Abstract
DgkB is a soluble diacylglycerol (DAG) kinase that is essential for membrane lipid homeostasis in many Gram-positive pathogens. Anionic phospholipids, like phosphatidylglycerol (PtdGro), were required for DgkB to recognize diacylglycerol embedded in a phospholipid bilayer. An activity-independent vesicle binding assay was used to determine the role of specific residues in DgkB-PtdGro interactions. Lys15 and Lys165 were required for DgkB to dock with PtdGro vesicles and flank the entrance to the DgkB active site. Mg2+ was required for vesicle binding. The compromised vesicle binding by mutants in the key asparate residues forming the structural Mg2+-aspartate-water network within the substrate binding domain revealed that interfacial binding of DgkB required a Mg2+-dependent conformational change. DgkB interaction with phospholipid vesicles was not influenced by the presence of ATP, but anionic vesicles decreased the Km of the enzyme for ATP. Arg100 and Lys15 are two surface residues in the ATP binding domain that were necessary for high affinity ATP binding. The key residues responsible for the structural Mg2+ binding site, the conformational changes that increase ATP affinity, and interfacial recognition of anionic phospholipids were identical in DgkB and the mammalian diacylglycerol kinase catalytic cores. This sequence conservation suggests that the mammalian enzymes also require a structural divalent cation and surface positively charged residues to bind phospholipid bilayers and trigger conformational changes that accelerate catalysis.
The diacylglycerol (DAG)2 kinase superfamily (Pfam00781) defines a group of soluble lipid kinases that phosphorylate membrane-associated DAG and function in regulating intracellular phospholipid metabolism and signaling. The mammalian family members share a conserved catalytic core domain that is associated with a variety of modules that confer intracellular membrane targeting specificity (for reviews, see Refs. 1 and 2). Staphylococcus aureus DgkB is a prototypical member of Pfam00781 that has the catalytic core of the DAG kinase superfamily, but lacks the ancillary domains characteristic of its mammalian cousins. DgkB proteins are essential in Gram-positive bacteria to re-introduce the large amount of DAG arising from lipoteichoic acid biosynthesis into the phospholipid biosynthetic pathway (3). DAG is exclusively found in the membrane phospholipid bilayer and the soluble DgkB must be capable of interfacial catalysis despite the absence of the membrane targeting modules present in its mammalian counterparts.
Interfacial activation is a key feature of soluble proteins that act on substrates embedded in a phospholipid bilayer and has been extensively studied in phospholipid hydrolases (4, 5). These enzymes can utilize monomeric substrates, but their activity increases significantly when substrate is presented in micelles or unilamellar vesicles. Electrostatic effects are crucial to interfacial recognition by the soluble phospholipase A2 proteins (4), and the structures of the anion-assisted phospholipase A2 dimer (for review see Ref. 5) reveal a series of lysine residues aligned along one aspect of the phospholipase A2 surface (the i-face) that bind anionic phospholipids and mediate docking to the bilayer. A second well studied mechanism is the association of phosphatidylinositol phospholipase C from Bacillus sp. with membranes via hydrophobic interactions involving tryptophan residues (6). These residues are located on the rim of a (βα)8-barrel flanking the entrance to the active site and function as interfacial anchors that selectively recognize phosphatidylcholine (PtdCho) (7, 8). X-ray studies reveal that the active sites of these enzymes are completely formed in solution (9-13) (7, 14). However, phospholipases A and C do undergo subtle conformational changes upon binding to the interface that cooperate in promoting bilayer association and processive catalysis (5, 7, 15-18). These conformational changes are important for the i-face to form a tight seal with the bilayer to exclude water from the active site channel and allow substrate entry, but appear to have a minimal effect on the configuration of the active site.
DgkB is a protein that is distinctly different from these examples of interfacial enzymes. DgkB activity toward monomeric dioctanoylglycerol substrate could not be detected (3) and the x-ray structure of the protein reveals that the active site exists in a catalytically inactive conformation in solution (19). DgkB is a dimer, and each monomer is divided into ATP-binding domain 1 and substrate-binding domain 2 (Fig. 1) (19). The ATP domain has an incompletely formed nucleotide binding site in which a crucial aspartate residue (Asp68) is unable to interact with ATP·Mg2+, and domain 2 is characterized by the presence of a structural Mg2+ cation binding site that must be occupied to support catalysis (19). DgkB must have a mechanism whereby it engages the bilayer to present the substrate to the active site, and it must also undergo a conformational change that promotes ATP binding. The goals of this study are to define the i-face of DgkB, to identify the molecular determinants on its surface that are required for interfacial binding, and to characterize the conformational changes associated with the activation of enzyme. We predict that these findings will extend to the entire superfamily of soluble DAG kinases.
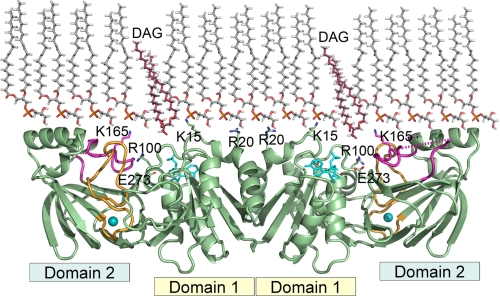
FIGURE 1.
Model depicting the interaction of DgkB with phospholipid bilayers. DgkB is a dimer that is shown docked to a PtdGro monolayer with its interfacial binding surface (i-face) consisting of Lys15, Arg20, and Lys165 facing the phospholipid bilayer. DAG is shown descending from the bilayer into the active site opening. Domain 1 is the ATP-binding module and domain 2 is the substrate-Mg2+-binding module. Catalysis occurs in the cavity between these two domains mediated by Glu273, the catalytic base. The key electropositive residues, Lys15, Arg20, and Lys165, protrude from the protein surface and are ideally positioned to interact with anionic phospholipids to orient the entrance to the active site toward the bilayer. ADP is shown as sticks, Mg2+ is depicted as a cyan ball, the β8-α6 loop is colored magenta, and the β10 -β11 loop is orange. The dotted magenta line indicates missing electron density in the β8-α6 loop in the x-ray structure. The model was drawn with PyMOL (DeLano) using the Mg2+·DgkB·ADP structure (Protein Data Bank accession 2QV7). The atomic coordinates of PtdGro and DAG were generated by Chem3D Pro 9.0 (CambridgeSoft). The intermolecular spacing within the PtdGro layer was based on a 45-Å2 molecular area experimentally observed in dioleoyl-PtdGro monolayer (41).
EXPERIMENTAL PROCEDURES
Materials—Sources of supplies were: Avanti Polar Lipids Inc., all phospholipids, 1,2-dioleoyl-sn-glycerol, 1,2-and di-O-octadecenyl-sn-glycerol; Sigma, 1,2-dioleoylethylene glycol; American Radiolabeled Chemicals Inc., 1,2-[14C]dioleoyl-sn-glycerol; Stratagene, QuikChange site-directed mutagenesis kit; PerkinElmer, the Nickel Chelate Histidine Detection Kit, The LB medium consisted of 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter. All other chemicals were of reagent grade or better.
