Abstract
Human immunodeficiency virus (HIV) persists in a latent form in infected individuals treated effectively with highly active antiretroviral therapy (HAART). In part, these latent proviruses account for the rebound in viral replication observed after treatment interruption. A major therapeutic challenge is to purge this reservoir. In this study, we demonstrate that suberoylanilide hydroxamic acid (SAHA) reactivates HIV from latency in chronically infected cell lines and primary cells. Indeed, P-TEFb, a critical transcription cofactor for HIV, is released and then recruited to the viral promoter upon stimulation with SAHA. The phosphatidylinositol 3-kinase/Akt pathway is involved in the initiation of these events. Using flow cytometry-based single cell analysis of protein phosphorylation, we demonstrate that SAHA activates this pathway in several subpopulations of T cells, including memory T cells that are the major viral reservoir in peripheral blood. Importantly, SAHA activates HIV replication in peripheral blood mononuclear cells from individuals treated effectively with HAART. Thus SAHA, which is a Food and Drug Administration-approved drug, might be considered to accelerate the decay of the latent reservoir in HAART-treated infected humans.
Highly active antiretroviral therapy (HAART)5 can reduce plasma human immunodeficiency virus (HIV) to undetectable levels indefinitely. Interruption of HAART, however, inevitably results in a rapid rebound of viremia, indicating that antiretroviral therapy-mediated viral suppression alone is unlikely to eradicate HIV. HIV DNA can integrate stably into the DNA of resting memory T cells where it will persist until these cells die (1). This major reservoir has an estimated half-life of 44 months, suggesting that 70 years of HAART treatment would be required for its eradication (2). Thus, designing new approaches to purge the reservoir of HIV represents a critical therapeutic challenge.
The establishment of viral latency in the pool of memory T cells is multifactorial (3, 4). The transcriptional repression of HIV plays a critical role. Several mechanisms have been identified, such as transcriptional interference (5), chromatin remodeling via acetylation or methylation (6–9), and lack of nuclear factor-κB (NF-κB) or other activators (10). Various approaches antagonizing these distinct transcription blocks have been used to reactivate HIV from latently infected cells (11). This reactivation should contribute to the clearance of otherwise hidden pro-viruses by the immune system and HAART. Ideally, because generalized immune activation also increases the number of targets for new infections and has significant toxic side effects, these strategies should induce HIV transcription without activating cells of the immune system. Indeed, administrations of IL-2 and anti-CD3 antibodies have resulted in significant side effects without having any apparent positive effect on the latent reservoir (12, 13). Various new activators have also been described, including IL-7 (14–16), prostratin (11, 17, 18), valproic acid (19), and hexamethylene bisacetamide (HMBA) (20, 21). These compounds stimulate HIV replication while having limited effects on the activation of T cells and remain under investigation.
Positive transcription elongation factor b (P-TEFb) is a critical component of the cellular transcriptional machinery that is also required for the elongation of HIV transcription. It is recruited to the HIV long terminal repeat (HIV LTR) mainly via the viral transactivator Tat. P-TEFb is composed of two subunits as follows: the catalytic subunit, cyclin-dependent kinase 9 (Cdk9), and the regulatory subunit, cyclin T1 (CycT1). It is present in two distinct complexes in cells, a small transcriptionally active complex (SC) and a large inactive complex (LC) (22). In the latter complex, it is associated with the inhibitory hexamethylene bisacetamide-induced protein 1 or 2 (HEXIM1 or -2) and the 7SK small nuclear RNA as well as with methyl phosphate capping enzyme (also called BCDIN3), La-related protein 7 (LARP7), and several other proteins (23, 24). In cells, an equilibrium is maintained between these complexes. However, various stimuli such as UV light, actinomycin D, or 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole can alter this equilibrium and release P-TEFb from the LC. P-TEFb is not recruited to the HIV LTR in latently infected T cells. Thus, releasing active P-TEFb from the LC should be an excellent strategy to activate viral replication in these cells. Indeed, UV light and actinomycin D activate HIV transcription via P-TEFb (25, 26). We and others recently demonstrated that HMBA also releases P-TEFb from the LC (20, 21, 27). However, these two compounds are not suitable for use in humans.
HMBA is the prototypic member of the family of hybrid polar compounds. These chemicals are characterized by their ability to induce terminal differentiation and/or apoptosis of various transformed cells (28, 29). Interestingly, HMBA stimulates viral replication in various latently infected cell lines as well as T cells from HAART-treated patients (21, 30–32). The PI3K/Akt pathway is involved in this release of P-TEFb. This pathway leads to the phosphorylation of threonine 270 and serine 278 in HEXIM1, which releases the SC. P-TEFb is required for the activation of HIV transcription by HMBA (21). Recent studies suggest that Sp1 could contribute to the recruitment of P-TEFb to the HIV promoter (20). In another study, the release of calcium also activates PP2B and PP1α, which contribute to this disruption of the LC in HeLa cells (27).
Another hybrid polar compound, suberoylanilide hydroxamic acid (SAHA), was recently approved for use as a treatment of cutaneous lymphoma, offering a potential clinically available approach for the clearance of the viral reservoir in HIV-infected patients treated effectively with HAART. Unlike HMBA, SAHA is a histone deacetylase (HDAC) inhibitor. However, like HMBA, it also activates the PI3K/Akt pathway transiently (33). Other studies demonstrated that the subsequent inhibition of this pathway contributes to the death of cancer cell lines (34).
In this study, we investigated the potential for SAHA to stimulate HIV replication from latently infected cells. Indeed, the PI3K/Akt pathway was involved in the activation of P-TEFb and its recruitment to the HIV LTR. Strikingly, SAHA was as potent as the powerful mitogen PHA in inducing HIV production from cultures of primary lymphocytes from patients treated effectively with HAART.
