Abstract
The Leishmania parasite is a widespread disease threat in tropical areas, causing symptoms ranging from skin lesions to death. Leishmania parasites typically invade macrophages but are also capable of infecting fibroblasts, which may serve as a reservoir for recurrent infection. Invasion by intracellular pathogens often involves exploitation of the host cell cytoskeletal and signaling machinery. Here we have observed a dramatic rearrangement of the actin cytoskeleton and marked modifications in the profile of protein tyrosine phosphorylation in fibroblasts infected with Leishmania major. Correspondingly, exposure to L. major resulted in degradation of the phosphorylated adaptor protein p130Cas and the protein-tyrosine phosphatase-PEST. Cellular and in vitro assays using pharmacological protease inhibitors, recombinant enzyme, and genetically modified strains of L. major identified the parasite protease GP63 as the principal catalyst of proteolysis during infection. A number of additional signaling proteins were screened for degradation during L. major infection as follows: a small subset was cleaved, including cortactin, T-cell protein-tyrosine phosphatase, and caspase-3, but the majority remained unaffected. Protein degradation occurred in cells incubated with Leishmania extracts in the absence of intact parasites, suggesting a mechanism permitting transfer of functional GP63 into the intracellular space. Finally, we evaluated the impact of Leishmania on MAPK signaling; unlike p44/42 and JNK, p38 was inactivated upon infection in a GP63- and protein degradation-dependent manner, which likely involves cleavage of the upstream adaptor TAB1. Our results establish that GP63 plays a central role in a number of hostcell molecular events that likely contribute to the infectivity of Leishmania.
Protozoans of the genus Leishmania cause a complex disease called leishmaniasis, whose clinical manifestations have been divided into three principal types, cutaneous, mucocutaneous, and visceral, exhibiting different degrees of severity and mortality (1, 2). This disease threatens over 350 million people in 88 countries in tropical, subtropical, and temperate regions (4).4 The development, multiplication, and transmission of Leishmania in the form of promastigotes between mammalian hosts are achieved by the sandfly insect vector (4).
Following inoculation into a vertebrate host, promastigotes are typically phagocytosed by macrophages where they differentiate into and multiply as amastigotes (1, 5). Heavily infected macrophages lyse and liberate amastigotes that will colonize other cells. In addition, both promastigotes and amastigotes of Leishmania major can be internalized by fibroblast cells (6). Despite their capability to synthesize nitric oxide, fibroblasts produce a much lower quantity of this microbicidal compound than macrophages (6). The limited capacity of fibroblast to eliminate parasites implies that these cells could act as a reservoir for long term infection (6). Nevertheless, little is known regarding the molecular events occurring in fibroblast cells upon contact with Leishmania parasites.
Several intracellular parasites hijack the actin cytoskeletal machinery to infiltrate and traffic inside their host cells (7, 8). Cellular proteins such as cortactin, Wiskott-Aldrich syndrome protein (WASP),5 Crk, and Crk-associated substrate (p130Cas) have been identified as targets of intracellular bacteria (9–12). Leishmania amastigotes induce activation of Cdc42 to re-organize the actin network and enter into Chinese hamster ovary (CHO) fibroblasts (13). Additionally, the activity of Cdc42 is involved in knitting a shell of actin around the internalized Leishmania parasite, a site at which other cytoskeletal regulators such as vinculin and WASP are also recruited (13, 14). Numerous biological processes, including those modulating the dynamics of actin cytoskeleton assembly, are controlled by the dual effects of protein-tyrosine kinases (PTKs) and protein-tyrosine phosphatases (PTPs). Leishmania can affect the state of tyrosine phosphorylation in macrophage cells by activating SHP-1 (Src homology-2 domain-containing phosphatase-1) (15, 16). However, the specific roles of other PTPs in this pathogenic process remain unclear. Interestingly, another nonreceptor PTP, PTP-PEST, has been extensively implicated in the regulation of WASP and p130Cas phosphorylation as well as in the modulation of vinculin-containing adhesion structure formation (17–20). These studies have established PTP-PEST as a critical regulator of actin remodeling and present this enzyme as a particularly interesting candidate target of Leishmania.
Downstream elements of cellular signal transduction such as members of the MAPKs have also been linked to the pathogenic outcome of Leishmania infection. The ability of promastigotes to manipulate and circumvent MAPK activation may represent a strategy to evade the macrophage-host cell defense mechanism. Incubation of macrophages with a p38 inhibitor prior to exposure to Leishmania donovani augmented their subsequent invasion by the parasite (21). Similarly, the anisomycin-mediated inhibition of L. donovani survival inside macrophages was dependent on p38 (21). Moreover, L. major down-regulates p38 to impair CD40-induced iNOS2 expression, inhibiting nitric oxide production and favoring survival within macrophages (22). By inhibiting p38, the parasite can also hijack another signal initiated by CD40 cross-linking, altering cytokine expression to its advantage; interleukin-12, a promoter of the host-protective T-helper type 1 (TH1) cell response, is reduced whereas interleukin-10, an inhibitor of TH1 cell and of NO production, is increased (23, 24). Although the interplay between p38 activity and Leishmania persistence is accepted, little is known regarding the parasitic elements involved in regulation of this MAPK.
Leishmania is coated by a characteristic glycocalyx, whose molecular components play a critical role in the initial contact between the parasite and its host environment. GP63, also referred to as major surface protease, leishmanolysin, or promastigote surface protease, is the most abundant protein covering Leishmania promastigotes (25, 26). Studies performed using different parasitic models demonstrated that GP63 plays a crucial role in complement fixation and processing, which protects Leishmania during its sojourn into mammalian hosts (27–29). Similarly, GP63 was recently shown to defend the parasite against antimicrobial peptides such as defensins and pexiganan (30). The abundance and diversity as well as the high catalytic activity at mammalian body temperature of this virulence factor (31–33) favor the dissemination of the parasite as it digests constituents of the extracellular matrix of the host such as collagen type IV, fibronectin, and laminin (34). Several species of Leishmania release proteolytically active GP63 in the surrounding milieu (35–38) presumably facilitating the propagation of the parasite. In addition, fragments from GP63-processed fibronectin can protect parasites within macrophages, as they attenuate production of reactive oxygen intermediates and favor amastigote proliferation (39). Furthermore, GP63 maximizes promastigote binding to and internalization in macrophages through its ability to interact with the α4/β1 integrin and to promote complement-dependent adhesion (40, 41). Moreover, similar to fibronectin, coating polystyrene surfaces with GP63 enhances in vitro spreading of fibroblasts (42). The expression of specific gp63 genes in the intracellular amastigote form (27, 43, 44) implies an intra-host cell function for this parasitic protease. Interestingly, the activity of GP63 was implicated in the protection of encapsulated proteins against phagolysosomal degradation as well as intra-macrophage survival of Leishmania mexicana amazonensis (45, 46). The identification of the myristoylated alanine-rich C kinase substrate-related protein (MRP), a cytosolic protein associated with the actin network of macrophages, as a substrate of GP63 reinforces the potential of this enzyme to modulate host cell activities within the intracellular space (47). Nonetheless, little else is known concerning its impact on host cell signal transduction or the existence of additional intracellular substrates for this parasitic protease.
Subverting normal cellular functions is a widespread strategy among intracellular parasites to take advantage of mammalian hosts. In this study, we describe distinctive effects of Leishmania infection on cell signaling in fibroblasts. During L. major infection, we found that the parasite manipulates cellular components, in part by altering the tyrosine phosphorylation state of several proteins. Upon contact with L. major, the adaptor Crk interacts with a truncated form of p130Cas, which further correlates with cleavage of two novel substrates, p130Cas and PTP-PEST, by GP63. Moreover, through GP63, L. major impacted the stability of additional proteins, including cortactin, TC-PTP (T-cell PTP), and caspase-3. Additionally, L. major was found to down-regulate p38 in a GP63-dependent manner. Direct activators of p38 include the MAPK kinase MKK3, MKK6, and the adaptor molecule TAB1 (TAK-1-binding protein-1), which interacts directly with p38 (48, 49). Interestingly, the inhibition of p38 occurred in concert with the GP63-mediated proteolysis of TAB1. This study reveals diverse and novel mechanisms by which Leishmania can monopolize different constituents of the fibroblast signal transduction machinery.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Plasmids—Chemicals were purchased at BioShop Canada Inc., Fisher, and Sigma. The PTP-PEST polyclonal antibodies (2528 and 2530) were described previously (19). Monoclonal antibodies specific for Crk, p130Cas, integrin-β1, Shc, and paxillin were from BD Transduction Laboratories. Antibodies against IκBα, STAT5 (C17), and JNK1 were from Santa Cruz Biotechnology. TC-PTP monoclonal antibodies (3E2) were described previously (50). Polyclonal rabbit antibodies against PTP-1B and focal adhesion kinase as well as monoclonal anti-cortactin (4F11) and phosphotyrosine (4G10) were from Upstate. Antibodies specific for AKT, caspase-3, phospho-Thr-202/Tyr-204 p44/42 MAPKs, p44/42 MAPKs, phospho-Thr-183/Tyr-185 JNK, phospho-Thr-180/Tyr-182 p38 MAPK, p38 MAPK, TAB1, MKK3, and MKK6 were from Cell Signaling Technology. The vector encoding GST-PTP-PEST (pEBG-PTP-PEST) has been described (51). cDNAs encoding p38 (image ID 4195842) and TAB1 (image ID 5356886) were obtained from American Type Culture Collection (ATCC). pET-28(c+)TAB1 was generated by PCR cloning of the TAB1 cDNA between the EcoRI and NotI sites. FLAG-TAB1-Myc-Myc WT and residues 1–418 (amino acid numbering) were made by inserting TAB1 cDNA amplified by PCR into pcDNA4/Myc-HisA between the EcoRI and NotI restriction sites. The sense oligonucleotide sequence contains the FLAG epitope, whereas the antisense oligonucleotides for both full-length and truncated TAB1 encodes an additional Myc epitope. The mammalian expression vector encoding GST-p38 was obtained by successively cloning the p38 PCR product into pDONR221 and pDEST27 using Gateway technology according to the manufacturer's instructions (Invitrogen). The integrity of the inserted DNA in each vector was verified by sequencing (Genome Quebec).
Cells and Parasites—Transient transfections were performed, and PTP-PEST clones (B14V, B15V, B11WT (wild type), and B118WT) were generated and maintained as described previously (51). Primary mouse embryonic fibroblasts (P-MEFs) were isolated from BALB/c embryos (The Jackson Laboratories) and grown as described (51). L. major A2 and gp63-null (L. majorgp63-/-) and rescued strains (in which gp63 gene 1 was re-introduced, L. majorgp63-/-rescued) (28) as well as L. donovani strain 2211, L. mexicana, Leishmania tarentolae, and Leishmania braziliensis 2249 promastigotes were all maintained at 25 °C in SDM-79 medium supplemented with 10% heat-inactivated FBS (Wisent) as described (52).
