Abstract
We have used site-directed spin labeling (SDSL) and electron paramagnetic resonance (EPR) to identify residues 17 and 137 as sites of interaction between the head domain and rod domain 1A of the intermediate filament protein vimentin. This interaction was maximal when compared with the spin labels placed at up- and downstream positions in both head and rod regions, indicating that residues 17 and 137 were the closest point of interaction in this region. SDSL EPR characterization of residues 120-145, which includes the site of head contact with rod 1A, reveals that this region exhibits the heptad repeat pattern indicative of α-helical coiled-coil structure, but that this heptad repeat pattern begins to decay near residue 139, suggesting a transition out of coiled-coil structure. By monitoring the spectra of spin labels placed at the 17 and 137 residues during in vitro assembly, we show that 17-137 interaction occurs early in the assembly process. We also explored the effect of phosphorylation on the 17-137 interaction and found that phosphorylation-induced changes affected the head-head interaction (17-17) in the dimer, without significantly influencing the rod-rod (137-137) and head-rod (17-137) interactions in the dimer. These data provide the first direct evidence for, and location of, headrod interactions in assembled intermediate filaments, as well as direct evidence of coiled-coil structure in rod 1A. Finally, the data identify changes in the structure in this region following in vitro phosphorylation.
The intermediate filament (IF)2 protein family is one of the largest in the human genome, with more than 65 members. Despite the limited primary sequence homology in the IF family, all cytoplasmic IF proteins are similar in predicted secondary structure, and have the ability to assemble into 8-12-nm IFs (1-3).
Vimentin is a Type III IF protein, common to many cells of mesenchymal origin (1, 4, 5). The vimentin monomer is a 53-kDa polypeptide, and like all IF proteins exhibits a conserved, predicted, tripartite domain structure, consisting of a central α-helical rod domain (∼310 amino acids long), flanked by non α-helical N- and C-terminal head and tail domains (6-10). The rod region contains a heptad repeat motif (abc-defg)n where almost 75% of the first and fourth (a and d) residues are hydrophobic. Because the a,d residues are aligned along one surface of the α-helix, they form a largely hydrophobic stripe along that surface, facilitating the assembly of two parallel helices into an α-helical coiled-coil dimer (11-15). Structure prediction programs suggest the central α-helical rod domain can be subdivided into extended regions where the coiled-coil prediction is very strong (coil domains 1A, 1B, 2A, and 2B), interrupted by short segments where this prediction weakens (linker domains) (11-13, 15) The head and tail domains that flank the central rod domain show great variation in both size and sequence and almost nothing is known of their structure or relationships in intact filaments. While the coiled-coil domain is clearly critical to filament assembly and filament structure, much evidence shows that the head domain plays a critical role in filament assembly as well (16-19).
Phosphorylation has been shown to contribute to the regulation of vimentin filament assembly and disassembly. Phosphorylation occurs exclusively in the head and tail domains, with nearly twenty phosphorylation sites identified, most of which are in the head domain. Several different kinases have been implicated (20-25).
IF proteins are generally insoluble in physiologic conditions, requiring chaotropes such as 8 m urea to achieve solubility. This has compounded the difficulty in achieving crystals for high-resolution structural determination. To date, no crystal structure of an intact IF protein or IF has been achieved, although great progress has been made using a divide and conquer approach of crystallizing IF fragments (26-29). Other very valuable data have been achieved through cross-linking studies (30)) circular dichroism (26, 31-33), small angle x-ray scattering (34) and cryo-electron tomography (35). However, very little experimental data are available for the structure of head region, and virtually no data on secondary structure are available to suggest any specific model of interaction between head and rod regions.
We have demonstrated that site-directed spin labeling and electron paramagnetic resonance (SDSL-EPR) can be used to study the secondary, tertiary, and quaternary structure of IF proteins and intact filaments under physiologic conditions and in real time. Using this approach we have demonstrated the presence of coiled-coil structure in the rod 1B and 2B regions, as well as identified points of overlap and orientation of adjacent dimers in intact filaments (36, 37). SDSL EPR studies on the linker 2 (L2) region have been productive as well, demonstrating that the Linker 2 is rigid, that the polypeptide back-bones of adjacent linkers in a dimer run in-parallel, and that linker-linker interactions occur very early in assembly, before coiled-coil structure is adopted (38). By placing spin labels at selected sites that report on monomer-monomer interaction, and different dimer-dimer interactions, we have also been able to identify structural changes that result from phosphorylation (39), as well as from mutations that cause human disease (40). These findings collectively provide direct tests of longstanding models of IF structure, confirming some elements of these models, and controverting others.