EXPERIMENTAL PROCEDURES
Patients—Samples from HIV-infected adults were obtained from the University of California, San Francisco SCOPE cohort. Cryopreserved PBMCs were obtained from subjects who had been treated without interruption with at least three different Food and Drug Administration-approved antiretroviral agents and who had plasma HIV RNA levels persistently below the limit of detection with commercially available assays (less than 50 or 75 copies/ml). Subjects were required to have undetectable plasma HIV RNA levels for at least 6 months but less than 5 years.
Cell Lines—The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: U1, ACH-2, J1.1 from Dr. Thomas Folks (35–37). JΔK cells were kindly provided by Arnold Rabson, The Cancer Institute of New Jersey, New Brunswick, and described previously (30). Jurkat cells were obtained from the ATCC. Jurkat, U1, ACH-2, and JΔK cells were grown in RPMI 1640 medium containing penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum at 37 °C with 5% CO2. TZM-Bl cells were cultured in Dulbecco's modified Eagle's medium containing the same supplements.
Immunoreagents and Chemicals—The anti-CycT1 (catalog number sc-8127) and anti-Cdk9 (catalog number sc-13130) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-HEXIM1 antibody was described previously (38). SAHA (synthesized by Prof. C. S. Chen) was resuspended in DMSO to obtain a stock solution of 20 mm. Chemical inhibitors, Akt inhibitor VIII (AI8), and LY 294002 (Calbiochem) were resuspended in DMSO.
Reagents Used for PhosFlow Analysis—The following antibodies were used for detection of cell surface markers in Phos-Flow: CD3 (clone SP34–2, Alexa700-conjugated at a dilution 1:100, purchased from Invitrogen); CD4 (clone S3.5, PE-Cy7, 1:100, Invitrogen); CD8 (clone 3B5, PE-Cy7, 1:1000, Invitrogen); CD27 (clone 0323, 1:100, APC-Alexa750, eBioscience); CD33 (PerCP-Cy5.5, 1:25, eBioscience); and CD45 (clone 2H4, ACD, 1:100, Beckman Coulter). Dead cells were stained with a violet-fluorescent fixable Live/Dead® amine-reactive dye (1:1000, Invitrogen). The phospho-specific antibodies (ERK1/2 (Thr(P)-202/Tyr(P)-204, Alexa488), Akt (Thr(P)-308, PE), both from BD Biosciences) were used at a dilution of 1:25. For mitogenic stimulation, PMA (100 ng/ml, Sigma) was used in combination with ionomycin calcium salt (1 μg/ml, Sigma). For fixation of cells, we used a final concentration of 2% paraformaldehyde (Electron Microscopy Sciences). Cells were permeabilized with 70% methanol (Fisher).
Activation of HIV Production in HIV Chronically Infected Cell Lines—U1 or JΔK cells were plated at 2 × 105 cells/ml in 24-well plates; supernatant was collected before stimulation with SAHA and then every day, and viral release in the supernatant was quantified by p24 ELISA (PerkinElmer Life Sciences). Base-line levels were 50–100 pg/ml in control cells. Pre-treatment with inhibitor was done 1 h before stimulation by 1 mm SAHA.
Luciferase Assay—TZM-Bl cells (106 cells) were lysed in 200 μl of luciferase reporter lysis buffer (catalog number E387A, Promega, Madison, WI). After centrifugation, cell debris was discarded. Half of the lysis solution was used for analysis using a luminometer.
Glycerol Gradient Sedimentation Analysis—Glycerol gradients (10–30%) were established by pipetting 2 ml of each of the glycerol fractions (10, 15, 20, 25, and 30% v/v) in buffer A (20 mm HEPES, pH 7.9, 0.3 m KCl, 0.2 mm EDTA, 0.1% Nonidet P-40) into centrifugation tubes (catalog number 331372, Beckman Instruments, Palo Alto, CA). Gradients were formed by standing for 6 h at 4 °C. Jurkat cells were lysed in 0.5 ml of buffer A containing 0.1% protease inhibitor and either 0.5% RNase inhibitor or RNase A (100 mg/ml) for 30 min at 4 °C. The lysates were centrifuged at 10,000 × g for 10 min, and the supernatants were loaded into tubes with the preformed glycerol gradients. Protein complexes were then fractionated by centrifugation in an SW 41Ti rotor (Beckman Instruments) at 38,000 rpm for 20 h. Ten fractions (1 ml) were collected from the top of the gradient, precipitated with trichloroacetic acid, and analyzed by immunoblotting with the appropriate antibodies (39).
Infection of PBMCs and Isolation of Resting CD4+ T Cells—PBMCs were isolated from buffy coats of healthy HIV-negative donors in a Ficoll-Hypaque density gradient (Pharmacia, Piscataway, NJ). PBMCs were then plated at 5 × 106 cells per ml in 24-well plates, using RPMI 1640 medium (10% human serum AB). After 30 min, nonadherent cells were isolated and cultured in complete RPMI 1640 medium (containing penicillin (100 IU/ml), streptomycin (100 μg/ml), and 10% fetal calf serum) with IL-2 (10 units/ml) and PHA (3 μg/ml). After 3 days, cells were treated or not with SAHA (1 mm) in the presence or absence of 1 mm Akt inhibitor 8 (AI8, Calbiochem, 30-min preincubation).
After isolation, 107 nonadherent cells were activated with PHA (5 μg/ml) and IL-2 (10 units/ml) for 3 days and infected with HIV strain HIV-1LAI (0.1 ng/ml p24). They were cultured in RPMI 1640 medium, 10% fetal calf serum, supplemented with IL-2 (10 units/ml). After 11 days, resting CD4+ T cells were isolated by negative selection using magnetic beads (Invitrogen). Cells were more than 95% pure as assessed by fluorescence-activated cell sorter. These cells were then cultured in RPMI 1640 medium, 10% fetal calf serum. These cells expressed neither CD25 nor HLA-DR.