Construction of Expression Plasmids and DgkB Purification—Construction of expression plasmid pAJ015 was based on a published procedure (3). Site-directed mutagenesis was performed with the QuikChange mutagenesis kit using 5 ng of plasmid and 125 ng of each complementary mutagenic primers between 36 and 48 bases in length, with melting temperatures lower than 78 °C, and with one or two G or C bases at the termini. Initial denaturation at 95 °C for 30 s was followed by 18 cycles of successive denaturation (95 °C), annealing (55 °C), and extension (68 °C) for 30 s, 1 min, and 8 min, respectively, using a thermal cycler (Applied Biosciences) equipped with a hot-top. DpnI-treated DNA was transformed into TOP10 Escherichia coli cells and the identity of the constructs were confirmed by DNA sequencing.
The E. coli BL21(DE3) strain (Invitrogen) harboring the His-tagged dgkB expression plasmid pAJ015 was grown in LB medium supplemented with 100 μg/ml carbenicillin at 37 °C until the A600 reached 0.5. Then, expression of DgkB was induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 400 μm and rotary shaking of the culture was continued at 25 °C for 16 h. Cells were harvested by centrifugation (6,000 × g for 15 min), resuspended in 20 mm Tris, pH 7.9, 500 mm NaCl, 1 mm β-mercaptoethanol, 10 mm imidazole, 10% (v/v) glycerol and protease inhibitor mixture (Roche). Bacterial cell disruption was achieved by a microfluidizer processor (Microfluidics) and the insoluble debris was removed by centrifugation (20,000 × g for 40 min). The cell-free extract was loaded onto a nickel-nitrilotriacetic acid affinity column (Qiagen). The resin was washed with 10 column volumes of 20 mm Tris, pH 7.9, 500 mm NaCl, 1 mm β-mercaptoethanol, 10 mm imidazole, and 10% (v/v) glycerol, followed by 10 column volumes of the same buffer containing 50 mm imidazole. DgkB was eluted from the column in the same buffer containing 500 mm imidazole. The protein was concentrated to 20 mg/ml using a centrifugal filter device (Amicon) and dialyzed against 20 mm MOPS, pH 7.0, 100 mm NaCl, 1 mm EDTA, and 1 mm β-mercaptoethanol. SDS-PAGE analysis of all mutants using Coomassie Blue staining revealed a single band corresponding to the DgkB monomer. Circular dichroism spectroscopy and size exclusion chromatography were used to confirm that the mutations in DgkB affected neither the secondary structure, nor the oligomeric state of proteins. The Bradford protein assay (20) was used for quantitation and protein samples were stored as 50% glycerol at -20 °C.
DAG Kinase Assays—DgkB activity was assessed by either a octyl glucoside:DAG mixed micellar assay performed as described previously (19), or a phospholipid vesicle assay that uses [1-14C]dioleoylglycerol incorporated into 100-nm unilamellar phospholipid vesicles composed of dioleoyl-PtdGro: DAG (90/10, mol %). Vesicles were prepared from dried PtdGro and DAG vortexed in 50 mm HEPES, pH 7.4, 150 mm LiCl at 40 °C for 30 min, followed by sonication of the aqueous mixture at 200 watts for 2 min, and passage (15 times) through a microextruder equipped with a 100-nm polycarbonate filter (Avanti Polar Lipids). The assay contained 50 mm HEPES, pH 7.4, 150 mm LiCl, 10 mm MgCl2, 5 mm ATP, lipid vesicles, and DgkB in a final volume of 50 μl. The reaction was initiated by addition of DgkB, incubated at 25 °C for 30 min, and then 40 μl was spotted onto a Whatmann DE81 filter disc, which was placed immediately in hexane:ethyl ether (1/1, v/v) and washed with 10 ml/disc solvent for 3 × 30 min. The bound [14C]phosphatidic acid was quantitated by liquid scintillation counting.
DgkB Vesicle Binding Assay—The AlphaScreen assay is a bead-based technology developed for assessing biomolecular protein-target interactions in a homogeneous microplate format (21, 22). AlphaScreen binding assays were developed to investigate protein-protein interactions (23), and we have developed an assay for DgkB-phospholipid interactions based on the same principles. The assay employed the Nickel Chelate Histidine Detection Kit (PerkinElmer) with amino-terminal histidine-tagged DgkB and biotinylated unilamellar phospholipid vesicles in 384-well microplate format. An interaction between DgkB and the phospholipid vesicle bring the donor (streptavidin) and acceptor (Ni2+-chelate) beads in close proximity, and upon excitation at 680 nm, the photosensitizer on the donor bead converts oxygen into singlet oxygen that interacts with chemiluminescent groups on the acceptor beads that are within 200 nm of the donor. The AlphaScreen assay is a sensitive method to reliably determine relative binding affinities, although the high avidity of the assay leads to an overestimate of the strength of the interaction by approximately a factor of 10 (24). Preliminary experiments established the range of DgkB and vesicle concentrations that were within the linear range of the assay. Vesicles at 20 mm total lipid concentration were prepared as outlined above under “DAG Kinase Assays” using lipid mixtures containing 0.2 mol % dipalmitoyl-PtdEtn-N-hexanoyl-biotin (Invitrogen). Reagents were diluted with buffer containing 50 mm HEPES, pH 7.4, 150 mm LiCl, and 1 mg/ml bovine serum albumin as follows: vesicles, to a stock concentration of 0.1 mm total lipid; DgkB, serial dilution between 3333 and 26 nm; streptavidin-coated donor beads and nickel chelate acceptor beads, to 0.1 mg/ml. The ratio of lipids in the vesicle preparations is always reported as mole %. Negative control was obtained by eliminating vesicles from the well mixture and the resulting signal to background ratio was ∼300:1.
Phosphatidylglycerol (PtdGro) and DAG in chloroform solutions containing 0.2 mol % dipalmitoyl-PtdEtn-N-hexanoyl-biotin were dried under nitrogen, mixed with 50 mm HEPES, pH 7.4, and 150 mm LiCl at a final concentration of 1 mm, vortexed at 40 °C for 30 min, followed by sonication of the aqueous mixture at 200 watts for 2 min, and extrusion of vesicles (15 times) through a microextruder furnished with a 100-nm polycarbonate filter (Avanti Polar Lipids). In a representative experiment, a 7.5-μl aliquot of the vesicle stock was mixed with 5 μl of donor beads, and 7.5 μl of protein aliquot was mixed with 5 μl of acceptor beads, followed by incubation in the dark at 25 °C for 1 h. The two solutions were then mixed under subdued light conditions and incubated for an hour, followed by data collection on a Fusion-Alpha plate reader (Packard). Each assay point was determined in triplicate with a 25-μl reaction volume per well at 30 μm (total lipid) final vesicle concentration, 20 μg/ml final donor- and acceptor-bead concentrations, and final DgkB concentrations between 0 and 1000 nm. His-tagged DgkB was required for this assay. The tag was located on the amino terminus on the opposite side of the protein from the i-face. Removal of the tag by proteolysis did not alter the activity of the protein (not shown), and the crystal structure was determined on the His-tagged protein (19), suggesting that the tag had no discernable effect on protein structure or function.
