Abstract
We have shown that branching morphogenesis of mammary ductal structures requires the action of the morphogen epimorphin/syntaxin-2. Epimorphin, originally identified as an extracellular molecule, is identical to syntaxin-2, an intracellular molecule that is a member of the extensively investigated syntaxin family of proteins that mediate vesicle trafficking. We show here that, although epimorphin/syntaxin-2 is highly homologous to syntaxin-1a, only epimorphin/syntaxin-2 can stimulate mammary branching morphogenesis. We construct a homology model of epimorphin/syntaxin-2 based on the published structure of syntaxin-1a, and we use this model to identify the structural motif responsible for the morphogenic activity. We identify four residues located within the cleft between helices B and C that differ between syntaxin-1a and epimorphin/syntaxin-2; through site-directed mutagenesis of these four amino acids, we confer the properties of epimorphin for cell adhesion, gene activation, and branching morphogenesis onto the inactive syntaxin-1a template. These results provide a dramatic demonstration of the use of structural information about one molecule to define a functional motif of a second molecule that is related at the sequence level but highly divergent functionally.
Branching morphogenesis is a developmental process involved in the formation of many organs, including mammary gland, lung, kidney, and salivary gland. In the mammary gland, branching morphogenesis allows the primitive anlage to develop into the highly ramified mammary ductal tree. Investigations of the signaling processes involved in mammary ductal branching have revealed that branching requires the presence of a growth factor and the morphogen epimorphin (1). Epimorphin binds to mammary epithelial cells through αv-integrins (2), activating morphogenic processes that are dependent upon the transcription factor CCAAT/enhancer-binding protein-β (C/EBPβ)5 (3), and mediating branching and invasion into the extracellular matrix through activation of matrix metalloproteinase-3 (MMP3) (4). Although epimorphin is required for branching morphogenic processes, overexpression of epimorphin can lead to pathological consequences, including ductal hyperplasia and mammary cancer (3, 5). Epimorphin plays a role in morphogenesis of other epithelial organs as well, including kidney (6), pancreas (7, 8), hair (9), intestine (10, 11), and lung (12).
Although epimorphin was first identified as an extracellular morphogen through the use of function-blocking antibodies in lung and skin organ culture assays (12, 13), the same molecule was later found to function in the cytoplasm as syntaxin-2, a member of the syntaxin family of proteins that controls vesicle fusion (14, 15). However, the idea that epimorphin/syntaxin-2 might have distinct roles dependent upon its location on the outside or inside of the plasma membrane is controversial (14–16), in part because epimorphin lacks a canonical peptide signal sequence to direct extracellular localization through the ER/Golgi pathway. Since the discovery of epimorphin/syntaxin-2, a number of molecules have been found that not only transit the plasma membrane in the absence of a signal sequence, but also have distinct functions in the cytosol and in the extracellular milieu (17). Although investigations of such bifunctional, bi-topological molecules are normally focused on either their intracellular or extracellular functions, the possibility that structural information about a molecule in one orientation could provide functional insight about the same molecule in the opposite orientation, where it binds to a completely different ligand or set of ligands, has remained largely unexplored.
Although the structure of epimorphin/syntaxin-2 has not been reported, structural data exist for other members of the syntaxin family (18). The conserved syntaxin structure is composed of an N-terminal autonomously folded 3-helix bundle domain (Habc), a flexible linker region, an α-helical SNARE domain, which participates in the coiled-coil assembly of the SNARE complex, and a C-terminal membrane anchor (Fig. 1, B and C) (19, 20). NMR and crystal structures have shown that some syntaxins partition between two conformations: the open conformation, in which the SNARE domain is able to participate in the multiprotein SNARE complex, and the closed conformation, in which the SNARE domain interacts with the b and c helices of Habc, to form an intramolecular antiparallel four-helix bundle (19–22). Deletion analyses have revealed that the functional domain of epimorphin can be distinguished from those of the syntaxins (1, 23), because the Habc domain alone is required for epimorphin morphogenic activity, whereas the SNARE domain is dispensable for morphogenesis (1) but absolutely necessary for syntaxin function (23, 24).
FIGURE 1.
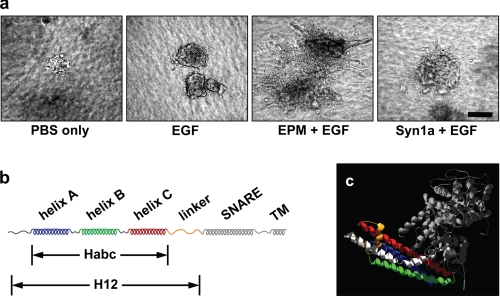
Epimorphin morphogenic activity resides in the Habc domain. a, epimorphin is necessary, and syntaxin-1a does not substitute for epimorphin in directing branching morphogenesis. Clusters of SCp2 mouse mammary epithelial cells were cultured for 10 days in three-dimensional collagen-I gels with the addition of: (a) phosphate-buffered saline only, (b) EGF, (c) EGF and the H12 domain of EPM (Hirai et al. (1)) and EGF, or (d) EGF and a fragment of syntaxin-1a (Syn1a) corresponding to H12 EPM. Scale bar, 100 μm. b, syntaxin motif structure. Syntaxins have three alpha helices (A, B, and C, comprising the Habc domain) connected by a linker to the SNARE helix, followed by a transmembrane domain. c, ribbon drawing of the closed conformation of Syntaxin1a (blue, green, and red, constituting the Habc domain, orange for the linker domain, and white for the SNARE domain) in complex with nSecA (gray); molecular coordinates are from PDB 3c98.
Here, we use structural and functional information about syntaxins to create an epimorphin homology model and to deduce the site of a key epimorphin ligand binding motif; we also demonstrate the specificity of the interactions formed by this motif using site-directed mutagenesis to create an active morphogen from the functionally inactive syntaxin-1a. These results identify precisely the minimal structural motif of epimorphin essential for its function as a morphogen and bring us closer to understanding its mode of interaction with critical physiological ligands.