Chromatin Immunoprecipitation (ChIP) Assays—ChIP was carried out essentially as described previously (40). Cross-linking was achieved by incubating 7 × 107 cells (U1 or JΔK) in 1% formaldehyde in medium for 10 min at room temperature. Cross-linking reactions were stopped by addition of glycine to a final concentration of 0.125 m. Cells were then pelleted in a conical tube and washed with cold phosphate-buffered saline. Cell pellets were then resuspended in 1 ml of Lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.0) for 10 min on ice and subjected to sonification to obtain DNA fragments averaging ∼200–500 bp in length. One-tenth of the total chromatin solution was used in each ChIP. Chromatin solutions were pre-cleared with protein A/G-Sepharose beads and then incubated with the appropriate antibody at 4 °C overnight. Protein A/G-Sepharose beads were then added, and the mixture was incubated for another 2 h. The beads were washed five times in TSE-150 (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, 150 mm NaCl), TSE-500 (like TSE-150 but with 500 mm NaCl), and buffer III (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.0), and twice in Tris/EDTA buffer. Immune complexes were eluted from the beads with elution buffer (1% SDS and 0.5% NaHCO3) for 15 min at room temperature. The DNA-protein complexes were then treated with proteinase K, followed by the reversion of cross-linking between DNA and protein at 65 °C for 4 h. DNA was extracted with phenol/chloroform, precipitated with ethanol, and dissolved in 30 μl of Tris/EDTA buffer. DNA (3 μl) was used with appropriate primer sets to amplify specific DNA fragments. Quantitative PCR was then performed using a Stratagene MX3000P real time PCR system. Primers used for LTR were described previously (7), and primers for Gag spanned the region between nucleotides 1144 and 1410. Standard curves for each primer pair were first obtained to check for their respective efficiency. Products were quantified using Brilliant SYBR Green QPCR (Stratagene, San Diego, CA), according to the manufacturer's directions. Relative intensity was calculated and normalized to the input. Fold induction was assessed as the immunoprecipitation with specific antibodies over isotype antibody control.
PhosFlow Analysis—Protein phosphorylation levels in different subsets of primary human PBMCs were analyzed by protein phosphorylation (41). In brief, frozen PBMCs were thawed in RPMI 1640 medium containing 5% fetal bovine serum, washed in phosphate-buffered saline containing 2% fetal bovine serum, and rested at 2 × 106 cells/ml in RPMI 1640 medium at 37 °C, 5% CO2 overnight. The following day, cells were washed with ice-cold phosphate-buffered saline and transferred to a 96-well V-bottom plate. Cells were stained for expression of the cell surface markers CD3, CD4, CD8, CD27, CD45RA, and CD33 for 30 min on ice. An amine-reactive dye was used as a dead cell marker. Subsequently, cells were stimulated with SAHA or PMA/ionomycin at 37 °C; stimulation was stopped after 15 min by immediate fixation with paraformaldehyde at a final concentration of 2%. After 20 min of fixation, cells were permeabilized in 70% ice-cold methanol for 20 min on ice. Cells were washed and stained with phospho-specific antibodies for 60 min on ice. Unstimulated control cells underwent the same manipulations. Cells were analyzed on a customized LSR II Flow Cytometer (BD Biosciences). Analyses of data were performed using FlowJo (Tree Star, Ashland, OR). Fold changes in phosphorylation were calculated as the ratio of median fluorescence intensity of stimulated cells over unstimulated cells.
RESULTS
SAHA Activates HIV Replication at the Level of Transcription—To characterize the ability of SAHA to stimulate viral replication, increasing concentrations of SAHA were used to stimulate chronically infected cell lines. We analyzed the production of viral particles using p24 ELISA. Increased HIV replication was detected in lymphocytic JΔK (Fig. 1A, lanes 2–5) and ACH-2 as well as monocytic U1 cell lines (data not shown) upon SAHA treatment. This increase was dose-dependent, reaching a 40-fold induction with 10 μm SAHA.
FIGURE 1.
SAHA activates HIV replication at the level of transcription. JΔK (A) or TZM-Bl cells (B) were stimulated with increasing concentrations of SAHA for 48 h. A, viral replication was measured in the supernatant of JΔK cells using p24 ELISA. B, cells were lysed, and luciferase activity was measured. All results are presented as fold induction compared with the unstimulated control. C, PBMCs from healthy donors were infected in vitro with HIV-1LAI, and resting CD4+ T cells were isolated after 14 days of culture in the absence of PHA. These isolated cells were cultured in the presence or absence of SAHA (500 nm) for 2 days, and viral replication was assessed using p24 ELISA. D and E, JΔK and TZM-Bl cells were preincubated or not with Akt and PI3K inhibitors, untreated or treated with SAHA (1 μm), and viral replication was assessed using p24 ELISA.
Previously, HMBA was demonstrated to stimulate HIV transcription (21, 32, 42). To determine whether SAHA stimulates viral replication by affecting gene expression from an HIV LTR, we used TZM-Bl cells that contain an integrated HIV LTR linked to the luciferase reporter gene in their genome. Indeed, SAHA induced a dose-dependent increase in luciferase activity, up to 30-fold compared with the unstimulated control in these cells (Fig. 1B, compare lanes 1 and 6).
Taken together, these results demonstrate that SAHA activates gene expression from the HIV LTR in cell lines. To assess the ability of SAHA to stimulate viral replication in primary cells, we used a primary cell model for latent infection, as described previously (21). Resting CD4+ T cells obtained using this protocol are quiescent with low levels of residual replication. Thus, any activation over this background is presented as fold induction. In these primary cells, SAHA induced a 5-fold increase in HIV replication (Fig. 1C, compare lanes 1 and 2). We conclude that SAHA stimulates expression from the HIV LTR and activates viral replication in latently infected cell lines as well as in primary cells.