A second method for assessing DgkB-vesicle interactions was performed essentially as described by Thomas and Glomset (25). Reagents were diluted with buffer containing 50 mm HEPES, pH 7.4, 150 mm LiCl, and 10 mm MgCl2 as follows: biotinylated PtdGro and PtdCho vesicles to a stock concentration of 5 and 20 mm, respectively; DgkB, serial dilution between 5000 and 40 nm. The DgkB stock (25 μl) solution was mixed with 25 μl of PtdGro vesicles and incubated at 25 °C for 1 h. The bound fraction of DgkB was precipitated by adding 10 μl of PtdCho vesicles, 5 μl of 7.5 mg/ml NeutrAvidin, and the cross-linked vesicles were removed by centrifugation at 21,000 × g for 5 min. DgkB in the supernatant was separated from unbound NeutrAvidin by SDS-gel electrophoresis, the gels were stained with Sypro Ruby and the amount of protein in each band was measured using a Typhoon 9200 PhosphorImager with excitation and emission wavelengths set at 532 and 610 nm, respectively. A standard curve of DgkB was linear between 0 and 1.25 μm DgkB. The amount of DgkB bound was calculated by subtracting the unbound DgkB in samples containing PtdGro vesicles from the DgkB in the control experiment containing only PtdCho vesicles.
Fluorescence Spectroscopy—Two methods were used to assess Mg2+-dependent conformational changes in DgkB in the absence of phospholipid vesicles. The fluorescence intensity change arising from the interaction between a protein and SYPRO Orange dye is related to the hydrophobic surface of the protein accessible to the dye and is a sensitive indicator of protein conformational changes (26). SYPRO Orange fluorescence was measured with excitation at 492 nm and emission at 580 nm. The cuvette held a 3-ml sample containing 0.15 μm DgkB, 1/10,000 dilution of SYPRO Orange DMSO stock (Invitrogen) in 50 mm HEPES, pH 7.4, 150 mm LiCl. Intrinsic tryptophan fluorescence intensity (27) was the second method of detecting conformational changes in DgkB. Because DgkB does not contain tryptophan, the DgkB[Y163W] mutant was constructed to place a tryptophan reporter residue on the β8-α6 loop adjacent to Lys165. Intrinsic tryptophan fluorescence was measured in 50 mm HEPES, pH 7.4, 150 mm LiCl. Excitation was at 280 nm and emission was recorded at 340 nm. All measurements were made using a FluoroLog-3 Horiba Jobin Yvon spectrofluorimeter in the absence or presence of Mg2+ and the difference in fluorescence was plotted as a function of Mg2+ concentration.
Data Analyses—All data analyses were performed using Prism 5 (GraphPad) statistical software. Dissociation constants were calculated from the binding isotherms measured by AlphaScreen assay. The triplicate data points were corrected for background counts obtained in a control reaction without vesicles, and values were plotted in the function of DgkB concentration. Data points between 0 and 250 nm were fitted to a single site binding hyperbola that yielded the dissociation constant and the maximum amount of protein bound. Dissociation constants for mutants were calculated based on the maximum binding of the wild-type protein. Data are reported as value ± S.E. The steady-state kinetic analysis used to assess the dependence of DgkB activity on ATP concentration was based on the vesicle and the mixed micellar assay described under “DAG Kinase Assays.” Initial velocities measured in triplicates at ATP concentrations between 0.78 and 3200 μm were plotted in the Michaelis-Menten plot and the data points were fitted to a single site hyperbola, V = Vmax × [ATP]/(Km + [ATP]) to determine Km. Negative cooperativity was observed for the dependence of DgkB and DgkB[K165A] activity on ATP concentration, thus Hill plots of V = Vmax × [ATP]h/(Kmh + [ATP]h) were used to determine the Hill coefficients. For proteins with Hill coefficients of 0.5, the Km of the low and high affinity ATP binding sites were calculated by fitting the data points to V = Vmax1 × [ATP]/(Km1 + [ATP]) + Vmax2 × [ATP]/(Km2 + [ATP]), a two-site binding hyperbola.
RESULTS
Working Hypothesis—DgkB lacks both exposed tryptophan residues and other hydrophobic surface features suggesting that ionic interactions dominate protein-phospholipid interactions. The crystal structure of the soluble Mg2+·DgkB·ADP ternary complex shows a cigar-shaped homodimer with the active site located between nucleotide-binding domain 1 and the substrate-Mg2+-binding domain 2. The DgkB surface is highly electronegative except for a distinct positive-charged patch due to the presence of Lys15, Arg20, and Lys165 whose side chains protrude from the same protein surface of the dimer that contains the paired active site entrances (Fig. 1). This structural analysis suggests that docking of DgkB to a phospholipid bilayer via these surface features would orient the protein with the active site entrance facing the phospholipid bilayer. Furthermore, the catalytic center of DgkB has an incompletely formed ATP binding site indicating that a conformational change to high-affinity ATP binding occurs upon docking of the protein to the bilayer.
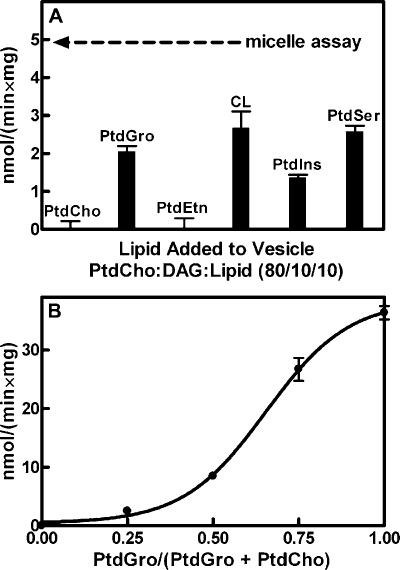
Activation of DgkB by Anionic Phospholipid Vesicles—DAG kinase activity is typically measured by presenting DAG to the enzyme in mixed micelles with a neutral detergent such as octyl glucoside (19, 28, 29). The detergent mixed micelle assay is a robust in vitro biochemical tool, but in the context of the cell, DAG is embedded in the phospholipid bilayer of cell membranes. Therefore, our first step was to develop a DAG kinase assay that presented DAG in phospholipid vesicles. DAG embedded in unilamellar dioleoyl-PtdCho vesicles was not a substrate for DgkB; however, doping these vesicles with 10 mol % of various lipids showed that the presence of anionic phospholipids, like dioleoyl-PtdGro, were activators of interfacial catalysis (Fig. 2A). PtdGro is a major component of S. aureus and other bacterial membranes (30). Increasing the proportion of PtdGro in the vesicles led to an increase in the specific activity of DgkB up to 90% PtdGro, 10% DAG (Fig. 2B). These data revealed that the presentation of DAG in the context of a negatively-charged phospholipid bilayer was required for DgkB activity. Neither PtdGro mixed with DAG in octyl glucoside-mixed micelles, nor the addition of soluble glycerolphosphate head groups in the detergent micelle assay affected DgkB activity (data not shown). We selected PtdGro for subsequent work because it is an abundant anionic phospholipid in S. aureus, which does not contain phosphatidylserine. Nonetheless, phosphatidylserine was capable of activating DkgB to the same extent as PtdGro (not shown). DgkB activity required the presence of a divalent cation. Mg2+ was the most potent cation, although catalysis was detected with Mn2+ and to a lesser extent with Ca2+, the same order of potency as found using the micelle assay (19).
FIGURE 2.
Anionic phospholipids activate DgkB. A, relationship between DgkB activity and the vesicle lipid composition. Vesicles were prepared with 80 mol % PtdCho, 10 mol % DAG, and 10 mol % of PtdGro, phosphatidylethanolamine (PtdEtn), cardiolipin (CL), phosphatidylinositol (PtdIns), or phosphatidylserine (PtdSer). The arrow indicates DgkB activity measured using the octyl glucoside mixed micellar assay. B, activation of DgkB catalysis by PtdGro. DAG substrate was presented in phospholipid vesicles containing 10 mol % DAG and 90 mol % PtdGro plus PtdCho at the indicated molar ratios. Assays were performed in triplicate, and error bars represent S.E. The vesicle assays contained 500 μm total lipid with 50 μm DAG and the micellar assay 50 μm total DAG dissolved in 100 mm octyl β-d-glucoside.