Cellular Treatment, Infection, and Immunoblotting—Stationary phase of Leishmania promastigotes was centrifuged at 2500 rpm (Allegra 6R Centrifuge, Beckman Coulter) for 5 min. Supernatant was removed, and pellets of parasites were washed with phosphate-buffered saline (PBS) and centrifuged at 2500 rpm for 5 min (Allegra 6R Centrifuge, Beckman Coulter). Parasites were resuspended in DMEM (containing the indicated concentration of heat-inactivated FBS) and added onto cells at the specified parasite:cell ratio and for the indicated duration. Cells were then rinsed on ice with PBS and lysed (100 mm Tris (pH 7.4), 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, 0.1% SDS, 50 mm NaF, 1 mm Na3VO4, Complete protease inhibitor (Roche Applied Science)). Cell extracts were cleared by centrifugation at 16,000 × g for 10 min at 4 °C, and the protein concentration of each sample was measured by the Bradford assay (Bio-Rad protein assay). Proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore), and immunoblotted as described (51).
The effect of pharmacological protease inhibitors on cellular protein degradation induced by L. major infection was determined as follows: caspase inhibitor (Z-VAD-fmk; Cedarlane), calpain inhibitor (PD150606; Calbiochem), proteasome inhibitor (lactacystin; Sigma), as well as calpain inhibitor and proteasome inhibitor combined together were added to cells 2 h prior to addition of parasites and co-incubated for an additional 15 min. Cells that were not exposed to any of the inhibitors were incubated with media containing DMSO as a vehicle control. Cells were harvested, lysed, and analyzed by immunoblotting as described above.
The effect on mammalian cells of parasite-conditioned medium and protein lysates prepared from cultured L. major was evaluated as follows. L. major cultures were centrifuged for 5 min at 2500 rpm (Allegra 6R Centrifuge, Beckman Coulter). Supernatants were carefully harvested, avoiding contact with the pellet and the wall of the tube. Conditioned SDM was centrifuged a second time for 10 min at 3500 rpm (Allegra 6R Centrifuge, Beckman Coulter) to remove any potentially residual parasites. Supernatant from cultured parasites was then diluted in normal parasite growth media to normalize the volume according to the parasite concentration in the original culture. Conditioned or fresh growth media were deposited on P-MEFs for 1 h, and cell lysates were prepared and analyzed as described above.
To prepare parasite lysates, L. major parasites were washed three times with PBS, resuspended in serum-free DMEM, and sonicated (Ultrasonic Processor, Sonics & Materials Inc.) two times for 10 s at an intensity of 50% at 4 °C with a 30-s incubation on ice between sonication steps. Lysates were then cleared by centrifugation for 2 min at 16,000 × g at 4 °C, and protein concentration was determined by the Bradford assay. P-MEFs were then incubated with serum-free DMEM supplemented with 333 μg/ml L. major protein extract for 1 h. The absence of parasites in both the conditioned media and L. major lysates was confirmed by microscopic examination (data not shown). Following the incubation, protein lysates were prepared from treated cells and analyzed by immunoblotting according to the procedures described above.
Incubation of Cellular Extracts and Recombinant Proteins with Parasite Lysate—The procedure for preparation of parasite extracts from L. major promastigotes (WT and gp63-/-) was based on a previously published protocol (47). L. major stationary phase promastigotes were washed with PBS and with TNB (10 mm Tris-HCl (pH 7.4), 100 mm NaCl, 0.4 mg/ml bovine serum albumin (Invitrogen), 5 μg/ml pepstatin, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin) before being resuspended in TNB. Parasites were then sonicated (Ultrasonic Processor, Sonics & Materials Inc.) two times at 4 °C for 5 s at an intensity of 50% with a 5-s incubation on ice between each sonication step. Samples were then centrifuged for 2 min at 16,000 × g at 4 °C, and supernatants were kept to perform the cleavage assay. PTP-PEST-/--WT expressing cells (clone B11WT) were rinsed with ice-cold PBS, collected in TNB, and lysed by three sonication steps of 15 s at an intensity of 50% at 4 °C, which were separated by 15-s incubations on ice. Samples were then centrifuged for 5 min at 16,000 × g at 4 °C; supernatants were collected, and protein concentrations were determined by the Bradford assay. Next, 200 μg of protein lysates from PTP-PEST-/--WT expressing cells was incubated with lysates made from 20 × 106 promastigotes (either from WT or gp63-/- L. major) or with TNB (control) at 37 °C for the indicated time. Samples were immediately placed on ice, and SDS sample buffer was added. Samples were boiled for 4 min, separated by SDS-PAGE, and analyzed by immunoblotting.
To obtain purified GST-PTP-PEST, protein extracts from transfected fibroblasts (51) were prepared in lysis buffer (100 mm Tris (pH 7.4), 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, Complete protease inhibitor (Roche Applied Science)). Cell lysates were cleared by centrifugation for 10 min at 16,000 × g at 4 °C and then incubated for 1 h at 4 °C with glutathione-Sepharose beads (Amersham Biosciences). The beads were then washed three times with lysis buffer and two times with TNB. Next, the beads were resuspended in TNB, incubated with the indicated quantity of L. major lysates, and prepared as described above for 15 min at 37 °C. Samples were immediately placed on ice, rinsed twice with ice-cold lysis buffer, and resuspended in SDS sample buffer. Samples were boiled for 4 min, separated by SDS-PAGE, and analyzed by immunoblotting.
Incubation of Recombinant Proteins with Recombinant GP63—GST-PTP-PEST was purified as described above. To produce His-TAB1, Escherichia coli cells transformed with pET-28(c+)TAB1 were induced for 2 h at 37 °C with 1 mm isopropyl β-d-1-thiogalactopyranoside and harvested by centrifugation. Bacterial cells were lysed (50 mm Tris (pH 7.5), 500 mm NaCl, 40 mm imidazole, 1% Triton X-100, and EDTA-free complete protease inhibitor), and recombinant proteins were isolated on nickel-Sepharose beads (Ni-Sepharose 6 Fast Flow, GE Healthcare) according to the manufacturer's instructions. Immobilized His-TAB1 was eluted in elution buffer (1 m imidazole, 500 mm NaCl, 50 mm Tris (pH 7.5), 1% Triton X-100, complete protease inhibitor) for 1 h at 4 °C, concentrated in storage buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 0.1 mm EGTA, 0.1 mm EDTA, 25% glycerol, 0.25 mm dithiothreitol, complete protease inhibitor) using Microcon filters (Millipore), and stored at -80 °C. Purified GST-PTP-PEST or His-TAB1 was incubated with recombinant GP63 (53) in TNB at 37 °C for 30 min. Samples were immediately placed on ice and resuspended in SDS sample buffer. Samples were boiled for 4 min, separated by SDS-PAGE, and analyzed by immunoblotting.
Immunofluorescence—Glass microscope coverslips were coated with 0.2% gelatin at 37 °C for 30 min and rinsed with PBS prior to seeding with P-MEFs. Infected P-MEFs were rinsed three times with PBS, fixed in 4% paraformaldehyde (diluted in PBS) for 20 min, treated with permeabilizing solution (4% paraformaldehyde, 0.1% Triton X-100 in PBS) for 20 min at 4 °C, and incubated for 45 min with 2% bovine serum albumin/PBS. Cells were washed with PBS and stained with a mixture of rhodamine-conjugated phalloidin (Molecular Probes) and 4′,6-diamidino-2-phenylindole (DAPI) (Roche Applied Science). Cells were washed three times with PBS and rinsed once with water, and coverslips were deposited on slides using Vectashield mounting medium (Vector Laboratories, Inc.). Random field images were acquired by confocal microscopy (Zeiss LSM 510-NLO).
Immunoprecipitation and Protein Complex Analysis—To analyze Crk interactions, serum-starved and L. major-infected cells were rinsed with ice-cold PBS and lysed (100 mm Tris (pH 7.4), 5 mm EDTA, 150 mm NaCl, 1% Triton X-100, 10 mm NaF, 1 mm Na3VO4, and Complete protease inhibitor (Roche Applied Science)). Cell extracts were centrifuged at 16,000 × g for 10 min at 4 °C, and the protein concentration of each sample was measured by the Bradford assay. Cleared protein lysates (2.4 mg) were then incubated at 4 °C for 2 h in the presence of 1.25 μg of anti-Crk monoclonal antibody (BD Transduction Laboratories), 25 μl of protein G-agarose beads (Invitrogen). Beads were then washed three times with lysis buffer, boiled in SDS-sample buffer, and analyzed by immunoblotting.
To analyze the interaction between p38 and TAB1, control and L. major-infected cells expressing FLAG-TAB1-Myc-Myc were rinsed with ice-cold PBS and lysed in interaction buffer (20 mm Tris (pH 7.5), 120 mm NaCl, 10% glycerol, 2 mm EDTA, 1% Triton X-100, 1 mm Na3VO4, 10 mm NaF, and Complete protease inhibitor (Roche Applied Science)). Cell extracts were centrifuged at 16,000 × g for 10 min at 4 °C, and the protein concentration of each sample was measured by the Bradford assay. Cleared protein lysates (2.4 mg) were then incubated at 4 °C for 2 h in presence of GST or GST-p38 immobilized on glutathione-Sepharose beads prepared as described below. Beads were then washed three times with interaction buffer, resuspended in SDS sample buffer, boiled, and analyzed by immunoblotting. To isolate GST fusion proteins (GST and GST-p38), transiently transfected HeLa cells (51) were rinsed with ice-cold PBS and lysed in interaction buffer, and protein extracts were cleared by centrifugation (10 min, 16,000 × g, 4 °C). Protein lysates were then incubated with glutathione-Sepharose beads for 1 h at 4 °C, washed three times with interaction buffer, then washed for 10 min in interaction buffer, and resuspended in interaction buffer.
RESULTS
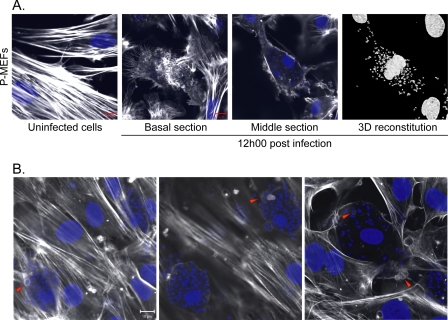
L. major Is Capable of Infecting Primary Embryonic Fibroblasts in Culture—Fibroblasts have been observed to be an alternative cell type to macrophages as hosts for the parasite Leishmania in animal models (6). Additionally, genetically manipulated fibroblasts generated by our laboratory have proved to be valuable models to investigate molecular mechanisms of signal transduction (20, 51, 54). To observe the infection of cultured fibroblasts, P-MEFs were incubated with L. major, fixed and stained, and examined for the presence of parasites by confocal microscopy. Under these experimental conditions, actin staining illustrated cellular morphology, and DAPI allowed the detection of both mammalian and parasitic nuclei as well as the kinetoplast of the parasite. Following 12 h of incubation in the presence of parasites, most cells became infected. In several cases, we observed that spread cells became rounded and exclusively exhibited F-actin cortically as well as in retracting filipodia at the cell-substratum interface (Fig. 1, A, basal section and middle section, and B). These detaching cells contained numerous parasites as seen by three-dimensional reconstitution of the DAPI signal (Fig. 1A). Fig. 1B illustrates another sample treated under similar conditions. Intriguingly, nuclear condensation, which is characteristic of apoptosis, was not observed in the infected cells, even those harboring a heavy load of parasites. In addition, we failed to detect increased caspase activity in cells exposed to L. major, even following 24 h of incubation (supplemental Fig. 1). This indicates that despite the magnitude of the stress induced by the parasite in its host cell, it does not activate apoptosis. Interestingly, we noticed the presence of parasites that appear to be surrounded by actin-rich structures similar to the actin cup described previously (Fig. 1B, arrowheads) (13). These observations imply communication between L. major and cytoskeletal regulators that could influence the infection process in fibroblasts.