SAHA-induced HIV Replication Is Dependent on the PI3K/Akt Signaling Pathway—The PI3K/Akt pathway is involved in HMBA-induced HIV replication (21). To determine whether this pathway is also activated by SAHA, we used PI3K and Akt inhibitors. Indeed, Akt and PI3K inhibitors decreased SAHA-induced viral replication by 70% (Fig. 1D, compare lanes 1–3). Importantly, these inhibitors had no significant effect on basal levels of HIV production. These results suggest that SAHA stimulates viral replication in cells in a PI3K/Akt-dependent manner.
SAHA Activates Akt in Primary Memory T Cell Subpopulation—To characterize this activation of PI3K/Akt under physiological conditions, we next used flow cytometry-based single cell analysis of protein phosphorylation to determine the protein phosphorylation status in different populations of primary T cells, as described previously (41). PI3K/Akt activation was followed using antibodies against the active, phosphorylated form of Akt. Antibodies against phospho-ERK1/2 were used as a control to follow the MAPK pathway. PBMCs from HIV-uninfected donors were analyzed for expression levels of specific cell surface markers on T cells (CD3, CD4, or CD8) and monocytes (CD3 and CD33). The maturation markers CD45RA and CD27 were used to subdivide CD3+CD4+ and CD3+CD8+ lymphocytes into subpopulations of naive (CD45RA+CD27+) and memory (CD45RA-CD27+) T cells. Stimulation with SAHA for 15 min induced the activation of Akt in all populations of T cells tested in a dose-dependent manner, reaching 2-fold induction of phosphorylation with 5 μm SAHA (Fig. 2A). Several time points were analyzed for activation with SAHA, and 15 min was found to be the best time point. Of interest, the positive control (PMA + ionomycin) also induced a 2.5-fold increase of Akt phosphorylation. Critically, Akt was activated in memory T cells, which contain most of the latent HIV proviruses in the blood (Fig. 2A, lane 4). Such stimulation had lesser effects in monocytes (Fig. 2A, lane 5). In contrast, whereas ERK1/2 MAPK was not phosphorylated upon stimulation with SAHA, PMA/ionomycin induced a more than 3-fold stimulation of this pathway in all populations analyzed (Fig. 2B). This suggests that SAHA does not induce global T cell activation, in agreement with previous studies (43). Overall, these results demonstrate that the PI3K/Akt pathway, which is involved in SAHA-dependent stimulation of HIV transcription, is activated by SAHA in PBMCs, including those in subpopulations of cells known to contain latent proviruses (3).
FIGURE 2.
PI3K/Akt pathway is stimulated by SAHA in primary memory T cells. Comparing stimulated over unstimulated cells, fold changes in phosphorylation was analyzed in T cells (CD3+), CD4+ T cells (CD3+CD4+), naive T cells (CD3+CD4+CD45RA+CD27+), memory T cells (CD3+CD4+CD45RA–), and monocytes/macrophages (CD4+CD33+) for PBMCs of healthy donors. Phosphorylation of Akt (A) and ERK1/2 MAPK (B) was analyzed. Data are representative of three independent experiments conducted with cells from three different donors.
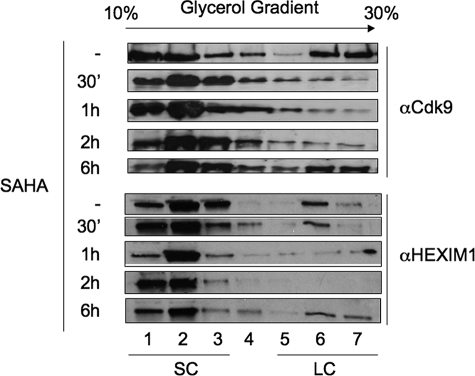
SAHA Releases P-TEFb from the Large Inactive Complex—To determine whether active P-TEFb is released from the LC upon SAHA treatment, we used glycerol gradient centrifugation (21) using Jurkat cells. Fractions were isolated and analyzed by Western blotting for the presence of Cdk9 subunit of P-TEFb (Fig. 3, upper panel). Upper fractions (Fig. 3, upper panel, lanes 1–3) contain the SC, free P-TEFb, whereas the lower fractions contain the inactive LC where HEXIM1 binds and inhibits P-TEFb (Fig. 3, upper panel, lanes 4–7). In untreated cells, Cdk9 was present in both the SC and LC (Fig. 3, upper panel). After 2 h of stimulation, Cdk9 completely disappeared from the LC. However, the complex reformed after 6 h of stimulation. Similar results were obtained with HEXIM1 (Fig. 3, lower panel). Indeed HEXIM1 was released from the LC at 2 h post-stimulation and reappeared in the LC after 6 h of stimulation. Of note, levels of CycT1 and CDK9 remained the same during 48 h (data not shown). Similar results were obtained in PBMCs from healthy donors (data not shown). In conclusion, SAHA induced a rapid but transient disruption of the LC in Jurkat cells.
FIGURE 3.
SAHA releases active P-TEFb. Jurkat T cells were untreated or treated with SAHA. At 30 min and 1, 2, or 6 h, cell lysates were subjected to glycerol gradient centrifugation. Fractions were collected and proteins precipitated before Western blotting with anti-HEXIM1 (bottom panels) and anti-Cdk9 (top panels) antibodies. Fractions 1–3 contain the active SC, whereas fractions 5–7 contain the inactive LC.