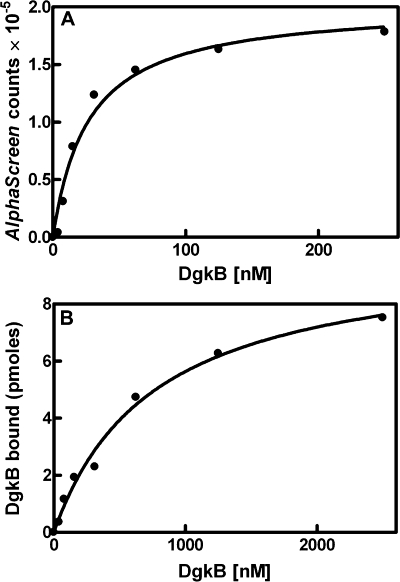
DgkB Binding to PtdGro Vesicles—The observation that DgkB catalysis was accelerated by the presentation of DAG in PtdGro vesicles suggested that the protein possessed an intrinsic affinity for anionic phospholipids. An AlphaScreen proximity assay was developed to detect and evaluate the molecular determinants required for the association between DgkB and 100-nm unilamellar phospholipid vesicles independent of catalysis. The AlphaScreen assay is a solution-based assay with a high signal to noise ratio that is described in detail under “Experimental Procedures.” This assay yielded an apparent Kd for DgkB of 34 ± 1 nm for PtdGro vesicles (Fig. 3A). The distinct advantage of the AlphaScreen assay is its reproducibility and sensitivity that allows a large number of DgkB mutants to be analyzed and the relative importance of specific residues and buffer components to be accurately compared. However, the high avidity of the AlphaScreen assay beads generally leads to apparent Kd values that are about 10-fold lower than are found using other assays. Therefore, we validated this concept by determining the DgkB Kd using the method developed by Thomas and Glomset (25) that measures the amount of DgkB bound to PtdGro vesicles using centrifugation to separate bound and free DgkB. The amount of DgkB bound was determined from the difference in the protein recovered in the supernatant in experiments performed with either PtdCho vesicles or PtdCho vesicles mixed with PtdGro vesicles using SDS-gel electrophoresis and staining with Sypro Ruby to detect the protein. Image analysis using the PhosphorImager and a standard DkgB protein curve was used to calculate the percent bound. This involved procedure yielded an apparent Kd of 760 ± 120 nm for DgkB (Fig. 3B). The apparent Kd determined by the centrifugation method was about 20-fold higher than the apparent Kd determined with AlphaScreen.
FIGURE 3.
Binding of DgkB to PtdGro vesicles. A, DgkB binding to PtdGro vesicles determined by the AlphaScreen assay. Measurements were made in triplicate and the mean ± S.E. are plotted. The apparent Kd was calculated as 34 ± 1 nm. B, DgkB binding to PtdGro vesicles determined from the difference in the recovery of soluble DgkB treated with PtdCho vesicles compared with the recovery of soluble DgkB in the presence of PtdCho plus PtdGro vesicles as measured by densitometry of stained SDS gels. The apparent Kd using this assay was 760 ± 120 nm. Details of these two assays are found under “Experimental Procedures.”
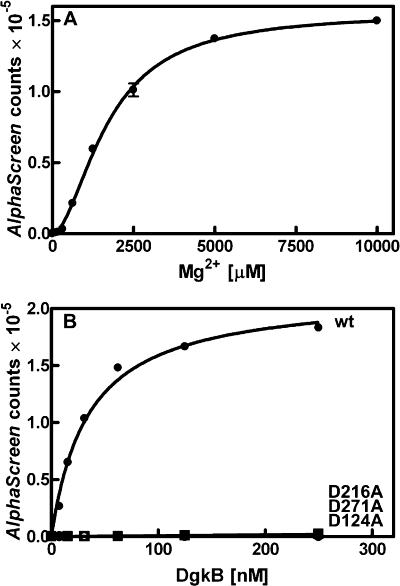
Mg2+-dependent DgkB Binding to Phospholipid Vesicles—DgkB binding to PtdGro vesicles was detected only in the presence of MgCl2 (Fig. 4A), meaning that the binding constant was higher than could be detected with the AlphaScreen assay. However, in the presence of MgCl2 DgkB-vesicle interactions were clearly evident (Table 1). Also, Mg2+ was required for the binding of DgkB to PtdGro/DAG vesicles. DgkB bound to PtdGro/DAG vesicles with an apparent Kd of 12 nm (Table 1), about 3-fold higher than to PtdGro alone. The binding of DgkB to PtdCho or PtdCho/DAG vesicles could not be demonstrated using this assay either in the presence or absence of Mg2+ (data not shown), underscoring the importance of anionic phospholipids for the association of DgkB with phospholipid bilayers. These data demonstrated that both PtdGro and Mg2+ were critical for vesicle binding.
FIGURE 4.
Mg2+ requirement for DgkB binding to PtdGro vesicles. DgkB binding to phospholipid vesicles was measured using the AlphaScreen proximity assay that signals the association of DgkB with 100-nm diameter phospholipid vesicles. Assays were conducted in triplicate and the dissociation constants calculated (value ± S.E.). A, phospholipid vesicles were composed of PtdGro and binding assays were performed in the presence of the indicated concentration of MgCl2. B, comparison of the binding of wild-type DgkB and three mutants in the aspartate residues that participate in the Mg2+·Asp·water network using the AlphaScreen assay in the presence of PtdGro vesicles and 10 mm Mg2+. The apparent Kd values are listed in Table 1. The symbols are: DgkB, •; DgkB[D216A], ▪; DgkB[D124A], ○; and DgkB[D271A], □. Error bars represent standard error.
TABLE 1.
Enzyme activity and phospholipid vesicle binding by DgkB and its mutant derivatives
|
DgkB
|
Activitya
|
Apparent Kd
(nm)b
|
|
|---|---|---|---|
| PtdGro | PtdGro/DAG | ||
| nmol/min/mg | |||
| Wild-type | 24.9 ± 0.1 | 34 ± 1 | 12 ± 1 |
| E273A | <0.4 | 37 ± 1 | 15 ± 1 |
| Mg2+ binding | |||
| D216A | 6.2 ± 0.1 | 170,000 ± 22,000 | 594 ± 33 |
| D124A | 2.4 ± 0.2 | 15,900 ± 920 | 13,640 ± 380 |
| D271A | <0.4 | 11,100 ± 516 | 6,730 ± 280 |
| Surface positive charge | |||
| K15A | 12.2 ± 0.2 | 114,320 ± 18274 | 231 ± 10 |
| R20A | 25.6 ± 0.4 | 444 ± 17 | 50 ± 5 |
| K165A | 13.9 ± 0.4 | 80,884 ± 9561 | 285 ± 17 |
| R100A | 16.8 ± 0.5 | 652 ± 32 | 22 ± 1 |
| K15A,K165A | 2.8 ± 0.4 | NDc | ND |
| ATP binding | |||
| D68A | <0.4 | 11 ± 1 | 12 ± 1 |
| T94A | 3.1 ± 0.1 | 31 ± 1 | 11 ± 1 |
| N96A | 2.3 ± 0.1 | 102 ± 1 | 18 ± 1 |
| D97A | 3.6 ± 0.2 | 25 ± 1 | 11 ± 1 |
The DgkB specific activities were determined by the vesicle assay at 30 μm total lipid vesicle concentration (27 μm PtdGro and 3 μm DAG).