FIGURE 1.
L. major parasites accumulate in P-MEFs and cause actin cytoskeleton rearrangements. P-MEFs were infected with the L. major parasite at a ratio of 1:20 (cells:parasite) for 12 h. Cells were then fixed, permeabilized, stained, and observed by confocal microscopy as described under “Experimental Procedures.” A, uninfected control cells show normal cytoskeletal organization. Basal section illustrates the cell-substratum interface. Detection of actin reveals the presence of numerous retracting filipodia in an L. major-infected cell. Section acquired across the infected cell shows the presence of several parasite nuclei (small blue dots) throughout the intracellular space delimited by the cortical actin cytoskeleton. Three-dimensional reconstitution of the detected DAPI signal illustrates the high number of parasites within the infected cell. B, L. major accumulates and forms actin cups inside fibroblasts. Arrowheads point to examples of actin cups. Experiment was performed as described for A. Scale bar, 10 μm.
Cellular Profile of Tyrosine Phosphorylation as well as p130Cas-Crk Complexes Are Modulated during L. major Infection—Functional modulation of numerous cytoskeletal proteins depends on their tyrosine phosphorylation. PTP-PEST has been recognized as an important phosphatase for the balanced activities of several of these molecules (55, 56). We used our PTP-PEST rescued fibroblast lines (51) to investigate the possible modulation of phosphotyrosine proteins in response to parasitic infection. In PTP-PEST-/- fibroblasts, re-expression of PTP-PEST correlates with a reduction of the phosphorylation level of some proteins (one near 250 kDa, another near 130 kDa, and one just less than 75 kDa) under serum-starved control conditions (Fig. 2A). Incubation of both cell lines (B14VPTP-PEST- and B11WTPTP-PEST+) with L. major parasites leads to dramatic changes in the phosphorylation state of numerous proteins. During the course of the infection, the phosphorylation content of proteins of ∼167, 128, 54, and 29 kDa decreased (Fig. 2A, black arrowheads), whereas those of ∼153, 89, 63, 49, and 33 kDa were augmented (Fig. 2A, white arrowheads). These results suggest that upon contact with the target cell, L. major can induce rapid modification of the tyrosine phosphorylation profile of cellular proteins, which undoubtedly impacts cellular signal transmission.
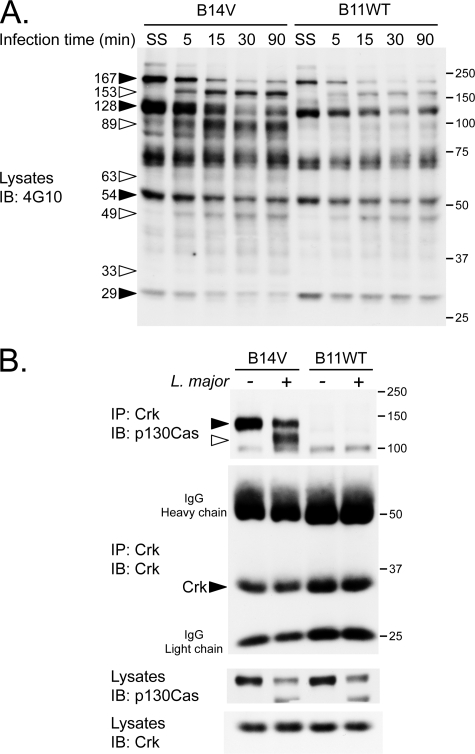
FIGURE 2.
L. major modulates fibroblast tyrosine phosphorylation profile and alters the interaction of p130Cas with Crk. A, PTP-PEST-/- EV (B14V) and PTP-PEST-/--expressing WT (B11WT) cells were serum-starved (SS) (0.05% heat-inactivated FBS DMEM) for 16 h and incubated in the absence or presence of L. major for the indicated time. Protein extracts were resolved by SDS-PAGE and analyzed by immunoblotting (IB) for protein tyrosine phosphorylation using an anti-phosphotyrosine antibody (4G10). B, B14V and B11WT were serum-starved and incubated for 15 min with or without L. major. Crk was immunoprecipitated (IP) from protein lysates. Immuno-isolated Crk and co-precipitated p130Cas were separated by SDS-PAGE and analyzed by immunoblotting. Filled arrowheads correspond to decreasing signals, and open arrowheads point to increasing signals corresponding to tyrosine-phosphorylated candidates detected under the different conditions. SS, serum-starved. The values on the right correspond to molecular mass in kDa.
Interestingly, the phosphorylation of a prominent band between 100 and 150 kDa significantly decreases during infection. An abundant phosphoprotein of this size, p130Cas, is a substrate of PTP-PEST (supplemental Fig. 2) (17, 54) that plays a role in the invasion of host cells by other microorganisms such as Salmonella typhimurium (57). To gain insight into the possible regulation of p130Cas signaling during infection with L. major, we investigated its interaction with Crk, an important signaling adaptor that binds to tyrosine-phosphorylated p130Cas (58). It was previously observed that expression of PTP-PEST decreased the formation of the p130Cas-Crk complex (18). In our cell system, we observed the assembly of this complex only in the absence of PTP-PEST (Fig. 2B, PTP-PEST-/- cells, B14V noninfected). Exposure of these cells (B14V) to L. major resulted in the interaction of Crk with a smaller form of p130Cas as demonstrated by co-immunoprecipitation (Fig. 2B) as well as in vitro binding to purified Crk Src homology-2 domain (supplemental Fig. 3). Also, in both cell lines (B14V and B11WT), we observed that the total amount of full-length p130Cas was diminished, whereas a smaller form appeared during the infection with Leishmania (Fig. 2B, bottom). The cellular content of Crk remained unchanged. These results suggest that p130Cas/Crk-mediated signaling events are modulated during infection by L. major.
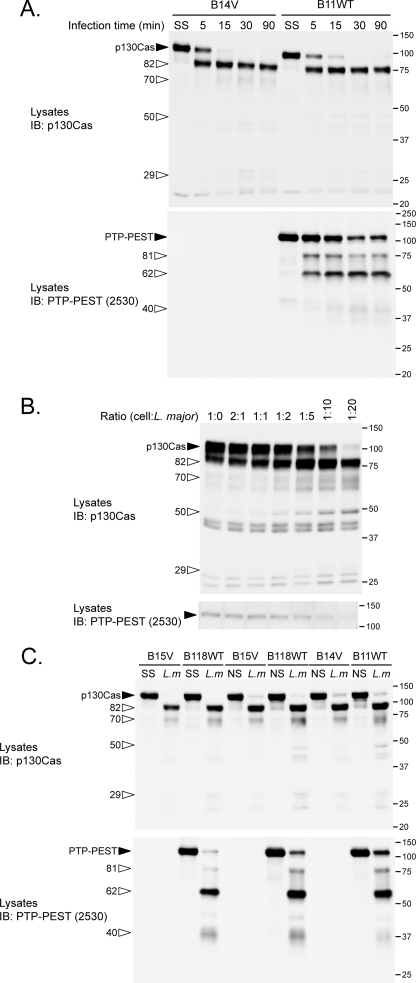
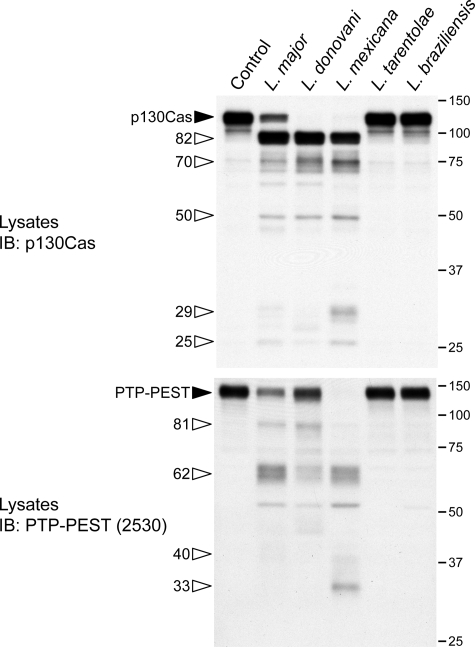
Cellular Exposure to Leishmania Leads to Proteolysis of p130Cas and PTP-PEST—The diminished amount of p130Cas and the binding of Crk to a smaller protein recognized by a p130Cas antibody were indicative of potential proteolysis of p130Cas in cells exposed to L. major. Therefore, we examined the consequence of incubating our PTP-PEST cell lines with L. major on p130Cas stability. Infection with L. major leads to cleavage of p130Cas in a time- and parasite concentration-dependent manner (Fig. 3, A and B). As infection progresses, p130Cas cleavage products accumulate at ∼82, ∼70, ∼50, and ∼29 kDa. To our surprise, we also found that PTP-PEST was cleaved during the infection, which yielded fragments of ∼81, ∼62, and ∼40 kDa. However, L. major does not require PTP-PEST expression to promote the proteolysis of p130Cas because it occurs in both PTP-PEST null and rescued clones (Fig. 3, A and C). Interestingly, in both primary and B11WT fibroblasts, p130Cas and PTP-PEST were degraded when cells were incubated with L. major, L. donovani, and L. mexicana but not with L. tarentolae nor L. braziliensis (Fig. 4 and data not shown). The absence of cleavage induced by L. tarentolae and L. braziliensis is not simply because of delayed kinetics, because extended incubation of fibroblasts with these species did not increase p130Cas or PTP-PEST proteolysis (supplemental Fig. 4). These data identify p130Cas and PTP-PEST as novel signaling targets of specific Leishmania species. To our knowledge, these observations include the first example of p130Cas and PTP-PEST cleavage occurring during parasitic infection.
FIGURE 3.
L. major infection induces proteolysis of p130Cas and PTP-PEST. A, fibroblasts positive and negative for PTP-PEST expression were starved for 16 h in 0.05% heat-inactivated FBS/DMEM and incubated in starvation medium without or with L. major at a ratio of 1:20 (cells:parasites) for the indicated duration. IB, immunoblot. B, PTP-PEST-/- cells expressing WT PTP-PEST (clone B11WT) were exposed to different ratios (cells:parasites) of L. major for 10 min. C, PTP-PEST-/- cells rescued with either empty vector (clones B14V and B15V) or with the WT enzyme (clones B11WT and B118WT) were maintained in starvation or regular media (nonstarved) 16 h prior to treatment. Indicated clones were incubated with or without L. major in the absence of serum or with media supplemented with 10% heat-inactivated-FBS. Cell lysates were prepared and analyzed by immunoblotting for p130Cas and PTP-PEST. Filled arrowheads identify intact proteins, and open arrowheads point to cleavage products. L.m, L. major; SS, serum-starved; NS, nonstarved or in presence of serum. The values on the right correspond to molecular mass in kDa.