P-TEFb Is Recruited to the HIV LTR upon SAHA Treatment—Because P-TEFb is released from HEXIM1 upon treatment with SAHA, we sought to determine whether P-TEFb was recruited to the HIV LTR and coding regions to stimulate HIV transcription. ChIP analyses were performed using antibodies against RNA polymerase II (RNAPII) and the CycT1 subunit of P-TEFb (Fig. 4). Recruitment to the promoter and coding regions was analyzed using the primers depicted in Fig. 4A. Indeed, CycT1 was recruited to the HIV promoter and coding region upon stimulation with SAHA (Fig. 4B, compare lanes 1 and 4 to lanes 2 and 5). Importantly, preincubation of cells with a PI3K inhibitor strongly reduced the recruitment of CycT1 (Fig. 4B, lanes 3 and 6). RNAPII was also recruited to the coding region upon stimulation with SAHA (Fig. 4B, compare lanes 10 and 11). This recruitment was dependent upon PI3K (Fig. 4B, lane 12). However, RNAPII was already present on the HIV promoter in basal conditions and only slightly increased after treatment with SAHA (Fig. 4B, compare lanes 7 and 8). These results are reminiscent of our previous studies using HMBA (21). Taken together, P-TEFb is recruited to the HIV LTR and coding region upon stimulation with SAHA. This recruitment leads to the appearance of RNAPII in the coding region, accounting for increased HIV transcription.
FIGURE 4.
SAHA stimulates the recruitment of RNAPII and P-TEFb to the HIV LTR. A, schematic representation of the HIV provirus integrated into the genome of JΔK cells. B, JΔK cells were preincubated or not for 1 h with the PI3K inhibitor, LY294002 (10 μm), and then stimulated or not with SAHA (1 μm). ChIP analyses were performed using antibodies against RNAPII and CycT1. The recruitment of these transcription factors to the promoter and coding regions was followed by PCR amplification using primer pairs 1 and 2, respectively.
SAHA Activates HIV Replication from Latency in PBMCs from HAART-treated Individuals—Most of the data in the published literature is based on data generated in HIV-infected cell lines. This experimental design may not be directly relevant to what is happening in patients for many reasons because of their transformed phenotype. We used primary cells for analyzing signaling pathways in Fig. 2. To address whether SAHA could be effective in vivo, we also analyzed the effect of SAHA on HIV replication using cells collected from 15 HAART-treated HIV-infected adults, all of whom had achieved durable viral suppression (plasma HIV RNA levels <75 copies/ml). At the time of our analysis, patients had been on HAART for a median of 21 months (interquartile range 15–33 months) and had evidence of stable viral suppression for a median of 13 months (interquartile range 9–18 months). The median CD4+ T cell count was 395 cells/mm3 (interquartile range 314–539). Cryopreserved PBMCs were stimulated with SAHA or with PHA for 6 and 24 h, respectively, washed, and then cultured in the presence of stimulated PBMCs from healthy donors for 21 days; viral replication was measured using p24 ELISA (Fig. 5). Unstimulated PBMCs were also analyzed.
FIGURE 5.
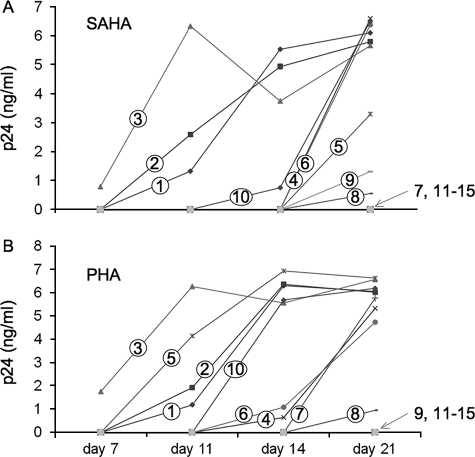
SAHA activates HIV replication in PBMCs from patients treated effectively with HAART. PBMCs from HAART-treated patients with undetectable viremia for more than 1 year were stimulated or not with SAHA at 500 nm for 6 h (A) or with PHA at 3μg/ml (B) for 24 h. Cells were then cultured in the presence of additional stimulated PBMCs, and viral replication was assessed by p24 ELISA over a period of 21 days. Results from 15 patients are presented. Patients were numbered according to their response to stimuli. Cells from patients 1–10 presented activation of HIV replication, whereas cells from patients 11–15 did not present any HIV replication upon the different stimuli used in these studies.
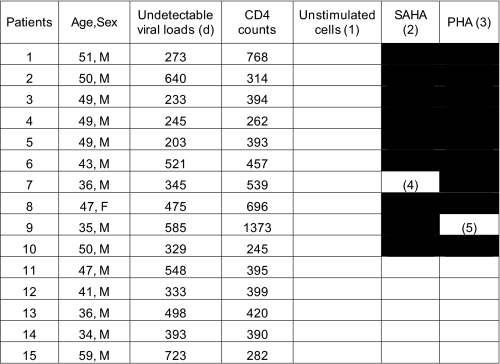
No replication was detected in unstimulated controls. PHA stimulated HIV replication in 9 of 15 samples (Table 1). Strikingly, SAHA also stimulated viral replication in 9 of 15 samples, at levels comparable with those observed with PHA. Noteworthy, the production of new virions in the supernatant was not detected simultaneously in all samples. Some patients responded to SAHA or PHA stimulation with slower kinetics (Fig. 5, A, patients 4–6 and 8–10, and B, patients 4, 6–8, and 10). In these studies, we used 107 cells per experiment. Because only one in a million cells is thought to contain integrated latent HIV (1), it is possible that the kinetic differences observed in different samples as well as the failure of PHA or SAHA to stimulate HIV in five samples may be due to partitioning of the few infected cells. In strong support to this hypothesis, two samples were reactivated by only one stimulus, i.e. SAHA (Fig. 5A, sample 9) or PHA (Fig. 5B, sample 7). Most probably, the number of infected cells was so low that they were present in only one of the two conditions tested. We did not observe any significant correlation between CD4 counts, length of undetectable viremia or age, and the potency of SAHA or PHA to reactivate HIV from latency (Table 1). Taken together, these results demonstrate that SAHA can reactivate HIV from latency in PBMCs from patients treated effectively with HAART, with efficiency comparable with that of the mitogen PHA.
TABLE 1.