The apparent Kd values for DgkB association with PtdGro vesicles were determined using the AlphaScreen assay at 30 μm total lipid vesicle (27 μm PtdGro and 3 μm DAG).
ND, no detectable binding.
Role of the Structural Mg2+ Site in DgkB Vesicle Binding—The Mg2+ requirement for DgkB docking to PtdGro vesicles (Fig. 4A) may be due to an electrostatic role for Mg2+ forming a bridge between DgkB and PtdGro. However, DgkB has a Mg2+-specific binding site that together with three key aspartate residues creates a network of ordered water molecules that organize the loop regions and connect the secondary structural elements in domain 2 (19). Site-directed mutagenesis of the three key asparate residues participating in the DgkB Mg2+·Asp·water network was employed to test whether this site was responsible for the observed Mg2+ dependence of DgkB binding to PtdGro vesicles (Fig. 4B). The dissociation constants for DgkB binding to PtdGro vesicles measured in the presence of 10 mm MgCl2 for DgkB[D124A], DgkB[D216A], and DgkB[D271A] mutants were orders of magnitude higher than wild-type DgkB, which had an apparent Kd in the AlphaScreen assay of 34 nm (Table 1). The membrane binding deficiency of these mutants was also examined by the method of Thomas and Glomset (25) using the assay conditions described in Fig. 3B. Each protein was tested at 2.5 μm total protein, which was saturating for the wild-type protein (Fig. 3B). The amount of vesicle-bound proteins in this experiment were as follows: DgkB, 7.5 ± 2.1 pmol; DgkB[D124A], 2.7 ± 0.9 pmol; DgkB[D216A], 0.7 ± 0.4 pmol; and DgkB[D271A], 1.2 ± 0.1 pmol in the presence of 10 mm MgCl2. In the absence of MgCl2, only 0.5 ± 0.3 pmol of DgkB was associated with PtdGro vesicles. Thus, the binding of the DgkB[D124A], DgkB[D216A], and DgkB[D271A] to PtdGro vesicles was significantly compromised in the presence of Mg2+ using this assay. Although the apparent Mg2+ requirement for vesicle binding was slightly higher in the AlphaScreen assay than the Kd for Mg2+ determined in solution (see below), these results established that the occupancy of the structural Mg2+ site in domain 2 underlies the Mg2+-dependent DgkB binding to PtdGro vesicles.
Mg2+-induced Conformational Change in DgkB—The analysis of the aspartate mutants suggested that a Mg2+-induced conformational change in DgkB was required to correctly orient key residues for vesicle interaction. The presence of Mg2+ stabilized DgkB to thermal denaturation as observed with the related YegS protein (31) (not shown). The contacts made by the Mg2+·Asp·water network involved loop regions of the protein (19), and the structural analysis suggested that Mg2+ was important for maintaining the organization of these loops rather than supporting the α-helical and β-sheet structural elements in the protein. Two fluorescence assays were used to determine whether there was a Mg2+-dependent conformational change in DgkB.
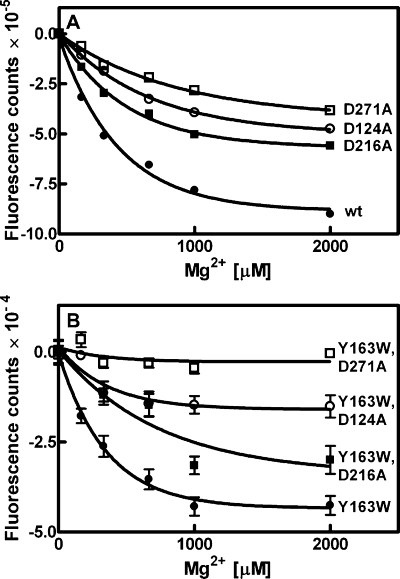
The intensity of the fluorescent signal emitted by the dye SYPRO Orange is related to the hydrophobicity of the protein surface (26). Titration of Mg2+ into a solution of DgkB and SYPRO Orange led to a decrease in the fluorescence intensity in the sample (Fig. 5A). This result indicated that Mg2+ binding to DgkB promoted a structural rearrangement that decreased the overall hydrophobicity of the DgkB surface. The apparent Kd for Mg2+ based on the Mg2+-induced change in SYPRO Orange fluorescence was 390 ± 13 μm. The same experiment was also performed with the panel of DgkB asparate mutants (Fig. 5A). These data suggested that the Mg2+-dependent conformational rearrangements reported by SYPRO Orange fluorescence in DgkB were compromised in all three aspartate mutants, which bound Mg2+ with lower affinities than wild-type DgkB. The rank order of importance for the aspartate residues in DgkB conformation change detected by SYPRO Orange was Asp271 > Asp124 > Asp216.
FIGURE 5.
Mg2+-induced conformational change in DgkB. Two methods were used to investigate potential conformational changes in DgkB triggered by Mg2+ binding in the absence of lipid. A, SYPRO orange fluorescent dye binding to DgkB decreased as a function of MgCl2 concentration. The Mg2+ dissociation constants calculated from these data were: DgkB, •, 390 ± 13 μm; DgkB[D216A], ▪, 490 ± 32 μm; DgkB[D124A], ○, 780 ± 61 μm; and DgkB[D271A], □, 1060 ± 106 μm. B, intrinsic tryptophan fluorescence of DgkB[Y163W] decreased as a function of MgCl2 concentration. The apparent dissociation constants for Mg2+ were: DgkB[Y163W] (•), 303 ± 72 μm; DgkB[Y163W,D216A] (▪), 845 ± 365 μm; DgkB[Y163W,D124A] (○), 424 ± 285 μm; and DgkB[Y163W,D271A] (□), not detected. Error bars represent S.E.
The second approach to investigate the connection between the Mg2+·Asp·water network and a conformational change in DgkB was to use site-directed mutagenesis to replace tyrosine at position 163 with a tryptophan (DgkB[Y163W]). Position 163 was selected because of its location on the β8-α6 loop and the fact that Tyr163 is not conserved in the DkgB protein family. DgkB[Y163W] introduced a single tryptophan residue into the β8-α6 loop that also contains Lys165, a residue important for DgkB docking to PtdGro vesicles (see below). The DgkB[Y163W] mutant protein exhibited slightly higher catalytic activity than the wild-type protein. The measurement of intrinsic tryptophan fluorescence is an indicator of any conformational change in the β8-α6 loop that alters the environment of the tryptophan and the adjacent Lys165. The observed change in fluorescence intensity as a function of MgCl2 concentration in DgkB[Y163W] indicated that binding of Mg2+ altered the environment of the fluorescent tryptophan probe located more than 20 Å from the structural Mg2+ site (Fig. 5B). Specifically, the observed quenching of the tryptophan fluorescent signal suggests that Trp163 was more solvent exposed in the presence of Mg2+ (32). The apparent Kd for Mg2+ in this analysis was 303 ± 72 μm. Mutations in each of the key asparatic acid residues were then introduced into DgkB[Y163W] and their Mg2+-dependent tryptophan fluorescence measured (Fig. 5B). Each of these mutants exhibited a compromised conformational change with the rank order of importance for the aspartates of Asp271 > Asp124 > Asp216, as noted in SYPRO Orange binding experiment. These data established that the binding of Mg2+ to DgkB promoted a conformational change in the β8-α6 loop region in domain 2.