FIGURE 4.
L. major, L. donovani, and L. mexicana, but neither L. tarentolae nor L. braziliensis, infections result in degradation of p130Cas and PTP-PEST. P-MEFs were exposed to the indicated species of Leishmania for 1 h at a ratio of 1:20. Protein lysates were harvested and analyzed by immunoblotting (IB) for p130Cas and PTP-PEST. Filled arrowheads identify intact proteins, and open arrowheads point to cleavage products. The values on the right correspond to molecular mass in kDa.
Leishmania Surface Metalloprotease GP63 Cleaves p130Cas and PTP-PEST—We next investigated whether factors, including host physiologic state as well as endogenous proteases, influence p130Cas and PTP-PEST proteolytic processing. The conditions under which cells are maintained contribute to signaling responses and may affect the activity of cellular proteases. To explore the possible impact of the cellular physiologic state on p130Cas and PTP-PEST stability, we compared the consequence of L. major infection among various clonal fibroblast lines (B14V, B15V, B11WT, and B118WT) as well as between serum-starved and growing cells (nonstarved). p130Cas and PTP-PEST from either serum-starved or non-starved cells were both cleaved at a similar rate following incubation with L. major (Fig. 3C). In addition, the proteolysis of p130Cas and PTP-PEST occurred similarly in all clones tested. p130Cas was previously identified as a substrate of both the calpain and caspase-3 proteases, whereas PTP-PEST was demonstrated to be cleaved by caspase-3 during apoptosis (51, 59, 60). We found that incubation of cells with protease inhibitors such as caspase inhibitor (Z-VAD-fmk), calpain inhibitor (PD150606), and proteasome inhibitor (lactacystin) failed to block p130Cas and PTP-PEST cleavage induced by L. major infection (Fig. 5A). Moreover, the pattern of cleavage products for p130Cas and PTP-PEST present following infection differs from the typical caspase-mediated profile observed during apoptosis (induced by TNF-α, Fig. 5B). Correlating with this observation, the characteristic cleavage of caspase-3, which activates this enzyme during apoptosis, was not induced by L. major infection. Instead, upon exposure to L. major, cellular caspase-3 undergoes distinct proteolytic processing leading to the emergence of a slightly smaller form detected by immunoblot (Fig. 5B). Also, it appears that some p130Cas fragments were targeted to the proteasome during the infection because an additional product was detected in cells treated with lactacystin (Fig. 5A, gray arrowhead). These results suggest that L. major-induced p130Cas and PTP-PEST cleavage occurs independently of host cell factors, including three important classes of proteases.
FIGURE 5.
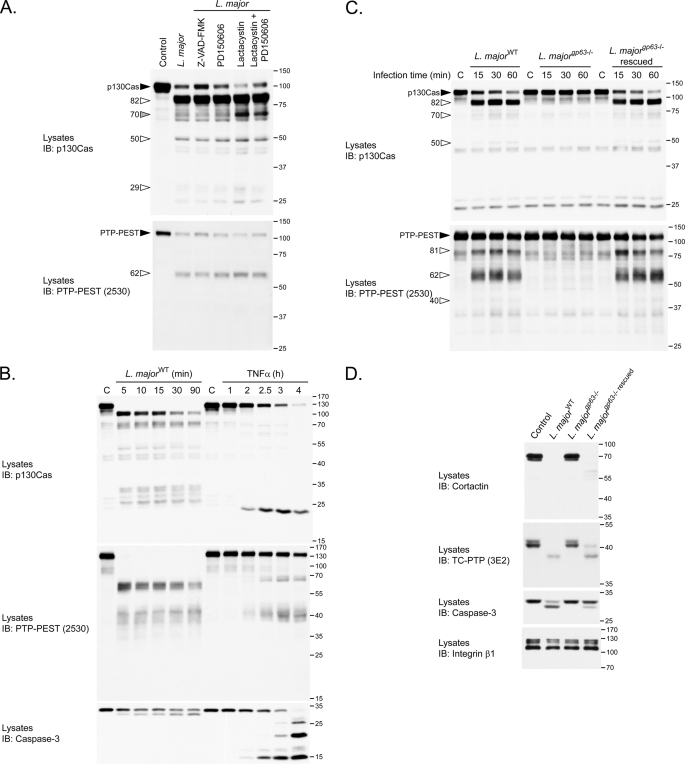
The parasitic protease GP63 is essential for L. major-induced degradation of p130Cas and PTP-PEST. A, to inhibit various endogenous cellular proteases, B11WT fibroblasts were preincubated with 100 μm Z-VAD-fmk, 100 μm PD150606, 10 μm lactacystin, or 100 μm PD150606 with 10 μm lactacystin for 2 h and subsequently exposed to L. major stationary promastigotes at a ratio of 1:20 (cells:parasites) in the presence of the same compounds for an additional 15 min. Lysates were then analyzed by immunoblotting (IB) for p130Cas and for PTP-PEST. B, B11WT cells were either treated with 10 μg/ml tumor necrosis factor-α in the presence of 10 μg/ml cycloheximide or with L. major at a ratio of 1:20 (cells:parasites) for the indicated times. Control cells for the tumor necrosis factor-α in the presence of treatment were incubated with media supplemented with 10 μg/ml cycloheximide only. C and D, B11WT cells were infected for the indicated times (C) or for 1 h 30 min (D) with L. majorwild type, L. majorgp63-/-, or L. majorgp63-/- rescue at a cell:parasite ratio of 1:20. Protein extracts were analyzed by immunoblotting for PTP-PEST (using 2530 or 2528 antibodies as indicated), p130Cas, caspase-3, cortactin, TC-PTP, and integrin-β1. C, control. Filled arrowheads identify intact proteins; the gray arrowhead points to a cleavage product specific to the lactacystin conditions, and open arrowheads point to other cleavage products. The values on the right correspond to molecular mass in kDa.
The inability of these protease inhibitors to block L. major-induced protein cleavage suggests that an enzyme of parasitic origin performs this proteolysis. The major surface metalloprotease, GP63, is the most abundant protein at the surface of the Leishmania parasite (25, 26). To determine whether GP63 plays a role in the degradation of p130Cas and PTP-PEST, we examined the consequence of incubating cells with parasites in which the genes encoding GP63 were ablated (28). In contrast to L. majorWT, L. majorgp63-/- parasites do not induce the cleavage of p130Cas and PTP-PEST (Fig. 5C). Importantly, re-introduction of gp63 into the -/- parasite (L. majorgp63-/-rescued) rescued this phenomenon. Similar observations were also obtained from primary embryonic fibroblasts placed in the presence of these different genotypes of L. major (data not shown).
Part of the infection processes of several intracellular parasites, including Leishmania, consists of taking control of specific cytoskeletal, tyrosine phosphorylation, and apoptotic modulators (13–16, 61, 62). Interestingly, we found that another cytoskeletal regulator, cortactin, and classical PTP, TC-PTP, as well as the apoptotic executioner caspase-3 were all degraded as a result of L. major infection in a GP63-dependent manner (Fig. 5D). In contrast, Crk, integrinβ1, STAT5, the MAPKs (ERK1/ERK2, JNK, and p38), Shc, focal adhesion kinase, paxillin, IκB, AKT, and PTP1B remained stable in cells incubated with L. major (Fig. 2 and Fig. 8 and supplemental Fig. 5). This underscores that the action of GP63 is specific to a subset of substrates. Together, these results identify GP63 as a novel regulator of p130Cas, PTP-PEST, cortactin, TC-PTP, and caspase-3 integrity.
FIGURE 8.
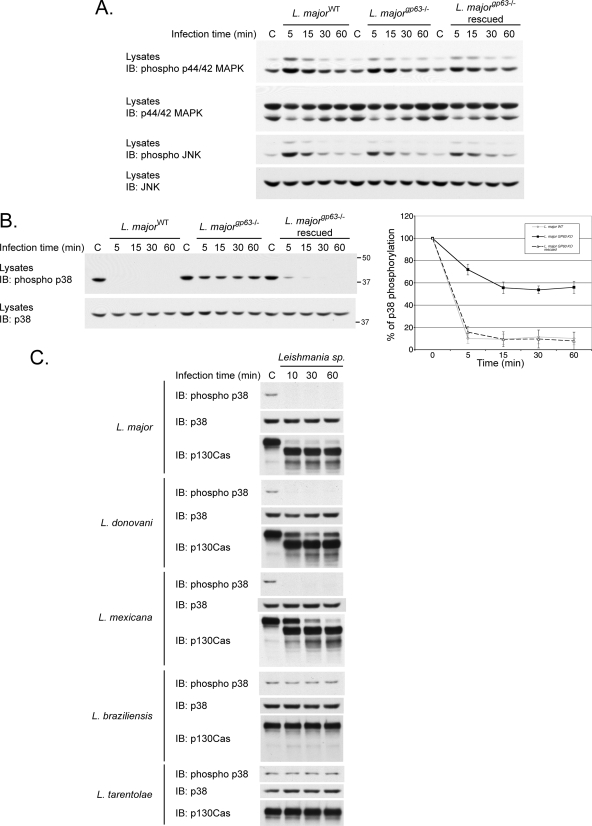
Leishmania infection modulates MAPKs and depends on GP63 activity to down-regulate p38. P-MEFs were left uninfected or incubated with L. majorWT (A, B, and C), with strains in which the GP63 gene was excised or re-introduced (L. majorgp63-/- or L. majorgp63-/- rescued) (A and B), or with L. donovani, L. mexicana, L. tarentolae, or L. braziliensis (C) for the indicated times. The activity of p44/42 (A), JNK (A), and p38 (B and C) MAPKs was measured by immunoblotting (IB) using phospho-specific antibodies for each protein. Total input of p44/42 (A), JNK (A), and p38 (B and C) as well as stability of p130Cas (C) was also measured. B, right panel, phosphorylated p38 levels were quantified and normalized to the total amount of p38 by densitometry. Values correspond to the means ± S.E. of three independent experiments. C, control.