SAHA activates HIV replication in PBMCs from patients treated effectively with HAART
Black and white rectangles indicate samples positive (black) and negative (white) for HIV replication. In column 5 (“Unstimulated cells (1)”), patients' PBMCs were not stimulated. In column 6 (“SAHA (2)”), patients' PBMCs were incubated with SAHA (500 nm for 6 h). In column 7 (“PHA (3)”), patients' PBMCs were stimulated with PHA (3 μg/ml for 24 h). Only PHA reactivated PBMCs from Patient 7 (4). Only SAHA reactivated PBMCs from Patient 9 (5).
DISCUSSION
In this study, we demonstrated that SAHA activates viral replication mainly at the level of transcription in chronically infected cells. The PI3K/Akt pathway is involved in this process. Indeed, both PI3K and Akt inhibitors decreased SAHA-induced HIV replication. In addition, SAHA activated PI3K/Akt in all populations of T cells, including memory T cells that harbor latent HIV. The critical transcription factor P-TEFb was released and recruited to the HIV LTR upon stimulation with SAHA, which is important for the reactivation of HIV from latency. Strikingly, we demonstrated that SAHA can activate viral replication in PBMCs from HAART-treated individuals as efficiently as PHA.
Previously, we characterized the role of HMBA in the reactivation of HIV from latency in chronically infected cell lines and PBMCs (21). In this study, we demonstrated that the mechanism of activation of HIV transcription by SAHA is very similar. The PI3K/Akt pathway is involved as demonstrated using both PI3K and Akt inhibitors. Others suggested that the release of calcium is also involved in the HMBA-induced release of P-TEFb (27). Thus, for HMBA, both pathways operate, especially in lymphocytic cell lines and PBMCs. However, this pathway is not involved in actinomycin D-induced release of P-TEFb (27). Moreover, in our study, inhibitors of the calcium pathway had little effect on the activation of HIV replication induced by SAHA (data not shown). Importantly, Akt was activated in all populations of primary T cells and monocytes. Moreover, SAHA did not activate the ERK1/2 pathway. Thus, SAHA acts on cells belonging to the major HIV latent reservoir by activating viral replication and does not result in global T cell activation. Further studies will be required to determine the mechanism of SAHA-induced activation of the PI3K/Akt pathway. SAHA inhibits a wide range of HDACs. HDACs are expressed in both nucleus and cytoplasm and affect many processes (44). One of their targets may be involved in triggering the PI3K/Akt pathway.
Following the stimulation of PI3K/Akt, SAHA increased transcription from the HIV LTR via P-TEFb. How does this transcription activation occur? SAHA, similar to HMBA, has a transient effect on the release of P-TEFb. It first releases P-TEFb from its inactive LC. This surge in SC is followed by the recruitment of P-TEFb to the HIV LTR. In quiescent cells, in which levels of key transcription factors involved in HIV transcription such as NF-κB, NF-AT, and others is rate-limiting, P-TEFb could be further recruited via bromodomain containing 4 (Bdr4), a protein known to be associated with active P-TEFb and bound to acetylated histones (45). Additionally, Sp1 could be involved (20). During this first phase of activation, productive elongation of HIV transcription occurs. In the second phase, ∼6 h after stimulation, P-TEFb returns to the LC. But the activation of HIV transcription is sustained over time for at least several days in chronically infected cell lines. This finding can be explained by the production of viral transcripts during the few hours of P-TEFb activation, which lead to the production of Tat. Tat is a powerful activator of HIV transcription that can use P-TEFb even from the LC (46). Thus once Tat is synthesized, P-TEFb no longer needs to be released from the LC for HIV transcription and efficient viral replication. Interestingly, other HDAC inhibitors (trichostatin A and sodium butyrate) were analyzed for reactivating latent DNA viruses such as Epstein-Barr virus (47), but they did not induce the expression of Epstein-Barr virus lytic genes. Most likely, although they require P-TEFb (48), these DNA viruses do not encode a transcriptional activator that could function analogously to Tat.
Previously, we demonstrated that HMBA could reactivate HIV from latency. However, clinical studies concluded that this compound is not suitable for use in human patients because of significant toxicities (49). However, SAHA also activates HIV replication. SAHA is a Food and Drug Administration-approved oral drug for the treatment of cutaneous lymphoma (50) and hence may be a reasonable choice for accelerating the decay of the latent reservoir in the context of HAART. In addition, SAHA neither activates nor induces the proliferation of PBMCs like IL-2 and PHA. SAHA also presents some growth inhibitory effects that could inhibit new infections. Indeed, concentrations of SAHA necessary to reactivate HIV from latency in primary cells are within clinically achievable concentrations (less than or equal to 1 μm) (51, 52). Moreover, these levels may only need to be achieved intermittently, thus suggesting the possibility of lower or pulsed administration and hence a better safety profile than that observed with SAHA in cancer patients (50). Importantly, in our studies, SAHA was only given once to PBMCs from HAART-treated patients. Thus, repeated administrations of SAHA to patients could have a huge effect on the reactivation of HIV from latency. However, given its multifactorial nature, combination therapies targeting different mechanisms of latency may also be required (5). Such additional agonists would be given together with intensified HAART to eradicate HIV and suppress residual replication. Recent integrase inhibitors could be important for the success of such an approach. Indeed, HIV decays faster when integration is inhibited (53).
In conclusion, our studies further demonstrate the importance of P-TEFb in HIV transcription, and our studies suggest new approaches to purge the viral reservoir in HAART-treated patients.
Acknowledgments
We thank Dr. Arnold B. Rabson for providing the JΔK cells and the plasmids coding for different forms of Akt. We are grateful to members of the Peterlin laboratory for stimulating discussions and continuous support.
