct (Fig. 2B, right upper panel). In contrast, constructs containing only CAP-Gly-1 with or without the serine-rich regions (H1-(1–155), H1-(58–155), and H1-(58–211)) were unable to either accumulate at the plus ends or colocalize with the MT network; in all cases they were diffusely localized regardless of expression level (Fig. 2C and data not shown). These observations are consistent with previous analysis of CLIP-170 fragments that led to the conclusion that two CAP-Gly motifs (either in cis or trans) are necessary for +TIP behavior (36).
However, our work with constructs containing CAP-Gly-2, but not CAP-Gly-1, indicates that this conclusion needs to be reassessed. Construct H1-(156–350), which contains CAP-Gly-2 and both serine-rich regions, displayed some labeling of the MT lattice without a strong preference for the plus end (Fig. 2D). Even more striking, the fragment containing CAP-Gly-2 and the third serine-rich region (H1-(203–350)) displayed clear plus-end tracking at low expression and obvious MT lattice decoration at higher expression levels (Fig. 2E and supplemental movie 3). In contrast, constructs containing CAP-Gly-2 and the second serine-rich region (H1-(122–300)), or CAP-Gly-2 alone (H1-(203–300)), failed to either track plus ends or bind along the MT lattice (note that MT lattice binding may be faintly visible in overexpressing cells near the periphery (Fig. 2F and data not shown)).
In summary, these experiments suggest that the proximal half of CLIP-170 cannot mediate either MT-plus-end tracking or MT lattice localization in vivo, consistent with previous reports (36). However, we also find that the distal half of the CLIP-170 head domain, missing the first CAP-Gly sequence, and consisting of the second CAP-Gly and the third serine-rich region, can both track plus ends and localize to MTs by itself.
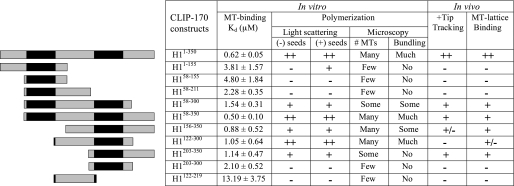
Interaction of CLIP-170 Fragments with Microtubules—To investigate the origins of these differences in activity, the ability of each of our CLIP-170 fragments to bind MTs was measured by cosedimentation assay (Table 1). As a control, the head domain H1-(1–350) was found to bind with a Kd value similar to that reported previously (25, 39). Examination of these data indicate the following. First, constructs containing single CAP-Gly domains bind MTs more weakly than those containing both CAP-Gly domains, with the second CAP-Gly having somewhat (∼2 times) higher affinity than the first. Second, the presence of any single serine-rich region improves the affinity, but only ∼2 times, with the second and third regions appearing to have a stronger effect than the first. Finally, the second serine-rich region by itself does have a detectable although weak affinity for MTs (we were not able to test the other serine-rich regions by themselves because these constructs were difficult to express).
TABLE 1.
Summary of the experimental data collected for the various CLIP-170 fragments
The affinity of each CLIP-170 fragment for taxol-MTs was measured by cosedimentation assay. Data are the average of three independent experiments. Values are ±S.D. The light scattering entries are a qualitative representation of the data presented in Fig. 3. The microscopy, tip tracking, and lattice binding entries are summaries of the images shown in Figs. 2 and 4 and supplemental Figs. 1 and 2, with data for some fragments not shown. Results are qualitatively summarized by the following symbols: ++, strong positive effect; +, moderate positive effect; +/–, borderline detectable positive effect; –, no effect detected.
Taken together, these MT-binding data indicate that each CAP-Gly and serine-rich region has some microtubule binding activity (the conclusion with regard to serine-rich regions 1 and 3 is tentative), but that the presence of a CAP-Gly motif and serine-rich region together is essential for efficient binding of CLIP-170 to MTs. Placing these paired domains in tandem increases the affinity even further.
