Abstract
Several lines of evidence indicate that polypeptide growth factors are important in articular cartilage homeostasis and repair. It is not yet clear how these growth factors are regulated. We tested the hypothesis that the growth factors responsible for regulating cartilage are themselves regulated by growth factors. We delivered insulin-like growth factor I (IGF-I), fibroblast growth factor-2 (FGF-2), and/or transforming growth factor-β1 (TGF-β1) to adult bovine articular chondrocytes in primary culture and measured the resulting changes in IGF-I, FGF-2, and TGF-β1 gene expression and protein production. These growth factors differentially regulated their own and each others gene expression and protein production. In concert, they regulated each other in an interactive fashion. Their interactions ranged from inhibitory to synergistic. The time course of the regulatory effects differed among the individual growth factors and combinations. Growth factor-induced changes in growth factor protein production by articular chondrocytes generally corresponded to the changes in gene expression patterns. These studies suggest that interactions among IGF-I, FGF-2, and TGF-β1 substantially modulate their regulatory functions. The results may help guide the application of growth factors to articular cartilage repair.
Damaged articular cartilage is responsible for considerable disability in the form of arthritis and joint trauma (1). Articular chondrocytes have a poor intrinsic repair capacity. Once articular cartilage substance is lost, the damage is generally permanent, and is often progressive (2). Polypeptide growth factors play a central role in articular chondrocyte function. Several anabolic and mitogenic factors have been identified that help maintain cartilage homeostasis in normal joints. These factors include insulin-like growth factor I (IGF-I),2 fibroblast growth factor-2 (FGF-2), and transforming growth factor-β1 (TGF-β1) (3).
IGF-I is considered a candidate for articular cartilage repair because it stimulates both cell proliferation and the synthesis of key matrix constituents, including aggrecan, type II collagen, and non-collagen proteins (4–9), and because it has been shown to promote repair in ex vivo (10) and animal models (11, 12). FGF-2 is a potent stimulus for chondrocyte proliferation (5, 13–15) and was among the earliest factors shown to promote the repair of osteochondral defects in vivo (16–18). TGF-β1 is a potent anabolic agent for chondrocytes (19–21). The effects of these factors are modulated by the presence of other factors (15, 22–26). However, little is known of the regulatory effects of these factors on each other.
We tested the hypothesis that growth factors known to be anabolic for articular chondrocytes regulate their own expression and that of other anabolic growth factors. We tested this hypothesis using IGF-I, FGF-2, and TGF-β1. We found that these growth factors, acting individually, differentially regulate each other. We further found that, in concert, they selectively interact to regulate each other both at the level of gene expression and protein production.
EXPERIMENTAL PROCEDURES
Cell Culture Reagents—Dulbecco's minimum essential medium (DMEM), fetal bovine serum, penicillin, streptomycin, and glutamine were from Invitrogen. Ascorbic acid and bovine serum albumin were from Sigma. Basal medium was prepared with DMEM, 50 μg/ml ascorbic acid, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine. Complete medium was prepared with basal medium supplemented with 10% fetal bovine serum. Human recombinant IGF-I was purchased from Peprotech Inc (Rocky Hill, NJ). Human recombinant FGF-2 and TGF-β1 were purchased from R&D Systems (Minneapolis, MN).
Chondrocyte Isolation and Culture—Bovine articular chondrocytes were isolated as previously described (27). Briefly, articular cartilage was harvested from fresh, 1-year-old bovine carpal joints and digested in complete medium supplemented with 0.1% collagenase type I (Worthington, Lakewood, NJ). After 16 h, the suspension was filtered through 100-μm mesh cell strainer (BD Biosciences, Bedford, MA), and isolated chondrocytes were washed twice and suspended in complete medium.
Chondrocytes were plated in 6-well plates at a density of 5 × 105 cells per well in 4 ml of complete medium. After 3 days, cells were washed, cultured in basal medium for 2 days, and then fresh basal medium for one additional day. The medium was then changed to basal medium containing 0.1% bovine serum albumin and the designated growth factor(s). Basal medium containing 0.1% bovine serum albumin without growth factors was used as control. Cells were cultured for treatment durations of 20 min to 5 days without growth factor or medium replacement.
Growth Factor Analysis by ELISA—Human IGF-I, human FGF-2, and human TGF-β1 DuoSet ELISA kits were purchased from R&D Systems. IGF-I and TGF-β1 were measured in cell culture medium. For TGF-β1 analysis, medium was pretreated with HCl to activate latent TGF-β1 and then neutralized with NaOH. No TGF-β1 was detected in medium without HCl pretreatment. No IGF-I was detected in the cell layer (data not shown). FGF-2 in the medium was less than 10% of the total FGF-2 production (data not shown) and was therefore measured in the cell layer by harvesting and sonicating cells in 0.5 ml of cell lysis buffer. The cell lysis buffer was prepared as previously described (28). Cell protein was determined by Bradford assay of the cell lysate.
RNA Isolation and Reverse Transcription—Total RNA was prepared from chondrocytes using the RNeasy Mini kit (Qiagen). Chondrocytes were lysed with lysis buffer RLT (RNeasy Mini kit, Qiagen) and homogenized by passing 6 times through a 20-gauge needle. On-column DNase digestion was performed to remove any residual DNA. RNA concentration and purity were determined by optical density at 260 nm and 280 nm. cDNA was prepared by reverse transcription of 2 μg of total RNA with a reaction volume of 50 μl for 2 h using the High-capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) and random primer. Reverse transcription was terminated by heating at 95 °C for 20 min. cDNA samples were diluted 1:20 for real-time PCR analysis, except for IGF-I mRNA analysis, for which cDNA samples were not diluted.