In the current study we have utilized SDSL-EPR to identify points of interaction between the head and rod 1A regions, and to characterize the secondary structure of the rod 1A region (residues 120-145). Finally we show how the headrod interactions are affected by both in vitro assembly, and phosphorylation.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis, Cloning, Overexpression, Purification, and Spin Labeling of Human Vimentin—Vimentin mutants were produced by site-directed mutagenesis, then purified and spin-labeled as described in detail previously (36, 37, 40).3 In short, site-directed mutagenesis was used to introduce cysteine residues at specific sites in a slightly modified vimentin expression construct (generously provided by Roy Quinlan, University of Durham, Durham, UK) using the Stratagene QuikChange kit. Coding sequences were confirmed by automated DNA sequencing. Mutant vimentin protein was produced by bacterial overexpression. Inclusion bodies were purified using lysozyme/DNase, high/low salt washes, and chromatography using fast protein liquid chromatography system (FPLC, Amersham Biosciences). Site-directed spin labeling was done by first treating the purified protein with 100 μm TCEP (Tris-(2-carboxyethyl) phosphine, hydrochloride; Molecular Probes, Eugene, OR) followed by spin labeling with 500 μm O-87500 ((1-oxyl-2,2,5,5-tetramethyl-_3-pyrroline-3-methyl) methanethiosulfonate-d15[MTSL-d15]; (Toronto Research Chemicals, Toronto, Canada) to make the EPR-active protein having the nitroxide spin label on the targeted cysteine residue. Because dithiothreitol used under phosphorylation reaction conditions would have released the methanethiosulfonate (MTSL) spin label from the cysteine, we coupled a 3-malemidoproxyl (MSL) spin label to the same position; this spin label is attached by alkylation of thiol, and is thus resistant to dithiothreitol. For phosphorylation experiments, the vimentin mutants were spin-labeled with 500 μm 3-malemidoproxyl (253375 Sigma Aldrich). The unincorporated label was separated from spin-labeled protein by chromatography over a CM-Sepharose column (FPLC, Amersham Biosciences). Protein concentrations were measured by the BCA method (BCA protein assay reagent; Pierce). All purified spin-labeled proteins were stored at -80 °C.
In Vitro Filament Assembly and Electron Microscopy—Filament assembly was generally conducted by dialyzing the spin-labeled protein from 8 m urea into filament assembly buffer, overnight. To identify the sequence of structural changes that occurred during in vitro assembly, dialysis was conducted in a stepwise manner, as described by Carter et al. (42). Electron microscopy of the negatively stained samples was performed to verify the filament assembly each time. In brief, after dialysis, ∼10 μl of the sample was removed and stained with 1% uranyl acetate on formvar-coated carbon grids and then observed with a Phillips CM-120 Electron Microscope operated at 80-kV acceleration voltage.
EPR Spectroscopy of Site-directed Spin Labels—EPR measurements of the spin-labeled proteins were conducted on a JEOL X-band spectrometer fitted with a loop-gap resonator (43). Spectra were collected from ∼5-7 μl of purified, spin-labeled, dialyzed protein, at a final protein concentration of 25-100 μm, loaded in a sealed, quartz capillary tube. Spectra were obtained by a single scan of 120 s over 100 G at a microwave power of 4 milliwatt at room temperature (unless otherwise specified). Modulation amplitude (0.125 mT) was optimized to the natural line width of the attached nitroxide as previously described (44). Normalization of the spectra to the same number of spins was done by normalizing each spectrum to the same integrated intensity/amplitude. To achieve the improvement in the fidelity of the calculation, each sample was double-integrated after its solubilization in 2% SDS.
In Vitro Phosphorylation of Vimentin Mutants by Protein Kinase A—Assembly of vimentin mutants into filaments was achieved by a single step dialysis against kinase reaction buffer. Following electron microscopy to verify the presence of filaments, samples were treated with protein kinase A (PKA; catalytic subunit from bovine heart, Sigma Aldrich) as previously reported (39). Briefly, protein (∼2 mg/ml) was dialyzed overnight against kinase reaction buffer/assembly buffer (20 mm Hepes, 60 mm NaCl, 2 mm MgCl2, 6 mm EGTA) at room temperature. Two identical sets of reaction aliquots were incubated at 30 °C for 3 h as described in Table 1. After the kinase treatment, the samples were analyzed by electron microscopy and by EPR spectroscopy.
TABLE 1.
Phosphorylation reaction conditions
| Control | ATP | Kinase | ATP+kinase | |
|---|---|---|---|---|
| μl | μl | μl | μl | |
| Protein sample | 20 | 20 | 20 | 20 |
| Kinase buffer | 10 | 5 | 5 | |
| ATP | 5 | 5 | ||
| Kinase | 5 | 5 |
RESULTS
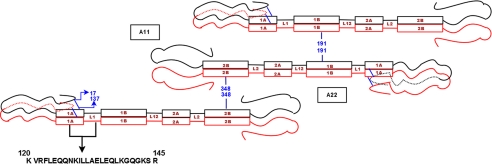
Fig. 1 shows a schematic of the domain structure of vimentin, emphasizing the predicted locations of the head, rod, linker, and tail regions. The figure diagrammatically shows the in-parallel, in-register nature of the dimer, as well as the A11 and A22 dimer-dimer relationships and their established points of overlap (centered on residues 191 and 348). The figure also shows the amino acid sequence in the rod 1A region from residues 120-145 that were analyzed in this report, and the schematized view of the head folding back on the rod domain.
FIGURE 1.
Schematic of the predicted domain structure of vimentin showing head, rod, linker, and tail regions. The placement of cysteine mutants spin labels in Rod 1A for EPR spectroscopy and the schematic view of A11 and A22 alignments of the tetramer with points of overlap at positions 191 and 348; also depicted in the figure is the folding of the head on the rod domain.
Identification of Head-Rod1A Interactions by SDSLEPR—It has been speculated that the vimentin head folds back onto the rod region (45), although no data, to our knowledge, have been presented. To directly test this model, we placed spin labels at multiple sites in both the head and rod domains, and mixed these samples, probing for evidence of spin-spin interaction. We found that the spin label at position 17 in the head region showed proximity to the spin label at residue 137, a “c” position in rod 1A. Note that the mixing of two samples of protein, one labeled at the 17 position, and one labeled at the 137 position, will yield a mix that is predicted to be 25% 17-17 interactions, 25% 137-137 interactions, and 50% 17-137 interactions. This dilution effect predicts a decrease in the spectral intensity reflective of 50% maximal interaction, a prediction that was observed to be true. The interaction seen between 17 and 137 was compared with control samples where 100% of the protein was labeled at residue 17, or at residue 137. In addition, we created double mutant (dm) protein, bearing spin labels at both 17 and 137 positions.