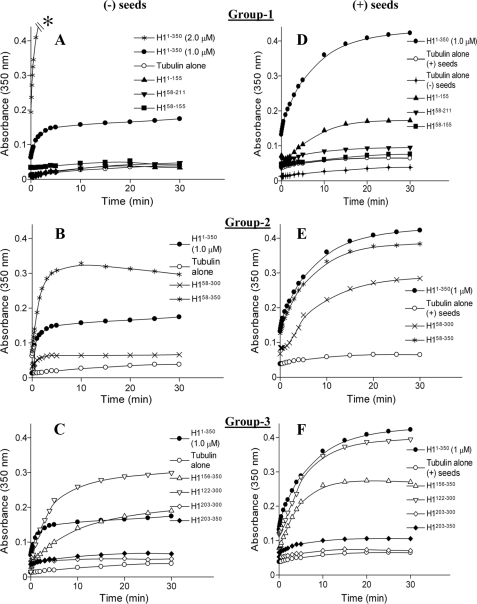
Effects of CLIP-170 Fragments on the Nucleation and Elongation Processes of Tubulin Polymerization—The CLIP-170 H1-(1–350) fragment has been shown to promote MT polymerization in vivo and in vitro and to induce MT nucleation in vitro (10, 36, 40). To determine which parts of the protein endow it with these activities, we tested the effect of the different CLIP-170 H1-(1–350) fragments on tubulin polymerization in vitro, utilizing the methods of light scattering, electron microscopy, and light microscopy. Two different conditions were used in these assays. In the first, tubulin (12 μm) was used without MT seeds or stabilizing factors. Because 12 μm tubulin is below the critical concentration for nucleation in this tubulin preparation, only proteins that nucleate MTs (or other polymers) are expected to give a light-scattering signal (Fig. 3, A–C). To test for polymerization promoting activity in the absence of nucleation activity, we utilized a second assay, in which preformed taxol-stabilized MT seeds were added to the polymerization assay along with the CLIP-170 fragments (Fig. 3, D–F). Finally, to ascertain whether the polymers detected by light scattering were indeed MTs or other non-MT structures, electron microscopy and light microscopy were used to look at the morphology of the tubulin polymers formed in the presence of the different CLIP-170 fragments (Fig. 4 and supplemental Figs. 1 and 2).
FIGURE 3.
Assembly promoting activity of CLIP-170 fragments. A–C, MT assembly in the presence of CLIP-170 fragments was monitored by change in the absorbance at 350 nm. Polymerization of tubulin (12 μm) was measured in the absence and presence of the different CLIP-170 fragments (2.0 μm) as shown. In this assay, H1-(1–350) (2.0 μm) causes strong bundling and thus produces an off-scale light scattering signal (A, asterisk). To minimize this effect and for comparisons with the other CLIP-170 fragments, H1-(1–350) was also used at 1.0 μm as shown. Data were extracted at different time points and plotted using the GraFit 6 program; the lines are provided as a guide. Each data point is the average from two independent experiments. D–F, polymerization with or without CLIP-170 fragments as shown was carried out in the presence of taxol seeds (1.0 μm) to observe the effects on microtubule elongation. The tubulin concentration was 12 μm and all CLIP-170 fragments were used at 1.0 μm concentration to minimize possible bundling effects.
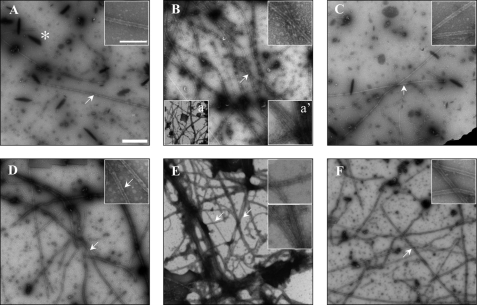
FIGURE 4.
Electron microscopy of the polymers assembled in the presence of tubulin and selected CLIP-170 fragments. Samples were prepared as described in Fig. 3, A–C. Large panels were acquired at ×10,000 magnification (scale bar, 1.0 μm). The insets show regions (indicated by arrows) of the same fields magnified to ×30,000 (scale bar, 0.2 μm). A, control sample in the absence of CLIP-170 fragments. This image shows one of the few MTs observed; the asterisk indicates dark precipitates of uranyl acetate crystals commonly seen in glutaraldehyde-fixed samples with low concentrations of polymerized tubulin. B, H1-(1–350), main panel shows a sample with reduced fragment concentration (1.0 μm versus the 2.0 μm used for other fragments) to allow visualization of individual MTs. The lower insets (a and a′, 10,000 and 30,000 magnification, respectively) show samples at 2.0 μm H1-(1–350), where bundling becomes strong. C, H1-(58–211), this image shows some of the few MTs observed in this sample. D, H1-(58–300), this is a representative image showing increase in the number of MTs with negligible bundling effects. E, H1-(122–300), representative image showing that this construct strongly promotes both MT formation and bundling. F, H1-(203–350), this representative image shows that this construct causes a mild increase in MT number with negligible bundling effects.