Real-time PCR Analysis—Chondrocyte FGF-2 mRNA, IGF-I mRNA, and TGF-β1 mRNA were measured by real-time PCR using a Prism 7000 Sequence Detector System and TaqMan Universal Master Mix (Applied Biosystems). Primers were synthesized by Invitrogen and probes were 5′-end FAM and 3′-end MGB nonfluorescent quencher (MGBNFQ) labeled and synthesized by Applied Biosystems (Table 1). The standard curve method was used to calculate the expression of target genes (IGF-I, FGF-2, and TGF-β1) and the content of 18S rRNA. At least five serial dilutions of cDNA were made to determine threshold cycle (CT) values, and CT values versus log of dilutions were used as the standard curve for each target gene and 18S rRNA. At all time points tested, IGF-I, FGF-2, and TGF-β1 did not alter 18S rRNA levels (data not shown). For this reason, gene expression data were normalized to 18S rRNA levels. At least three independent experiments were performed using different batches of articular chondrocytes obtained from different bovine joints at different times. Fold changes of target gene expression are expressed as the ratio of growth factor-treated samples to the untreated controls. Data are presented as the average of fold changes.
TABLE 1.
Primers and probes used for real-time PCR
| Gene | ACC. no. | Primer | Probe |
|---|---|---|---|
| 18S | AF176811 | Forward: 5′-AGAAACGGCTACCACATCCAA-3′ | 5′-AAGGCAGCAGGCGC-3′ |
| Reverse: 5′-GGGTCGGGAGTGGGTAATTT-3′ | |||
| FGF-2 | NM_174056 | Forward: 5′-TGGTATGTGGCACTGAAACGA-3′ | 5′-CTGGGCAGTATAAACT-3′ |
| Reverse: 5′-TTCTGCCCAGGTCCTGTTTT-3′ | |||
| IGF-I | AY277405 | Forward: 5′-CATCCTCCTCGCATCTCTTCTATC-3′ | 5′-CCTGTGCTTGCTCG-3′ |
| Reverse: 5′-CGTGGCAGAGCTGGTGAA-3′ | |||
| TGF-β1 | M36271 | Forward: 5′-TGAGCCAGAGGCGGACTACT-3′ | 5′-CAAGGAGGTCACCCGC-3′ |
| Reverse: 5′-TGCCGTATTCCACCATTAGCA-3′ |
Statistical Analysis—The effects of growth factor and time were evaluated using analysis of variance (ANOVA). The ANOVA used terms for growth factor, time, and the growth factor-by-time interaction, as well as a random effect to correlate data from the same experimental run. A second analysis, also using ANOVA, directly tested whether combining growth factors lead to synergistic or inhibitory effects. Synergistic effects were those for which the value of the combined growth factors was significantly greater than the sum of the effects of the individual growth factors. Inhibitory effects were those for which the value of the combined growth factors was significantly less than the sum of the effects of the individual growth factors. p values less than 0.05 were considered statistically significant. To simplify the figures, p values are given in the text.
RESULTS
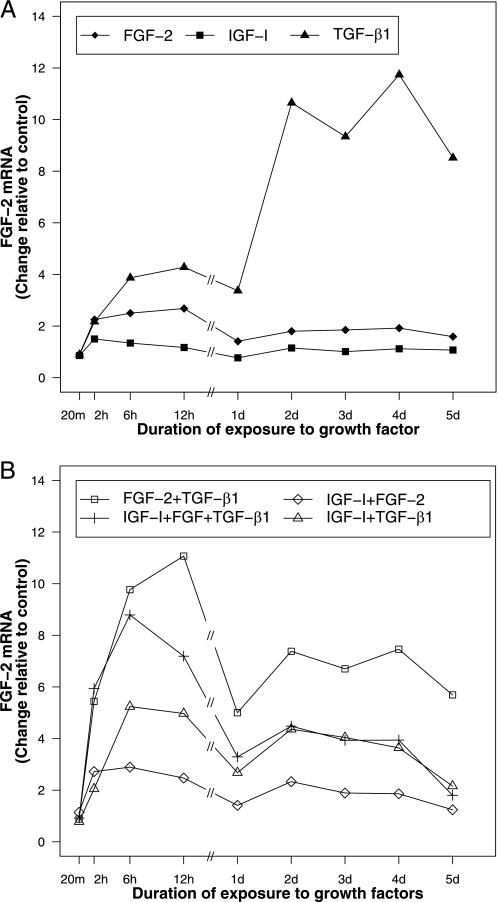
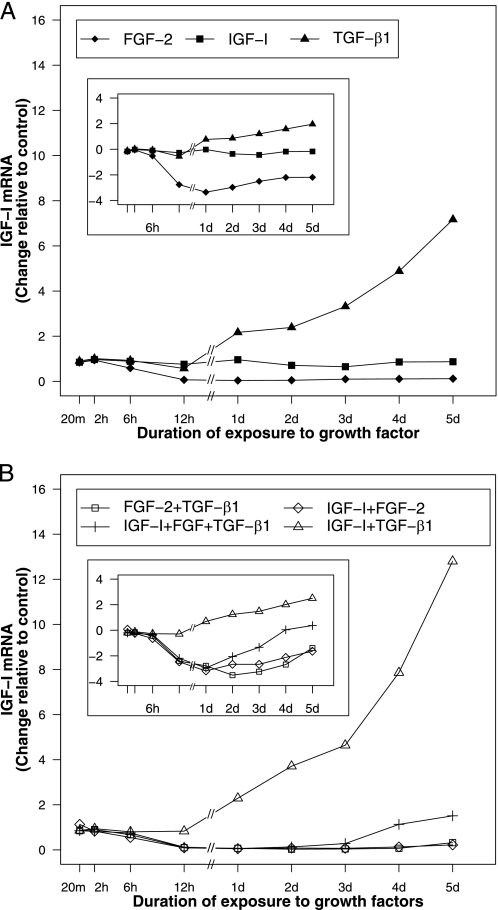
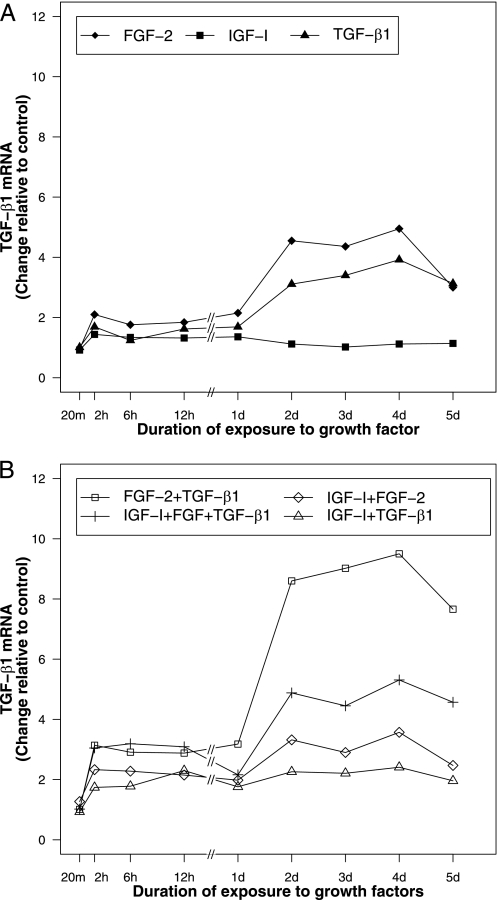
FGF-2, IGF-I, and TGF-β1 Differentially Regulate Their Own Gene Expression—FGF-2 increased FGF-2 transcripts to a maximum of 2.7-fold (p < 0.001) at 12 h and then maintained a stimulation of 1.4–1.9-fold through the 5-day duration of these studies (p ≤ 0.022) (Fig. 1A). IGF-I had little effect on IGF-I expression (Fig. 2A). TGF-β1 increased its own gene expression < 1.7-fold from 2 h through 1 day (p ≤ 0.013 for all time points) and then produced a further time-dependent stimulation to 3.9-fold at 4 days (p = 0.016) (Fig. 3A). Detailed data are given in supplemental Table S1.
FIGURE 1.