This work was supported, in whole or in part, by National Institutes of Health Grants AI49104 and AI058708 (to B. M. P.) and R01 AI40312 and AI47062 (to J. M. M.). This work was also supported by the University of California, San Francisco, Center for AIDS Research Grants P30 AI027763, P30 MH59037, and CC99-SF-001 and the University of California, San Francisco, Clinical and Translational Research Institute Grant UL1 RR024131, a component of the National Institutes of Health Roadmap for Medical Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HAART, highly active antiretroviral therapy; HIV, human immunodeficiency virus; SAHA, suberoylanilide hydroxamic acid; PI3K, phosphatidylinositol 3-kinase; PBMC, peripheral blood mononuclear cell; PMA, phorbol 12-myristate 13-acetate; LTR, long terminal repeat; ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; HMBA, hexamethylene bisacetamide; RNAPII, RNA polymerase II; SC, small complex; LC, large complex; IL, interleukin; MAPK, mitogen-activated protein kinase.
References
- 1.Chun, T. W., Finzi, D., Margolick, J., Chadwick, K., Schwartz, D., and Siliciano, R. F. (1995) Nat. Med. 1 1284–1290 [DOI] [PubMed] [Google Scholar]
- 2.Finzi, D., Blankson, J., Siliciano, J. D., Margolick, J. B., Chadwick, K., Pierson, T., Smith, K., Lisziewicz, J., Lori, F., Flexner, C., Quinn, T. C., Chaisson, R. E., Rosenberg, E., Walker, B., Gange, S., Gallant, J., and Siliciano, R. F. (1999) Nat. Med. 5 512–517 [DOI] [PubMed] [Google Scholar]
- 3.Lassen, K., Han, Y., Zhou, Y., Siliciano, J., and Siliciano, R. F. (2004) Trends Mol. Med. 10 525–531 [DOI] [PubMed] [Google Scholar]
- 4.Contreras, X., Lenasi, T., and Peterlin, B. M. (2006) Future Virol. 1 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenasi, T., Contreras, X., and Peterlin, B. M. (2008) Cell Host Microbe 4 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcello, A. (2006) Retrovirology 3 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams, S. A., Chen, L. F., Kwon, H., Ruiz-Jarabo, C. M., Verdin, E., and Greene, W. C. (2006) EMBO J. 25 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bednarik, D. P., Cook, J. A., and Pitha, P. M. (1990) EMBO J. 9 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishida, T., Hamano, A., Koiwa, T., and Watanabe, T. (2006) Retrovirology 3 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganesh, L., Burstein, E., Guha-Niyogi, A., Louder, M. K., Mascola, J. R., Klomp, L. W., Wijmenga, C., Duckett, C. S., and Nabel, G. J. (2003) Nature 426 853–857 [DOI] [PubMed] [Google Scholar]
- 11.Williams, S. A., Chen, L. F., Kwon, H., Fenard, D., Bisgrove, D., Verdin, E., and Greene, W. C. (2004) J. Biol. Chem. 279 42008–42017 [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., Engel, D., Mizell, S. B., Hallahan, C. W., Fischette, M., Park, S., Davey, R. T., Jr., Dybul, M., Kovacs, J. A., Metcalf, J. A., Mican, J. M., Berrey, M. M., Corey, L., Lane, H. C., and Fauci, A. S. (1999) Nat. Med. 5 651–655 [DOI] [PubMed] [Google Scholar]
- 13.Stellbrink, H. J., van Lunzen, J., Westby, M., O'Sullivan, E., Schneider, C., Adam, A., Weitner, L., Kuhlmann, B., Hoffmann, C., Fenske, S., Aries, P. S., Degen, O., Eggers, C., Petersen, H., Haag, F., Horst, H. A., Dalhoff, K., Mocklinghoff, C., Cammack, N., Tenner-Racz, K., and Racz, P. (2002) AIDS 16 1479–1487 [DOI] [PubMed] [Google Scholar]
- 14.Nunnari, G., and Pomerantz, R. J. (2005) Expert Opin. Biol. Ther. 5 1421–1426 [DOI] [PubMed] [Google Scholar]
- 15.Scripture-Adams, D. D., Brooks, D. G., Korin, Y. D., and Zack, J. A. (2002) J. Virol. 76 13077–13082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, F. X., Xu, Y., Sullivan, J., Souder, E., Argyris, E. G., Acheampong, E. A., Fisher, J., Sierra, M., Thomson, M. M., Najera, R., Frank, I., Kulkosky, J., Pomerantz, R. J., and Nunnari, G. (2005) J. Clin. Investig. 115 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korin, Y. D., Brooks, D. G., Brown, S., Korotzer, A., and Zack, J. A. (2002) J. Virol. 76 8118–8123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkosky, J., Culnan, D. M., Roman, J., Dornadula, G., Schnell, M., Boyd, M. R., and Pomerantz, R. J. (2001) Blood 98 3006–3015 [DOI] [PubMed] [Google Scholar]
- 19.Lehrman, G., Hogue, I. B., Palmer, S., Jennings, C., Spina, C. A., Wiegand, A., Landay, A. L., Coombs, R. W., Richman, D. D., Mellors, J. W., Coffin, J. M., Bosch, R. J., and Margolis, D. M. (2005) Lancet 366 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary, S. K., Archin, N. M., and Margolis, D. M. (2008) J. Infect. Dis. 197 1162–1170 [DOI] [PubMed] [Google Scholar]
- 21.Contreras, X., Barboric, M., Lenasi, T., and Peterlin, B. M. (2007) PLoS Pathog. 3 1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlin, B. M., and Price, D. H. (2006) Mol. Cell 23 297–305 [DOI] [PubMed] [Google Scholar]