In control polymerization assays with tubulin alone, negligible polymerization was seen without seeds (Fig. 3, A–D). Control experiment with seeds (1 μm) showed a mild increase in light scattering (polymerization) as a function of time (Fig. 3, D–F). In contrast, assays with H1-(1–350) showed robust polymerization in both assays (Fig. 3, all panels). These observations confirm the strong effects of H1-(1–350) on tubulin polymerization as reported previously (40).
The fragments containing only a single CAP-Gly without a serine-rich domain (H1-(58–155) and H1-(203–300)) or a serine-rich domain alone (H1-(122–219)) had negligible effects on light scattering in either assay, suggesting that these domains in isolation are incapable of promoting either MT nucleation or polymerization (Fig. 3 and data not shown). However, different results were obtained with certain combinations of CAP-Gly and serine-rich regions.
More specifically, a construct containing CAP-Gly-1 together with serine-rich region 1 (H1-(1–155)) could not promote polymerization in the absence of exogenous MT seeds (Fig. 3A), consistent with previous reports (36). However, we were surprised to see that this construct promoted polymerization when MT seeds were provided (Fig. 3D). Even more striking, a construct containing serine-rich region 2 and CAP-Gly-2 (H1-(122–300)) induced polymerization in both the presence and absence of seeds (Fig. 3, C and F). Similar but less dramatic activity was seen with a construct containing CAP-Gly-2 and two serine-rich regions (H1-(156–350)), whereas a construct consisting of CAP-Gly-2 and the third serine-rich region (H1-(203–350)) had low but detectable activity in both assays (Fig. 3, C and F). These observations suggest that these constructs promote both nucleation and polymerization.
To complement the above experiments and test whether the observed changes in light scattering are due to formation of MTs or non-MT aggregates, we examined by electron microscopy the morphology of the polymerization products produced in the absence of MT seeds. As expected, few MTs were observed in control experiments containing tubulin alone; addition of the full-length head domain (H1-(1–350)) increased the number of MTs dramatically and induced some bundling (Fig. 4, A and B, Table 1, and supplemental Figs. 1 and 2).
In all cases where light scattering was observed to be higher than with the tubulin alone control, more MTs were observed than in this control (Figs. 3 and 4, Table 1, and supplemental Fig. 2), verifying the idea that CAP-Gly-2 can indeed induce MT formation as long as it placed in conjunction with one of the adjacent serine-rich regions. However, we cannot rule out the possibility that these constructs also induce other less regular structures. It should also be noted that there was a strong correlation between activity in the light scattering assay and MT bundling activity as observed in EM and light microscopy (supplemental Figs. 1 and 2 and data not shown), reminding us that increased light scattering signals cannot be simply equated to increased polymerization.
DISCUSSION
The main focus of this work was to define the minimal plus-end tracking unit of CLIP-170 and relate its plus-end tracking behavior to the MT binding and polymerization activity observed in vitro. It has already been established that the initial 350 amino acids of CLIP-170 contain the entire region responsible for MT binding, and this fragment displays plus-end tracking behavior similar to that of the full-length CLIP-170 in vivo (10, 25). Our approach was to further break down this MT binding domain into different subdomains and then characterize the activities of these subdomains in vivo and in vitro.
We find that a fragment consisting of the distal half of the CLIP-170 head domain (containing only CAP-Gly-2 and serine-rich region 3) has all of the activities normally associated with the full head domain as follows: +TIP behavior in vivo as well as microtubule nucleation and elongation promoting activities in vitro. However, removal of the serine-rich region abrogates all of these activities. These observations demonstrate that a single CAP-Gly motif can be sufficient for these activities if it is in the right context. They also suggest that the combination of a CAP-Gly and serine-rich region is necessary for these activities. Consistent with the results of Ref. 36, we find that removal of either the first or third serine-rich regions leaves intact both CLIP-170 +TIP behavior and its tubulin nucleation and elongation activities, demonstrating that the +TIP activity of fragment H1-(203–350) cannot simply be a function of the third serine-rich region.
In contrast, the proximal half of the CLIP-170 head domain, which also contains a CAP-Gly and serine-rich regions, lacks the +TIP and nucleation activities. However, it does have the ability to promote MT elongation in the presence of MT seeds (Fig. 3D). This activity argues against the idea that the proximal domain lacks function simply because of folding problems. In addition, we have observed that both the first and second CAP-Gly motifs can activate EB1, but that the proximal fragment has greater EB1 activating activity.3 These observations demonstrate that the two halves of CLIP-170 are not functionally equivalent. The full-length head domain does have considerably stronger +TIP and polymerization promoting activities than does the distal half by itself, indicating that the proximal half does contribute in a significant way to the microtubule directed activities of CLIP-170.