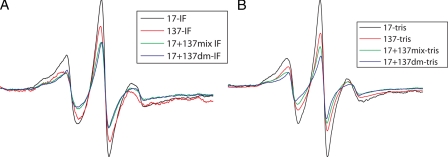
The resulting spectra are shown in Fig. 2, A and B. The narrower shape of the spectra for the single mutants (17-17 and 137-137) implies that there is more mobility and less dipolar interaction as compared with broadening of the spectra for the mixture that has both mutants together (17 + 137). The double mutant (17 + 137(dm)) showed greater broadening of spectra as compared with single-labeled mutants suggesting interaction between these positions in the head and rod, respectively. It is also notable that the line broadening is accompanied by the reduction in the amplitude of the double mutants, which is an indication of strong interaction.
FIGURE 2.
Comparison of EPR spectra for single mutants (17-17 and 137-137), 17 + 137 mixture, and 17 + 137 double mutant showing the mobility of the spin labels and dipolar interaction. The double mutant (17 + 137 dm) showed greater broadening of spectra emphasizing the strong interaction at these positions in head and rod domain. A, EPR spectra scanned when spin-labeled proteins were dialyzed against IF assembly buffer and against low ionic strength Tris buffer (B).
To further substantiate this observation, we calculated d1/d scores from spectra obtained from frozen samples as shown in Table 2. The double mutant (17 + 137) showed significant broadening (d1/d = 0.45) in comparison to that of single-labeled mutants (position 17 (d1/d = 0.39) and position 137 (d1/d = 0.38)) providing direct evidence of interaction between head and rod at these sites. To identify the point of closest proximity in this head-rod 1A region, we placed spin labels both up- and downstream from each site to determine whether the interaction increased or decreased. The specific combinations tested and the resulting d1/d values are shown in Table 2. When position 16 was mixed with positions 130, 136, 137, and 139, or when position 22 was mixed with 130, 136, and 137, there was much weaker interaction than that observed for residue 17. This established that the highest d1/d value was achieved for the 17-137 interaction, suggesting that this is point of closest apposition of head and rod1A in the region tested, and is consistent with the head folding back on the rod region.
TABLE 2.
d1/d values for head rod interactions
| Residue | d1/d |
|---|---|
| 16 | 0.40 |
| 17 | 0.39 |
| 22 | 0.38 |
| 130 | 0.36 |
| 136 | 0.38 |
| 139 | 0.41 |
| 16 + 130 | 0.37 |
| 16 + 136 | 0.38 |
| 16 + 137 | 0.39 |
| 16 + 139 | 0.37 |
| 17 + 130 | 0.39 |
| 17 + 136 | 0.39 |
| 17 + 137 mix | 0.42 |
| 17 + 137 dm | 0.45 |
| 17 + 139 | 0.39 |
| 22 + 130 | 0.36 |
| 22 + 136 | 0.38 |
| 22 + 137 | 0.37 |
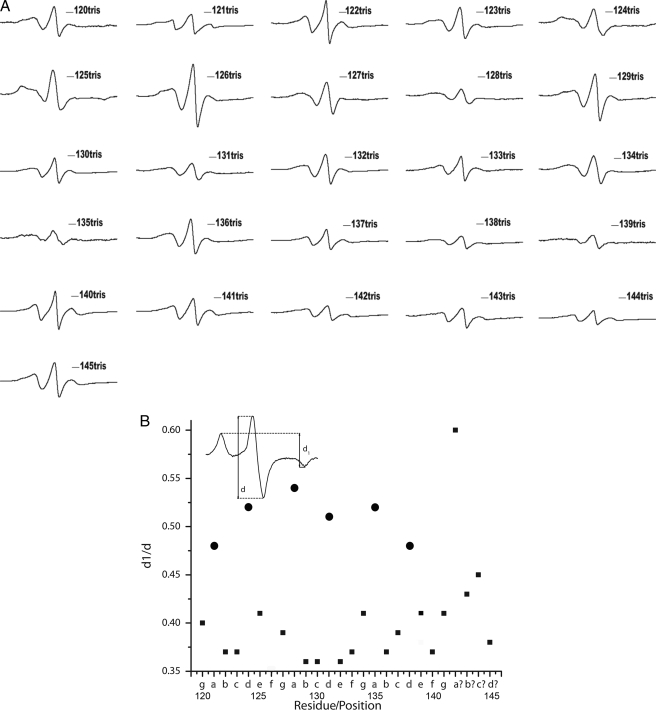
Structural Analysis of the Rod 1A Region—To explore the structure of the rod 1A region (residues 120-145), we introduced spin labels at each residue, confirmed by electron microscopy that each mutant was assembly competent, and collected EPR spectra. Fig. 3A shows the normalized EPR spectra of 26 vimentin mutants within this region. Within a coiled-coil dimer, the a and d positions reside at the interface between two monomers, and are thus closer to one another. The b, c, e, f, and g positions, in contrast, reside on the external surface of the dimer, and are therefore more distant from one another. Our previous EPR characterization of rod 2B was able to differentiate between the a,d and non-a,d positions both qualitatively by appearance of the spectra, and quantitatively through the calculation of d1/d values from spectra of frozen samples. Residues 121, 124, 128, 131, 135, 138 are predicted to be a,d positions, and therefore on the interface between the 2 helices. Room temperature EPR analysis show broadening of spectra which is an indication of strong magnetic dipolar interaction between spin labels, supporting the assignment of these positions as a and d positions. In contrast, the other residues in this region are predicted to be on the external surface of the dimer, and to have reduced dipolar interactions. The spectra from each of these other residues is consistent with a non-a,d position except for position 139. Room temperature spectra from 138 and 139 are similar, but characterization at low temperature provides additional insight, discussed below. Collectively, these spectra provide direct evidence that is consistent with an α-helical, coiled-coil structure, similar to what we have demonstrated in our earlier work on rod 2B domain where the pattern of coiled-coil structure by EPR spectroscopy was established (37).