EXPERIMENTAL PROCEDURES
Reagents—Bradford protein dye reagent was from Bio-Rad. Restriction enzymes, T4 DNA ligase, and alkaline phosphatase were from New England Biolabs (Ipswich, MA). Isopropyl-1-thio-β-d-galactopyranoside and protease inhibitor were from Calbiochem. Coomassie Brilliant Blue R-250 dye and TaqDNA Polymerase were from Sigma. Purified oligonucleotides used for mutagenesis and PCR were from Bio-Synthesis (Lewisville, TX). Deoxynucleotide triphosphates and epidermal growth factor were from Roche Applied Science (Indianapolis, IN). Prof. Frederick M. Hughson (Dept. of Molecular Biology, Princeton University) provided samples of syntaxin-1a protein lacking the transmembrane domain.
Three-dimensional Homology Modeling—Homology modeling was carried out using the SWISS-MODEL/PROMOD II server using the “first approach mode” (25). In brief, the amino acid sequence of syntaxin-2 was submitted to the server, and suitable templates with a sequence identity of more than 25% were selected. Five template structure coordinates (3c98B.pdb, 1ez3B.pdb, 1ez3C.pdb, 1ex3A.pdb, and 1br0A.pdb) were superimposed, and a structural and local pairwise alignment of the target sequence to the main template structures was generated. The positions of the backbone atoms of the template structure were averaged, and the best loops were selected using a method that accounts for force field energy, steric hindrance, and favorable interactions. Starting with conserved residues, the model side chains were built by isosteric replacement of template structure side chains. Deviations in the model were energy-minimized using the GROMOS96 force field.
Generation of Recombinant Epimorphin—Expression constructs were generated by PCR amplification using cDNA for mouse epimorphin or human syntaxin-1 as template. Habc-epimorphin (EPM) lacks the N-terminal 26 amino acids, the linker, SNARE helix, and transmembrane domains and was PCR-amplified with oligonucleotides as follows: HSHisF, CCGCGCCATATGCACCATCACCATCACCATGGCGGGGATCATTTCATGGACGGTTTCTTCCAT, and HSHisR, CGCGCGAAGCTTTTATTATTTGCTTCGCTCCCGGAACAGGAT.
Recombinant syntaxin-1a (Habc-1a) is derived from the homologous three-helix bundle domain of syntaxin-1a and was PCR-amplified using the following oligonucleotides: HS1AHis, CCGCGCCATATGCACCATCACCATCACCATGGCGGGGACCGCTTCATGGATGAGTTCTTTGAA, and HS1AHisR, CGCGCGAAGCTTTTATTATTTGCAGCGTTCTCGGTAGTCTGA.
Both Habc-EPM and 3-Hlx-1A were cloned as Nde1 and HindIII fragments into a pET-27b(+) (Novagen, San Diego, CA) expression vector leading to a N-terminal fusion of His6 tag to the protein fragment. Recombinant Habc-1→2 is derived from Habc-1A with the proposed ligand binding site residues mutated using the QuikChange™ site-directed mutagenesis kit from Stratagene. All mutations were introduced by PCR amplification of the entire expression plasmid using two mutated oligonucleotide primers. Two complementary primers carrying the mutation were used for the substitution mutations. The sequences of the sense primers used for the substitution of the amino acids indicated were the following, with the modified codons inbold and the nucleotide changed indicated in bold as follows. To create the 1→2 protein: M79N, 5′-AGGAACTGGAGGAGCTCAACTCGGACATTAAGAAGACAG-3′; E101D, 5′-CGAGCAGAGCATCGACCAGGAGGAAGGTC-3′; C145S, 5′-GACTACCGAGAACGCAGCAAATAATAAAAGCTTGCGG-3′; and Y141F, 5′-CCACTCAGTCAGACTTCCGAGAACGCAGC-3′. The sequences of the primers used to generate the 2→1 protein were: D79M, 5′-AAGAAGAGCTGGAGGACCTGATGAAAGAGATCAAGAAAACTGCTAAC-3′; D101E, 5′-ATTGAGCAGAGCTGTGAACAGGACGAGAATGGG-3′; S145C, 5′-CCGGGAGCGATGCAAAGGCCGCATC-3′; and F141Y, 5′-GCGCAGATCCTGTACCGGGAGCGATGC-3′. The procedures used for protein production are in the supplemental Materials and Methods.
Cell Culture and Cell Assays—The mouse mammary epithelial cell lines SCp2 and EpH4 were maintained in growth medium (Dulbecco's modified Eagle's medium/F-12 supplemented with 2% fetal bovine serum, 5 μg/ml insulin, and 50 μg/ml gentamicin); the human breast epithelial cell line MCF10A was grown in Dulbecco's modified Eagle's medium/F-12 supplemented with 5% horse serum, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 μg/ml insulin. Adhesion assays were carried out essentially as previously described (1) with the following modifications. Wells of a 24-well polystyrene plate (bacterial-grade, Falcon) were coated overnight at 4 °C with 250 μl/well of recombinant EPM at a concentration of 25 μg/ml. The wells were air-dried, then blocked with a 0.1% solution of pluronic F108 (BASF) in phosphate-buffered saline at room temperature for 30 min, and then washed twice with phosphate-buffered saline. Cells (100,000 cells/well) were plated onto the protein-coated substrata and incubated at 37 °C for 2 h. The wells were then washed three times with phosphate-buffered saline, and the remaining cells attached to the plate were trypsinized and counted. Wells coated with fibronectin were used as a positive control for adhesion. Blocking antibodies were: α1 (Ha31/8, 25 μg/ml, BD Biosciences); α5 (5H10-27, 25 μg/ml, BD Biosciences); αv (H9.2B8, 25 μg/ml, BD Biosciences); β1 (Ha2/5, 25 μg/ml, BD Biosciences); and β3 (2C9.G2, 25 μg/ml, BD Biosciences).