Role of Lys15 and Lys165 in DgkB Binding to PtdGro—The specific binding of DgkB to anionic phospholipid vesicles suggested that ionic interactions between positively charged residues on the surface of the protein and the anionic phospholipid was responsible for docking DgkB to the bilayer. The inclusion of 500 mm NaCl in the DgkB PtdGro vesicle assay inhibited its activity by 75%, further supporting the idea that electrostatic interactions contributed to the ability of DgkB to recognize PtdGro vesicles (not shown). The analysis of the DgkB structure shows that its surface is largely electronegative, except for a patch of positively charged residues (Lys15, Arg20, and Lys165) that lie along one aspect of the protein (Fig. 1) (19). Site-directed mutagenesis was used to prepare a series of DgkB mutants with alanine substituted for each of these positively charged residues and their vesicle binding properties were determined (Table 1) in experiments performed as illustrated in Fig. 4B. Lys15 and Lys165 were the key residues required for DgkB docking to both the PtdGro and PtdGro/DAG bilayers (Table 1). The DgkB[R20A] and DgkB[R100A] mutants exhibited about an order of magnitude decrease in PtdGro vesicle binding, but only had 4- and 2-fold lower affinities, respectively, in the presence of PtdGro/DAG vesicles. The association of the DgkB[K15A,K165A] double mutant with either PtdGro or PtdGro/DAG vesicles was not detected by AlphaScreen, and this double mutant was more catalytically compromised than the single mutants (Table 1). These data showed that Lys15 and Lys165 were the key residues required for DgkB docking to phospholipid vesicles.
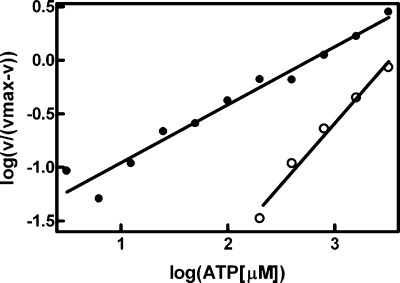
ATP Affinity and Vesicle Binding—A key feature of the interfacial activation of DgkB was a significant change in the kinetic parameters related to ATP. The mutation of four residues involved in the ATP·Mg2+ binding site (Asp68, Thr94, Asn96, and Asp97) compromised catalytic activity, but had either small (DgkB[T94A]) or no change in the affinity of DgkB for PtdGro or PtdGro/DAG vesicles (Table 1) indicating that ATP binding had little effect on the association of DgkB with phospholipid vesicles. The ATP Km in the mixed micelle assay was 3.7 ± 0.4 mm (Table 2) and there was no indication of cooperative behavior based on a Hill plot (Fig. 6). However, DgkB exhibited high negative cooperativity for ATP in the vesicle assay with a Hill coefficient of 0.54 (Fig. 6). The cooperative behavior was so pronounced that two apparent Km values could be calculated by fitting the data to a 2-site binding model (Table 2). The high affinity site exhibited a Km of 32 ± 12 μm, whereas the low affinity site exhibited a Km of 2.1 mm. The importance of key residues to the ATP Km change was investigated by kinetic analysis of mutant proteins (Table 2). These data showed that DgkB[K165A] had the same ATP kinetic properties as DgkB with distinct high and low affinity sites. However, neither DgkB[K15A] nor DgkB[R100A] mutants exhibited cooperative behavior and lacked the high affinity ATP binding mode. These data suggested that the interaction of these two residues with the PtdGro bilayer was responsible for a conformational change that promoted ATP binding. The cooperative behavior of DgkB indicated a communication between the two subunits of the dimer. Therefore, the ATP kinetics in the DgkB[R20A] mutant was analyzed because this residue lies on either side of the dimer interface, and may function as a membrane anchor that prevents the free rotation of domains 1 and 2 in both monomers when the dimer is bound to phospholipid bilayers. DgkB[R20A] was not a cooperative enzyme, but rather exhibited only the low Km for ATP (Table 2). These data demonstrated that Arg20 was critical to support the negative cooperative behavior of DgkB. These data were consistent with a model in which the binding of DgkB to the anionic phospholipid vesicles triggered a conformational change in one monomer of DgkB that significantly increased its affinity for ATP, whereas the other monomer had an ATP Km that remained similar to that found in the mixed micelle assay.
TABLE 2.
Kinetic constants for DgkB and selected DgkB mutants
| Assay | DgkB | Hill coefficientsa | Km1; Km2b |
|---|---|---|---|
| μm | |||
| Vesicle | Wild-type | 0.54 ± 0.02 | 32 ± 12; 2,180 ± 760 |
| K15A | 0.96 ± 0.06 | 402 ± 62 | |
| R20A | 0.90 ± 0.12 | 74 ± 15 | |
| R100A | 0.95 ± 0.10 | 750 ± 165 | |
| K165A | 0.57 ± 0.03 | 40 ± 6; 2,500 ± 770 | |
| Micelle | Wild-type | 1.14 ± 0.04 | 3,700 ± 400 |
Hill coefficients for DgkB and its mutants obtained from the slope of the Hill plots.
ATP Km for DgkB and its mutants; to assess ATP binding of the non-identical catalytic sites of DgkB and DgkB[K165A], data points of the Michaelis-Menten plot were fitted to a two-site binding hyperbola as discussed under “Experimental Procedures.”
FIGURE 6.
Vesicle binding decreases the Km for ATP. Kinetic constants for ATP were determined using either the vesicle (PtdGro/DAG, 90/10, mol %) (•) or octyl glucoside-mixed micelle (○) assays. The Hill plots indicated strong negative cooperativity for ATP binding in the vesicle assay with a Hill coefficient (slope) of 0.54. In contrast, the micellar assay did not exhibit cooperative behavior and the Hill coefficient was 1.14.
DISCUSSION
Our experiments support the model for DgkB docking to phospholipid bilayers depicted in Fig. 1, and define the electropositive surface of DgkB as the i-face of DgkB. Lys15 and Lys165 are the two most important amino acids required for electrostatic association of DgkB with anionic phospholipid bilayers. Lys15 (domain 1) and Lys165 (domain 2) are located on opposite sides of the active site entrance and are appropriately positioned to both dock the protein to the phospholipid surface and facilitate access of DAG to the active site. The docking mode depicted in Fig. 1 also creates a hydrophobic channel to the active site that allows the apolar substrate to descend from the bilayer into the catalytic pocket. Exactly how DAG is recognized in the bilayer remains to be determined. However, we note that the loop between residues 146 and 157 (dotted line) is mobile and not visible in the electron density maps of the soluble protein, and this region is positioned to act as a substrate recognition motif following the docking of DgkB to the bilayer.