FIGURE 3.
A, EPR spectra of cysteine spin-labeled mutants in Rod 1A region from 120-145 at room temperature. Spectra are normalized to the same number of spins using double integration after adding 2% of SDS to each sample. B, dipolar broadening ratio (d1/d) versus the residue position in the heptad is plotted. All the a,d positions are indicated as circles, and are clustered near 0.5 value indicating that these positions are located within 1.5 nm of each other. Inset: line-width ratio d1/d from spectra acquired at -100 °C reflects the dipolar interaction strength.
A particularly valuable feature of the EPR approach is the ability to determine the distances between spin labels (46). Freezing the protein prior to the collection of the spectra eliminates the residual motion due to the side chains and tumbling of the protein, which in turn allows for discrimination between motion of the spin label, and proximity between two spin labels. Thus, to further evaluate each sample we collected the spectra from frozen samples, and used the spectra to calculate spin-spin distance using the d1/d ratio (depicted in the inset, Fig. 3B). The dipolar broadening ratio (d1/d) versus the residue position was plotted to visually highlight the difference between a,d and non-a,d positions as shown in Fig. 3B. All of the a and d positions, highlighted as asterisks, are clustered near the 0.5 value, evidence that the spin labels at these positions are located within 1.5 nm of each other. Thus, the d1/d score for the various spectra clearly show the close proximity of spin labels placed at a,d positions as compared with non-a,d positions of the heptad, in what constitutes a “coiled-coil EPR signature.” Table 3 shows the periodicity of the distance separation along the positions of the heptad. These data provide evidence that is consistent with α-helical coiled-coil structure.
TABLE 3.
Heptad position and d1/d values for residues 120-145 in rod 1A of vimentin
| Residue/heptad position | d1/d |
|---|---|
| 120-g | 0.40 |
| 121-a | 0.48 |
| 122-b | 0.37 |
| 123-c | 0.37 |
| 124-d | 0.52 |
| 125-e | 0.41 |
| 126-f | 0.35 |
| 127-g | 0.39 |
| 128-a | 0.54 |
| 129-b | 0.36 |
| 130-c | 0.36 |
| 131-d | 0.51 |
| 132-e | 0.36 |
| 133-f | 0.37 |
| 134-g | 0.41 |
| 135-a | 0.52 |
| 136-b | 0.37 |
| 137-c | 0.38 |
| 138-d | 0.48 |
| 139-e | 0.41 |
| 140-f | 0.37 |
| 141-g | 0.41 |
| 142 | 0.60 |
| 143 | 0.43 |
| 144 | 0.45 |
| 145 | 0.38 |
Calculation of d1/d for positions 138 and 139 reveals that position 139 exhibits characteristics of being a typical non-a,d position. Position 138 is an a,d position with a typical d1/d close to 0.5. The next predicted a,d residue, position 142, exhibits larger d1/d value (0.6), which is higher than the values a,d positions would normally show. This, combined with the observation that the heptad repeat pattern is interrupted at position 142, suggests a departure from the α-helix coiled-coil pattern in this region.
These data collectively point to a coiled-coil structure within rod 1A from position 120-141, with a deviation from coiled-coil structure occurring near position 142 similar to what has been predicted (47). Thus, EPR data are able to confirm a long-standing prediction, namely that at most of rod 1A forms a coiled-coil in intact IFs.
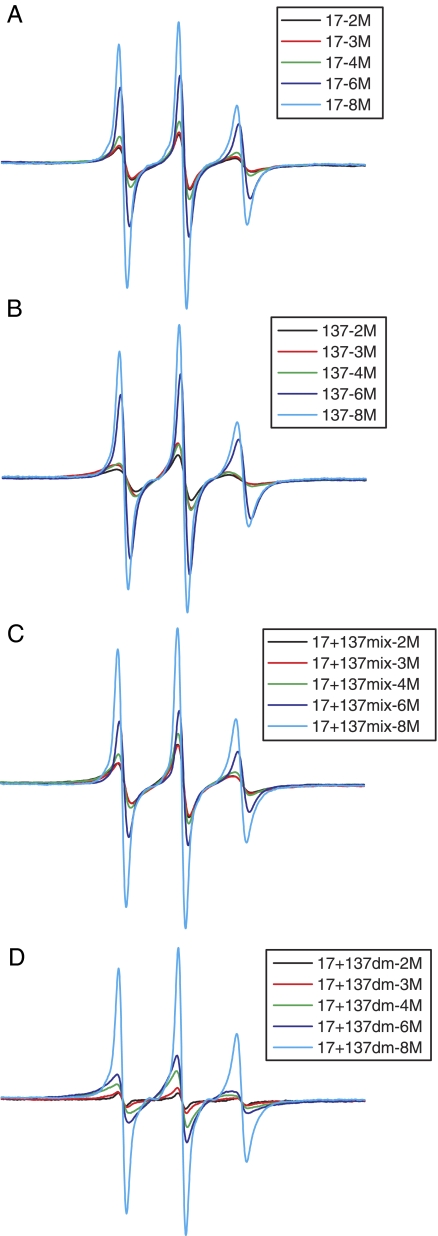
Sequencing the Stages of in Vitro Filament Assembly—One unique advantage of SDSL EPR is that the emergence and progression of spin-spin interactions can be monitored during in vitro assembly of filaments, by acquiring spectra from samples that are at different stages of assembly. It has been previously shown by SDSL EPR that the initiation of α-helix formation starts when the protein is dialyzed from 8 m to 6 m urea, followed by coiled-coil formation at 4 m urea, and tight packing of A11 and A22 tetramers at 2 m urea (36, 37). We used this same approach to explore the progression of 17-137 interactions during in vitro assembly. Spectra collected from samples harvested from dialysis against different concentrations of urea are shown in Fig. 4. In the 17 + 137 double mutant, spectral broadening starts at about 6 m urea, with tight association achieved at 4 m, beyond which little further change is seen. This suggests that the specific association between head and rod domains occurs at an early stage in the in vitro assembly process. It is also interesting to note that the interaction is stronger and more pronounced in the double mutant than in the mixture of single spin labels, suggesting that the head of one vimentin chain folds back along the rod of the same polypeptide. These results suggest that the interaction between the head and the Rod 1A occurs very early in the process of assembly formation, even before the vimentin dimer is formed, which would result in residues 17-17 and 137-137 coming into close proximity.