Transcriptional analysis was performed using RNA from SCp2 cells grown for 2 days in growth medium (control) or growth medium supplemented with 25 μg/ml Habc-EPM, Habc-Syn1a, or Habc-1→2. Isolated total RNA was labeled and hybridized to Affymetrix mouse 430_2 gene expression chips at the Mayo Clinic microarray core facility. Processing, normalization, and background correction were carried out using the GCRMA function of Genespring. Quantitative real-time PCR was performed on reverse-transcribed cDNA, using ABI 8900 fast cycler instruments, following manufacturer protocols. TaqMan assays used were purchased from ABI and were: MMP3 (Mm01168406_g1), Fas (Mm01204974_m1), Mgp (Mm00485009_m1), Aqp3 (Mm01208559_m1), and Krt1–16 (Mm00492979_g1). Branching morphogenesis assays were performed using modifications of previously published protocols (3, 4), as described in the supplemental Materials and Methods.
RESULTS
Our previous studies had shown that extracellular epimorphin, when presented in combination with a growth factor such as EGF, is sufficient to direct mammary epithelial branching morphogenesis; we had further localized the morphogenic activity of epimorphin to a domain of the molecule contained within the first 187 amino acids, a fragment that we designated as H12 (1, 3). To determine whether this property is unique to epimorphin among the syntaxin family of proteins, we used a three-dimensional collagen branching assay to compare the morphogenic capacity of this fragment of EPM with a fragment of syntaxin-1a that also lacks the transmembrane domain (Syn1a). We found that cells exposed to EPM in combination with EGF showed abundant branching morphogenesis, whereas cells treated with EGF alone or with Syn1a and EGF showed no branching (Fig. 1A).
The H12 domain of epimorphin consists of an N-terminal region, the three helix Habc bundle, and a linker sequence (Fig. 1B). Crystal and NMR structural analyses of syntaxins suggest that the N-terminal 28 amino acids and the linker region have little defined structure, and thus that the Habc domain is the only structured element of H12-EPM (Fig. 1C) (20, 26–29). We hypothesized that a minimal fragment containing only the Habc bundle would possess all of the functional activity of the complete H12 domain. We expressed the Habc domains of epimorphin (Habc-EPM) and syntaxin-1a (Habc-Syn1a) and found that only Habc-EPM was sufficient to reproduce the effects of H12 in branching morphogenesis (not shown). We therefore used the Habc proteins of EPM and Syn1a in all subsequent experiments.
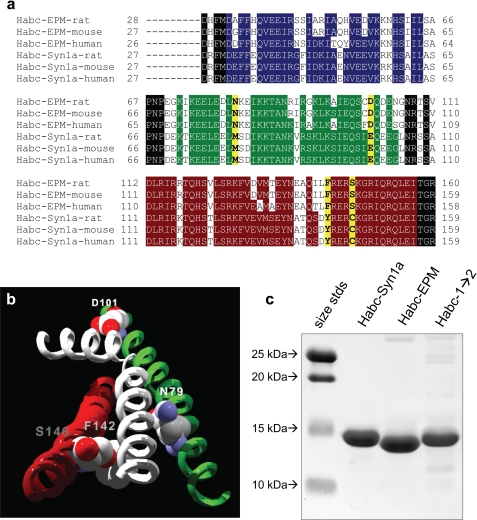
Epimorphin and syntaxin-1a show a very high level of sequence similarity, with 68% identity and 82% homology for human epimorphin versus human syntaxin-1a when compared within the Habc homologous domain (Fig. 2A). That epimorphin and syntaxin-1a proteins showed very different effects in the morphogenesis assay (Fig. 1A) suggested that the morphogenic activity of epimorphin was critically dependent upon presentation of a highly specific ligand binding interface, in which these two proteins were likely to differ by a just a few amino acids. Structural studies of syntaxins in the closed conformation have revealed that, within the N-terminal Habc bundle, the interface between the second and third helices forms a groove that binds to the SNARE helix in the closed conformation of the intact molecule to form a four-helix structure (20, 26–29). We hypothesized that epimorphin binds to its receptor through a comparable interaction, that is, that there exists a helix on the epimorphin receptor that serves as an alternative epimorphin ligand, binding to the cleft between the second and third helices of the Habc domain. We constructed an epimorphin homology model by threading the sequence of epimorphin through the published crystal structure of syntaxin-1a (Fig. 2B). We used this model to identify the amino acid residues of the epimorphin Habc bundle with side chains pointing toward the cleft formed by the second and third helices.
FIGURE 2.
Design and production of Habc-1→2. a, sequence alignment of the Habc domain of rat, mouse, and human epimorphin with rat, mouse, and human syntaxin 1a. Residues highlighted yellow are those that differ between the two molecules and are predicted by the epimorphin homology model to face inwards toward the cleft between helices B and C. Homologous residues in helices A, B, and C are colored blue, green, and red, respectively. b, location of the key functional residues on the epimorphin homology model. Helices B and C are depicted as green and red, respectively, and the white helix represents a hypothetical helix that aligned along the long groove formed by the helices B and C. The space-filling molecules are the residues that are oriented toward the cleft and that differ between syntaxin1a and EPM. c, image of Coomassie-stained gel of recombinant proteins.
Of the residues in the homology model that were predicted to be in contact with the hypothetical fourth helix, four were found to differ between syntaxin-1a and epimorphin and to be oriented toward the predicted binding cleft (syn1a/epimorphin: D79E, D101E, F142Y, and S146C). It is noteworthy that, although the α-helical domains of the Habc bundle are generally conserved between epimorphin and syntaxin-1a, these four residues that differ between the two molecules are conserved between species in mouse, rat, and human (Fig. 2A). We used site-directed mutagenesis to generate a mutant construct of Habc-Syn1a in which these residues were mutated to match the corresponding positions on epimorphin (Habc-1→2, Fig. 2, B and C). We used similar methods to generate a mutant construct of Habc-EPM in which the residues were mutated to match the corresponding positions on syntaxin-1a (Habc-2→1, not shown).