FGF-2 gene expression in articular chondrocytes treated with FGF-2, IGF-I, and TGF-β1 individually (A) and in combination (B). Articular chondrocytes were incubated with 50 ng/ml FGF-2, 200 ng/ml IGF-I, or/and 100 ng/ml TGF-β1, or no growth factor (control) and harvested after the indicated time periods. Data are expressed as fold change compared with control.
FIGURE 2.
IGF-I gene expression in articular chondrocytes treated with FGF-2, IGF-I, and TGF-β1 individually (A) and in combination (B). Articular chondrocytes were incubated with 50 ng/ml FGF-2, 200 ng/ml IGF-I, or/and 100 ng/ml TGF-β1, or no growth factor (control) and harvested after the indicated time periods. Data are expressed as fold change compared with control. The same data are expressed as the natural logarithm of fold change of IGF-I mRNA (inset).
FIGURE 3.
TGF-β1 gene expression in articular chondrocytes treated with FGF-2, IGF-I, and TGF-β1 individually (A) and in combination (B). Articular chondrocytes were incubated with 50 ng/ml FGF-2, 200 ng/ml IGF-I, or/and 100 ng/ml TGF-β1, or no growth factor (control) and harvested after the indicated time periods. Data are expressed as fold change compared with control.
FGF-2, IGF-I, and TGF-β1 Differentially Regulate Each Others Gene Expression—Each growth factor was delivered individually to articular chondrocytes and gene expression of the other growth factors was assessed.
FGF-2 Expression—IGF-I increased FGF-2 transcripts 1.5-fold by 2 h (p = 0.009). FGF-2 transcripts returned to baseline by 1 day. TGF-β1 stimulated a progressive, time-dependent increase of FGF-2 transcripts to 11.7-fold at 4 days (p = 0.008) (Fig. 1A).
IGF-I Expression—FGF-2 decreased IGF-I transcripts in a progressive, time-dependent fashion to 4% of the baseline value by 1 day (p < 0.0001). This inhibition persisted at ∼10% of baseline for the remainder of the 5-day experiments (p < 0.001 for all time points). TGF-β1 exerted little effect during the first 12 h of exposure, but subsequently increased IGF-I expression to a maximum of 7.2-fold at 5 days (p = 0.009) (Fig. 2A).
TGF-β1 Expression—FGF-2 increased TGF-β1 transcripts through 2 h, exerted little further effect through 1 day, and then further increased TGF-β1 transcripts to 4.6-fold at 2 days (p < 0.001). IGF-I produced a small (1.3–1.4-fold) increase in TGF-β1 expression from 6 h through 1 day (p ≤ 0.008), that returned to baseline thereafter (Fig. 3A). Detailed data are given in supplemental Table S1).
Delivery of a Second Growth Factor Demonstrates Interactive Regulation of Growth Factor Gene Expression—Two growth factors were delivered simultaneously, and growth factor gene expression was compared with that generated by individual growth factors.
FGF-2 Expression—FGF-2 and TGF-β1 synergistically stimulated FGF-2 expression from 2 h through 1 day. The effect of FGF-2 and TGF-β1 together was 1.3–1.9-fold greater than the sum of the effects of the two factors individually from 2 h through 1 day (p ≤ 0.015 for all time points). However, from 2 days to 5 days, FGF-2 reduced the stimulatory effect of TGF-β1 on FGF-2 expression by 28–36% (p ≤ 0.005 for all time points). The addition of IGF-I to TGF-β1 modestly and transiently increased TGF-β1 stimulation of FGF-2 expression (1.4-fold) at 6 h (p = 0.005) and then markedly reduced the stimulatory effect of TGF-β1 on FGF-2 expression (57–75%) from 1 day through 5 days (p < 0.0001 for all time points). IGF-I did not substantially alter the effect of FGF-2 on FGF-2 transcript levels (Fig. 1B).