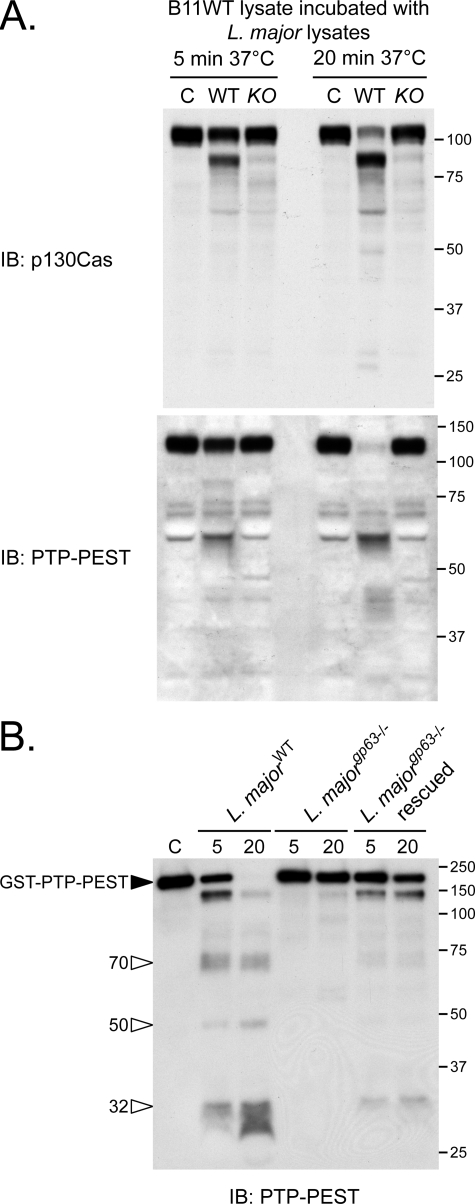
To obtain further insight into the regulation of p130Cas and PTP-PEST protein stability by GP63, we incubated fibroblast lysates or purified GST-PTP-PEST with protein extracts prepared from different L. major genotypes. As shown in Fig. 6A, the addition of extracts from WT L. major parasites to a total cell lysate induced the degradation of p130Cas and PTP-PEST following 5 and 20 min of incubation. The profile of bands observed in this in vitro assay was quite similar to that obtained when fibroblast cells were exposed to live parasites (Fig. 3). In contrast, no significant cleavage was detected, neither after 5 nor 20 min of incubation, when proteins extracted from gp63-/- L. major were added to the total cell lysate. These observations imply that the integrity of both parasitic and host cells is not essential for the cleavage reaction to occur. Importantly, purified PTP-PEST incubated with L. major lysates was also degraded when GP63 was present (Fig. 6B), confirming that cellular proteins are dispensable for the cleavage of PTP-PEST triggered by the parasite. Finally, recombinant GP63 efficiently cleaved purified GST-PTP-PEST (supplemental Fig. 6). These experiments point to GP63 as a prerequisite for p130Cas and PTP-PEST degradation and strongly suggest that these cellular signaling proteins are genuine substrates of GP63.
FIGURE 6.
L. major lysates depend on parasite expression of GP63 to induce the proteolysis of p130Cas and PTP-PEST in vitro. A, extracts from B11WT cells were incubated with lysates from stationary phase wild-type (WT) or gp63-/- (KO) L. major promastigotes or with lysis buffer (control, C) for the indicated times. B, GST-PTP-PEST was isolated from transfected PTP-PEST-/- cells, incubated under control conditions, or with either 5 or 20 μg of parasite lysates prepared from the identified L. major genotypes for 15 min. Samples were analyzed by immunoblotting (IB) for p130Cas (A) and PTP-PEST (2530 antibody) (A and B). Filled arrowheads point to intact proteins, and open arrowheads identify cleavage products. The values on the right correspond to molecular mass in kDa.
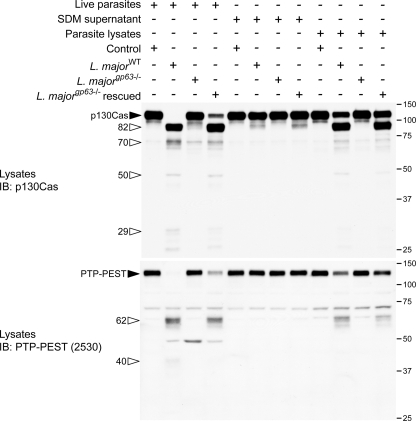
The cleavage of p130Cas and PTP-PEST occurs rapidly following the cellular exposure to the parasite. This suggests the existence of a mechanism facilitating entry of GP63 into target cells, before internalization of the protozoan commences. To test this hypothesis, we examined the impact of challenging P-MEFs with either live parasite, supernatant from promastigote cultures, or parasite lysates. As expected, incubation of P-MEFs with live parasites leads to the cleavage of both p130Cas and PTP-PEST. Interestingly, detectable levels of p130Cas cleavage products were present following exposure of the cells to media in which L. major was growing (Fig. 7, SDM supernatant). Importantly, incubation of cells with Leishmania lysates that do not contain intact parasites induced pronounced cellular p130Cas and PTP-PEST cleavage. Under all these conditions, the occurrence of cellular protein degradation was strictly dependent on the capacity of the protozoan to synthesize GP63. These findings imply a process that promotes the transfer of GP63 from the parasite to its target cells.
FIGURE 7.
Exposure of P-MEFs to L. major-conditioned medium and to parasite lysates induces protein fragmentation in a GP63-dependent manner. P-MEFs were exposed to live parasites, centrifuged culture supernatant, or lysates of L. major for 1 h. Control cells for the live parasite and parasite lysate groups were incubated with serum-free DMEM during the course of the experiment, whereas the supernatant control was incubated in fresh SDM. Fibroblast protein integrity was analyzed by immunoblotting (IB) using antibodies specific for p130Cas and PTP-PEST (2530). Filled arrowheads point to intact proteins, and open arrowheads identify cleavage products. The values on the right correspond to molecular masses in kDa.
L. major Infection Modulates MAPKs and Causes GP63-dependent Inactivation of p38—The phosphorylation of p130Cas favors JNK (63) and p44/42 MAPK activation (64), whereas the expression of PTP-PEST in B-cells interferes with Ras-mediated p44/42 phosphorylation (65), and promotes the activation of p38 in fibroblasts stimulated with anisomycin (supplemental Fig. 7). To evaluate the impact of GP63 on the activity of host MAPKs, we measured the phosphorylation levels of three members of the MAPKs (p44/42, JNK, and p38) in P-MEFs exposed to different strains of L. major. Immunoblotting using a phospho-specific antibody shows an increase in p44/42 MAPK phosphorylation in cells incubated with L. major for 5 min, which is followed by a gradual decrease as the infection progressed (Fig. 8A). This transient ERK activation occurred similarly in cells infected with parasites both positive and negative for GP63 (WT, gp63-/-, or gp63-/- rescued). Likewise, JNK was up- and down-regulated independently of gp63 genotype. In contrast, a dramatic dephosphorylation of p38 occurred following L. major infection (Fig. 8B). Interestingly, parasites lacking GP63 induced only a partial dephosphorylation of p38 (Fig. 8B). Noteworthy, re-introduction of gp63 in the L. majorgp63-/- strain (rescued) was sufficient to restore its capacity to inactivate p38 (Fig. 8B). Because our results, presented in Fig. 4, revealed that the ability to provoke p130Cas and PTP-PEST cleavage is species-specific, we sought to verify the possible correlation between the proteolytic capacity of certain species and the modulation of p38 phosphorylation. Treatment of P-MEFs with L. major, L. donovani, and L. mexicana but not with L. tarentolae nor L. braziliensis caused the disappearance of the phosphorylated form of p38 (Fig. 8C). This inactivation of p38 correlated with the ability of the parasite to induce protein cleavage as detected by p130Cas degradation. The reduced level of phosphorylated p38 was not due to its degradation since the total amount of p38 remained constant throughout all tested infections (Fig. 8, B and C). Altogether, these results reveal a novel mechanism of p38 regulation dependent on protein cleavage in which GP63 appears to play a key role.
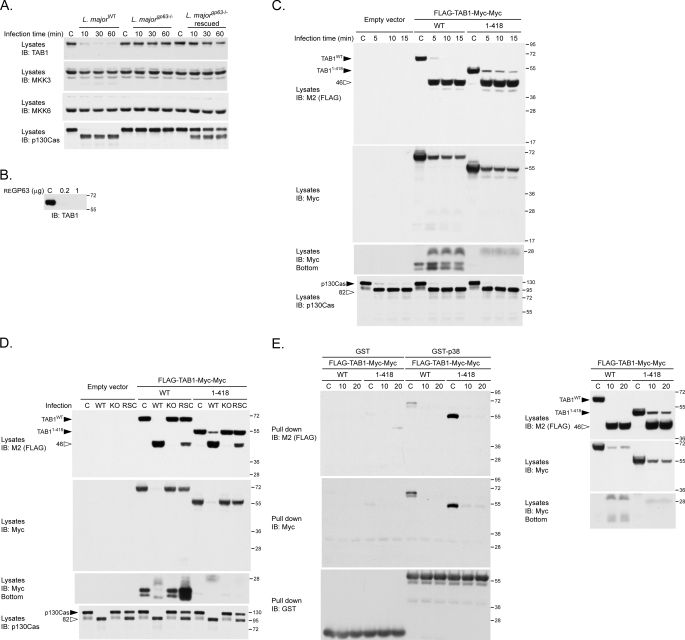
GP63 Cleaves the p38 Regulator TAB1 during L. major Infection, Generating Products Unable to Bind p38—The modulation of MAPKs observed during the infection with L. major prompted us to analyze molecules that lie upstream of p38. Even though PTP-PEST expression regulates p38 in certain contexts, we did not observe differences in p38 modulation between PTP-PEST-/- and re-expressing fibroblasts treated with L. major (data not shown). As direct regulators of p38, the MAPK kinase (MKK) MKK3 and MKK6 can phosphorylate its activation loop, whereas binding of the scaffolding protein TAB1 induces its autophosphorylation (48, 49). L. major infection induced a marked disappearance of cellular TAB1, whereas MKK3 and MKK6 levels remained constant (Fig. 9A). The diminished TAB1 levels correlated with the presence of GP63 on L. major and paralleled p130Cas degradation. Moreover, recombinant his-TAB1 is degraded by purified GP63 (Fig. 9B). These data identify TAB1 as a novel substrate of GP63.
FIGURE 9.
L. major regulates p38/TAB1 binding via GP63-mediated proteolysis of TAB1. A, P-MEFs were incubated with L. majorWT, L. majorgp63-/-, or L. majorgp63-/- rescued for the indicated duration. Protein lysates were collected, and the stability of the p38 regulators TAB1, MKK3, and MKK6 was verified by immunoblotting (IB). B, His-TAB1 purified from expressing bacteria was incubated under control conditions (C) or with 0.2 or 1 μg of recombinant GP63 for 30 min. The stability of TAB1 was then analyzed by immunoblotting using anti-TAB1 antibodies. C, PTP-PEST-/- cells transfected with an empty vector or with vectors encoding either FLAG-TAB1 (WT)-Myc-Myc or FLAG-TAB1-(1–418)-Myc-Myc were incubated under control conditions (C) or with L. majorWT at a ratio of 1:20 (cells:parasites) for the indicated time. Protein lysates were harvested and analyzed by immunoblotting using the indicated antibodies. D, PTP-PEST-/- cells transfected with an empty control vector or with vectors encoding FLAG-TAB1 (WT)-Myc-Myc or FLAG-TAB1-(1–418)-Myc-Myc were incubated under control conditions (C) or with L. majorWT, or L. majorgp63-/- (KO), or L. majorgp63-/--rescue (RSC) at a ratio of 1:20 (cells:parasites) for 15 min. Protein lysates were harvested and analyzed by immunoblotting using the indicated antibodies. E, PTP-PEST-/- cells expressing either FLAG-TAB1 (WT)-Myc-Myc or FLAG-TAB1-(1–418)-Myc-Myc were incubated under control conditions (C) or with L. majorWT at a ratio of 1:20 (cells:parasites) for the indicated time. Lysates extracted from infected and noninfected cells were incubated with either GST or GST-p38 immobilized on glutathione-Sepharose beads, and the ability of the GST fusion proteins to interact with various forms of TAB1 was investigated by immunoblotting for the FLAG and Myc epitopes. The quantity of GST fusion proteins used in each assay was assessed by immunoblot against GST. The presence of TAB1 proteins in each extracts was also verified by immunoblotting with the indicated antibody (E, right panel). The values on the right correspond to molecular mass in kDa.