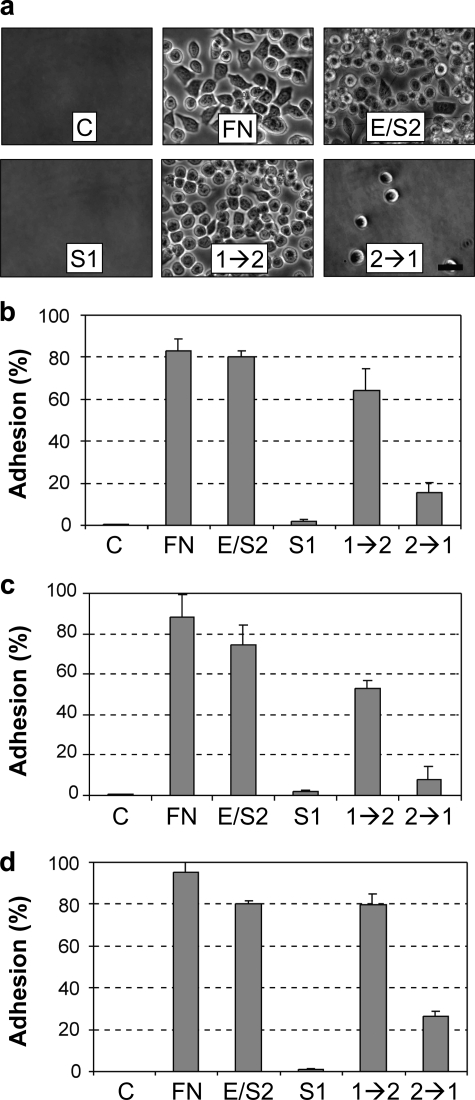
We determined the activity of the quadruple mutant Habc-1→2 and Habc-2→1 proteins in assays that measure binding to epithelial cells, alteration of gene expression, and induction of mammary branching morphogenesis. Association of epimorphin with cells can be assessed through cell binding assays in which proteins are adsorbed to the surface of culture plates and then overlaid with cells in suspension (1, 2). We found that the SCp2 cells adhered well to the positive control fibronectin, to Habc-EPM, and to Habc-1→2 protein, but not to Habc-Syn1a or to Habc-2→1 (Fig. 3, A and B). We found similar results with other mammary epithelial cell lines, as well, including mouse EpH4 cells (Fig. 3C) and human MCF10A cells (Fig. 3D). These results show that the mutation of the four amino acids within the cleft formed by the second and third helices of syntaxin-1a reconstitutes the cell binding activity of epimorphin/syntaxin-2. As we had found greatly reduced binding of the mammary epithelial cells to the Habc-2→1 mutant, we used the Habc-Syn1a mutant as control for further experiments.
FIGURE 3.
Cell adhesion to Habc-1→2. a, images of SCp2 cells to uncoated plates (C), or to plates coated with fibronectin (FN), Habc-EPM (E/S2), Habc-Syn1a (S1), Habc-1→2(1→2), or Habc-2→1(2→1); scale bar, 25 μm. b, quantification of SCp2 cell adhesion (C, untreated control). c, quantification of EpH4 cell adhesion. d, quantification of MCF10A cell adhesion. For b–d, quantification is expressed as means ± S.E., p < 0.01 for C, S1, or 2→1 versus FN, E/S2, or 1→2 for all three cell lines.
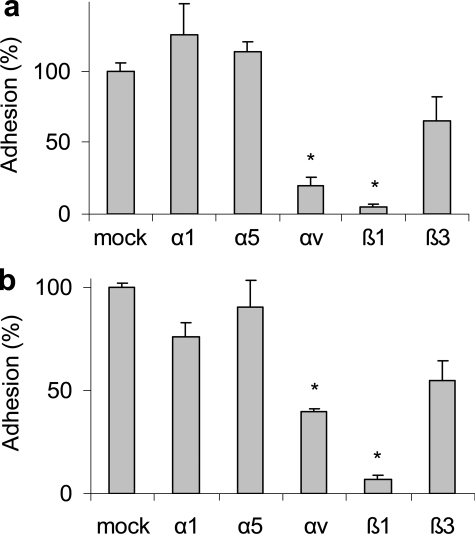
We had previously found that epimorphin binds to mammary epithelial cells through adhesion to αv-integrins (2), so we tested whether anti-integrin antibodies blocked binding of SCp2 cells to Habc-EPM and Habc-1→2. We found that antibodies that blocked integrins αv and β1 also blocked cell association with Habc-EPM (Fig. 4A) and Habc-1→2 (Fig. 4B), but that antibodies targeting integrins α1, α5, and β3 had little effect. These results show that cell binding to Habc-EPM and Habc-1→2 occurs through the same receptors.
FIGURE 4.
Cell adhesion to Habc-EPM and Habc-1→2 is mediated by integrins. Adhesion of SCp2 cells to plates coated with Habc-EPM (a) or Habc-1→2(b) is blocked by antibodies against integrin-αv or integrin-β1, but not by antibodies against integrins-α1, -α5, or -β3. Quantification is expressed as means ± S.E., *, p < 0.005.