- 23.Jeronimo, C., Forget, D., Bouchard, A., Li, Q., Chua, G., Poitras, C., Therien, C., Bergeron, D., Bourassa, S., Greenblatt, J., Chabot, B., Poirier, G. G., Hughes, T. R., Blanchette, M., Price, D. H., and Coulombe, B. (2007) Mol. Cell 27 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krueger, B. J., Jeronimo, C., Roy, B. B., Bouchard, A., Barrandon, C., Byers, S. A., Searcey, C. E., Cooper, J. J., Bensaude, O., Cohen, E. A., Coulombe, B., and Price, D. H. (2008) Nucleic Acids Res. 36 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casse, C., Giannoni, F., Nguyen, V. T., Dubois, M. F., and Bensaude, O. (1999) J. Biol. Chem. 274 16097–16106 [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, V. T., Kiss, T., Michels, A. A., and Bensaude, O. (2001) Nature 414 322–325 [DOI] [PubMed] [Google Scholar]
- 27.Chen, R., Liu, M., Li, H., Xue, Y., Ramey, W. N., He, N., Ai, N., Luo, H., Zhu, Y., Zhou, N., and Zhou, Q. (2008) Genes Dev. 22 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richon, V. M., Webb, Y., Merger, R., Sheppard, T., Jursic, B., Ngo, L., Civoli, F., Breslow, R., Rifkind, R. A., and Marks, P. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5705–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel, D. S., Zhang, X., Feinman, R., Teitz, T., Zelenetz, A., Richon, V. M., Rifkind, R. A., Marks, P. A., and Michaeli, J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoni, B. A., Rabson, A. B., Kinter, A., Bodkin, M., and Poli, G. (1994) Virology 202 684–694 [DOI] [PubMed] [Google Scholar]
- 31.Klichko, V., Archin, N., Kaur, R., Lehrman, G., and Margolis, D. (2006) J. Virol. 80 4570–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vlach, J., and Pitha, P. M. (1993) J. Gen. Virol. 74 2401–2408 [DOI] [PubMed] [Google Scholar]
- 33.Liu, Y., Denlinger, C. E., Rundall, B. K., Smith, P. W., and Jones, D. R. (2006) J. Biol. Chem. 281 31359–31368 [DOI] [PubMed] [Google Scholar]
- 34.Rahmani, M., Yu, C., Reese, E., Ahmed, W., Hirsch, K., Dent, P., and Grant, S. (2003) Oncogene 22 6231–6242 [DOI] [PubMed] [Google Scholar]
- 35.Clouse, K. A., Powell, D., Washington, I., Poli, G., Strebel, K., Farrar, W., Barstad, P., Kovacs, J., Fauci, A. S., and Folks, T. M. (1989) J. Immunol. 142 431–438 [PubMed] [Google Scholar]
- 36.Folks, T. M., Justement, J., Kinter, A., Dinarello, C. A., and Fauci, A. S. (1987) Science 238 800–802 [DOI] [PubMed] [Google Scholar]
- 37.Perez, V. L., Rowe, T., Justement, J. S., Butera, S. T., June, C. H., and Folks, T. M. (1991) J. Immunol. 147 3145–3148 [PubMed] [Google Scholar]
- 38.Li, Q., Price, J. P., Byers, S. A., Cheng, D., Peng, J., and Price, D. H. (2005) J. Biol. Chem. 280 28819–28826 [DOI] [PubMed] [Google Scholar]
- 39.Yik, J. H., Chen, R., Pezda, A. C., and Zhou, Q. (2005) J. Biol. Chem. 280 16368–16376 [DOI] [PubMed] [Google Scholar]
- 40.Nissen, R. M., and Yamamoto, K. R. (2000) Genes Dev. 14 2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweneker, M., Favre, D., Martin, J. N., Deeks, S. G., and McCune, J. M. (2008) J. Immunol. 180 6490–6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, N., Pezda, A. C., and Zhou, Q. (2006) Mol. Cell. Biol. 26 7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, N., Zhao, D., Kirschbaum, M., Zhang, C., Lin, C. L., Todorov, I., Kandeel, F., Forman, S., and Zeng, D. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4796–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minucci, S., and Pelicci, P. G. (2006) Nat. Rev. Cancer 6 38–51 [DOI] [PubMed] [Google Scholar]
- 45.Yang, Z., Yik, J. H., Chen, R., He, N., Jang, M. K., Ozato, K., and Zhou, Q. (2005) Mol. Cell 19 535–545 [DOI] [PubMed] [Google Scholar]
- 46.Barboric, M., Yik, J. H., Czudnochowski, N., Yang, Z., Chen, R., Contreras, X., Geyer, M., Matija Peterlin, B., and Zhou, Q. (2007) Nucleic Acids Res. 35 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Countryman, J. K., Gradoville, L., and Miller, G. (2008) J. Virol. 82 4706–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bark-Jones, S. J., Webb, H. M., and West, M. J. (2006) Oncogene 25 1775–1785 [DOI] [PubMed] [Google Scholar]
- 49.Andreeff, M., Stone, R., Michaeli, J., Young, C. W., Tong, W. P., Sogoloff, H., Ervin, T., Kufe, D., Rifkind, R. A., and Marks, P. A. (1992) Blood 80 2604–2609 [PubMed] [Google Scholar]
- 50.Duvic, M., Talpur, R., Ni, X., Zhang, C., Hazarika, P., Kelly, C., Chiao, J. H., Reilly, J. F., Ricker, J. L., Richon, V. M., and Frankel, S. R. (2007) Blood 109 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fantin, V. R., Loboda, A., Paweletz, C. P., Hendrickson, R. C., Pierce, J. W., Roth, J. A., Li, L., Gooden, F., Korenchuk, S., Hou, X. S., Harrington, E. A., Randolph, S., Reilly, J. F., Ware, C. M., Kadin, M. E., Frankel, S. R., and Richon, V. M. (2008) Cancer Res. 68 3785–3794 [DOI] [PubMed] [Google Scholar]
- 52.Ramalingam, S. S., Parise, R. A., Ramanathan, R. K., Lagattuta, T. F., Musguire, L. A., Stoller, R. G., Potter, D. M., Argiris, A. E., Zwiebel, J. A., Egorin, M. J., and Belani, C. P. (2007) Clin. Cancer Res. 13 3605–3610 [DOI] [PubMed] [Google Scholar]
- 53.Sedaghat, A. R., Dinoso, J. B., Shen, L., Wilke, C. O., and Siliciano, R. F. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4832–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]