IGF-I Expression—The addition of FGF-2 to TGF-β1 reversed the stimulatory effect of TGF-β1 on IGF-I gene expression. In combination, FGF-2 and TGF-β1 produced a time-dependent inhibition of IGF-I expression that closely paralleled the inhibitory action of FGF-2 alone. In contrast, the addition of IGF-I to TGF-β1 increased IGF-I transcripts synergistically after 1 day (p < 0.0001 for all time points). By 5 days, the effect of IGF-I and TGF-β1 together was 1.8-fold (p = 0.017) greater than the sum of the effects of the two factors individually. The inhibitory effect of FGF-2 on IGF-I expression was partially reversed by the addition of IGF-I to FGF-2, but only at 5 days (p = 0.015) (Fig. 2B).
TGF-β1 Expression—The addition of IGF-I to FGF-2 produced a 1.3-fold increase (p = 0.003) in TGF-β1 transcripts at 6 h. The effect of IGF-I then became inhibitory, reducing the stimulatory effect of FGF-2 by 27–33% from 2 days through 4 days (p ≤ 0.004 for all time points). Thus, the combination of IGF-I and FGF-2 generated TGF-β1 transcript levels that were intermediate to those generated by each factor individually. Similarly, the addition of IGF-I to TGF-β1 increased TGF-β1 transcripts 1.4-fold from 6 h through 12 h (p < 0.001 for these two time points). Then, from 2 days through 5 days, IGF-I reduced the stimulatory effect of TGF-β1 on TGF-β1 gene expression by 27–39% (p ≤ 0.004 for all time points). The combination [IGF-I + TGF-β1], as for the combination [IGF-I + FGF-2], generated TGF-β1 transcript levels that were intermediate to those generated by IGF-I and TGF-β1 individually.
The combination [FGF-2 + TGF-β1] synergistically stimulated TGF-β1 expression at all time points after 1 day (Fig. 3B). Specifically, the effect of [FGF-2 + TGF-β1] was 1.2–1.5-fold greater than the sum of the effects of the individual factors from 2 days through 5 days (p ≤ 0.001 for all time points). Detailed data are given in supplemental Tables S2 and S3.
Delivery of a Third Growth Factor Modulates the Effect of Growth Factor Pairs—All three growth factors were delivered simultaneously and growth factor gene expression was compared with that generated by growth factor pairs.
FGF-2 Expression—The addition of IGF-I to the combination [FGF-2 + TGF-β1] inhibited FGF-2 expression in a time-dependent fashion from 12 h through 5 days (p ≤ 0.001 for all time points). By 5 days, IGF-I had reduced by 68% the stimulatory effect of [FGF-2 + TGF-β1] on FGF-2 expression (p < 0.0001). In contrast, the addition of TGF-β1 to [IGF-I + FGF-2] increased FGF-2 transcript levels at all time points tested (p ≤ 0.009) except 20 m. The addition of FGF-2 to the combination [IGF-I + TGF-β1] augmented the stimulatory effect of [IGF-I + TGF-β1] at all time points from 2 h through 12 h (p ≤ 0.006), but this effect was transient and disappeared by 1 day (Fig. 1B).
IGF-I Expression—The addition of IGF-I to [FGF-2 + TGF-β1] gradually, but completely, overcame the inhibition of IGF-I expression by [FGF-2 + TGF-β1]. This restoration of IGF-I expression began at 1 day and achieved baseline levels at 4 days. The addition of TGF-β1 to [FGF-2 + IGF-I] also increased IGF-I expression at all time points from 2 days to 5 days (p ≤ 0.008). In contrast, the addition of FGF-2 to [IGF-I + TGF-β1] produced a marked downward shift in IGF-I transcript levels, overcoming the stimulatory effect of [IGF-I + TGF-β1] until day 4 when IGF-I transcript levels returned to baseline (Fig. 2B). Taken together, these data suggest that FGF-2 and TGF-β1 exert opposing effects on IGF-I gene expression.
TGF-β1 Expression—The addition of IGF-I to [FGF-2 + TGF-β1] markedly reduced TGF-β1 gene expression at all time points from 1 day to 5 days (p < 0.001). This reflects a partial abrogation by IGF-I of the synergistic actions of FGF-2 and TGF-β1 on TGF-β1 expression. Conversely, the addition of FGF-2 to [IGF-I + TGF-β1] increased TGF-β1 expression at all time points (p ≤ 0.023) except 20 m and 1 day. The addition of TGF-β1 to [IGF-I + FGF-2] caused a similar, though less marked, increase in TGF-β1 transcripts at all time points (p ≤ 0.015) except 20 m and 1 day (Fig. 3B). Detailed data are given in supplemental Tables S2 and S3.
The Time Course of Growth Factor Regulation of Growth Factor Expression Differs Among Growth Factors—The effect of duration of growth factor treatment on growth factor gene expression was determined for individual and combinations of growth factors for treatment durations of 20 min to 5 days.
FGF-2 Expression—In all groups, FGF-2 expression increased by 2 h and peaked between 2 and 12 h before declining until 1 days. After 1 day, FGF-2 expression again increased in all groups except the IGF-I treatment group, in which FGF-2 expression remained at a baseline level after 12 h. Only TGF-β1 produced a higher delayed peak (11.7-fold, p = 0.008) than early peak (4.3-fold, p = 0.0001) of FGF-2 expression. By 5 days, the second peak of FGF-2 expression had begun to decline in all groups (Fig. 1, A and B).