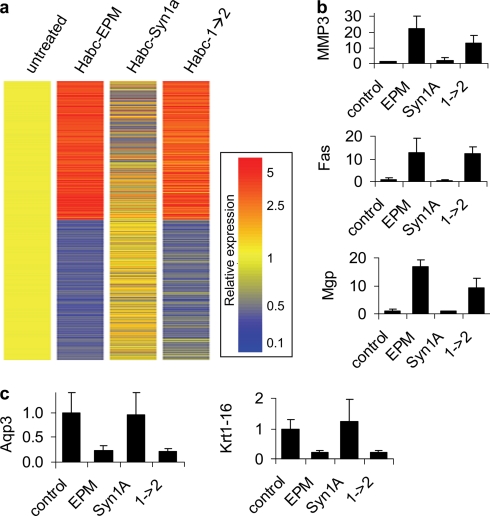
We next assessed global transcriptional alterations of cells treated with the Habc constructs. When expression levels were normalized to untreated cells, 1166 genes were found to be regulated >2-fold in at least one of the treatment conditions, and many of these showed common pattern of regulation between Habc-EPM and Habc-1→2 (Fig. 5A; data for all genes is in supplemental Table S1). We validated the microarray results for selected genes relevant to epimorphin function and mammary branching morphogenesis. Epimorphin has been shown to induce MMP3 (4), a molecule involved in mammary ductal side branching (30), and we found that both Habc-EPM and Habc-1→2 (but not Habc-Syn1a) induced MMP3 (Fig. 5B). We also found that Habc-EPM and Habc1→2 increased gene expression of a number of additional transcripts encoding molecules known to be involved in branching morphogenesis, including Fas (31) and matrix GLA protein (32) (Fig. 5B). Both Habc-EPM and Habc-1→2 also selectively down-regulated transcription of a number of mammary luminal epithelial cell markers, including aquaporin-3 (33) and keratin-16 (34) (Fig. 5C), consistent with the loss of luminal morphology during branching morphogenesis (35). In all of these cases, transcript level alterations induced by Habc-EPM were also found in Habc-1→2, but not in Habc-Syn1a.
FIGURE 5.
Gene expression in cells treated with Habc-EPM, Habc-Syn1a, and Habc-1→2. a, gene expression fingerprint of SCp2 cells treated with Habc-EPM, Habc-Syn1a, and Habc-1→2, normalized to expression levels in untreated cells. Heat map displays expression for transcripts regulated by >2-fold relative to untreated cells. b, real-time quantitative PCR of MMP3, Fas, and matrix gla protein (Mgp) transcripts, which are activated by Habc-EPM (EPM) and Habc-1→2(1→2) relative to Habc-Syn1a (Syn1a). c, real-time QPCR of aquaporin-3 (Aqp3) and keratin-16 (Krt1–16) transcripts, which are inhibited by Habc-EPM and Habc-1→2. All comparisons for b and c are p < 0.05 for Habc-EPM and Habc-1→2 versus Habc-Syn1a.
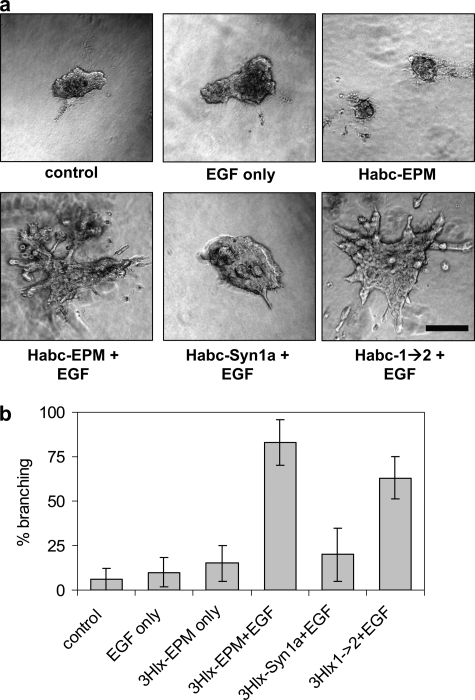
We assessed the ability of the recombinant proteins to induce mammary branching morphogenesis of SCp2 cell clusters. We found that treatment with EGF, or with EGF and Habc-Syn1a, led to growth of the cell clusters, but little or no branching, whereas those treated with Habc-EPM and EGF or Habc-1→2 and EGF exhibited robust branching activity (Fig. 6, A and B). These results show that, by mutating four amino acids, we have reconstituted the epimorphin morphogenic activity using an inactive syntaxin-1a template.
FIGURE 6.
Branching morphogenesis directed by Habc-EPM and by Habc-1→2. SCp2 cell clusters were embedded in collagen-I gels and exposed for 4 days to 50 ng/ml EGF and/or 25 μg/ml of the indicated recombinant proteins. a, representative images of branching assay. Scale bar, 200 μm. b, quantification of branching expressed as means ± S.E. More than 25 randomly selected cell clusters were measured for each condition, and positive branching was assessed when clusters displayed two or more cell projections of greater length than the central cell cluster.
DISCUSSION
In this study, we used structural information from the crystal structure of the homologous protein syntaxin-1a to develop a hypothesis about a potential ligand binding motif of epimorphin/syntaxin-2 and to identify amino acids essential for the function of this motif. This strategy relied upon the fact that epimorphin and syntaxin-2 have identical sequences but distinct behavior and functional domains, reflective of their different localizations: epimorphin is present on the extracellular surface of the plasma membrane (1), whereas syntaxin-2 is found on the cytoplasmic face. It is noteworthy that epimorphin/syntaxin-2 has no canonical signal sequence, and as such, extracellular localization of epimorphin apparently occurs without transit through the endoplasmic reticulum and Golgi apparatus. A variety of proteins lacking canonical signal sequences, including fibroblast growth factors 1 and 2 and interleukin-1β, have been found to exit cells using nonclassic protein secretion pathways (36), but epimorphin/syntaxin-2 is further distinguished in that it has distinct extracellular and intracellular functions. Inside the cell, syntaxin-2 mediates membrane fusion, whereas outside the cell, epimorphin acts as a morphogen. There are other examples of proteins with topologically distinct functions (17). For example, phosphohexose isomerase functions inside the cell in glycolysis and gluconeogenesis, but the same molecule has been extensively investigated as autocrine motility factor in the extracellular space (37, 38), and HMGB1 acts as an architectural DNA-binding protein in the nucleus, but the same molecule is known as amphoterin outside the cell, where it acts as a pro-inflammatory cytokine (39). Our strategy to identify functional domains of epimorphin through structural analysis of the homologous syntaxin-1a could potentially be applied to other proteins with distinct extracellular and intracellular functions, or to homologous pairs of proteins with distinct functions in different subcellular compartments or different extracellular localizations.