To examine further the mode of TAB1 proteolysis, we exposed cells expressing different forms of TAB1 (WT or 1–418), flanked by FLAG (N terminus) and Myc (C terminus) epitopes, to L. major. Truncation of the C-terminal portion of TAB1 was previously found to generate a form of the protein (TAB1 1–418) that displays increased affinity for p38 and contains the residues essential for their interaction (49). Both versions of TAB1 were degraded in infected cells and produced the same N-terminal fragment (46 kDa) detected by antibodies against FLAG (Fig. 9C). The C-terminal cleavage products detected via the Myc tag were substantially less abundant and therefore appear only upon longer exposure (Fig. 9C, IB: myc). In addition, profile of these C-terminal fragments differed according to the version of TAB1 expressed (WT or 1–418) (Fig. 9C, IB: myc, bottom). In all cases, the efficient generation of cleavage products depended on the parasitic expression of GP63 (Fig. 9D). Thus, GP63-mediated TAB1 proteolysis generates a stable N-terminal fragment and other smaller C-terminal products, suggesting that GP63 cleaves TAB1 at multiple sites. Importantly, L. major infection impaired p38 binding to TAB1 (both WT and 1–418) (Fig. 9E). Specifically, none of the TAB1 cleavage products (FLAG- or Myc-tagged) generated during L. major infection were able to interact with GST-p38. Together, these results show that GP63 processes TAB1, which alters the formation of TAB1-p38 complexes and could thereby modulate p38 signaling.
DISCUSSION
Upon inoculation into a mammalian organism, Leishmania parasites must adapt to this foreign environment and take advantage of their immediate surrounding components and cells. During this process, molecules covering the parasite undoubtedly play a critical role as they are directly in contact with their extracellular milieu. Herein, we demonstrate that the modulation of multiple mammalian signaling molecules occurs early during infection by Leishmania. Importantly, GP63 appears to be a critical player in disrupting the state of several signaling proteins.
Even though macrophages are believed to be the primary target of Leishmania, this parasite is capable of colonizing other cell types (6, 66–69). Fibroblasts are abundant in the immediate environment where promastigotes are inoculated, can harbor parasites in animal models, and were proposed to play an important role for long term infection (6). Our data confirm that L. major is capable of infecting cultured fibroblasts isolated from mouse embryos. The accumulation of parasites within them gave rise to heavily infected spheroids similar to those typically observed in spleen biopsies from infected individuals (2). Interestingly, no nuclear condensation or caspase activation characteristic of apoptotic cells was detected in highly infected fibroblasts despite the intense stress undoubtedly induced by the presence of parasites. Correspondingly, Leishmania mediates a delay in programmed cell death induction in neutrophils and macrophages (70, 71). This implies that the parasite induces modifications in the cellular signaling machinery early during the infectious process to avoid activation of the apoptotic program. Besides the involvement of the mannose receptors in the attachment of Leishmania to fibroblasts and the implication of Cdc42 in parasitic internalization (13, 72), little else is known about the molecular events occurring within fibroblasts during Leishmania infection. Here we have shown that the interaction between Leishmania and fibroblasts induces several additional modifications of host signaling proteins.
Leishmania provoked important changes in the tyrosine phosphorylation level of several proteins (Fig. 2). The effect of inhibition of PTKs illustrates their requirement for the internalization of L. donovani by macrophages (73). On the other hand, the modulation of tyrosine phosphorylation content in macrophages infected with L. donovani was associated with the general activation of PTPs, including SHP-1 (15). Our results identify another PTP, PTP-PEST, that is proteolyzed in cells that encounter Leishmania. This post-translational modification of PTP-PEST could modulate its enzymatic properties, as the cleavage of PTP-PEST was shown to augment its catalytic activity (51). Additionally, Leishmania induced GP63-dependent cleavage of TC-PTP likely near its C-terminal nuclear localization signal (supplemental Fig. 8). Removal of the nuclear localization signal could allow the phosphatase to access additional substrates and also enhance its catalytic activity (74–76). The altered tyrosine phosphorylation profile found in cells infected with Leishmania may also be due to the cleavage of highly phosphorylated proteins such as p130Cas. The phosphorylation of p130Cas fragments was likely maintained because they remained capable of binding Crk. Thus, the combined regulation of PTKs, PTPs, and protein stability is likely responsible for the dramatic changes in phosphoprotein levels in Leishmania-infected cells.
Proteolysis of cellular proteins, including p130Cas and PTP-PEST during the Leishmania infective process, could be a maneuver used by the parasite to take control of the cellular machinery. PTP-PEST and p130Cas cleavage occurred independently of growth conditions or the presence of various cellular protease inhibitors, which implies that the host cell proteolytic apparatus is not activated during L. major infection. In contrast, in in vivo and in vitro analysis, taking advantage of L. major strains positive or negative for gp63 gene expression supports the identification of p130Cas and PTP-PEST as novel, genuine substrates of the parasite protease GP63. Additionally, the in vitro cleavage of PTP-PEST by recombinant GP63 indicates that it is directly responsible for this proteolytic event. Interestingly, cells exposed to L. major, L. mexicana, or L. donovani all exhibited p130Cas and PTP-PEST proteolysis, whereas those incubated with L. braziliensis or L. tarentolae did not. Genetic analysis based on gp63 gene organization and sequences has grouped members of the Viannia subgenus (which includes L. braziliensis) in a separate cluster from other Leishmania species (77). Divergence in the composition of L. braziliensis gp63 genes could limit their access to intracellular substrates or modify their specificity, thereby explaining the limited cleavage observed in our experiments. On the other hand, the lizard parasite L. tarentolae possesses a variant GP63 lacking enzymatic activity (78). Experiments using genetically modified strains of L. major led to the identification of additional substrates of GP63 as follows: cortactin, TC-PTP, caspase-3, and TAB1. Even though GP63 is targeting several substrates, we believe that this enzyme does not cause general protein degradation because the majority of signaling proteins tested were resistant to its presence. The specific contributions of each of these individual cleavage events to Leishmania pathology remain unresolved. Nonetheless, the GP63 substrates we have identified are implicated in multiple physiologic functions, including cytoskeletal rearrangement, cell proliferation, and apoptosis, pointing to several potential avenues by which the parasite may take advantage of its host.
The secretion and transfer of virulence factors into host cells is of paramount importance for the pathogenic processes of several microorganisms (79). Here we have provided evidence for the rapid cleavage of p130Cas and PTP-PEST upon initial contact between L. major and fibroblast cells, presumably before internalization of the parasite. The previously reported cleavage of the intracellular protein MRP (47) as well as the activity of GP63 found on amastigotes (39) are also indicative of an important role for GP63 inside mammalian cells. The capability of the parasite to induce protein degradation before it enters into its host cells implies a mechanism allowing the transfer of GP63 to the intracellular space of its host. Clinical isolates causing cutaneous (Leishmania tropica) or visceral (Leishmania infantum) leishmaniasis as well as L. amazonensis, L. major, L. mexicana, and L. donovani were observed to release proteolytically active GP63 in culture supernatants (36–38). Recent investigations performed with Leishmania chagasi indicated that incubation of stationary phase promastigotes under parameters reproducing the extracellular mammalian host environment (37 °C and Matrigel) stimulated the secretion of internal GP63 (35). Furthermore, in amastigotes, the majority of GP63 is localized in the flagellar pocket of the parasite, which is the principal site of exocytosis (44), indicating a role for the liberation of GP63 inside the host cell. During the course of our experiments, we noticed that throughout the initial contact between the protozoan and the fibroblasts, a large proportion of the microorganisms presented their flagellum (and flagellar pocket) toward the mammalian cells.6 This behavior could allow parasites to concentrate their secretion products, including GP63, in the direction of their targeted cell. Remarkably, incubation of fibroblasts with L. major culture supernatant and even parasite lysates led to the appearance of degradation products, which correlated with GP63 protein expression. These observations reveal that L. major secretes or contains all the components necessary for GP63 entry and that this phenomenon does not require parasitic integrity. Interestingly, a recent study on L. donovani reported the presence of microvesicles budding from the flagellar pocket and identified a set of 151 distinct proteins secreted by the parasite (80). We postulate that during the Leishmania/mammalian cell initial interaction, release of GP63 in close vicinity to the host cell surface, possibly with other secreted transport effectors, facilitates its entry into the cytosolic space, allowing it to reach additional substrates.
Reorganization of the actin cytoskeleton plays a central role in the internalization of many intracellular parasites. Interestingly, heat killing of L. amazonensis amastigotes, a process that also abrogates GP63 activity (45), prevent their internalization in CHO cells, supporting a unique property of live amastigotes to accomplish entry into cells (13). The small Rho-GTPase Cdc42 is also necessary for Leishmania entry into CHO cells (13). Cdc42 was previously shown to signal toward WASP and N-WASP to stimulate actin nucleation (81). During intracellular trafficking of Leishmania, WASP, vinculin, Arp2/3, and other cytoskeletal regulators gather with actin filaments around engulfed parasites to form an actin cup structure thought to protect the foreign parasite from phagolysosomal digestion (13, 14, 61). Cortactin, another actin regulator that interacts with Arp2/3 and N-WASP, is similarly recruited to actin-rich structures exploited by other intracellular microbes (9). The assembly of the PTP-PEST-PSTPIP-WASP complexes was previously shown to allow PTP-PEST to dephosphorylate WASP (19) and to inhibit WASP-induced actin polymerization (82). It was recently demonstrated that caspase-3-mediated cleavage of PTP-PEST dissociates its phosphatase domain from PSTPIP and was expected to prevent it from dephosphorylating and inhibiting WASP (51, 83). Importantly, we show that GP63 mediates the degradation of PTP-PEST during Leishmania infection, which could function to affect parasite-induced actin cytoskeleton remodeling. Moreover, we observed that p130Cas and cortactin were cleaved in cells exposed to parasites expressing GP63. MRP, another identified substrate of GP63, is associated with actin filaments in macrophages cells (47). GP63 was also shown to interact with fibronectin receptor (integrin) and to stimulate the internalization of Leishmania parasites (40). Thus, we propose that GP63 will act from both outside and inside the cell to alter the activity of host cell signaling molecules. This modulation may assist parasite engulfment through the formation of actin fibers and membrane protrusions. Future investigations will attempt to decipher the precise role of GP63 in modulating the dynamic remodeling of the cytoskeleton.