FIGURE 4.
EPR spectra showing the sequence of in vitro assembly of intermediate filaments. The spectra were recorded from the samples undergoing stepwise assembly at 8 m urea (light blue), 6 m urea (dark blue), 4 m urea (green), 3 m urea (red), and 2 m urea (black) and are normalized for the protein concentration in each sample. Spectra of assembly intermediates taken from single mutants 17 (A), 137 (B), mixture of 17-137 (C), and double mutant 17 + 137 (D) are shown.
Effect of in Vitro Phosphorylation by Protein Kinase A—Phosphorylation of the head domain regulates assemby/disassembly of vimentin IFs in vivo during mitosis. To explore the effect of phosphorylation on the head-rod interaction, we spin labeled the mutants (17, 137, and 17 + 137 dm) with the MSL (3-malemidoproxyl) spin label (selected because it is not affected by the dithiothreitol used in the kinase reaction buffer, explained under “Experimental Procedures”). Spin-labeled proteins were assembled by overnight dialysis. The intact filaments were treated with protein kinase A. Controls consisted of samples lacking ATP, or lacking the protein kinase. Filament status was assessed by electron microscopy, establishing that filament disassembly occurred only in samples treated with the protein kinase and ATP, as predicted, and not in the controls.
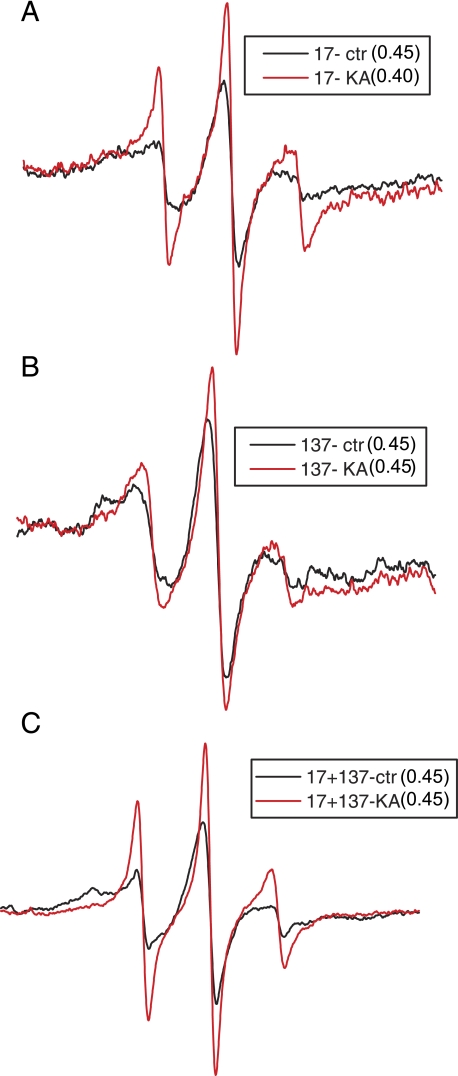
The phosphorylation of the IF resulted in a broadening of the spectrum of spin label at residue 17 (see Fig. 5A). The kinase-treated spectrum has a narrower line showing a marked increase in motion. The same change in the spectral shape can be observed when the spin label was placed at residue 137, and at residues 17 and 137 in the double mutant, but to a lesser extent in the latter. However, the decrease in the line broadening and increase in the spectral amplitude can be influenced either by the decreased contact between the dimers or by the increased distance between the spins. To discriminate between changes due to motion and changes due to distance, we repeated the study, but eliminated the motional component by measuring spectra at -100 °C. The d1/d ratio, the semi-quantitative measure of the distance between spins, was calculated from the EPR spectra of frozen samples (Fig. 5). The d1/d value for the pre- and post-kinase spectra for protein spin-labeled at residue 17 shows decrease in value from 0.45 to 0.40 after kinase treatment. This confirms that the spins become spatially more distant as a result of phosphorylation. There was no change in the d1/d ratio for spins at the 137 position, nor for the 17 + 137 double mutant. This suggests that the phosphorylation-induced change in the spin label motion at position 17 was significant, implying partial disruption of head-head interactions between monomers in a dimer. However, there was no effect on either rod-rod or head-rod interaction as the distance between the spin labels was not influenced.
FIGURE 5.