IGF-I Expression—IGF-I expression responded more slowly than FGF-2 or TGF-β1 expression to growth factor treatment. The inhibitory effect of FGF-2, and of [FGF-2 + IGF-I], on IGF-I expression first appeared at 6 h. The stimulatory effect of TGF-β1, and of [IGF-I + TGF-β1] did not appear until 1 day and the addition of TGF-β1 to FGF-2 delayed the inhibitory effect of FGF-2 (Fig. 2B). Unlike FGF-2 alone (Fig. 2A), the inhibition by [FGF-2 + TGF-β1] and by [FGF-2 + IGF-I] showed a trend toward restoration by 5 days. In response to [IGF-I + FGF-2 + TGF-β1], IGF-I expression followed an inhibitory time course until 1 day, then progressively recovered to baseline at 4 days (p = 0.582) and became stimulatory to 1.5-fold of baseline at 5 days (p = 0.003) (Fig. 2B). Thus, the addition of IGF-I or TGF-β1 as a third regulator to [FGF-2 + TGF-β1] or [FGF-2 + IGF-I], accelerated the recovery of IGF-I gene expression from its inhibition by FGF-2 (Fig. 2B).
TGF-β1 Expression—TGF-β1 expression increased by 2 h for every growth factor and growth factor combination tested except one (p ≤ 0.013), followed by a plateau through 1 day and by a second increase in TGF-β1 expression over the second 24-h period (Fig. 3, A and B). The exception was cells treated with IGF-I. IGF-I produced only a small (1.3–1.4-fold), transient (6 h through 1 day, p ≤ 0.008) increase (Fig. 3A). The second peaks of TGF-β1 expression, in contrast to those of FGF-2 expression, achieved similar or higher magnitude than the corresponding first peaks in all treatment groups except the IGF-I treatment group. This was especially pronounced for [FGF2 + TGF-β1], which generated a first peak of 2.9–3.2-fold from 2 h through 1 day (p ≤ 0.013) and a second of 7.7–9.5-fold from 2 days through 5 days (p ≤ 0.021) (Fig. 3B). Detailed data are given in supplemental Table S4.
Relation between Regulatory Effects on Growth Factor Gene Expression and Growth Factor Protein Production—The effects of growth factors on the production of other growth factors were assessed by ELISA and compared with their effects on gene expression.
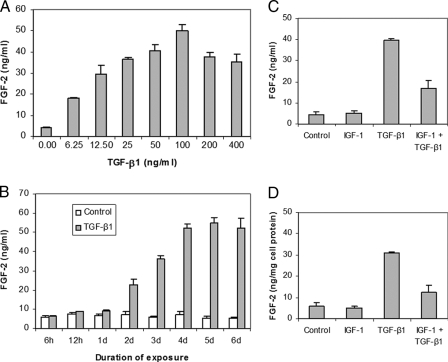
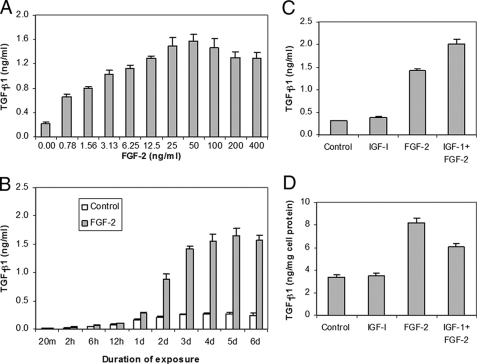
FGF-2 Production—In dose response studies, IGF-I failed to increase FGF-2 production (data not shown). TGF-β1 increased FGF-2 in a dose-dependent manner to 12.2-fold at 100 ng/ml TGF-β1(p < 0.0001) (Fig. 4A). In time course studies, TGF-β1 increased FGF-2 after 12 h to a plateau of 7.4-fold at 4 days (p < 0.0001) (Fig. 4B). The addition of IGF-I strongly inhibited TGF-β1-induced FGF-2 production (Fig. 4, C and D). These patterns of protein production closely parallel the patterns of FGF-2 transcripts in the gene expression studies.
FIGURE 4.
Regulation of FGF-2 production by articular chondrocytes. Dose response to TGF-β1 at 5 days (A). Time course in response to 100 ng/ml TGF-β1 (B). Response at 5 days to IGF-I 200 ng/ml and TGF-β1 100 ng/ml expressed as FGF-2 content (C). Response at 5 days to IGF-I 200 ng/ml and TGF-β1 100 ng/ml expressed as FGF-2 content normalized to cell protein content (D).
IGF-I Production—IGF-I in the medium was less than 200 pg/ml at 20 m and decreased with time in control and chondrocytes treated with FGF-2, 0–400 ng/ml or TGF-β1, 0–400 ng/ml (data not shown). This is consistent with the decrease in IGF-I transcripts caused by FGF-2. The increase in IGF-I transcripts generated by TGF-β1 was not reflected by an increase in IGF-I protein.
TGF-β Production—In dose-response studies, IGF-I did not affect TGF-β1 production (data not shown). Thus, the small (1.3–1.4-fold), transient increase in TGF-β1 transcripts elicited by IGF-I was not reflected in a change in TGF-β1. FGF-2 increased TGF-β1 7.2-fold at 50 ng/ml FGF-2 (p < 0.001) (Fig. 5A), consistent with its effects on TGF-β1 transcripts (Fig. 3A). In time-course studies, FGF-2 increased TGF-β1 6.6-fold at 5 days (p < 0.001) (Fig. 5B). This corresponds to an increase in TGF-β1 transcripts of >4.4-fold from 2 days to 4 days. The addition of IGF-I and FGF-2 generated an apparent additive effect on TGF-β1 production (Fig. 5C), but reduced TGF-β1 protein production (p = 0.002) when normalized to cell protein content (Fig. 5D) consistent with the reduction in TGF-β1 transcripts in the gene expression studies.
FIGURE 5.
Regulation of TGF-β1 production by articular chondrocytes. Dose response to FGF-2 at 5 days (A). Time course in response to 50 ng/ml FGF-2 (B). Response at 5 days to IGF-I 200 ng/ml and FGF-2 50 ng/ml expressed as TGF-β1 content (C). Response at 5 days to IGF-I 200 ng/ml and FGF-2 50 ng/ml expressed as TGF-β1 content normalized to cell protein content (D).