Investigations of structurally homologous but mechanistically divergent families of proteins have prompted suggestions that new protein functions may have evolved through an opportunistic process termed “recruitment,” wherein the pre-existing structural features of an active site or ligand binding site are exploited for a new purpose (40, 41). Analysis of protein sequence data suggests that protein speciation can proceed through intermediates with promiscuous functionality that can bind multiple ligands and facilitate multiple biological processes (40). For a molecule such as epimorphin/syntaxin 2, the acquisition of dual topology would provide an additional mechanism to acquire multifunctionality (17). For a syntaxin originally evolved to carry out an intracellular function through the selective binding of a particular ligand from among the array of potential intracellular binding partners, extracellular localization would result in exposure to a novel pool of potential ligands, allowing conscription of a pre-existing protein binding site for new functional interactions.
For epimorphin, we hypothesized that the binding cleft between helices b and c, which accommodates the SNARE helix in the closed form of syntaxin 1a, could also be used as a molecular interface for binding to the extracellular receptor for epimorphin. Initially we envisioned this alternative ligand as another α-helix capable of forming an intermolecular four-helix bundle with the epimorphin three-helix domain. Although this is certainly an attractive model, the structural nature of the interaction between epimorphin and its receptor remains to be elucidated, and one could envision alternative models that would also be consistent with the recruitment of the epimorphin binding site that we have identified here. A striking example of the diversity of protein-protein interactions that can be mediated by a three-helix bundle is found in the Golgi-localized, g-adaptin ear-containing, ARF-binding (GGA) protein family. The GAT domain conserved within this family belongs to the syntaxin trihelical bundle fold (42). The GAT domain has been shown to represent a polyfunctional module that can interact with a wide variety of accessory proteins, using overlapping but distinct sites of molecular interaction (43). GAT domain binders include the coiled-coil domain of endosome fusion mediator Rabaptin-5 (44, 45), which likely forms an interaction resembling the four-helix bundle of the closed form of syntaxin. However, other GAT binders such as ubiquitin are recognized through alternative modes of interaction not involving helical epitopes (46, 47). Thus, although a variety of possibilities exist, the precise mode of interaction of epimorphin with its receptor remains to be experimentally determined.
We have recently identified the αv, β1, and β5 integrin subunits as proteins that directly bind to epimorphin and mediate its morphogenic effects (2). Integrin ectodomains are known to bind to a wide variety of extracellular matrix, cell surface, and soluble protein and glycoprotein ligands (48, 49); however, only a handful of structural studies of integrins bound to their ligands have been reported, and the variety of binding interactions that must contribute to specificity among the integrins remains to be uncovered. The nature of the interaction between epimorphin and cognate integrins will be an important area of future investigation.
The discovery of the active site of epimorphin may have applicability to investigations of pathologies of the breast and other organs. The transcription factor C/EBPβ is a key downstream mediator of the effects of epimorphin in mammary epithelial cells (3). C/EBPβ functions as a homo- or heteromeric dimer of its two constituent isoforms: LAP (liver activating protein) and LIP (liver inhibiting protein) (50–52). LAP and LIP are mutually antagonistic, and changes in their relative ratio can lead to dramatically altered cellular properties (53–55), including malignant transformation in the mammary gland: transgenic mice with increased LIP expression in mammary epithelial cells develop hyperplasias that can spontaneously progress to neoplasia and invasive carcinoma (56), and increased LIP levels are associated with development of human breast cancer (56–60). We have found that epimorphin increases the ratio of LIP to LAP in cultured mammary epithelial cells and in the mammary glands of Whey Acidic Protein promoter-epimorphin transgenic mice (3); these mice develop mammary tumors as they age (5). Because misregulation of epimorphin can contribute to mammary tumor development, inhibiting epimorphin binding may have potential as a cancer preventative or therapeutic approach in specific cancer cell types. The identification of the epimorphin binding cleft may facilitate development of inhibitors capable of blocking the binding of epimorphin to its cell surface receptors. Moreover, because epimorphin has been implicated in normal and pathological development of a number of organs in addition to the breast, including intestine (10, 61), lung (62, 63), pancreas (7), liver (64, 65), cartilage (66), and hair (9, 67), our identification of a key epimorphin interaction domain may be applicable to investigations of these organs in both normal development and in neoplasia.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA64786 (to M. J. B.), CA57621 (to M. J. B. and Zena Werb), CA122086 (to D. C. R.), and CA128660 (to C. M. N. and D. C. R.) from NCI. This work was also supported by the Office of Biological and Environmental Research of the Dept. of Energy (Grant DE-AC03-76SF00098 and a Distinguished Fellow Award to M. J. B.) and the Breast Cancer Research Program (BCRP) of the Dept. of Defense (an Innovator Award to M. J. B.; Grant W81XWH-04-1-0582 to C. M. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Table S1.
Footnotes
The abbreviations used are: C/EBPβ, CCAAT/enhancer-binding protein-β; MMP3, matrix metalloproteinase-3; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; LAP, liver-activating protein; LIP, liver-inhibitory protein; EPM, epimorphin; Habc, N-terminal autonomously folded 3-helix bundle domain; EGF, epidermal growth factor.