Some inconsistencies are present in the literature regarding the response of the p38 MAPK to Leishmania infection. Several reports suggest that the parasite either actively inhibits or avoids activating p38 (22, 23, 84), whereas others indicate that p38 is induced during infection (21, 85–87). Despite the discrepancy, most reports affirm that activation of p38 is detrimental to parasite survival. Here we report that activating phosphorylation of p38 is diminished following exposure of fibroblasts to L. major and correlates with the capability of GP63 to cleave intracellular substrates. Moreover, the ability to down-regulate p38 was specific to certain Leishmania species; L. major, L. mexicana, and L. donovani caused complete inactivation of p38, a task which L. tarentolae and L. brasiliensis failed to perform. These results underscore that Leishmania requires functional GP63 to achieve inactivation of p38. Importantly, this GP63-dependent cellular response was unique to p38. In contrast, the p44/42 and JNK MAPKs reacted similarly in response to infection, regardless of gp63 genotype. Leishmania did not entirely monopolize the host signaling machinery because infected cells maintained the ability to activate MAPKs (p44/42) and STAT-5 following growth hormone stimulation as well as MAPKs (p44/42) and AKT downstream of lysophosphatidic acid stimulation (data not shown). Interestingly, another pathogenic microorganism, Bacillus anthracis, also secretes a metalloproteinase termed lethal factor to inhibit MAPK signaling (88). In this case, lethal factor cleaves the MKKs, preventing efficient MAPK activation (88). Thus, we verified the integrity of p38 upstream regulators in cells submitted to L. major. Although we did not notice any effect on MKK3 nor MKK6, the adaptor molecule TAB1 was depleted upon infection. We then showed that TAB1 is cleaved in a GP63-dependent manner, generating fragments unable to interact with p38. As the p38/TAB1 interaction was previously shown to modulate p38 activity (49), we believe that alterations in the stability of this complex caused by GP63 could contribute to the decreased p38 phosphorylation occurring during Leishmania infection.
As the most abundant protein covering the surface of Leishmania (25, 26), GP63 undoubtedly plays fundamental roles in signals initiated upon contact with host cells. In the extracellular milieu within mammalian hosts, parasites are significantly protected from complement-mediated lysis by GP63 (27, 29, 89). Nevertheless, the vulnerability of GP63-deficient L. major to the effect of complement may not be solely responsible for their reduced infectivity in an animal model (28). Indeed, additional functions, including the promotion of host cell attachment and internalization, have been attributed to GP63 (40, 41). In addition, the GP63-mediated degradation of extracellular matrix components such as fibronectin facilitates parasite dissemination and inhibits the activation of the protective response of infected macrophages (34, 39). A limited number of intracellular functions have also been ascribed to GP63. For example, two intracellular substrates of GP63, MRP and NF-κB p65RelA, have been identified (47, 52). Also, expression and activity of GP63 are important to protect engulfed parasites during phagolysosomal transition (45, 46). Our results expand significantly the number of known targets of GP63 and emphasize its importance in the parasitic program that remodels intracellular signaling networks of host cells in its proximity.
The intrusion of virulence factors and the exploitation of cellular components are crucial strategies for host invasion by a wide array of pathogenic microorganisms. In this study, we have uncovered a series of intracellular effects of Leishmania in fibroblasts, a potentially important target cell of this parasite. Our results point to the importance of the metalloprotease GP63 in regulating several important signaling proteins, contributing to downstream changes in global protein tyrosine phosphorylation levels as well as a specific effect on p38 MAPK activation. In addition, proteins modulating apoptosis and the actin cytoskeleton are over-represented among the identified GP63 targets. Thus, our results suggest novel mechanisms by which GP63 could actively participate in the conditioning of host cells through the modulation of both signaling and structural regulators. If these changes are prerequisite for efficient infection by Leishmania, our results could contribute to the development of drugs that would impair host cell invasion by this virulent parasite.
Supplementary Material
Acknowledgments
We thank Dr. Greg Matlashewski, Dr. Laurent Lessard, and Karen Doody for critical reviews of the manuscript. We thank Jacynthe Laliberté for advice on confocal microscopy and Dr. Serge Hardy for help with apoptosis assays.
This work was supported by Canadian Institutes of Health Research Grant MOP-12466, National Cancer Institute of Canada Grant 015200, and operating grants (to M. L. T.). The M. O. laboratory is supported by Canadian Institutes of Health Research Grant MOP-12671. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8.
Footnotes
World Health Organization (2008) Leishmaniasis, The Disease and Its Epidemiology, http://www.who.int/leishmaniasis/disease_epidemiology/en/index.html.
The abbreviations used are: WASP, Wiskott-Aldrich syndrome protein; CHO, Chinese hamster ovary; p130Cas, Crk-associated substrate; P-MEFs, primary mouse embryonic fibroblast cells; PTP, protein-tyrosine phosphatase; PTK, protein-tyrosine kinase; TC-PTP, T-cell PTP; WT, wild type; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; FBS, fetal bovine serum; GST, glutathione S-transferase; Z, benzyloxycarbonyl; fmk, fluoromethyl ketone; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; MKK, MAPK kinase; P-MEF, primary mouse embryonic fibroblast; MRP, myristoylated alanine-rich C kinase substrate-related protein; JNK, c-Jun N-terminal kinase; PEST, Pro-Glu-Ser-Thr.
M. Hallé, M. Olivier, and M. L. Tremblay, unpublished observations.
References
- 1.Cunningham, A. C. (2002) Exp. Mol. Pathol. 72 132-141 [DOI] [PubMed] [Google Scholar]
- 2.Roberts, L. S., Janovy, J., and Schmidt, G. D. (2000) in Gerald D. Schmidt & Larry S. Roberts' Foundations of Parasitology (Roberts, L. S., and Janovy, J., Jr., eds) 7th Ed., pp. 61-88, McGraw-Hill, New York
- 3.Deleted in proof
- 4.Kamhawi, S. (2006) Trends Parasitol. 22 439-445 [DOI] [PubMed] [Google Scholar]
- 5.Hepburn, N. C. (2000) Clin. Exp. Dermatol. 25 363-370 [DOI] [PubMed] [Google Scholar]
- 6.Bogdan, C., Donhauser, N., Doring, R., Rollinghoff, M., Diefenbach, A., and Rittig, M. G. (2000) J. Exp. Med. 191 2121-2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens, J. M., Galyov, E. E., and Stevens, M. P. (2006) Nat. Rev. Microbiol. 4 91-101 [DOI] [PubMed] [Google Scholar]
- 8.Rottner, K., Lommel, S., Wehland, J., and Stradal, T. E. (2004) J. Pathol. 204 396-406 [DOI] [PubMed] [Google Scholar]
- 9.Selbach, M., and Backert, S. (2005) Trends Microbiol. 13 181-189 [DOI] [PubMed] [Google Scholar]
- 10.Weidow, C. L., Black, D. S., Bliska, J. B., and Bouton, A. H. (2000) Cell Microbiol. 2 549-560 [DOI] [PubMed] [Google Scholar]
- 11.Gouin, E., Welch, M. D., and Cossart, P. (2005) Curr. Opin. Microbiol. 8 35-45 [DOI] [PubMed] [Google Scholar]
- 12.Stamm, L. M., Pak, M. A., Morisaki, J. H., Snapper, S. B., Rottner, K., Lommel, S., and Brown, E. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14837-14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morehead, J., Coppens, I., and Andrews, N. W. (2002) Infect. Immun. 70 4571-4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodge, R., and Descoteaux, A. (2005) Cell Microbiol. 7 1647-1658 [DOI] [PubMed] [Google Scholar]
- 15.Blanchette, J., Racette, N., Faure, R., Siminovitch, K. A., and Olivier, M. (1999) Eur. J. Immunol. 29 3737-3744 [DOI] [PubMed] [Google Scholar]
- 16.Nandan, D., Yi, T., Lopez, M., Lai, C., and Reiner, N. E. (2002) J. Biol. Chem. 277 50190-50197 [DOI] [PubMed] [Google Scholar]
- 17.Garton, A. J., Flint, A. J., and Tonks, N. K. (1996) Mol. Cell. Biol. 16 6408-6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garton, A. J., and Tonks, N. K. (1999) J. Biol. Chem. 274 3811-3818 [DOI] [PubMed] [Google Scholar]
- 19.Côté, J. F., Chung, P. L., Théberge, J. F., Hallé, M., Spencer, S., Lasky, L. A., and Tremblay, M. L. (2002) J. Biol. Chem. 277 2973-2986 [DOI] [PubMed] [Google Scholar]
- 20.Angers-Loustau, A., Côté, J. F., Charest, A., Dowbenko, D., Spencer, S., Lasky, L. A., and Tremblay, M. L. (1999) J. Cell Biol. 144 1019-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junghae, M., and Raynes, J. G. (2002) Infect. Immun. 70 5026-5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Awasthi, A., Mathur, R., Khan, A., Joshi, B. N., Jain, N., Sawant, S., Boppana, R., Mitra, D., and Saha, B. (2003) J. Exp. Med. 197 1037-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathur, R. K., Awasthi, A., Wadhone, P., Ramanamurthy, B., and Saha, B. (2004) Nat. Med. 10 540-544 [DOI] [PubMed] [Google Scholar]
- 24.Kima, P. E. (2007) Int. J. Parasitol. 37 1087-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao, C., Donelson, J. E., and Wilson, M. E. (2003) Mol. Biochem. Parasitol. 132 1-16 [DOI] [PubMed] [Google Scholar]