Normalized EPR spectra of phosphorylated spin-labeled mutants (A) 17, (B) 137, and (C) double mutant 17 + 137. For each indicated vimentin mutant, the red line corresponds to protein kinase A + ATP (KA)-treated sample and black line as buffer control sample (Control). The change in the spectral line reveals the degree of broadening and dipolar interaction, and is calculated by the d1/d ratio. (17-Control = 0.45 & 17-KA = 0.40, 137-Control = 0.45 & 137-KA = 0.45 and 17 + 137 dm-Control = 0.45 & 17 + 137 dm-KA = 0.44).
DISCUSSION
Crystallization has not been achieved for either the IF or an IF protein, limiting the ability to characterize the different domains and their molecular architecture in intact filaments. However, incremental advances in our understanding of IF structure have been achieved through a variety of approaches. Similarly, changes in the structure that occur during physiological processes such as kinase-mediated disassembly are also known only to a limited degree.
Rod 1A, specifically an R-C mutation in the highly conserved LNDR motif, is one of the most common sites of human disease-causing mutations among IF proteins, and thus an understanding of the structure at this site, and the impact of mutations on this structure is of particular interest(8, 48-50). Algorithms that predict secondary structure suggest the onset of α-helical coiled-coil structure somewhere near residue 100 (51, 52). The strength of the prediction begins to weaken around residue 139, leading to the hypothesis that the region near 139 is a transition to non-α-helical coiled-coil linker (47). Recent x-ray crystallographic data produced data, which suggest that this region (fragment 1A) exists as a single α-helix and not coiled-coil dimer (27). However, based on the curve of the helix and dimeric nature of the rod 1A peptide in solution, the authors concluded that within intact filaments, rod 1A most likely forms a coiled-coil. The SDSL EPR data reported here support this hypothesis.
In a previous study, we reported a pattern of EPR spectra that constitute an EPR “signature” for the coiled-coil structure (36, 37). We report here the same pattern for residues 120-141, direct evidence in support of the coiled-coil motif. It is possible that positions 139-142 represent a transition from coiled-coil structure to another structure and that the d1/d value calculated for position 142 is just the first indication of this transition. Relatively low d1/d values for positions 139, 140, and 141 could be consistent with coiled-coil non-a,d positions, or they could be consistent with a transition from coiled-coil to an as yet not determined linker structure. We previously identified Rod 1B region 169-193 as coiled-coil, but have no data concerning the resumption of coiled-coil structure between positions 145 and 169.
Previous reports have shown the importance of the head domain in the filament assembly and stability (17, 53, 54), and data have been presented concerning the affinity of head domain sequences for sequences in the C terminus of rod 2B (55-57). It has been hypothesized that the head domain folds back on the rod, but the experimental evidence supporting the hypothesis has been lacking (1, 16, 41). We reasoned that such an interaction would take place between positions in the head, and positions on the external surface of the rod 1A coiled-coil dimer. Thus, by positioning spin labels at various non-a,d positions of rod 1A and mixing these one at a time with spin-labeled head domain mutants, we were able to identify an interaction between the head (residue 17) and rod 1A (residue 137). Spin labels placed upstream and downstream from both residues, showed reduced levels of spin-spin interaction, suggesting that the 17-137 residues were at, or near the peak of this interaction. Further, by following the spectral changes that occurred during stepwise assembly, we showed tight packing was achieved between residues 17 and 137 earlier than seen for 137-137 or 17-17. This indicates that the head rod interaction within a polypeptide chain occurs very early in assembly, prior to the interaction between the chains at either the 17 or 137 sites. However, it must be underscored that this is in vitro assembly, a process that may/may not replicate in vivo assembly.
Having established in an earlier report that phosphorylation-induced changes in IF structure can be followed by SDSL EPR (39), we explored whether phosphorylation affected the 17-137 interaction. As the head domain has been shown to be the main target of phosphorylation, and phosphorylation has been shown to reduce the affinity of the head for rod 2B (23), we expected to see major effects on the 17-137 interaction. However the spectra suggest that no major change occurs in the proximity between residues 17 and 137 as calculated by the d1/d ratios. Similar results were seen for the 137-137 interaction in the rod region. However, the data indicate that the dimer head-head interaction near residue 17 was disrupted as a consequence of phosphorylation. As we recently demonstrated, phosphorylation of vimentin produces A11 tetramers (39). The data presented here extend this picture to reveal that the head of each vimentin chain is folded back along the rod 1A of the same chain. However, phosphorylation of the head interferes with the interaction between the head domains at position 17.
SDSL EPR provides a focused snapshot of local structural features near the site of the spin label, as well as changes in structural properties that can be induced by phosphorylation. Assembly of a comprehensive view of a protein, or in this case the interaction between multiple proteins, requires integration of many such snapshots. In a previous report we used SDSL EPR to establish that residue 191 is the point of overlap between two adjacent dimers in intact filaments (Ref. 36, and see the A11 tetramer in Fig. 1). The same study also established that these dimers are arranged in an antiparallel orientation. These findings place residue 137 opposite linker 2 and rod 2 in the A11 tetramer. In the current report we establish that the head domain folds back on the rod, bringing residues 17 and 137 into close proximity. This now places the head, and its multiple phosphorylation sites, adjacent to the linker 2 and rod domain 2 in the A11 dimer as well.