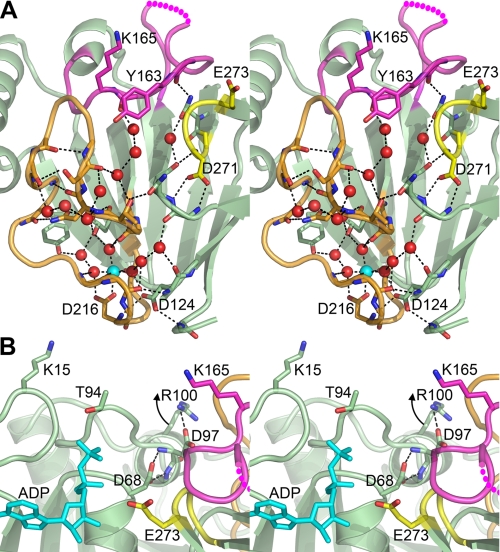
The fundamental importance of the structural Mg2+·Asp·water network in domain 2 to the global organization of the DgkB substrate binding domain (domain 2) is illustrated in Fig. 7A. Our previous work showed that this network mediates the positioning of Glu273, the catalytic base (19). Our current spectroscopic experiments extend the importance of the Mg2+ binding site to encompass a more global conformational change required for DgkB binding to phospholipid vesicles. Site-directed mutagenesis of the aspartates forming the network verify that the occupancy of the structural Mg2+ site underlies the Mg2+-dependent docking to anionic phospholipid vesicles. A major component of the Mg2+ effect can be attributed to a conformational change in the loop containing Lys165.
FIGURE 7.
Structural interpretation of the role for Mg2+ and Arg100 in DgkB binding to anionic phospholipids and ATP. A, binding to PtdGro vesicles is dependent on the structural Mg2+ site that affects the orientation of Lys165, a key membrane recognition element. The stereo view is a cross-section of the DgkB structure at the domain interface (looking toward domain 2) depicting the conserved Mg2+·Asp·water network and its connection to Tyr163 and Lys165 (colored magenta) via the β10 -β11 loop (colored orange). The color code is: Mg2+, cyan sphere; water, red spheres; β8-α6 loop, magenta; β10 -β11 loop, orange; loop, β15-β16 turn, yellow; hydrogen bonds are indicated as dashed black lines; and the dashed magenta line indicates the missing electron density from the β8-α6 loop. B, a view of the DgkB active site showing the essential Asp68 residue hydrogen bonded to the backbone amides in helix α4 containing Arg100. The data indicate that Arg100 interacts with anionic phospholipids to move helix α4 upward toward the bilayer breaking the connection between Asp68 and helix α4. This movement allows the rotation of the Asp68 side chain toward the reader to interact with the ATP γ-phosphate and Mg2+ at the P-loop. ADP is shown in blue sticks, Glu273 (the catalytic base) is shown in yellow, and the structural elements are colored as in A. The models were drawn with PyMOL (DeLano) using the Mg2+·DgkB·ADP structure (PDB accession 2QV7).
The interfacial activation of ATP binding is central to DgkB catalysis. The DgkB active site depicted in Fig. 7B represents a catalytically incompetent conformation of the enzyme in solution with an incompletely formed ATP binding pocket in domain 1 and Glu273, the catalytic base in domain 2, displaced from its required location (19). Two conformational changes are therefore necessary for catalysis to proceed. The first is a rotation of Asp68, an essential catalytic residue involved in coordinating ATP·Mg2+ bound at the P-loop (19). In solution, this movement is constrained by the hydrogen bonding of the Asp68 to the backbone amides of Asp97-Phe98 in helix α4 and also by a helix dipole interaction (Fig. 7B). Thus, a conformational change that breaks the connection between helix α4 and Asp68 is required before Asp68 can rotate to interact with ATP·Mg2+. Lys15 and Arg100 are both required for the conformational change that alters the ATP affinity, but whereas Lys15 is a key residue for binding to phospholipid vesicles, Arg100 is not. This suggests a two-step model for the conformational change. Lys15 first binds to PtdGro bringing the more buried Arg100 closer to the bilayer, and then Arg100 engages the anionic phospholipid surface moving helix α4 away from the active site. This model for the positioning of Asp68 is supported by the structure of a similar protein, NAD kinase in complex with ATP (33), in which helix α4 is replaced by a loop positioned further away from the P-loop. The second conformational change involves a hinge-like motion between domains 1 and 2 to bring Glu273 to the active site (19). Similar movements are commonly observed in kinases following ATP binding to the apoenyzme (34, 35), and we have predicted that this occurs in DgkB based on the structure (19).
A second feature of DgkB catalysis on PtdGro/DAG vesicles is that the ATP binding kinetics exhibit strong negative cooperativity which, in its limiting case, means half-of-the-sites binding stoichiometry (36). This effect in DgkB is so pronounced that both the high and low affinity ATP binding sites are clearly distinguished. This behavior generally arises from ligand-induced structural asymmetry in multisubunit enzymes and the model suggests an interaction between the 2 monomers of DgkB such that the conformational change associated with the high affinity binding of ATP to one monomer prevents the same conformational change from occurring in the other monomer. Alternatively, the activated conformation of one monomer tightly bound to the bilayer may sterically prevent the binding of the partner active site. Such communication would occur across the dimer interface, which is flanked by Arg20 with its side chain extended toward the anionic phospholipid bilayer (Fig. 1). The elimination of negative cooperativity and the presence of only a high affinity ATP binding mode in the DgkB[R20A] mutant reveals that the anchoring of the dimer interface to the bilayer via Arg20 prevents a conformation change to high affinity ATP from simultaneously occurring in both subunits.