DISCUSSION
To our knowledge, this is the first systematic analysis of growth factor regulation of growth factor expression in articular chondrocytes. Prior studies have reported that IGF-I expression is stimulated (29) and inhibited (30) by FGF-2. In the present studies, FGF-2 reduced IGF-I transcripts to 7% by 12 h. In contrast, TGF-β1 increased IGF-I transcripts to 7.2-fold baseline values by 5 days. Thus, FGF-2 and TGF-β1 appear to be potent counter-regulators of IGF-I expression by these cells. When TGF-β1 and FGF-2 were given together, FGF-2 almost completely eliminated the marked stimulatory action of TGF-β1 on IGF-I gene expression. This suggests an additional level of regulation by TGF-β1 and FGF-2, one that determines the growth factor priority. In this case, the effect of FGF-2 took priority over the effect of TGF-β1. The finding that, by 5 days, TGF-β1 appears to have begun to override this inhibition by FGF-2, suggests that the interaction between FGF-2 and TGF-β1 may be time-limited. TGF-β1 has been previously reported to inhibit IGF-I expression (29, 31). This difference among studies may reflect the well-known sensitivity of TGF-β to details of experimental conditions (19, 32, 33), but may also reflect the relationships among these growth factors. The seemingly discrepant results are consistent with the observation that TGF-β1 stimulates FGF-2 and FGF-2 inhibits IGF-I. Taken together, the data indicate that the regulation of IGF-I by TGF-β1 and FGF-2 is complex.
IGF-I has been reported to increase its own gene expression (34, 35). In the current studies, IGF-I alone had no consistent effect on IGF-I gene expression, but augmented IGF-I gene expression in the presence of TGF-β1. Indeed the synergistic interaction of IGF-I with TGF-β1 generated the highest IGF-I transcript levels observed in these studies. IGF-I, like TGF-β1, rescued IGF-I gene expression from FGF-2 inhibition. Just as the rescue by TGF-β1 required the presence of IGF-I, the rescue by IGF-I required the presence of TGF-β1. This suggests the presence of a positive interaction between IGF-I and TGF-β1 that negatively modulates the pathway by which FGF-2 regulates IGF-I gene expression.
Consistent with prior studies (30), FGF-2 increased its own expression. Indeed, it generated the most rapid self-regulatory action observed in these studies, increasing its own transcript level 2.7-fold by 12 h. The largest magnitude stimulation by a single growth factor observed in these studies was the nearly 12-fold increase in FGF-2 transcripts generated by TGF-β1. Further, TGF-β1 acted in concert with FGF-2 to synergistically increase FGF-2 expression at early time points and by accelerating the onset of this increase. The peak effect of TGF-β1 alone occurred at 4 days, while that of TGF-β1 and FGF-2 occurred at 12 h. In contrast, after 1 day, FGF-2 reduced TGF-β1 stimulation of FGF-2 expression. Although IGF-I alone did not substantially alter FGF-2 expression or the effect of FGF-2 on FGF-2 expression at any time point, it markedly depressed the stimulatory effect of TGF-β1 after 1 day. Taken together, these data suggest that the signaling networks employed by these three factors interact in a complex fashion to regulate FGF-2 expression and do so by modulating both the magnitude and time course of expression.
TGF-β1 increased its own transcripts by nearly 4-fold. This was the most pronounced self regulation observed in these studies. Consistent with prior data (30), FGF-2 also increased TGF-β1 transcript levels. Together, their effect was initially additive and then synergistic (Fig. 3). IGF-I reduced the stimulatory effect of FGF-2, TGF-β1, and [FGF-2 + TGF-β1], but only after 1 day. Taken together, these results suggest that FGF-2 and TGF-β1 employ distinct pathways in regulating TGF-β1 expression. The results further suggest that these pathways share elements that augment TGF-β1 expression and are not shared by IGF-I.
The data suggest that differences in the actions and the interactions of these growth factors are due, in part, to differing time courses. The regulation of IGF-I expression is a case in point. Acting individually, FGF-2 inhibition of IGF-I expression began between 6 h and 12 h. TGF-β1 stimulation of IGF-I expression began between 12 h and 1 day. Consistent with this, FGF-2, TGF-β1, and IGF-I together were initially inhibitory and subsequently stimulatory. The finding that 5 days were required for TGF-β1 and IGF-I to fully rescue the cells from the inhibitory effect of FGF-2 indicates that these factors modulate each others effects over a relative protracted time period. The different time courses, as well as the different actions observed for FGF-2 and TGF-β1 suggest that IGF-I gene expression involves a variety of competing regulatory elements.
FGF-2 and TGF-β1 additively increased TGF-β1 gene expression from 2 h through 1 day and synergistically after 1 day (Fig. 3). This contrasts with their regulation of FGF-2 gene expression. Together, FGF-2 and TGF-β1 synergistically stimulated FGF-2 expression from 2 h through 1 day. After 1 day, FGF-2 inhibited the stimulatory effect of TGF-β1 on FGF-2 expression (Fig. 1). Thus, for both target genes, the interaction between FGF-2 and TGF-β1 is similar during the early time course, but becomes opposite later in the time course. As for IGF-I expression, the long duration of the observed growth factor actions suggests that a single administration of growth factor, or growth factors, may generate effects that persist beyond the time frame of the initial dose. The observation that TGF-β1, with or without FGF-2, augments the expression of both FGF-2 and TGF-β1 suggests the presence of a feed-forward loop that may help sustain their expression over time. Because TGF-β1 produced by the cells required HCl treatment for detection, there may exist additional positive and negative regulatory components such as TGF-β1 activators or TGF-β1 binding proteins that modulate TGF-β1 function. Further studies will be required to elucidate the role of such modulators in articular cartilage and to determine whether a single dose of TGF-β is sufficient to sustain a feed-forward loop.