Our observations conflict with an earlier report concluding that two CAP-Gly domains are required for CLIP-170 +TIP and polymerization promoting activities (36). The difference in conclusion between our work and that of Ref. 36 most likely results from a focus (36) on constructs containing CAP-Gly-1 (they did not examine constructs containing only the distal half of the protein). Our observations are consistent with their report that fragments containing CAP-Gly-1 and missing CAP-Gly-2 fail to show plus-end tracking or promote tubulin nucleation.
Because the two CAP-Gly domains are very similar in structure, one might have expected that if one CAP-Gly has an activity, the other would have it as well. What is the origin of these differences in plus-end tracking and nucleation abilities? One possibility comes from the affinity analysis. The constructs containing CAP-Gly-2 (H1-(156–350), H1-(122–300), H1-(203–350), and H1-(203–300)) bind MTs with higher affinity than the constructs with CAP-Gly-1 (Table 1). These differences in affinity, although small, might endow CAP-Gly-2 with greater plus-end tracking and tubulin nucleation/elongation activities. The affinity difference may originate in the fact that CAP-Gly-2 has a more basic tubulin-binding groove, as shown in recent crystal structure analysis (32). In addition, our data indicate that serine-rich regions contribute to these activities (Table 1). It is not clear whether the differences between constructs that have the same CAP-Gly motif and different serine-rich regions result more from differences in the characteristics of the serine-rich regions or from differences in the position of these regions relative to the CAP-Gly motifs.
Serine-rich regions are present in most CAP-Gly proteins and have been proposed to be linkers between the different domains of the protein, sites of regulation by phosphorylation, or additional tubulin binding domains (28, 34, 41). In the CLIP family of proteins, these regions are poorly conserved at the primary sequence level and have characteristics of unstructured sequences (34). These attributes are consistent with passive linkers, but our data together with previous analysis of CLIP-115 and p150Glued (28, 42) indicate that the serine-rich regions have more active and specific roles. Moreover, the data suggest that the serine-rich regions of CLIP-170 act as additional and perhaps independent MT binding domains, similar to what has been proposed for p150Glued (42). Indeed, one might expect that a second binding site in addition to the CAP-Gly would be required for a fragment such as H1-(122–300) to be able to nucleate MTs. The poor sequence conservation of these domains and the weak affinity observed for the isolated serine-rich region 2 suggests that this binding is likely relatively nonspecific. However, we cannot rule out the possibility that the serine-rich regions enhance CAP-Gly activity in other ways, for example by altering the CAP-Gly structure.
It is intriguing that +TIP behavior and polymerization-promoting ability (both nucleation and elongation) correlate well with each other and with increased MT binding affinity (Table 1), but it is important to note that the correlation is neither complete nor simple. For example, H1-(122–300) has relatively high affinity and strong polymerization promoting ability in vitro, but in vivo this construct fails to track MT tips and localizes to the MT lattice only weakly. In contrast, H1-(203–350) has less dramatic effects on polymerization in vitro, but both track MT plus ends and bind to the MT lattice robustly (Table 1 and Fig. 2E). In addition, one construct containing the first CAP-Gly (H1-(1–155)) failed to show any activity in vivo or in normal (unseeded) light scattering assays but did induce MT growth in the presence of MT seeds (Fig. 3 and Table 1). The physiologic role and mechanism of this difference in activity are not yet clear, but they underscore the distinction between elongation and nucleation and the importance of considering how each may be altered to regulate MT dynamics.
Given that constructs containing only CAP-Gly-2 and a serine-rich region have all of the MT-related activities normally associated with CLIP-170, the question then arises as to the role of CAP-Gly-1. One possibility is that the two halves of the MT binding domain act cooperatively to provide CLIP-170 with more efficient +TIP and polymerization promoting activity. A second possibility (not mutually exclusive with the first) is that the two CAP-Gly domains endow CLIP-170 with the ability to bind simultaneously to tubulin and other CAP-Gly-binding proteins. One such protein is EB1 (18, 35). Interestingly, it has been reported that EB1 has a higher affinity for CAP-Gly-1 than CAP-Gly-2 (33). Given the observation that CAP-Gly-2 has higher affinity for MTs, this leads to the speculation that when both EB1 and tubulin are present, perhaps the first CAP-Gly binds to EB1 and the second to tubulin. Such a model could explain how CLIP-170 and EB1 can each recruit the other to MTs upon overexpression (17), and may ultimately prove important in explaining the cooperative effects of EB1 and CLIP-170 on tubulin polymerization.