- 26.Bouvier, J., Etges, R. J., and Bordier, C. (1985) J. Biol. Chem. 260 15504-15509 [PubMed] [Google Scholar]
- 27.Joshi, P. B., Sacks, D. L., Modi, G., and McMaster, W. R. (1998) Mol. Microbiol. 27 519-530 [DOI] [PubMed] [Google Scholar]
- 28.Joshi, P. B., Kelly, B. L., Kamhawi, S., Sacks, D. L., and McMaster, W. R. (2002) Mol. Biochem. Parasitol. 120 33-40 [DOI] [PubMed] [Google Scholar]
- 29.Brittingham, A., Morrison, C. J., McMaster, W. R., McGwire, B. S., Chang, K. P., and Mosser, D. M. (1995) J. Immunol. 155 3102-3111 [PubMed] [Google Scholar]
- 30.Kulkarni, M. M., McMaster, W. R., Kamysz, E., Kamysz, W., Engman, D. M., and McGwire, B. S. (2006) Mol. Microbiol. 62 1484-1497 [DOI] [PubMed] [Google Scholar]
- 31.Chaudhuri, G., and Chang, K. P. (1988) Mol. Biochem. Parasitol. 27 43-52 [DOI] [PubMed] [Google Scholar]
- 32.Yao, C., Leidal, K. G., Brittingham, A., Tarr, D. E., Donelson, J. E., and Wilson, M. E. (2002) Mol. Biochem. Parasitol. 121 119-128 [DOI] [PubMed] [Google Scholar]
- 33.Yao, C., Luo, J., Storlie, P., Donelson, J. E., and Wilson, M. E. (2004) Mol. Biochem. Parasitol. 135 171-183 [DOI] [PubMed] [Google Scholar]
- 34.McGwire, B. S., Chang, K. P., and Engman, D. M. (2003) Infect. Immun. 71 1008-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, C., Donelson, J. E., and Wilson, M. E. (2007) Eukaryot. Cell 6 1905-1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis, M., Sharma, D. K., Hilley, J. D., Coombs, G. H., and Mottram, J. C. (2002) J. Biol. Chem. 277 27968-27974 [DOI] [PubMed] [Google Scholar]
- 37.McGwire, B. S., O'Connell, W. A., Chang, K. P., and Engman, D. M. (2002) J. Biol. Chem. 277 8802-8809 [DOI] [PubMed] [Google Scholar]
- 38.Jaffe, C. L., and Dwyer, D. M. (2003) Parasitol. Res. 91 229-237 [DOI] [PubMed] [Google Scholar]
- 39.Kulkarni, M. M., Jones, E. A., McMaster, W. R., and McGwire, B. S. (2008) Infect. Immun. 76 1738-1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brittingham, A., Chen, G., McGwire, B. S., Chang, K. P., and Mosser, D. M. (1999) Infect. Immun. 67 4477-4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, D. Q., Kolli, B. K., Yadava, N., Lu, H. G., Gilman-Sachs, A., Peterson, D. A., and Chang, K. P. (2000) Infect. Immun. 68 80-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizvi, F. S., Ouaissi, M. A., Marty, B., Santoro, F., and Capron, A. (1988) Eur. J. Immunol. 18 473-476 [DOI] [PubMed] [Google Scholar]
- 43.Voth, B. R., Kelly, B. L., Joshi, P. B., Ivens, A. C., and McMaster, W. R. (1998) Mol. Biochem. Parasitol. 93 31-41 [DOI] [PubMed] [Google Scholar]
- 44.Hsiao, C. H., Yao, C., Storlie, P., Donelson, J. E., and Wilson, M. E. (2008) Mol. Biochem. Parasitol. 157 148-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhuri, G., Chaudhuri, M., Pan, A., and Chang, K. P. (1989) J. Biol. Chem. 264 7483-7489 [PubMed] [Google Scholar]
- 46.Seay, M. B., Heard, P. L., and Chaudhuri, G. (1996) Infect. Immun. 64 5129-5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corradin, S., Ransijn, A., Corradin, G., Roggero, M. A., Schmitz, A. A., Schneider, P., Mauel, J., and Vergeres, G. (1999) J. Biol. Chem. 274 25411-25418 [DOI] [PubMed] [Google Scholar]
- 48.Zarubin, T., and Han, J. (2005) Cell Res. 15 11-18 [DOI] [PubMed] [Google Scholar]
- 49.Ge, B., Gram, H., Di Padova, F., Huang, B., New, L., Ulevitch, R. J., Luo, Y., and Han, J. (2002) Science 295 1291-1294 [DOI] [PubMed] [Google Scholar]
- 50.Ibarra-Sanchez, M. J., Wagner, J., Ong, M. T., Lampron, C., and Tremblay, M. L. (2001) Oncogene 20 4728-4739 [DOI] [PubMed] [Google Scholar]
- 51.Hallé, M., Liu, Y. C., Hardy, S., Théberge, J. F., Blanchetot, C., Bourdeau, A., Meng, T. C., and Tremblay, M. L. (2007) Mol. Cell. Biol. 27 1172-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gregory, D. J., Godbout, M., Contreras, I., Forget, G., and Olivier, M. (2008) Eur. J. Immunol. 38 1071-1081 [DOI] [PubMed] [Google Scholar]
- 53.Button, L. L., Wilson, G., Astell, C. R., and McMaster, W. R. (1993) Gene (Amst.) 134 75-81 [DOI] [PubMed] [Google Scholar]
- 54.Côté, J. F., Charest, A., Wagner, J., and Tremblay, M. L. (1998) Biochemistry 37 13128-13137 [DOI] [PubMed] [Google Scholar]
- 55.Sastry, S. K., Rajfur, Z., Liu, B. P., Cote, J. F., Tremblay, M. L., and Burridge, K. (2006) J. Biol. Chem. 281 11627-11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jamieson, J. S., Tumbarello, D. A., Hallé, M., Brown, M. C., Tremblay, M. L., and Turner, C. E. (2005) J. Cell Sci. 118 5835-5847 [DOI] [PubMed] [Google Scholar]
- 57.Shi, J., and Casanova, J. E. (2006) Mol. Biol. Cell 17 4698-4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Defilippi, P., Di Stefano, P., and Cabodi, S. (2006) Trends Cell Biol. 16 257-263 [DOI] [PubMed] [Google Scholar]
- 59.Shim, S. R., Kook, S., Kim, J. I., and Song, W. K. (2001) Biochem. Biophys. Res. Commun. 286 601-608 [DOI] [PubMed] [Google Scholar]
- 60.Kook, S., Shim, S. R., Choi, S. J., Ahnn, J., Kim, J. I., Eom, S. H., Jung, Y. K., Paik, S. G., and Song, W. K. (2000) Mol. Biol. Cell 11 929-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lerm, M., Holm, A., Seiron, A., Sarndahl, E., Magnusson, K. E., and Rasmusson, B. (2006) Infect. Immun. 74 2613-2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carmen, J. C., and Sinai, A. P. (2007) Mol. Microbiol. 64 904-916 [DOI] [PubMed] [Google Scholar]
- 63.Kyaw, M., Yoshizumi, M., Tsuchiya, K., Kagami, S., Izawa, Y., Fujita, Y., Ali, N., Kanematsu, Y., Toida, K., Ishimura, K., and Tamaki, T. (2004) Mol. Pharmacol. 65 832-841 [DOI] [PubMed] [Google Scholar]
- 64.Stuart, L. M., Bell, S. A., Stewart, C. R., Silver, J. M., Richard, J., Goss, J. L., Tseng, A. A., Zhang, A., El Khoury, J. B., and Moore, K. J. (2007) J. Biol. Chem. 282 27392-27401 [DOI] [PubMed] [Google Scholar]
- 65.Davidson, D., and Veillette, A. (2001) EMBO J. 20 3414-3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woelbing, F., Kostka, S. L., Moelle, K., Belkaid, Y., Sunderkoetter, C., Verbeek, S., Waisman, A., Nigg, A. P., Knop, J., Udey, M. C., and von Stebut, E. (2006) J. Exp. Med. 203 177-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Zandbergen, G., Klinger, M., Mueller, A., Dannenberg, S., Gebert, A., Solbach, W., and Laskay, T. (2004) J. Immunol. 173 6521-6525 [DOI] [PubMed] [Google Scholar]
- 68.Belle, E. A. (1958) Can. Med. Assoc. J. 79 726-728 [PMC free article] [PubMed] [Google Scholar]
- 69.Oliveira, S. H., Fonseca, S. G., Romao, P. R., Figueiredo, F., Ferreira, S. H., and Cunha, F. Q. (1998) Parasite Immunol. 20 405-412 [DOI] [PubMed] [Google Scholar]
- 70.Aga, E., Katschinski, D. M., van Zandbergen, G., Laufs, H., Hansen, B., Muller, K., Solbach, W., and Laskay, T. (2002) J. Immunol. 169 898-905 [DOI] [PubMed] [Google Scholar]
- 71.Moore, K. J., and Matlashewski, G. (1994) J. Immunol. 152 2930-2937 [PubMed] [Google Scholar]
- 72.Hespanhol, R. C., de Nazare, C. S. M., Meuser, M. B., de Nazareth, S. L. M. M., and Corte-Real, S. (2005) J. Histochem. Cytochem. 53 35-44 [DOI] [PubMed] [Google Scholar]
- 73.Ghosh, D., and Chakraborty, P. (2002) Biosci. Rep. 22 395-406 [DOI] [PubMed] [Google Scholar]
- 74.Zhao, Z., Zander, N. F., Malencik, D. A., Anderson, S. R., and Fischer, E. H. (1992) Anal. Biochem. 202 361-366 [DOI] [PubMed] [Google Scholar]
- 75.Cool, D. E., Tonks, N. K., Charbonneau, H., Fischer, E. H., and Krebs, E. G. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 7280-7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hao, L., Tiganis, T., Tonks, N. K., and Charbonneau, H. (1997) J. Biol. Chem. 272 29322-29329 [DOI] [PubMed] [Google Scholar]
- 77.Victoir, K., Arevalo, J., De Doncker, S., Barker, D. C., Laurent, T., Godfroid, E., Bollen, A., Le Ray, D., and Dujardin, J. C. (2005) Parasitology 131 207-214 [DOI] [PubMed] [Google Scholar]
- 78.Campbell, D. A., Kurath, U., and Fleischmann, J. (1992) FEMS Microbiol. Lett. 75 89-92 [DOI] [PubMed] [Google Scholar]
- 79.Coburn, B., Sekirov, I., and Finlay, B. B. (2007) Clin. Microbiol. Rev. 20 535-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silverman, J. M., Chan, S. K., Robinson, D. P., Dwyer, D. M., Nandan, D., Foster, L. J., and Reiner, N. E. (2008) Genome Biol. 9 R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higgs, H. N., and Pollard, T. D. (2001) Annu. Rev. Biochem. 70 649-676 [DOI] [PubMed] [Google Scholar]
- 82.Badour, K., Zhang, J., Shi, F., Leng, Y., Collins, M., and Siminovitch, K. A. (2004) J. Exp. Med. 199 99-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hallé, M., Tremblay, M. L., and Meng, T. C. (2007) Cell Cycle 6 2773-2781 [DOI] [PubMed] [Google Scholar]
- 84.Privé, C., and Descoteaux, A. (2000) Eur. J. Immunol. 30 2235-2244 [DOI] [PubMed] [Google Scholar]
- 85.Liu, L., Wang, L., Zhao, Y., Wang, Y., Wang, Z., and Qiao, Z. (2006) Parasitol. Res. 99 189-193 [DOI] [PubMed] [Google Scholar]
- 86.Balaraman, S., Singh, V. K., Tewary, P., and Madhubala, R. (2005) Mol. Biochem. Parasitol. 139 117-127 [DOI] [PubMed] [Google Scholar]
- 87.Ruhland, A., Leal, N., and Kima, P. E. (2007) Cell Microbiol. 9 84-96 [DOI] [PubMed] [Google Scholar]
- 88.Turk, B. E. (2007) Biochem. J. 402 405-417 [DOI] [PubMed] [Google Scholar]
- 89.Thiakaki, M., Kolli, B., Chang, K. P., and Soteriadou, K. (2006) Microbes Infect. 8 1455-1463 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.