The effect of these growth factors on growth factor protein production generally corresponded closely with their effects on gene expression. As an apparent exception, IGF-I reduced the stimulation by FGF-2 of TGF-β1 transcripts in gene expression studies, and increased the stimulation by FGF-2 of TGF-β1 in growth factor production studies. To assess this inconsistency, we determined the effect of [IGF-I + FGF-2] on total cell protein and normalized TGF-β1 protein to total cell protein. We found that, consistent with the gene expression data, IGF-I reduced the stimulatory effect of FGF-2 on TGF-β1 protein/cell protein (Fig. 5, C and D). These data suggest that in the presence of FGF-2, IGF-I augments cellular protein synthesis to a greater degree than it augments TGF-β1 protein synthesis. To determine whether normalization to total cell protein similarly influences the interpretation of other growth factor interactions, we determined the effect of [IGF-I + TGF-β1] on total protein production and normalized FGF-2 protein to total cell protein. No change in the inhibitory effect of IGF-I, or the relation between gene expression and protein production was observed (Fig. 4, C and D).
The time course of TGF-β1 action on FGF-2 protein production (Fig. 4B) and of FGF-2 action on TGF-β1 protein production (Fig. 5B), were similar to their gene expression time courses (Figs. 1A and 3A), but the protein curves showed a small right shift compared with the transcript curves. The decrease in FGF-2 or TGF-β1 transcripts at 5 days was not seen for FGF-2 or TGF-β1 protein. This difference may reflect the right shift in protein production, or a potentially longer half-life of the protein than the mRNA. Further studies will be required to separate the respective roles of transcriptional, translational and post-translational mechanisms in the regulation of these growth factors.
Because many chondrocyte functions are involved in articular cartilage homeostasis and repair, the optimization of growth factor therapy for cartilage disorders will likely necessitate the use of more than one growth factor. The present data demonstrate that potentially therapeutic growth factors regulate each other, in some instances to a substantial degree. This means that delivery of one growth factor will likely generate not only the effects of that factor, but of other growth factors as well. Furthermore, the delivery of more than one factor may be expected to result in modulated effects that reflect growth factor interactions with each other. These effects may be in addition to previously identified interactions among effector signaling pathways activated by these growth factors (36–39). IGF-I acts on chondrocytes through a heterodimeric tyrosine kinase receptor that employs phosphoinositol-3 (PI3) kinase and mitogen-activated protein (MAP) kinase cascades to generate its cellular actions (reviewed in Ref. 40). FGF-2 employs a different family of tyrosine kinase receptors, but also uses MAP kinase and PI3 kinase downstream pathways (reviewed in Ref. 41). TGF-β1 activates serine/threonine kinase receptors and post-receptor pathways that are distinct from those of FGF-2 and IGF-I in using Smad protein-mediated signaling pathways. However, all three growth factor pathways interact at several levels (reviewed in Refs. 40, 41). The many shared elements within these growth factor signaling pathways may serve as the basis for involvement of these growth factors in a signaling network. The current data do not identify the sites of interaction among these signal transduction mechanisms. They do however indicate that these mechanisms provide a rich field for further study. Finally, the time course data suggest that growth factor regulation of other growth factors may be expected to modulate the actions these factors in a therapeutic setting.
The results of these studies may serve to guide the selection of individual growth factors and of combinations of growth factors for application to articular cartilage repair. In this capacity, the results indicate that regimens for growth factor-based therapeutics will include a variety of options. Taken together, the present data suggest that the therapeutic application of growth factors for articular cartilage are perhaps more complex than previously appreciated. Further studies will be required to elucidate the specific mechanisms by which these growth factors regulate each other.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AR047702 from NIAMS (to S. B. T.) and the Department of Veterans Affairs. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4.
Footnotes
The abbreviations used are: IGF, insulin-like growth factor; FGF, fibroblast growth factor; TGF, transforming growth factor; ANOVA, analysis of variance; ELISA, enzyme-linked immunosorbent assay.
References
- 1.Praemer, A., Furner, S., and Rice, D. P. (1999) Musculoskeletal Conditions in the United States, pp. 3–39, American Academy of Orthopaedic Surgery, Rosemont, IL
- 2.Buckwalter, J. A., and Mankin, H. J. (1998) Arthritis Rheum. 41 1331–1342 [DOI] [PubMed] [Google Scholar]