Supplementary Material
Acknowledgments
We thank Jose Chaverri for help with cloning and the members of the Goodson laboratory for insightful discussions and critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM065420 (to H. V. G.) and R01 GM076177 (to A. J. H.). This work was also supported by American Heart Association Postdoctoral Fellowship 0825871G (to K. K. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2, Movies S1–S3, and additional references.
Footnotes
The abbreviations used are: MT, microtubule; PIPES, 1,4-piperazinediethanesulfonic acid; GFP, green fluorescent protein.
K. K. Gupta, Z. Zhu, and H. V. Goodson, manuscript in preparation.
References
- 1.Kirschner, M., and Mitchison, T. (1986) Cell 45 329–342 [DOI] [PubMed] [Google Scholar]
- 2.Desai, A., and Mitchison, T. J. (1997) Annu. Rev. Cell Dev. Biol. 13 83–117 [DOI] [PubMed] [Google Scholar]
- 3.Gundersen, G. G., and Cook, T. A. (1999) Curr. Opin. Cell Biol. 11 81–94 [DOI] [PubMed] [Google Scholar]
- 4.Ross, J. L., Ali, M. Y., and Warshaw, D. M. (2008) Curr. Opin. Cell Biol. 20 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walczak, C. E. (2000) Curr. Opin. Cell Biol. 12 52–56 [DOI] [PubMed] [Google Scholar]
- 6.Gundersen, G. G. (2002) Nat. Rev. Mol. Cell Biol. 3 296–304 [DOI] [PubMed] [Google Scholar]
- 7.Howard, J., and Hyman, A. A. (2003) Nature 422 753–758 [DOI] [PubMed] [Google Scholar]
- 8.Morrison, E. E. (2007) Cell. Mol. Life Sci. 64 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhmanova, A., and Steinmetz, M. O. (2008) Nat. Rev. Mol. Cell Biol. 9 309–322 [DOI] [PubMed] [Google Scholar]
- 10.Diamantopoulos, G. S., Perez, F., Goodson, H. V., Batelier, G., Melki, R., Kreis, T. E., and Rickard, J. E. (1999) J. Cell Biol. 144 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ligon, L. A., Shelly, S. S., Tokito, M., and Holzbaur, E. L. (2003) Mol. Biol. Cell 14 1405–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirnauer, J. S., Grego, S., Salmon, E. D., and Mitchison, T. J. (2002) Mol. Biol. Cell 13 3614–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galjart, N. (2005) Nat. Rev. Mol. Cell Biol. 6 487–498 [DOI] [PubMed] [Google Scholar]
- 14.Akhmanova, A., Hoogenraad, C. C., Drabek, K., Stepanova, T., Dortland, B., Verkerk, T., Vermeulen, W., Burgerering, B. M., De Zeeuw, C. I., Grosveld, F., and Galjart, N. (2001) Cell 104 923–935 [DOI] [PubMed] [Google Scholar]
- 15.Sharp, D. J., Mennella, V., and Buster, D. W. (2005) Cell Cycle 4 1482–1485 [DOI] [PubMed] [Google Scholar]
- 16.Honnappa, S., Okhrimenko, O., Jaussi, R., Jawhari, H., Jelesarov, I., Winkler, F. K., and Steinmetz, M. O. (2006) Mol. Cell 23 663–671 [DOI] [PubMed] [Google Scholar]
- 17.Goodson, H. V., Skube, S. B., Stalder, R., Valetti, C., Kreis, T. E., Morrison, E. E., and Schroer, T. A. (2003) Cell Motil. Cytoskeleton 55 156–173 [DOI] [PubMed] [Google Scholar]
- 18.Komarova, Y., Lansbergen, G., Galjart, N., Grosveld, F., Borisy, G. G., and Akhmanova, A. (2005) Mol. Biol. Cell 16 5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coquelle, F. M., Caspi, M., Cordelières, F. P., Dompierre, J. P., Dujardin, D. L., Koifman, C., Martin, P., Hoogenraad, C. C., Akhmanova, A., Galjart, N., de Mey, J. R., and Reiner, O. (2002) Mol. Cell. Biol. 22 3089–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierre, P., Scheel, J., Rickard, J. E., and Kreis, T. E. (1992) Cell 70 887–900 [DOI] [PubMed] [Google Scholar]