The residues involved in interfacial recognition in DgkB are conserved in the catalytic cores of the mammalian DAG kinases (19), and this leads to the conclusion that the mammalian enzymes bind to phospholipid bilayers and undergo the same conformational changes as DgkB. This is supported by the observation that the catalytic activities of mammalian DAG kinases are also stimulated by a variety of anionic phospholipids (25, 29, 37-40). Thomas and Glomset (25) studied the binding of DAG kinase to phospholipid vesicles independent of its catalysis and made two key observations. First, DAG kinase does not bind to PtdCho vesicles unless an anionic phospholipid like phosphatidylserine, phosphatidylinositol, or PtdGro is present. Second, vesicle binding is absolutely dependent on Mg2+ and Mg2+ increased the thermal stability of the enzyme. The Mg2+ concentration required for 50% binding is about 300 μm, which closely matches the measurements made with DgkB. The three key aspartate residues (Asp124, Asp216, and Asp271) that coordinate Mg2+ in domain 2 are completely conserved in the mammalian DAG kinases (19), and site-directed mutagenesis of these conserved aspartates in pig DGKα show that they are all required for catalysis (29). These data indicate that the mammalian DAG kinase has a structural Mg2+ site like the one illustrated in Fig. 7A that governs protein stability, catalysis, and membrane interactions. It is generally thought that the membrane binding properties of mammalian DAG kinases is controlled by the presence of structural modules that lie outside the catalytic core that dock the kinases to the membrane (1, 2). Although these accessory modules are important in targeting the DAG kinases to selected intracellular membrane systems and integrating their spatiotemporal activation to signaling events, the phospholipid recognition properties and conformation of the catalytic cores of the DAG kinases described in this report are also critical to the function of all members of this protein family regardless of their intracellular location or ancillary activation mechanisms.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34496. This work was also supported by Cancer Center Support Grant CA21765 and the American Lebanese Syrian Associated Charities. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DAG, diacylglycerol; DgkB, S. aureus diacylglycerol kinase; PtdGro, phosphatidylglycerol; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Merida, I., Avila-Flores, A., and Merino, E. (2008) Biochem. J. 409 1-18 [DOI] [PubMed] [Google Scholar]
- 2.Sakane, F., Shin-ichi, I., Masahiro, K., Satoshi, Y., and Hideo, K. (2007) Biochim. Biophys. Acta 1771 793-806 [DOI] [PubMed] [Google Scholar]
- 3.Jerga, A., Lu, Y.-J., Schujman, G. E., de Mendoza, D., and Rock, C. O. (2007) J. Biol. Chem. 282 21738-21745 [DOI] [PubMed] [Google Scholar]
- 4.Winget, J. M., Pan, Y. H., and Bahnson, B. J. (2006) Biochim. Biophys. Acta 1761 1260-1269 [DOI] [PubMed] [Google Scholar]
- 5.Jain, M. K., and Berg, O. G. (2006) Curr. Opin. Chem. Biol. 10 473-479 [DOI] [PubMed] [Google Scholar]
- 6.Feng, J., Wehbi, H., and Roberts, M. F. (2002) J. Biol. Chem. 277 19867-19875 [DOI] [PubMed] [Google Scholar]
- 7.Shao, C., Shi, X., Wehbi, H., Zambonelli, C., Head, J. F., Seaton, B. A., and Roberts, M. F. (2007) J. Biol. Chem. 282 9228-9235 [DOI] [PubMed] [Google Scholar]
- 8.Zhang, X., Wehbi, H., and Roberts, M. F. (2004) J. Biol. Chem. 279 20490-20500 [DOI] [PubMed] [Google Scholar]
- 9.Scott, D. L., White, S. P., Otwinowski, Z., Yuan, W., Gelb, M. H., and Sigler, P. B. (1990) Science 250 1541-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White, S. P., Scott, D. L., Otwinowski, Z., Gelb, M. H., and Sigler, P. B. (1990) Science 250 1560-1563 [DOI] [PubMed] [Google Scholar]
- 11.Brunie, S., Bolin, J., Gewirth, D., and Sigler, P. B. (1985) J. Biol. Chem. 260 9742-9749 [PubMed] [Google Scholar]
- 12.Thunnissen, M. M., Ab, E., Kalk, K. H., Drenth, J., Dijkstra, B. W., Kuipers, O. P., Dijkman, R., de Haas, G. H., and Verheij, H. M. (1990) Nature 347 689-691 [DOI] [PubMed] [Google Scholar]
- 13.Scott, D. L., Otwinowski, Z., Gelb, M. H., and Sigler, P. B. (1990) Science 250 1563-1566 [DOI] [PubMed] [Google Scholar]
- 14.Heinz, D. W., Ryan, M., Bullock, T. L., and Griffith, O. H. (1995) EMBO J. 14 3855-3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatulian, S. A., Biltonen, R. L., and Tamm, L. K. (1997) J. Mol. Biol. 268 809-815 [DOI] [PubMed] [Google Scholar]
- 16.van den Berg, B., Tessari, M., Boelens, R., Dijkman, R., de Haas, G. H., Kaptein, R., and Verheij, H. M. (1995) Nat. Struct. Biol. 2 402-406 [DOI] [PubMed] [Google Scholar]
- 17.Guo, S., Zhang, X., Seaton, B. A., and Roberts, M. F. (2008) Biochemistry 47 4201-4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, J., Bradley, W. D., and Roberts, M. F. (2003) J. Biol. Chem. 278 24651-24657 [DOI] [PubMed] [Google Scholar]
- 19.Miller, D. J., Jerga, A., Rock, C. O., and White, S. W. (2008) Structure 16 1036-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 21.Ullman, E. F., Kirakossian, H., Singh, S., Wu, Z. P., Irvin, B. R., Pease, J. S., Switchenko, A. C., Irvine, J. D., Dafforn, A., Skold, C. N., and Wagner, D. B. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 5426-5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullman, E. F., Kirakossian, H., Switchenko, A. C., Ishkanian, J., Ericson, M., Wartchow, C. A., Pirio, M., Pease, J., Irvin, B. R., Singh, S., Singh, R., Patel, R., Dafforn, A., Davalian, D., Skold, C., Kurn, N., and Wagner, D. B. (1996) Clin. Chem. 42 1518-1526 [PubMed] [Google Scholar]
- 23.Stokka, A. J., Gesellchen, F., Carlson, C. R., Scott, J. D., Herberg, F. W., and Tasken, K. (2006) Biochem. J. 400 493-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazar, G. A., Dang, W., Karki, S., Vafa, O., Peng, J. S., Hyun, L., Chan, C., Chung, H. S., Eivazi, A., Yoder, S. C., Vielmetter, J., Carmichael, D. F., Hayes, R. J., and Dahiyat, B. I. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 4005-4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas, W. E., and Glomset, J. A. (1999) Biochemistry 38 3320-3326 [DOI] [PubMed] [Google Scholar]
- 26.Niesen, F. H., Berglund, H., and Vedadi, M. (2007) Nat. Protoc. 2 2212-2221 [DOI] [PubMed] [Google Scholar]
- 27.Anderson, K. S., Sikorski, J. A., and Johnson, K. A. (1988) Biochemistry 27 1604-1610 [DOI] [PubMed] [Google Scholar]
- 28.Walsh, J. P., and Bell, R. M. (1986) J. Biol. Chem. 261 6239-6247 [PubMed] [Google Scholar]
- 29.Abe, T., Lu, X., Jiang, Y., Boccone, C. E., Qian, S., Vattem, K. M., Wek, R. C., and Walsh, J. P. (2003) Biochem. J. 375 673-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White, D. C., and Frerman, F. E. (1967) J. Bacteriol. 94 1854-1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nichols, C. E., Lamb, H. K., Lockyer, M., Charles, I. G., Pyne, S., Hawkins, A. R., and Stammers, D. K. (2007) Proteins 68 13-25 [DOI] [PubMed] [Google Scholar]
- 32.Royer, C. A. (2006) Chem. Rev. 106 1769-1784 [DOI] [PubMed] [Google Scholar]
- 33.Liu, J., Lou, Y., Yokota, H., Adams, P. D., Kim, R., and Kim, S. H. (2005) J. Mol. Biol. 354 289-303 [DOI] [PubMed] [Google Scholar]
- 34.Suhre, K., and Sanejouand, Y. H. (2004) Nucleic Acids Res. 32 W610-W614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, S. S., Buechler, J. A., and Yonemoto, W. (1990) Annu. Rev. Biochem. 59 971-1005 [DOI] [PubMed] [Google Scholar]
- 36.Koshland, D. E., Jr. (1970) in The Enzymes (Boyer, P. D., ed) pp. 341-396, Academic Press, New York
- 37.Fanani, M. L., Topham, M. K., Walsh, J. P., and Epand, R. M. (2004) Biochemistry 43 14767-14777 [DOI] [PubMed] [Google Scholar]
- 38.Sakane, F., Kai, M., Wada, I., Imai, S., and Kanoh, H. (1996) Biochem. J. 318 583-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thirugnanam, S., Topham, M. K., and Epand, R. M. (2001) Biochemistry 40 10607-10613 [DOI] [PubMed] [Google Scholar]
- 40.Tu-Sekine, B., Ostroski, M., and Raben, D. M. (2007) Biochemistry 46 924-932 [DOI] [PubMed] [Google Scholar]
- 41.Saccani, J., Castano, S., Beaurain, F., Laguerre, M., and Desbat, B. (2004) Langmuir 20 9190-9197 [DOI] [PubMed] [Google Scholar]