Phosphorylation has been shown to induce disassembly of the vimentin IF into tetrameric-sized subunits. Using SDSL EPR we established that the A11 interaction remains intact after phosphorylation-induced disassembly. The same study also established that that phosphorylation induces measurable structural changes in the linker 2 and rod 2 domains of vimentin, despite the fact that the majority of phosphorylation occurs in the head domain, and no phosphorylation occurs in the rod domain(39). We demonstrate in the current report that the 17-17 distances are increased upon phosphorylation, indicating that the head domains at this site are driven apart as a result of phosphorylation, presumably by negative charge repulsion. The demonstration that phosphorylation induces structural changes in the head domain, and that the head domain is adjacent to the linker 2-rod 2 regions begins to suggest a model of how head phosphorylation is capable of inducing structural changes in linker 2 and rod 2 of an adjacent dimer.
Collectively, the data presented here provide another step forward in determining the relationships both within and between proteins in the intact IFs, under physiologic conditions, as well as changes in IF structure that occur with phosphorylation, a physiologic trigger for IF disassembly.
This work was supported, in whole or in part, by National Institutes of Health Grants EY017575 (to P. G. F.) and P30 EY012576 and AG029246 (to J. C. V.). Work was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-12088-01 from the National Center for Research Resources, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IF, intermediate filament; EPR, electron paramagnetic resonance; SDSL, site-directed spin labeling.
J. Hess, M. S. Budamagunta, J. Voss, and P. Fitzgerald, submitted manuscript.
References
- 1.Herrmann, H., and Aebi, U. (2004) Annu. Rev. Biochem. 73 749-789 [DOI] [PubMed] [Google Scholar]
- 2.Omary, M. B., and Coulombe, P. (2004) Intermediate Filament Cytoskeleton, Elsevier, New York
- 3.Oshima, R. G. (2007) Exp. Cell Res. 313 1981-1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrmann, H., and Harris, J. R. (1998) Intermediate Filaments, Plenum, New York, NY
- 5.Herrmann, H., Bar, H., Kreplak, L., Strelkov, S. V., and Aebi, U. (2007) Nat. Rev. Mol. Cell. Biol. 8 562-573 [DOI] [PubMed] [Google Scholar]
- 6.Albers, K., and Fuchs, E. (1992) Int. Rev. Cytol. 134 243-279 [DOI] [PubMed] [Google Scholar]
- 7.Conway, J. F., and Parry, D. A. D. (1988) Int. J. Biol. Macromol. 10 79-98 [Google Scholar]
- 8.Fuchs, E. (1994) J. Cell Biol. 125 511-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, E., and Hanukoglu, I. (1983) Cell 34 332-334 [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, E., and Weber, K. (1994) Annu. Rev. Biochem. 63 345-382 [DOI] [PubMed] [Google Scholar]
- 11.Parry, D., and Fraser, R. D. (1985) in Cellular and Molecular Biology of Intermediate Filaments (Goldman, R. D. and Steinert, P., eds), Plenum Press, New York
- 12.Parry, D. A., and Steinert, P. M. (1992) Curr. Opin. Cell Biol. 4 94-98 [DOI] [PubMed] [Google Scholar]
- 13.Parry, D. A., Steven, A. C., and Steinert, P. M. (1985) Biochem. Biophys. Res. Commun. 127 1012-1018 [DOI] [PubMed] [Google Scholar]
- 14.Parry, D. A., Strelkov, S. V., Burkhard, P., Aebi, U., and Herrmann, H. (2007) Exp. Cell Res. 313 2204-2216 [DOI] [PubMed] [Google Scholar]
- 15.Parry, D. A. D., and Steinert, P. M. (1995) Intermediate Filament Structure, R. G. Landes Company, Austin, TX
- 16.Traub, P., Scherbarth, A., Wiegers, W., and Shoeman, R. L. (1992) J. Cell Sci. 101 363-381 [DOI] [PubMed] [Google Scholar]
- 17.Traub, P., and Vorgias, C. E. (1983) J. Cell Sci. 63 43-67 [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann, E., Weber, K., and Geisler, N. (1985) J. Mol. Biol. 185 733-742 [DOI] [PubMed] [Google Scholar]
- 19.Quinlan, R. A., and Franke, W. W. (1983) Eur. J. Biochem. 132 477-484 [DOI] [PubMed] [Google Scholar]
- 20.Celis, J. E., Larsen, P. M., Fey, S. J., and Celis, A. (1983) J. Cell Biol. 97 1429-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans, R. M., and Fink, L. M. (1982) Cell 29 43-52 [DOI] [PubMed] [Google Scholar]
- 22.Fey, S. J., Larsen, P. M., and Celis, J. E. (1983) FEBS Lett. 157 165-169 [DOI] [PubMed] [Google Scholar]
- 23.Inagaki, M., Nishi, Y., Nishizawa, K., Matsuyama, M., and Sato, C. (1987) Nature 328 649-652 [DOI] [PubMed] [Google Scholar]
- 24.Omary, M. B., Ku, N. O., Tao, G. Z., Toivola, D. M., and Liao, J. (2006) Trends Biochem. Sci 31 383-394 [DOI] [PubMed] [Google Scholar]
- 25.Eriksson, J. E., He, T., Trejo-Skalli, A. V., Harmala-Brasken, A. S., Hellman, J., Chou, Y. H., and Goldman, R. D. (2004) J. Cell Sci. 117 919-932 [DOI] [PubMed] [Google Scholar]