References
- 1.Hirai, Y., Lochter, A., Galosy, S., Koshida, S., Niwa, S., and Bissell, M. J. (1998) J. Cell Biol. 140 159-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirai, Y., Nelson, C. M., Yamazaki, K., Takebe, K., Przybylo, J., Madden, B., and Radisky, D. C. (2007) J. Cell Sci. 120 2032-2043 [DOI] [PubMed] [Google Scholar]
- 3.Hirai, Y., Radisky, D., Boudreau, R., Simian, M., Stevens, M. E., Oka, Y., Takebe, K., Niwa, S., and Bissell, M. J. (2001) J. Cell Biol. 153 785-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simian, M., Hirai, Y., Navre, M., Werb, Z., Lochter, A., and Bissell, M. J. (2001) Development 128 3117-3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bascom, J. L., Fata, J. E., Hirai, Y., Sternlicht, M. D., and Bissell, M. J. (2005) Cancer Res. 65 8617-8621 [DOI] [PubMed] [Google Scholar]
- 6.Horikoshi, S., Yoshikawa, M., Shibata, T., Takahashi, K., Shirato, I., and Tomino, Y. (2001) Exp. Nephrol. 9 412-419 [DOI] [PubMed] [Google Scholar]
- 7.Lehnert, L., Lerch, M. M., Hirai, Y., Kruse, M. L., Schmiegel, W., and Kalthoff, H. (2001) J. Cell Biol. 152 911-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tulachan, S. S., Doi, R., Hirai, Y., Kawaguchi, Y., Koizumi, M., Hembree, M., Tei, E., Crowley, A., Yew, H., McFall, C., Prasadan, K., Preuett, B., Imamura, M., and Gittes, G. K. (2006) Dev. Growth Differ. 48 65-72 [DOI] [PubMed] [Google Scholar]
- 9.Takebe, K., Oka, Y., Radisky, D., Tsuda, H., Tochigui, K., Koshida, S., Kogo, K., and Hirai, Y. (2003) FASEB. J. 17 2037-2047 [DOI] [PubMed] [Google Scholar]
- 10.Fritsch, C., Swietlicki, E. A., Lefebvre, O., Kedinger, M., Iordanov, H., Levin, M. S., and Rubin, D. C. (2002) J. Clin. Investig. 110 1629-1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, Y., Wang, L., Iordanov, H., Swietlicki, E. A., Zheng, Q., Jiang, S., Tang, Y., Levin, M. S., and Rubin, D. C. (2006) J. Clin. Investig. 116 1535-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirai, Y., Takebe, K., Takashina, M., Kobayashi, S., and Takeichi, M. (1992) Cell 69 471-481 [DOI] [PubMed] [Google Scholar]
- 13.Gumbiner, B. M. (1992) Cell 69 385-387 [DOI] [PubMed] [Google Scholar]
- 14.Bennett, M. K., Garcia-Arraras, J. E., Elferink, L. A., Peterson, K., Fleming, A. M., Hazuka, C. D., and Scheller, R. H. (1993) Cell 74 863-873 [DOI] [PubMed] [Google Scholar]
- 15.Pelham, H. R. (1993) Cell 73 425-426 [DOI] [PubMed] [Google Scholar]
- 16.Spring, J., Kato, M., and Bernfield, M. (1993) Trends Biochem. Sci. 18 124-125 [DOI] [PubMed] [Google Scholar]
- 17.Radisky, D. C., Hirai, Y., and Bissell, M. J. (2003) Trends Cell Biol. 13 426-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ungar, D., and Hughson, F. M. (2003) Annu. Rev. Cell Dev. Biol. 19 493-517 [DOI] [PubMed] [Google Scholar]
- 19.Chen, X., Lu, J., Dulubova, I., and Rizo, J. (2008) J. Biomol. NMR 41 43-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misura, K. M., Scheller, R. H., and Weis, W. I. (2000) Nature 404 355-362 [DOI] [PubMed] [Google Scholar]
- 21.Antonin, W., Dulubova, I., Arac, D., Pabst, S., Plitzner, J., Rizo, J., and Jahn, R. (2002) J. Biol. Chem. 277 36449-36456 [DOI] [PubMed] [Google Scholar]
- 22.Dulubova, I., Sugita, S., Hill, S., Hosaka, M., Fernandez, I., Sudhof, T. C., and Rizo, J. (1999) EMBO J. 18 4372-4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weimbs, T., Low, S. H., Chapin, S. J., Mostov, K. E., Bucher, P., and Hofmann, K. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3046-3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieber, J. J., Willig, K. I., Heintzmann, R., Hell, S. W., and Lang, T. (2006) Biophys. J. 90 2843-2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez, I., Ubach, J., Dulubova, I., Zhang, X., Sudhof, T. C., and Rizo, J. (1998) Cell 94 841-849 [DOI] [PubMed] [Google Scholar]
- 27.Poirier, M. A., Xiao, W., Macosko, J. C., Chan, C., Shin, Y. K., and Bennett, M. K. (1998) Nat. Struct. Biol. 5 765-769 [DOI] [PubMed] [Google Scholar]
- 28.Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998) Nature 395 347-353 [DOI] [PubMed] [Google Scholar]
- 29.Lerman, J. C., Robblee, J., Fairman, R., and Hughson, F. M. (2000) Biochemistry 39 8470-8479 [DOI] [PubMed] [Google Scholar]
- 30.Wiseman, B. S., Sternlicht, M. D., Lund, L. R., Alexander, C. M., Mott, J., Bissell, M. J., Soloway, P., Itohara, S., and Werb, Z. (2003) J. Cell Biol. 162 1123-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, J., Sapi, E., Brown, W., Nilsen, J., Tartaro, K., Kacinski, B. M., Craft, J., Naftolin, F., and Mor, G. (2000) J. Clin. Investig. 106 1209-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert, K. A., and Rannels, S. R. (2004) Am. J. Physiol. 286 L1179-L1187 [DOI] [PubMed] [Google Scholar]
- 33.Matsuzaki, T., Machida, N., Tajika, Y., Ablimit, A., Suzuki, T., Aoki, T., Hagiwara, H., and Takata, K. (2005) Histochem. Cell Biol. 123 501-512 [DOI] [PubMed] [Google Scholar]
- 34.Tsubura, A., Okada, H., Senzaki, H., Hatano, T., and Morii, S. (1991) Histopathology 18 517-522 [DOI] [PubMed] [Google Scholar]