- 3.Trippel, S. B. (1997) Instr. Course Lect. 46 473–476 [PubMed] [Google Scholar]
- 4.Bonassar, L. J., Grodzinsky, A. J., Frank, E. H., Davila, S. G., Bhaktav, N. R., and Trippel, S. B. (2001) J. Orthop. Res. 19 11–17 [DOI] [PubMed] [Google Scholar]
- 5.Sah, R. L., Chen, A. C., Grodzinsky, A. J., and Trippel, S. B. (1994) Arch. Biochem. Biophys. 308 137–147 [DOI] [PubMed] [Google Scholar]
- 6.Trippel, S. B., Corvol, M. T., Dumontier, M. F., Rappaport, R., Hung, H. H., and Mankin, H. J. (1989) Pediatr. Res. 25 76–82 [DOI] [PubMed] [Google Scholar]
- 7.McQuillan, D. J., Handley, C. J., Campbell, M. A., Bolis, S., Milway, V. E., and Herington, A. C. (1986) Biochem. J. 240 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luyten, F. P., Hascall, V. C., Nissley, S. P., Morales, T. I., and Reddi, A. H. (1988) Arch. Biochem. Biophys. 267 416–425 [DOI] [PubMed] [Google Scholar]
- 9.Sandell, L. J., and Dukek, E. J. (1988) Orthop. Trans. 12 377–378 [Google Scholar]
- 10.Madry, H., Zurakowski, D., and Trippel, S. B. (2001) Gene Ther. 8 1443–1449 [DOI] [PubMed] [Google Scholar]
- 11.Madry, H., Kaul, G., Cucchiarini, M., Stein, U., Zurakowski, D., Remberger, K., Menger, M. D., Kohn, D., and Trippel, S. B. (2005) Gene Ther. 12 1171–1179 [DOI] [PubMed] [Google Scholar]
- 12.Nixon, A. J., Fortier, L. A., Williams, J., and Mohammed, H. (1999) J. Orthop. Res. 17 475–487 [DOI] [PubMed] [Google Scholar]
- 13.Trippel, S. B. (1995) J. Rheumatol. Suppl. 43 129–132 [PubMed] [Google Scholar]
- 14.Henson, F. M., Bowe, E. A., and Davies, M. E. (2005) Osteoarthritis Cartilage 13 537–544 [DOI] [PubMed] [Google Scholar]
- 15.Kato, Y., Iwamoto, M., and Koike, T. (1987) J. Cell Physiol. 133 491–498 [DOI] [PubMed] [Google Scholar]
- 16.Cucchiarini, M., Madry, H., Ma, C., Thurn, T., Zurakowski, D., Menger, M. D., Kohn, D., Trippel, S. B., and Terwilliger, E. F. (2005) Mol. Ther. 12 229–238 [DOI] [PubMed] [Google Scholar]
- 17.Wellmitz, G., Petzold, E., Jentzsch, K. D., Heder, G., and Buntrock, P. (1980) Exp. Pathol. (Jena) 18 282–287 [DOI] [PubMed] [Google Scholar]
- 18.Fujimoto, E., Ochi, M., Kato, Y., Mochizuki, Y., Sumen, Y., and Ikuta, Y. (1999) Arch. Orthop. Trauma Surg. 119 139–145 [DOI] [PubMed] [Google Scholar]
- 19.Rosier, R. N., O'Keefe, R. J., Crabb, I. D., and Puzas, J. E. (1989) Connect. Tissue Res. 20 295–301 [DOI] [PubMed] [Google Scholar]
- 20.Morales, T. I., and Roberts, A. B. (1988) J. Biol. Chem. 263 12828–12831 [PubMed] [Google Scholar]
- 21.Redini, F., Galera, P., Mauviel, A., Loyau, G., and Pujol, J. P. (1988) FEBS Lett. 234 172–176 [DOI] [PubMed] [Google Scholar]
- 22.Prins, A. P., Lipman, J. M., McDevitt, C. A., and Sokoloff, L. (1982) Arthritis Rheum. 25 1228–1238 [DOI] [PubMed] [Google Scholar]
- 23.Osborn, K. D., Trippel, S. B., and Mankin, H. J. (1989) J. Orthop. Res. 7 35–42 [DOI] [PubMed] [Google Scholar]
- 24.Guerne, P. A., Sublet, A., and Lotz, M. (1994) J. Cell Physiol. 158 476–484 [DOI] [PubMed] [Google Scholar]
- 25.Chopra, R., and Anastassiades, T. (1998) J. Rheumatol. 25 1578–1584 [PubMed] [Google Scholar]
- 26.Horton, W. E., Jr., Higginbotham, J. D., and Chandrasekhar, S. (1989) J. Cell Physiol. 141 8–15 [DOI] [PubMed] [Google Scholar]
- 27.Madry, H., and Trippel, S. B. (2000) Gene Ther. 7 286–291 [DOI] [PubMed] [Google Scholar]
- 28.Fanning, P. J., Emkey, G., Smith, R. J., Grodzinsky, A. J., Szasz, N., and Trippel, S. B. (2003) J. Biol. Chem. 278 50940–50948 [DOI] [PubMed] [Google Scholar]
- 29.Elford, P. R., and Lamberts, S. W. (1990) Endocrinology 127 1635–1639 [DOI] [PubMed] [Google Scholar]
- 30.Shida, J. I., Jingushi, S., Izumi, T., Ikenoue, T., and Iwamoto, Y. (2001) J. Orthop. Res. 19 259–264 [DOI] [PubMed] [Google Scholar]
- 31.Tsukazaki, T., Usa, T., Matsumoto, T., Enomoto, H., Ohtsuru, A., Namba, H., Iwasaki, K., and Yamashita, S. (1994) Exp. Cell Res. 215 9–16 [DOI] [PubMed] [Google Scholar]
- 32.Carrington, J. L., and Reddi, A. H. (1990) Exp. Cell Res. 186 368–373 [DOI] [PubMed] [Google Scholar]
- 33.van der Kraan, P. M., Vitters, E. L., and van den Berg, W. B. (1992) Ann. Rheum. Dis. 51 643–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nixon, A. J., Saxer, R. A., and Brower-Toland, B. D. (2001) J. Orthop. Res. 19 26–32 [DOI] [PubMed] [Google Scholar]
- 35.Froger-Gaillard, B., Hossenlopp, P., Adolphe, M., and Binoux, M. (1989) Endocrinology 124 2365–2372 [DOI] [PubMed] [Google Scholar]
- 36.Loeser, R. F., Pacione, C. A., and Chubinskaya, S. (2003) Arthritis Rheum. 48 2188–2196 [DOI] [PubMed] [Google Scholar]
- 37.Loeser, R. F., Chubinskaya, S., Pacione, C., and Im, H. J. (2005) Arthritis Rheum. 52 3910–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wroblewski, J., and Edwall-Arvidsson, C. (1995) J. Bone Miner. Res. 10 735–742 [DOI] [PubMed] [Google Scholar]
- 39.Bradham, D. M., in der, W. B., Precht, P., Balakir, R., and Horton, W. (1994) J. Cell Physiol. 158 61–68 [DOI] [PubMed] [Google Scholar]
- 40.Danielpour, D., and Song, K. (2006) Cytokine Growth Factor Rev. 17 59–74 [DOI] [PubMed] [Google Scholar]
- 41.Eswarakumar, V. P., Lax, I., and Schlessinger, J. (2005) Cytokine Growth Factor Rev. 16 139–149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.