- 26.Strelkov, S. V., Herrmann, H., Geisler, N., Lustig, A., Ivaninskii, S., Zimbelmann, R., Burkhard, P., and Aebi, U. (2001) J. Mol. Biol. 306 773-781 [DOI] [PubMed] [Google Scholar]
- 27.Strelkov, S. V., Herrmann, H., Geisler, N., Wedig, T., Zimbelmann, R., Aebi, U., and Burkhard, P. (2002) EMBO J. 21 1255-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strelkov, S. V., Kreplak, L., Herrmann, H., and Aebi, U. (2004) Methods Cell Biol. 78 25-43 [DOI] [PubMed] [Google Scholar]
- 29.Strelkov, S. V., Schumacher, J., Burkhard, P., Aebi, U., and Herrmann, H. (2004) J. Mol. Biol. 343 1067-1080 [DOI] [PubMed] [Google Scholar]
- 30.Steinert, P. M., Marekov, L. N., Fraser, R. D., and Parry, D. A. (1993) J. Mol. Biol. 230 436-452 [DOI] [PubMed] [Google Scholar]
- 31.Delacourte, A., Dousti, M., and Loucheux-Lefebvre, M. H. (1982) Biochim. Biophys. Acta 709 99-104 [DOI] [PubMed] [Google Scholar]
- 32.Geisler, N., Heimburg, T., Schunemann, J., and Weber, K. (1993) J. Struct. Biol. 110 205-214 [DOI] [PubMed] [Google Scholar]
- 33.Traub, P., Kuhn, S., and Grub, S. (1993) J. Mol. Biol. 230 837-856 [DOI] [PubMed] [Google Scholar]
- 34.Sokolova, A. V., Kreplak, L., Wedig, T., Mucke, N., Svergun, D. I., Herrmann, H., Aebi, U., and Strelkov, S. V. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16206-16211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldie, K. N., Wedig, T., Mitra, A. K., Aebi, U., Herrmann, H., and Hoenger, A. (2007) J. Struct. Biol. 158 378-385 [DOI] [PubMed] [Google Scholar]
- 36.Hess, J. F., Budamagunta, M. S., Voss, J. C., and FitzGerald, P. G. (2004) J. Biol. Chem. 279 44841-44846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hess, J. F., Voss, J. C., and FitzGerald, P. G. (2002) J. Biol. Chem. 277 35516-35522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess, J. F., Budamagunta, M. S., Shipman, R. L., FitzGerald, P. G., and Voss, J. C. (2006) Biochemistry 45 11737-11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittenger, J. T., Hess, J. F., Budamagunta, M. S., Voss, J. C., and Fitzgerald, P. G. (2008) Biochemistry 47 10863-10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hess, J. F., Budamagunta, M. S., FitzGerald, P. G., and Voss, J. C. (2005) J. Biol. Chem. 280 2141-2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., Parry, D. A., Jones, L. N., Idler, W. W., Marekov, L. N., and Steinert, P. M. (2000) J. Cell Biol. 151 1459-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter, J. M., Hutcheson, A. M., and Quinlan, R. A. (1995) Exp. Eye Res. 60 181-192 [DOI] [PubMed] [Google Scholar]
- 43.Hubbell, W. L., Froncisz, W., and Hyde, J. S. (1987) Rev. Sci. Instrum. 58 1879-1886 [Google Scholar]
- 44.Chomiki, N., Voss, J. C., and Warden, C. H. (2001) Eur. J. Biochem. 268 903-913 [DOI] [PubMed] [Google Scholar]
- 45.Bar, H., Strelkov, S. V., Sjoberg, G., Aebi, U., and Herrmann, H. (2004) J. Struct. Biol. 148 137-152 [DOI] [PubMed] [Google Scholar]
- 46.Likhtenshtein, G. I. (1993) Biophysical Labeling Methods in Molecular Biology, pp. 57-62, Cambridge University Press, New York
- 47.Conway, J. F., and Parry, D. A. (1990) Int. J. Biol. Macromol. 12 328-334 [DOI] [PubMed] [Google Scholar]
- 48.Coulombe, P. A., and Fuchs, E. (1993) Semin. Dermatol. 12 173-190 [PubMed] [Google Scholar]
- 49.Coulombe, P. A., Hutton, M. E., Vassar, R., and Fuchs, E. (1991) J. Cell Biol. 115 1661-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuchs, E., Coulombe, P., Cheng, J., Chan, Y. M., Hutton, E., Syder, A., Degenstein, L., Yu, Q. C., Letai, A., and Vassar, R. (1994) J. Investig. Dermatol. 103 25S-30S [DOI] [PubMed] [Google Scholar]
- 51.Lupas, A., Van Dyke, M., and Stock, J. (1991) Science 252 1162-1164 [DOI] [PubMed] [Google Scholar]
- 52.Berger, B., Wilson, D. B., Wolf, E., Tonchev, T., Milla, M., and Kim, P. S. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 8259-8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herrmann, H., Haner, M., Brettel, M., Muller, S. A., Goldie, K. N., Fedtke, B., Lustig, A., Franke, W. W., and Aebi, U. (1996) J. Mol. Biol. 264 933-953 [DOI] [PubMed] [Google Scholar]
- 54.Herrmann, H., Hofmann, I., and Franke, W. W. (1992) J. Mol. Biol. 223 637-650 [DOI] [PubMed] [Google Scholar]
- 55.Gohara, R., Tang, D., Inada, H., Inagaki, M., Takasaki, Y., and Ando, S. (2001) FEBS Lett. 489 182-186 [DOI] [PubMed] [Google Scholar]
- 56.Beuttenmuller, M., Chen, M., Janetzko, A., Kuhn, S., and Traub, P. (1994) Exp. Cell Res. 213 128-142 [DOI] [PubMed] [Google Scholar]
- 57.Mucke, N., Wedig, T., Burer, A., Marekov, L. N., Steinert, P. M., Langowski, J., Aebi, U., and Herrmann, H. (2004) J. Mol. Biol. 340 97-114 [DOI] [PubMed] [Google Scholar]