- 21.Perez, F., Diamantopoulos, G. S., Stalder, R., and Kreis, T. E. (1999) Cell 96 517–527 [DOI] [PubMed] [Google Scholar]
- 22.Fukata, M., Watanabe, T., Noritake, J., Nakagawa, M., Yamaga, M., Kuroda, S., Matsuura, Y., Iwamatsu, A., Perez, F., and Kaibuchi, K. (2002) Cell 109 873–885 [DOI] [PubMed] [Google Scholar]
- 23.Pierre, P., Pepperkok, R., and Kreis, T. E. (1994) J. Cell Sci. 107 1909–1920 [DOI] [PubMed] [Google Scholar]
- 24.Scheel, J., Pierre, P., Rickard, J. E., Diamantopoulos, G. S., Valetti, C., van der Goot, F. G., Haner, M., Aebi, U., and Kreis, T. E. (1999) J. Biol. Chem. 274 25883–25891 [DOI] [PubMed] [Google Scholar]
- 25.Folker, E. S., Baker, B. M., and Goodson, H. V. (2005) Mol. Biol. Cell 16 5373–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riehemann, K., and Sorg, C. (1993) Trends Biochem. Sci. 18 82–83 [DOI] [PubMed] [Google Scholar]
- 27.Hayashi, I., Plevin, M. J., and Ikura, M. (2007) Nat. Struct. Mol. Biol. 14 980–981 [DOI] [PubMed] [Google Scholar]
- 28.Hoogenraad, C. C., Akhmanova, A., Grosveld, F., De Zeeuw, C. I., and Galjart, N. (2000) J. Cell Sci. 113 2285–2297 [DOI] [PubMed] [Google Scholar]
- 29.Lallemand-Breitenbach, V., Quesnoit, M., Braun, V., El Marjou, A., Pous, C., Goud, B., and Perez, F. (2004) J. Biol. Chem. 279 41168–41178 [DOI] [PubMed] [Google Scholar]
- 30.Li, S., Finley, J., Liu, Z. J., Qiu, S.-H., Chen, H., Luan, C.-H., Carson, M., Tsao, J., Johnson, D., Lin, G., Zhao, J., Thomas, W., Nagy, L. A., Sha, B., DeLucas, L. J., Wang, B.-C., and Luo, M. (2002) J. Biol. Chem. 277 48596–48601 [DOI] [PubMed] [Google Scholar]
- 31.Bartolini, F., Tian, G., Piehl, M., Cassimeris, L., Lewis, S. A., and Cowan, N. J. (2005) J. Cell Sci. 118 1197–1207 [DOI] [PubMed] [Google Scholar]
- 32.Mishima, M., Maesaki, R., Kasa, M., Watanabe, T., Fukata, M., Kaibuchi, K., and Hakoshima, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10346–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisbrich, A., Honnappa, S., Jaussi, R., Okhrimenko, O., Frey, D., Jelesarov, I., Akhmanova, A., and Steinmetz, M. O. (2007) Nat. Struct. Mol. Biol. 14 959–967 [DOI] [PubMed] [Google Scholar]
- 34.Miller, R. K., D'Silva, S., Moore, J. K., and Goodson, H. V. (2006) Curr. Top. Dev. Biol. 76 49–87 [DOI] [PubMed] [Google Scholar]
- 35.Watson, P., and Stephens, D. J. (2006) J. Cell Sci. 119 2758–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slep, K. C., and Vale, R. D. (2007) Mol. Cell 27 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 38.Gaskin, F., Cantor, C. R., and Shelanski, M. L. (1974) J. Mol. Biol. 89 737–755 [DOI] [PubMed] [Google Scholar]
- 39.Ligon, L. A., Shelly, S. S., Tokito, M. K., and Holzbaur, E. L. (2006) FEBS Lett. 580 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnal, I., Heichette, C., Diamantopoulos, G. S., and Chretien, D. (2004) Curr. Biol. 14 2086–2095 [DOI] [PubMed] [Google Scholar]
- 41.Sim, K. L., and Creamer, T. P. (2002) Mol. Cell. Proteomics 1 983–995 [DOI] [PubMed] [Google Scholar]
- 42.Culver-Hanlon, T. L., Lex, S. A., Stephens, A. D., Quintyne, N. J., and King, S. J. (2006) Nat. Cell Biol. 8 264–270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.