- 35.Fata, J. E., Werb, Z., and Bissell, M. J. (2004) Breast Cancer Res. 6 1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prudovsky, I., Mandinova, A., Soldi, R., Bagala, C., Graziani, I., Landriscina, M., Tarantini, F., Duarte, M., Bellum, S., Doherty, H., and Maciag, T. (2003) J. Cell Sci. 116 4871-4881 [DOI] [PubMed] [Google Scholar]
- 37.Haga, A., Niinaka, Y., and Raz, A. (2000) Biochim. Biophys. Acta 1480 235-244 [DOI] [PubMed] [Google Scholar]
- 38.Sun, Y. J., Chou, C. C., Chen, W. S., Wu, R. T., Meng, M., and Hsiao, C. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5412-5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lotze, M. T., and Tracey, K. J. (2005) Nat. Rev. Immunol. 5 331-342 [DOI] [PubMed] [Google Scholar]
- 40.Glasner, M. E., Gerlt, J. A., and Babbitt, P. C. (2006) Curr. Opin. Chem. Biol. 10 492-497 [DOI] [PubMed] [Google Scholar]
- 41.Redfern, O. C., Dessailly, B., and Orengo, C. A. (2008) Curr. Opin. Struct. Biol. 18 394-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suer, S., Misra, S., Saidi, L. F., and Hurley, J. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4451-4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattera, R., Puertollano, R., Smith, W. J., and Bonifacino, J. S. (2004) J. Biol. Chem. 279 31409-31418 [DOI] [PubMed] [Google Scholar]
- 44.Mattera, R., Arighi, C. N., Lodge, R., Zerial, M., and Bonifacino, J. S. (2003) EMBO J. 22 78-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhai, P., He, X., Liu, J., Wakeham, N., Zhu, G., Li, G., Tang, J., and Zhang, X. C. (2003) Biochemistry 42 13901-13908 [DOI] [PubMed] [Google Scholar]
- 46.Prag, G., Lee, S., Mattera, R., Arighi, C. N., Beach, B. M., Bonifacino, J. S., and Hurley, J. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2334-2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akutsu, M., Kawasaki, M., Katoh, Y., Shiba, T., Yamaguchi, Y., Kato, R., Kato, K., Nakayama, K., and Wakatsuki, S. (2005) FEBS Lett. 579 5385-5391 [DOI] [PubMed] [Google Scholar]
- 48.Luo, B. H., Carman, C. V., and Springer, T. A. (2007) Annu. Rev. Immunol. 25 619-647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnaout, M. A., Goodman, S. L., and Xiong, J. P. (2007) Curr. Opin. Cell Biol. 19 495-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ossipow, V., Descombes, P., and Schibler, U. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 8219-8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Descombes, P., and Schibler, U. (1991) Cell 67 569-579 [DOI] [PubMed] [Google Scholar]
- 52.Buck, M., Turler, H., and Chojkier, M. (1994) EMBO J. 13 851-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hata, K., Nishimura, R., Ueda, M., Ikeda, F., Matsubara, T., Ichida, F., Hisada, K., Nokubi, T., Yamaguchi, A., and Yoneda, T. (2005) Mol. Cell. Biol. 25 1971-1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, C. N., Slack, J. M., and Tosh, D. (2000) Nat. Cell Biol. 2 879-887 [DOI] [PubMed] [Google Scholar]
- 55.Tang, Q. Q., Otto, T. C., and Lane, M. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 850-855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zahnow, C. A., Cardiff, R. D., Laucirica, R., Medina, D., and Rosen, J. M. (2001) Cancer Res. 61 261-269 [PubMed] [Google Scholar]
- 57.Bundy, L. M., and Sealy, L. (2003) Oncogene 22 869-883 [DOI] [PubMed] [Google Scholar]
- 58.Dearth, L. R., Hutt, J., Sattler, A., Gigliotti, A., and DeWille, J. (2001) J. Cell. Biochem. 82 357-370 [DOI] [PubMed] [Google Scholar]
- 59.Eaton, E. M., Hanlon, M., Bundy, L., and Sealy, L. (2001) J. Cell. Physiol. 189 91-105 [DOI] [PubMed] [Google Scholar]
- 60.Milde-Langosch, K., Loning, T., and Bamberger, A. M. (2003) Breast Cancer Res. Treat. 79 175-185 [DOI] [PubMed] [Google Scholar]
- 61.Shirasaka, T., Iizuka, M., Yukawa, M., Hirai, Y., Horie, Y., Itou, H., Kon-No, S., Fukushima, T., and Watanabe, S. (2003) J. Gastroenterol. Hepatol. 18 570-577 [DOI] [PubMed] [Google Scholar]
- 62.Terasaki, Y., Fukuda, Y., Suga, M., Ikeguchi, N., and Takeya, M. (2005) Respir. Res. 6 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Terasaki, Y., Fukuda, Y., Ishizaki, M., and Yamanaka, N. (2000) Am. J. Respir. Cell Mol. Biol. 23 168-174 [DOI] [PubMed] [Google Scholar]
- 64.Segawa, D., Miura, K., Goto, T., Ohshima, S., Mikami, K., Yoneyama, K., Shibuya, T., Watanabe, D., Kataoka, E., Yoshino, R., and Watanabe, S. (2005) J. Gastroenterol. Hepatol. 20 1769-1780 [DOI] [PubMed] [Google Scholar]
- 65.Miura, K., Nagai, H., Ueno, Y., Goto, T., Mikami, K., Nakane, K., Yoneyama, K., Watanabe, D., Terada, K., Sugiyama, T., Imai, K., Senoo, H., and Watanabe, S. (2003) Biochem. Biophys. Res. Commun. 311 415-423 [DOI] [PubMed] [Google Scholar]
- 66.Oka, Y., Sato, Y., Tsuda, H., Hanaoka, K., Hirai, Y., and Takahashi, Y. (2006) Dev. Biol. [DOI] [PubMed]
- 67.Akiyama, M., Amagai, M., Smith, L. T., Hashimoto, K., Shimizu, H., and Nishikawa, T. (1999) Br. J. Dermatol. 141 447-452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.